Abstract
Bioprinting of engineered bacteria is of great interest for applications of synthetic biology in the context of living biomaterials, but so far, only a few viable approaches are available for the printing of gels hosting live Escherichia coli bacteria. Here, we develop a gentle extrusion-based bioprinting method based on an inexpensive alginate/agarose ink mixture that enables printing of E. coli into three-dimensional hydrogel structures up to 10 mm in height. We first characterize the rheological properties of the gel ink and then study the growth of the bacteria inside printed structures. We show that the maturation of fluorescent proteins deep within the printed structures can be facilitated by the addition of a calcium peroxide-based oxygen generation system. We then utilize the bioprinter to control different types of interactions between bacteria that depend on their spatial position. We next show quorum-sensing-based chemical communication between the engineered sender and receiver bacteria placed at different positions inside the bioprinted structure and finally demonstrate the fabrication of barrier structures defined by nonmotile bacteria that can guide the movement of chemotactic bacteria inside a gel. We anticipate that a combination of 3D bioprinting and synthetic biological approaches will lead to the development of living biomaterials containing engineered bacteria as dynamic functional units.
Keywords: synthetic biology, bacteria, living biomaterials, bioprinting, bacterial communication, chemotaxis
1. Introduction
Additive manufacturing of biocompatible scaffold structures and 3D printing of gel-encapsulated mammalian cells have become popular approaches for the generation of spatially differentiated cell cultures and models of living tissues, which promise to have a wide range of biomedical applications.1−4 In the present work, we focus on bioprinting of bacterial systems, which is comparatively less developed but is expected to be of similar importance for the engineering of spatially organized bacterial communities and could ultimately lead to the realization of living, bacteria-based biomaterials.5−8 Spatial arrangement and confinement of bacterial communities also allows the study and control of their interactions with their environment, their surrounding scaffold matrix,9 or other communities under well-defined experimental conditions. This is of particular interest for applications in synthetic biology, which involve spatially distributed synthetic cell–cell communication and division of labor between different types of genetically engineered bacteria.
The ability to arrange engineered bacteria in general 3D geometries allows us to precisely set the initial and boundary conditions that dictate the evolution of genetically programmed dynamical systems in a spatial context. In particular, printing in 3D enables topologies that are impossible to achieve in 2D—e.g., core–shell geometries or 3D networks that avoid crossing of lines. While structured bacteria-laden thin 2D hydrogels are homogeneously influenced by environmental parameters such as gas supply, nutrients, or stress factors, structures extended in 3D can be designed (and can develop into) systems with richer internal structure and heterogeneity. As in naturally occurring systems, environmental parameters exhibit noticeable gradients over thicker biofilm structures, which can also provide positional information10 and therefore facilitate autonomous differentiation within the printed structures.
Functional biomaterials with biosensing11,12 or biocatalytic capabilities13 have been previously realized by immobilizing bacteria within various hydrogel-based support matrices, which in some cases also allowed a rough positioning of the bacteria in 3D. At a more macroscopic scale, hydrogel matrices were loaded with bacterial cultures to enable printing of biofilms for the degradation of pollutants,14 the realization of self-healing structures,6 and even bacterial electrodes.15
More recently, 3D bioprinting techniques were utilized to achieve better spatial control over bacteria-laden structures. A chaotic advection technique was developed that allowed continuous printing of multilayered filaments with a very precise definition of the interface between different live bacterial communities.16 A stereolithographic approach was used to structure biofilms into precisely defined, regular 3D-grid patterns17 with a potential application in environmental sensing. Laser-based lithography was utilized to contain bacterial cells within picoliter-sized gel compartments, which enabled the observation of cell–cell interactions and bacterial signaling between small populations.18 Bacteria were also encapsulated in a Pluronic F127 diacrylate-containing ink followed by UV cross-linking, which was used for the realization of printable bacteria-based biosensors.19 In a different approach,5 printed Bacillus subtilis spores were mixed with liquid agarose that started new bacterial colonies upon solidification of the gel and incubation after printing. The use of bacterial spores allowed printing at temperatures as high as 70 °C, which would not have been possible with living bacteria. An alternative approach20,21 comprises extrusion of homogeneous Escherichia coli biofilms using a custom-built 3D printer that is loaded with an alginate ink, which solidifies upon contact with a calcium-containing substrate. While this approach does not require high temperatures, only a few layers can be printed as diffusion of calcium ions is required for the solidification of the alginate gel. The majority of these examples resulted in homogeneous bacterial structures containing only a single bacterial clone.
Printing of living cells generally requires a compromise between printability and biocompatibility of the materials used for printing. Most approaches employed to print functional bacterial gels involve a curing step after printing, either using a chemical crosslinker,6,15,20,21 photo-cross-linking,18,19 or submersion in a CaCl216,22 or BaCl2 bath.23 The hydrogels used to encapsulate and print the bacteria had to be chosen specifically14 to facilitate printing at sufficiently low temperatures, which ensures cell viability and prevents sporulation of spore-forming bacteria.
Importantly, previous approaches discussed above were not used to execute engineered genetic functions within the bacteria and did not utilize their spatial organization within arbitrary voxel-defined 3D hydrogel structures. Such a capability is of major interest, however, as E. coli is the most developed genetic “workhorse” with a broad range of tools available for engineering biology.
To address this need, in the present work, we developed a gentle approach to print living E. coli bacteria embedded in a nontoxic hydrogel environment composed of an inexpensive mixture of alginate and agarose, which allowed extrusion-based printing at moderate temperatures and without the need for chemical cross-linking. The addition of calcium peroxide and the enzyme catalase24−26 to the gel ink ensured a continuous supply of the bacteria with oxygen within the printed gel over a time course of at least 24 h.
Using this method, we were able to sequentially print three different bacterial clones in a single layer and express fluorescent proteins at the respective locations. Dual extrusion of bacteria-free and bacteria-laden hydrogels enabled printing of bacteria at programmed positions within 3D structures of heights of up to 10 mm.
Bacterial bioprinting opens up the possibility to precisely define the boundary and initial conditions of dynamical systems composed of bacteria with genetically programmed interactions,27−31 which potentially allows combining the advantages of top-down patterning with self-organized spatiotemporal differentiation for the realization of living biomaterials.
To demonstrate this capability, we applied our bioprinting approach to control quorum-sensing-based bacterial communication between engineered sender and receiver bacteria placed at precisely defined locations. We further demonstrate that a bacteria-ink containing nonmotile bacteria can be used to create living boundaries that cannot be crossed by chemotactic bacteria and thus can be used to guide chemotaxis within a hydrogel.
2. Results and Discussion
2.1. Encapsulation of Living E. coli Bacteria in Printable Hydrogel Matrices
To establish a reliable biofabrication procedure that enables reproducible printing of living bacteria into any desired target structure, several challenges have to be addressed: first, the printing process is required to induce only minimal stress on the bacteria, which favors printing procedures that work at low temperatures, low pressure, and do not involve radical crosslinkers. Second, a nontoxic hydrogel composition has to be found that has good printability with fast solidification and high structural fidelity. Lastly, after extrusion, the printed bacterial gels have to be incubated for recovery and growth of the bacteria. To this end, the printed structures need to be embedded in a supportive environment that prevents shrinking, drying, or depletion of nutrients.
We first sought to find a printable hydrogel composition compatible with a moderate-temperature printing process using a custom-built gel extrusion printer (Figure 1A and Experimental Section),32 in which the highest continuously applied temperature does not exceed ≈42 °C, and which allows for fast postprint solidification by cooling to room temperature (i.e., not requiring a cross-linking step). E. coli bacteria are known to display strongly reduced cell growth after exposure to elevated temperatures during growth,33,34 with only some strains sustaining temperatures of up to 48.5 °C.35 Higher temperature generally results in bacterial cell death or—in the case of spore-forming bacteria such as B. subtilis(5)—sporulation, which precludes a direct extrusion of living cells at elevated temperatures.
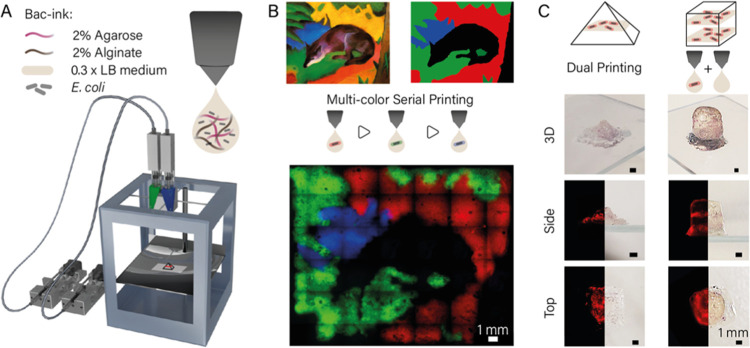
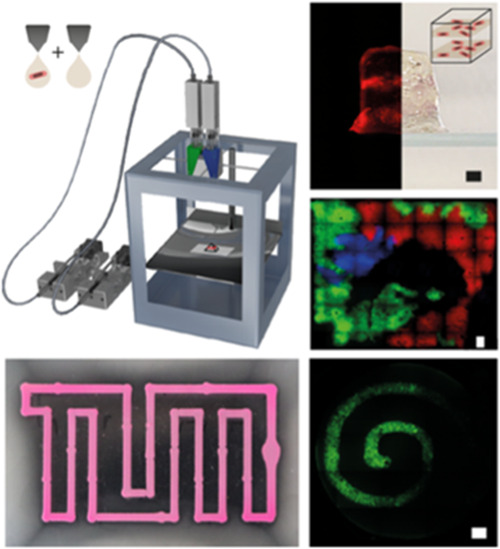
Figure 1.
(A) Schematic image of a custom-built dual extrusion gel printer32 used for bacteria printing. Two syringe pumps were hydraulically connected to a printhead holding two syringes, which were filled with bacteria-ink. The printhead was heated to 42 °C, and the gel quickly solidified upon cooling on the base plate. (B) Bacteria (E. coli DH5α) expressing either mRFP, mVenus, or mTurquoise were printed sequentially into hydrogel layers and incubated overnight to represent the painting “Blue Fox” by the Munich expressionist Franz Marc. The image demonstrates the resolution of ink deposition and the reproducibility of printing at specific locations. (C) Using dual extrusion with blank and bacteria-ink, three-dimensional gel objects were printed that contained single layers of bacteria. Images of the 3D structures (top and side view) were taken using a Canon 80D camera; fluorescence images were obtained with a Nikon Ti2-E microscope from the bottom and from slices of the structures.
We found a mixture of 2% agarose and 2% alginate to meet our requirements for a bacteria-ink, which are both hydrogels proven to be compatible with cell culture conditions.36 While alginate works as a viscosity enhancer, agarose provides fast solidification after extrusion and a good spatial resolution. To reduce osmotic stress, the bacteria-ink was prepared with 0.3 × LB medium, which was found to be an LB concentration compatible with printing at 42 °C.
Viscosity measurements indicate that the LB content has an appreciable influence on the temperature-dependent viscosity of the bacteria-ink mixture, which shows a steep decline in the temperature range from 20 to 35 °C (Figures S1 and S2). The bacteria themselves were found to have only a minor influence on the viscosity and printability of the gel ink (Figure S3).
During printing, the bulk of the gel is enclosed in the printhead, which works as a heat block at 42 °C and fixes the temperature of the bacteria-ink to 42 ± 1.5 °C within the sample syringe. By contrast, the tip of the nozzle is continuously exposed to room temperature, cooling the gel in this area to 36 ± 1.5 °C during the printing procedure. The temperature of the extruded gel droplets thus exactly matches the gelation temperature, while the main E. coli gel at 42 °C provides tolerable conditions as well as a comparatively low viscosity, which improves printability and reduces the strain on the cells. At higher LB concentrations, alginate cross-linking by the residual calcium in the medium results in gels with a larger variability in viscosity at the printing temperature and lower viscosities at room temperature (Figures S1 and S2). With bacteria-ink containing 0.3 × LB medium, however, we were able to print structures up to 30 mm in size in x-y and 10 mm in the z dimension. The final storage modulus of the solid gel was found to be on the order of 40 kPa after cooling to 20 °C. The printed gels showed moderate swelling (on the order of 10% in volume after 1 day, reduced to only 5% after 1 week) when submerged in media (Figure S4).
Printing of larger structures was only limited by longer printing times, which resulted in the drying of parts of the hydrogel during the printing process. As explained in more detail below, the expression and maturation of fluorescent proteins inside the printed gel were supported by an oxygen-generating system composed of calcium peroxide and catalase.
2.2. Bioprinting of Living Bacteria in 2D and 3D
We assessed the printing performance of the bacteria-ink using various test structures. We first investigated the spatial position of the bacteria over time by imaging a printed triangular structure (Figure S5) with 4x, 10x, and 60x magnification at a single position directly after printing and after 24 h of incubation. The total fluorescence signal increased considerably due to the growth of the mRFP-expressing bacteria. Printed bacteria appeared to remain at or close to their initial positions over time (Figure S5).
To demonstrate the spatial resolution and reproducibility of the printing procedure in 2D, bacteria that constitutively expressed mRFP, mVenus, and mTurquoise, respectively, were printed sequentially in the same plane (Figure 1B). After incubation at 37 °C for 24 h, the gel image is clearly visible in the three fluorescent channels.
Single voxels have a diameter of 770 ± 5 μm (corresponding to 120% of the outer nozzle diameter) and thus show a good print resolution for the type of nozzles employed. For the art print individual, sequentially printed color channels showed overlapping signals from two fluorescence channels only in the border regions between subsequent print layers. The overlaps were at widths of 50–500 μm, indicating the precision of the alignment of repeated printing steps with respect to each other (for comparison, the x-y precision of the custom print body is 12.5 μm for a one-step print job).
We further used the dual extrusion capability of our printer to generate a pyramid with one and a cuboid with two distinct bacterial bands within a single print job (Figure 1C). Ink containing mRFP producing bacteria and ink without bacteria were prepared and loaded into two separate glass syringes. The gels were printed alternatingly from the two separate syringes to form the 3D shapes with a base area of 6 mm × 6 mm each and a height of 5 mm (pyramid) and 10 mm (cuboid), respectively. Each bacterial band consisted of two layers printed with bacteria-containing gel. After 24 h of incubation at 37 °C in LB medium in a 10% Pluronic F127 sealed chamber, the printed structures were released from the Pluronic by dissolving the gel after 15 min storage at 4 °C. The bacterial bands show high RFP production and are clearly colored red. Photography and microscopy images from the bottom and side of the prints demonstrate the precise deposition of the bacterial layers within the complete gel structure. Sealing with Pluronic was used to prevent swelling of the gel prints (Figure S4), which was found to lead to a loss of bacteria at the boundaries of the printed structures.
2.3. Growth Conditions in the Hydrogel Matrix
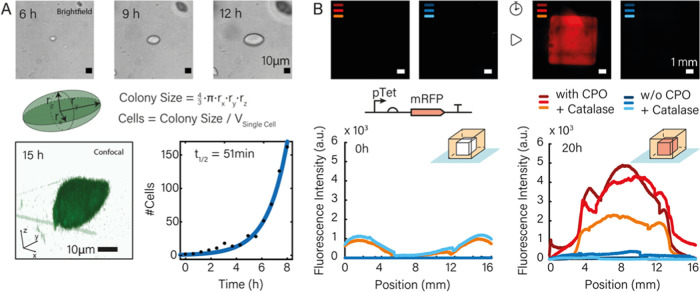
To gain a better understanding of the bacterial growth conditions inside the printed gels, we studied size and shape of the printed colonies for the first 8–15 h after extrusion using bright-field and confocal fluorescence microscopy (Figure 2). The colonies typically exhibited ellipsoidal, in some cases spherical shapes, whose volume grew roughly exponentially over time (Figure 2A). This finding is consistent with previous studies on the growth of bacteria gels, which found the formation of lenticular colonies in dense gel matrices such as 1% agar. The formation of such colonies is attributed to the splitting of the gel due to the pressure exerted by the growing bacteria along a fault line and a compression of the agar perpendicular to it.39 In the case of our gel mixture, a pore size of several 100 nm is expected,40 i.e., on the order of or slightly below the diameter of an E. coli bacterium.
Figure 2.
(A) Bacterial growth observed by bright-field and confocal fluorescence microscopy. Top row: Brightfield images of a microcolony at t = 6, 9, 12 h. The size of the colonies appeared roughly ellipsoidal in shape, which was confirmed by confocal fluorescence microscopy (bottom row, left). Cell numbers were determined for each time point by studying colony sizes in printed hydrogels for different time points (cf. Supporting Information 3.3). These numbers were used to determine in situ growth rates of the bacteria by fitting an exponential to the data (bottom row, right: data from confocal microscopy; cf. Figure S6 for data from bright-field microscopy). The bacteria displayed exponential growth over at least 8 h with a doubling time t1/2 around 50 min (experiments were performed in the presence of CPO/catalase). (B) For a good fluorescence readout, a continuous oxygen supply in the gel is necessary as the maturation of the majority of fluorescent proteins depends on oxygen.15,37,38 We casted two cuboid structures and sealed both in LB-ink to mimic the procedure applied for printed structures. With the addition of 0.1% calcium peroxide and 20 μg/mL of the enzyme catalase (final values), the fluorescence signal strongly increased during 20 h of growth at 37 °C, while in the absence of any oxygen generator, no fluorescence could be detected. Experimental triplicates are indicated by different shadings of the same color (red: with CPO + catalase, blue: without CPO and catalase). The graphs on the bottom show fluorescence intensity measured along a cross section of the printed gel cubes. The broadening of the profile of the cross section is caused by gel swelling.
We converted the volume of the colonies into a cell count assuming a single cell volume of 1.5 μm3, which we also experimentally confirmed directly after the printing process. Exponential fits to the cell number as a function of time resulted in doubling times around 50 min (cf. Figure S6 for experimental replicates).
In previous work on bacterial bioprinting,5 expression of fluorescent proteins was only observed at gel–air interfaces, which is explained by the lack of oxygen—required for FP maturation—inside the gel. As our studies required fluorescence monitoring also of bacteria deeply embedded in the gel, we utilized a calcium peroxide-based oxygen generation system, which is known as an additive from conventional cell culture.24−26 Within the gel, CPO degrades to hydrogen peroxide, whose conversion to oxygen and water is then catalyzed by the enzyme catalase. As shown in Figure 2B, we casted (6 mm)3-sized hydrogel cubes with bacteria containing the plasmid pTet-mRFP and induced the expression of mRFP via the addition of anhydrotetracycline (aTc) directly before mixing with bacteria-ink either with or without the addition of CPO and catalase. While after 20 h of incubation, the sample without CPO did not provide an appreciable fluorescence signal, the gels containing the oxygen-generating system showed a clear fluorescence signal in all triplicates. As a tradeoff, we found that the CPO/catalase mixture negatively affects bacterial cell growth, resulting in an increase of the doubling time of the bacteria by a factor of ≈2 compared to media without CPO/catalase (Figure S7), which most likely is caused by the oxidative stress imposed by the mixture. If fluorescence observation of the bacteria is not required, the addition of CPO/catalase can be omitted, leading to better growth conditions in the gel matrix.
2.4. Diffusion of Genetic Inducers
We next studied the diffusion of inducer molecules and the resulting spatiotemporal gene expression response of bacteria embedded in the bioprinted gel, for which we conducted experiments with aTc-inducible DH5αZ1 cells carrying a plasmid that encodes mRFP under the control of a pTet promoter (Figure 3A,B). The cells were printed into a circular shape with a diameter of 12 mm. Directly after the printing process, a 2 μL voxel containing aTc (a 1:1 mixture of 0.2 mg/mL aTc and gel ink, resulting in a final concentration of ≈0.2 μg/mL (or 0.43 μM) in the ≈ 1 mL reaction chamber) was pipetted into the center of the structure using a positive displacement pipette. The printed structure was then sealed and imaged continuously for 12 h at 37 °C. After incubation for 3–4 h, the fluorescence signal started to rise in the center while small colonies became visible in the bright-field images. After incubation for 10 h, large bacterial colonies were visible in the bright-field channel and the fluorescent protein was highly expressed by the bacteria.
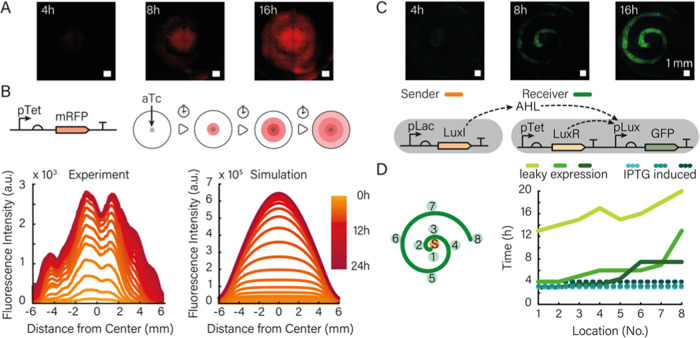
Figure 3.
(A) Diffusion of the genetic inducer aTc from the center of a printed gel and subsequent expression of mRFP by bacteria contained in the gel (scale bar 1 mm). (B) mRFP was expressed under the control of a pTet promoter and thus responds to aTc diffusion. The graph on the left shows the fluorescence intensity measured along the diameter of the bioprint for different time points. The observed dips in the graph are stitching artifacts. Simulations (right) that model aTc diffusion, bacterial growth, and gene expression are in good agreement with the experimental observations. (C) Fluorescence channel of 4× microscopy images of a sender–receiver system at 4, 8, and 16 h after the start of the experiment. The increase in fluorescence intensity is synchronous (time intervals of 10 min showed no differences) upon induction of the sender bacteria with IPTG and consequently with high AHL production. The corresponding gene circuit is shown below the micrographs. (D) Fluorescence time traces (right) corresponding to the locations indicated on the left for an experiment, in which the sender bacteria were not induced, and thus only a small amount of AHL is produced by leaky expression of LuxI from the Lac promoter.
The observed spatiotemporal dynamics of the fluorescence signal in the gel result from a combination of the diffusion of the inducer from the center into the gel print, bacterial cell growth throughout the print, and inducer concentration-dependent gene expression in the bacteria. As shown in Figure 3B, the rise of the fluorescence signal can be well recapitulated in a numerical simulation of these processes using experimentally realistic parameters for bacterial growth rate and expression dynamics (cf. Figures S6, S8, S9, and Supporting Information Section 4.1 on the numerical model). Screening of the value of the diffusion coefficient for aTc in simulations suggests a diffusivity on the order of 200 μm2/s within the gel (Figure S14).
2.5. Bacterial Communication in a Bioprinted Environment
Bioprinting with bacterial inks opens up the possibility to arrange different types of communicating or otherwise interacting bacteria into well-defined spatial relations. This is of particular interest for applications in synthetic biology, which require the precise control of the boundary conditions affecting the execution of spatiotemporal gene circuits27−30 by engineered bacteria. As an example for this type of application, we studied the dynamics of a synthetic bacterial sender–receiver system embedded in a bioprinted gel structure. The sender cells are DH5α cells hosting a plasmid coding for the 3OC6-HSL (AHL) synthase LuxI under the control of a Lac promoter (cf. materials and methods). Upon induction with IPTG, the cells express LuxI that consecutively produces the quorum-sensing signal AHL that can freely diffuse out of the bacteria and through the gel. The corresponding receiver cells contain a plasmid for the constitutive production of the AHL-dependent transcriptional activator LuxR, which controls the expression of GFP via the pLux promoter. The basic function of the sender–receiver system was first checked in bulk experiments using a plate reader (Figure S10).
To illustrate the spatial organization of the sender–receiver system, we then printed the receiver cells in the form of an Archimedean spiral (defined by r(ϕ)= aϕ, where r and ϕ are polar coordinates and a is a constant) and positioned the sender cells at its center (r = 0).
As expected, AHL produced by the senders activates the production of GFP by the receivers along the spiral (Figure 3C, Supporting Information Video 1). The signal typically increased over 10–16 h until nutrient depletion set in, followed by a decay in GFP fluorescence.
The behavior of the sender–receiver system differs from that of the aTc-inducer system due to the much higher sensitivity of LuxR activation to AHL (with a Kd on the order of only 10 nM). We therefore performed experiments both with fully induced senders and with uninduced senders that generated AHL via leaky expression of LuxI (and thus had a correspondingly lower “sender strength”41).
We found considerable variability among the sender–receiver experiments (Figure S11), which is likely related to the sensitivity of the dynamics with respect to variations in bacterial growth and the very low AHL concentration required for induction (cf. discussion in Section 4.2 of the Supporting Information). At low sender strength, in some cases, there was a clear differentiation in spatial response of the bacteria, with the closer receiver bacteria responding faster than the more remote ones (Figure 3D). In other cases, there was no clear spatial order or the complete receiver spiral started to produce GFP synchronously (Figure S11). Simulations indicate that in this case the AHL concentration throughout the gel had already reached nanomolar concentrations before the receivers started to grow appreciably (Figures S15 and S16).
2.6. Guiding Chemotaxis with Printed Boundaries
As a second example for spatial control over bacterial dynamics provided by the bacterial bioprinter, we investigated the motion of chemotactic bacteria42 in engineered environments. Chemotactic bacteria can move on top of (and to a certain extent also underneath) the surface of soft agar gels,43 propelling themselves toward areas of more favorable environmental conditions. When motile bacteria are inoculated in a spot in the middle of a soft agar plate, they concentrically migrate over the plate.39,42−44 The patterns are the result of an interplay of mass transport, chemotaxis, and metabolism44—the bacteria metabolize chemicals in the gel, creating a gradient of these chemicals, and chemotactically move in response to the gradient.
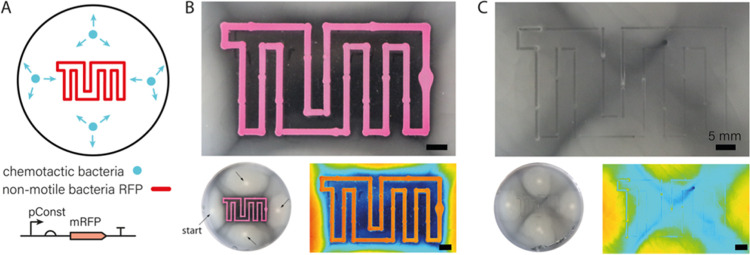
For our experiments, we prepared soft agar plates at agar concentrations between 0.22 and 0.24% (w/v) denser gels such as those used for 3D printing do not result in sufficient chemotactic movement of the bacteria. We then printed artificial boundaries in the shape of our university logo formed by RFP-producing nonmotile E. coli cells mixed with 1% alginate as printable ink into the soft agar (Figure 4A). The plates were inoculated with 2 μL aliquots of a suspension of a chemotactic E. coli strain (MG1655) placed at approximately equal distances from the outline of the logo. Within 20 h, the chemotactic cells grew into a thick biofilm over the agar surface, avoiding only the area enclosed by and between the printed borders. Apparently, the presence of the printed bacterial border region prevented the chemotactic bacteria from propagating into the interior of the logo, which was found to be only scarcely populated (Figures 4B and S12).
Figure 4.
(A) Artificial boundaries in the form of a TUM logo were printed into soft agar using nonmotile bacterial cells constitutively expressing RFP for better visualization. Directly after printing, chemotactic cells were inoculated at four locations (indicated with blue dots) and grown at 37 °C. (B) After 20 h of incubation, the printed boundaries showed a high intensity of RFP fluorescence. The chemotactic cells successfully spread over the whole surface outside of the boundaries—visible through the milky appearance of the gel—but do not enter the interior of the logo. Bottom left: The small image shows a view of the whole plate; the four bright inoculation points are indicated with arrows. Bottom right: High-contrast representation of the apparent bacterial density in the image above in heatmap coloring. Blue corresponds to low bacterial density, red to high density. (C) For comparison, the TUM logo was printed with gel ink not containing any bacteria. In this case, the bacteria spread over the whole plate without hindrance. Bottom left: View of the whole plate as in (B). Bottom right: High-contrast representation in heatmap coloring as in (B).
As for conventional chemotaxis experiments on soft agar plates,42,44 we assume that the chemotactic bacteria do not enter the border region due to the unfavorable nutrient conditions generated by the border bacteria, potentially combined with the physical obstruction caused by the nonmotile bacteria themselves, which cannot migrate into the agar deeply. Motile bacteria approaching the border either have to enter into the gel, below the nonmotile colony, or move sideways (in parallel to the printed border) on top of the agar. As the latter movement occurs with higher mobility, the bacteria preferentially spread in this direction. Our printed barrier structures thus guide the movement of the motile bacteria by shaping the concentration gradient to which they respond.
We found that the effectiveness of chemotactic guiding depends on the distance and topology of inoculated and printed colonies. For instance, when chemotactic bacteria are inoculated too close to a barrier structure, they reach the printed border before the nonmotile bacteria could grow sufficiently and deplete the agar around them, leading to penetration of the barrier. We also observe that chemotactic bacteria can move through a printed border when they are printed inside of a barrier structure (Figure S13). In this case, the motile bacteria must cross the barrier by moving into the soft agar below the barrier, as there is no alternative way to choose. In control experiments, in which a bacteria-free alginate boundary was printed, the chemotactic bacteria were able to completely fill the whole agar plate (Figures 4C and S12).
2.7. Discussion and Outlook
We anticipate that the spatial organization of live bacteria by 3D bioprinting will enable the generation of soft living biomaterials that are capable of simple forms of differentiation and other types of information processing.27,28 In such materials, bacteria can sense their mutual presence as well as gradients of environmental inputs and produce molecules in response to the information conveyed by these inputs in a spatially differentiated manner.
Communication between bacteria further supports spatial organization and division of labor between the bacteria. Systems of communicating bacteria with distributed functions benefit from our dual extrusion bioprinting approach, which enables the spatial positioning of bacteria of the same species containing different types of genetic circuits or also the realization of spatially organized consortia of different species.
Apart from applications in spatially distributed biocomputing,28,45 artificial pattern formation and the development of self-differentiating bacteria-based materials,27,29,30,46 bioprinted bacterial structures are expected to be of interest in a variety of other contexts such as environmental sensing and bioremediation.14,17 Printed structures may be used to model naturally occurring bacterial niches and study ecological interactions under controlled conditions.16 Similar systems might be useful to investigate the efficacy of antimicrobial agents under realistic conditions. To this end, the used biomaterials can be easily modified and the influence of 3D structure and geometry can be addressed by custom hydrogel patterning during the printing process.
Extrusion of different types of bacteria could even be used for the spatial organization of cocultures that cooperatively perform more complex biosynthetic tasks. This would allow coculturing of bacterial species with conflicting growth requirements—e.g., E. coli and cyanobacteria—and to optimize sizes and distances between the cooperators to optimize metabolic flux through such systems. It is also conceivable to develop bioprinting approaches that enable the simultaneous printing of mammalian and microbial cells, which is of interest for the study of host–pathogen interactions and screening for adequate cures.
One of the major challenges for the realization of “living” biomaterials will be to supply the printed bacteria with nutrients and remove waste products efficiently. Submersion into liquid media can provide nutrients by diffusion into the gels, at least for small structures, but will result in swelling and a potential loss of bacteria from the gel to the surroundings. Using an encapsulation strategy to contain and stabilize the printed gels as in the present work will inevitably limit the lifetime of the system. To generate much larger, spatially organized structures, dedicated supply lines embedded into the 3D-printed gels might be required that ensure cell viability for extended periods of time.
3. Conclusions
In conclusion, we have demonstrated a simple approach toward extrusion-based 3D printing of living hydrogel materials using a bacteria-laden ink based on a mixture of agarose and alginate. This inexpensive gel mixture has several advantages for applications in synthetic biology—alginate and agarose are nontoxic compounds commonly available in molecular biology labs. Gel formation does not require chemical or photo-cross-linking, and the gel mix is compatible with mild low-temperature extrusion conditions that ensure the survival of the bacteria throughout the whole printing process and thereafter. Specifically, E. coli cells were shown to grow into extended microcolonies within the gel for at least 10–15 h. Importantly, the addition of an oxygen-generating system based on calcium peroxide and catalase enabled the expression of fluorescent proteins deep within the gel, an advantage that has to be weighed against an observed reduction in cell growth caused by these compounds.
We showed the possibility of dual extrusion of hydrogels into arbitrary, voxel-defined 3D shapes and sequential extrusion of hydrogels containing different types of bacteria, both with a voxel size of 770 μm at 50–500 μm spatial resolution. Notably, the ability to print living E. coli bacteria opens up the possibility to spatially organize genetically engineered, communicating, and interacting bacteria in a programmable and reproducible manner, which is of great interest for applications in synthetic biology. We demonstrated this capability by controlling spatiotemporal gene expression dynamics using diffusing inducer molecules and in the context of a bacterial sender–receiver system. We also showed that bacteria-ink loaded with nonmotile bacteria can be used to define and grow living boundaries, which influence the chemotactic movement of motile bacteria coembedded in the hydrogel. We anticipate that similar systems that combine 3D bioprinting approaches with synthetic biology could be developed into living biomaterials that contain engineered bacteria as functional units for sensing, actuation, and bioproduction.
4. Experimental Section
4.1. Bioprinter
Bioprinting of bacteria-containing hydrogels was performed using a custom 3D printer setup based on a commercially available Ultimaker Original+ modified with custom-built components.32 Hydrogel is extruded from heated printheads via hydraulically coupled syringe pumps. To increase the variety of structures that can be manufactured, the printer was equipped with two extruders. The extruders can be loaded with any pair of hydrogels that can be printed at the same temperature. Both extruders are directly controlled by the Ultimaker board. Executable GCODEs for all structures were generated by our Python-based software (available on https://github.com/julia-mueller/bioprinter/).
4.2. Bacterial Strains and Culture Conditions
For all experiments, E. coli strains (DH5α, DH5αZ1, BL21, MG1655) with different plasmids were cultured from glycerol stocks and grown overnight in LB medium containing selective antibiotic (carbenicillin, Carb) in an incubator shaker (Innova44, New Brunswick) at 37 °C and 250 rpm. On the following day, cells were diluted 1:100 in LB medium containing carbenicillin and cultivated to an OD of 0.6. Prior to printing, the cells were centrifuged at 2000 rcf at 4 °C for 2 min, and the pellet was resuspended in fresh LB medium containing carbenicillin. The chemotactic bacteria used in Figure 4 were MG1655, while the nonmotile bacteria were DH5α.
4.3. Plasmids Coding for Fluorescent Proteins
High copy number plasmids (iGEM part pSB1A3) containing a constitutive promoter (iGEM part J23106) and the coding sequence for mVenus (YFP), or mTurquoise-2 (CFP), were transformed to DH5α cells for constitutive protein expression. The coding sequence for mRFP under a pTet promoter (iGEM part I13521) on a high copy number plasmid (iGEM part pSB1A3) was transformed to DH5alpha, which does not express a TetR, for “constitutive” and to DH5αZ1 cells for aTc-inducible protein expression.
4.4. Sender–Receiver Plasmids
The sender plasmid (iGEM part K116638) is a high copy number plasmid (iGEM part pSB1A2) and contains a pLac promoter (iGEM part R0010) controlling the AHL synthase LuxI (iGEM part C0061) and was transformed into DH5α cells for IPTG-inducible AHL expression. Leaky expression of LuxI and thus the production of AHL can occur as there are small amounts of Lactose present in LB medium. The receiver plasmid is a high copy number plasmid (iGEM part pSB1A3) that contains an expression cassette coding for the transcription factor LuxR (C0062) regulated by a Tet promoter (iGEM part R0040), and a second cassette coding for GFP (iGEM part E0040) regulated by a AHL inducible Lux promoter (pLux). We transformed this plasmid to BL21(DE3)pLysS.
4.5. Bacteria-Ink
For our bacteria-ink, we used final concentrations of 2% agarose and 2% alginate in 0.3 × LB medium containing (final) concentrations of 0.1% calcium peroxide (CPO) and 20 μg/mL catalase. The gels formed without the addition of calcium (in addition to the CPO).
4.6. Bioprinting of Bacteria
Mixed bacteria-ink was prepared in a microwave oven and kept at 70 °C for 15 min in an oven prior to mixing with bacteria culture. One milliliter of a liquid bacterial culture at OD 0.6 was mixed with 2 mL of bacteria-ink that was enriched with 60 μL of catalase (end concentration 20 μg/mL) and 200 μL of CPO (end concentration 0.1% w/v). The mixed bacteria-ink was loaded into the printhead that was preheated to the printing temperature. Extrusion on glass slides (microscopy slides 76 mm × 26 mm × 1.5 mm, VWR, Germany, and Menzel slides 76 mm x 26 mm, #1.5, Thermo Scientific, Germany) was performed with nozzle No. G23 (ID 330 μm, OD 640 μm) (Vieweg, Germany). For dual extrusion, a second gel was prepared and loaded into a second glass syringe in the printhead. Prints on agar plates were performed with a mixture of 500 μL of liquid bacterial culture at OD 0.6 and 500 μL 2% (w/v) alginate loaded to the printhead and extruded at 37 °C with nozzle No. G25 (ID 250 μm, OD 510 μm) (Vieweg, Germany).
After printing, the extruded structures were sealed with a custom 3D-printed plastic frame, LB-bacteria-ink, and closed with a glass lid. Directly after sealing, imaging with an epifluorescence microscope (Nikon Ti2-E) was started. For different magnifications, a 4×, 10×, and 60× objectives were employed. For larger structures (>1 mm), multipoint imaging with 4× magnification and consecutive stitching with 0% overlap of single images was performed.
4.7. Sample Sealing
All samples contain between 0.1 and 0.3 mL of bacteria-ink extruded in thin lines and are thus extremely prone to drying and shrinking during incubation at 37 °C. As drying of the hydrogel leads to cell death, drying and consecutive shrinking (cf. Figure S4B) was prevented using custom 3D-printed plastic chambers (polylactic acid, BASF, Germany) and glass cover slides (round 18 mm diameter and square 15 mm, #1.5, Carl Roth, Germany) that are attached to the cover slides with instant glue.
4.8. Large-Sample Stabilization
For stabilization during incubation of larger structures (pyramid and cuboid with bacterial layers), the chambers were filled up with 10% Pluronic F127 (Sigma-Aldrich, Germany) in LB medium. This inverse thermostable polymer stabilizes higher structures as its viscosity enhances extremely while incubating at 37 °C and can be removed as it liquefies after 15 min at 4 °C. As shown in early work on bacteria cultures on Pluronic F127 as agar substitute, bacteria are able to grow on this polymer.47,48 However, higher concentrations of Pluronic lead to cell death if the cells are exposed to Pluronic for several days (this polymer is also used as an antifouling agent used for long-term antibiofilm surface treatment49). With a maximum of 24 h incubation of bacteria-ink structures using low concentrations of this stabilizing agent, we ensured cell growth on the printed hydrogel structure without contamination of the surrounding auxiliary structure by washed-out cells.50
4.9. Nutrient and Oxygen Supply
To prevent drying and to ensure a continuous supply with nutrients, we embedded all printed structures in bacteria-ink in 1 × LB medium, which provides enough nutrients to feed the enclosed bacteria for at least 24 h. Sealing of the structures reduces the supply of the samples with oxygen, which is required, however, for the maturation of the fluorescent proteins used for optical readout,38,51 in particular for red fluorescent proteins.37 We therefore augmented our bacteria-containing ink with 0.1% (w/v) calcium peroxide (CPO) and 20 μg/mL catalase, which has been found sufficient to supply cell cultures with oxygen for over a week.26,52 CPO slowly decays into hydrogen peroxide at room temperature, while catalase promotes the conversion of H2O2 to oxygen and water.
Acknowledgments
The authors would like to thank Benedikt Buchmann (Prof. A. Bausch) and Maria Bauer (Prof. O. Lieleg) for their help with rheology measurements and confocal imaging.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.1c20836.
Experimental protocols, description of bacterial strains and gene sequences, additional experimental data including rheology measurements (Figures S1–S4); cell growth characterization (Figures S5–S7), gene expression in gels (Figures S8 and S9); sender–receiver communication (Figures S10 and S11); guided chemotaxis (Figures S12 and S13); details on modeling of the gene circuits (Figures S14–S16); Supporting Information references (PDF)
Author Contributions
F.C.S. directed the research; J.M, E.F. and F.C.S. conceived of the idea and designed the research. J.M., J.R., and A.J. performed 3D printing: J.M. and J.R. for diffusion studies, fluorescence experiments, and screening of printing parameters; J.M. for colony growth and sender–receiver experiments; and A.J. for experiments with dual extrusion and with chemotactic cells with the help of M.E. E.F. carried out cloning of plasmids for fluorescent readout. J.M. and A.J. developed the python software to generate printable GCODE. J.M., E.F., J.R., A.J., M.E., and F.C.S. analyzed the data and discussed the results. J.M and F.C.S. wrote the manuscript with the help of E.F. and A.J.
This work was funded by the European Research Council (project AEDNA, grant no. 694410). The authors gratefully acknowledge funding by the Bavarian Ministry of Science and the Arts through the ONE MUNICH Project “Munich Multiscale Biofabrication.”
The authors declare no competing financial interest.
Supplementary Material
References
- Murphy S. V.; Atala A. 3d Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785. 10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- Dey M.; Ozbolat I. T. 3d Bioprinting of Cells, Tissues and Organs. Sci. Rep. 2020, 10, 14023 10.1038/s41598-020-70086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B. S.; Das S.; Jang J.; Cho D.-W. Decellularized Extracellular Matrix-Based Bioinks for Engineering Tissue- and Organ-Specific Microenvironments. Chem. Rev. 2020, 120, 10608–10661. 10.1021/acs.chemrev.9b00808. [DOI] [PubMed] [Google Scholar]
- Kim H.; Kang B.; Cui X.; Lee S. H.; Lee K.; Cho D. W.; Hwang W.; Woodfield T. B. F.; Lim K. S.; Jang J. Light-Activated Decellularized Extracellular Matrix-Based Bioinks for Volumetric Tissue Analogs at the Centimeter Scale. Adv. Funct. Mater. 2021, 31, 2170231 10.1002/adfm.202170231. [DOI] [Google Scholar]
- González L. M.; Mukhitov N.; Voigt C. A. Resilient Living Materials Built by Printing Bacterial Spores. Nat. Chem. Biol. 2020, 16, 126–133. 10.1038/s41589-019-0412-5. [DOI] [PubMed] [Google Scholar]
- Huang J.; Liu S.; Zhang C.; Wang X.; Pu J.; Ba F.; Xue S.; Ye H.; Zhao T.; Li K.; Wang Y.; Zhang J.; Wang L.; Fan C.; Lu T. K.; Zhong C. Programmable and Printable Bacillus Subtilis Biofilms as Engineered Living Materials. Nat. Chem. Biol. 2019, 15, 34–41. 10.1038/s41589-018-0169-2. [DOI] [PubMed] [Google Scholar]
- Balasubramanian S.; Aubin-Tam M. E.; Meyer A. S. 3d Printing for the Fabrication of Biofilm-Based Functional Living Materials. ACS Synth. Biol. 2019, 8, 1564–1567. 10.1021/acssynbio.9b00192. [DOI] [PubMed] [Google Scholar]
- Gilbert C.; Tang T.-C.; Ott W.; Dorr B. A.; Shaw W. M.; Sun G. L.; Lu T. K.; Ellis T. Living Materials with Programmable Functionalities Grown from Engineered Microbial Co-Cultures. Nat. Mater. 2021, 20, 691–700. 10.1038/s41563-020-00857-5. [DOI] [PubMed] [Google Scholar]
- Kandemir N.; Vollmer W.; Jakubovics N. S.; Chen J. Mechanical Interactions between Bacteria and Hydrogels. Sci. Rep. 2018, 8, 10893 10.1038/s41598-018-29269-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuis J. O.; Tkacik G.; Wieschaus E. F.; Gregor T.; Bialek W. Positional Information, in Bits. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 16301–8. 10.1073/pnas.1315642110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettaieb F.; Ponsonnet L.; Lejeune P.; Ouada H. B.; Martelet C.; Bakhrouf A.; Jaffrezic-Renault N.; Othmane A. Immobilization of E. coli Bacteria in Three-Dimensional Matrices for Isfet Biosensor Design. Bioelectrochemistry 2007, 71, 118–25. 10.1016/j.bioelechem.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Shin H. J. Agarose-Gel-Immobilized Recombinant Bacterial Biosensors for Simple and Disposable on-Site Detection of Phenolic Compounds. Appl. Microbiol. Biotechnol. 2012, 93, 1895–904. 10.1007/s00253-011-3700-x. [DOI] [PubMed] [Google Scholar]
- Trelles J. A.; Bentancor L.; Schoijet A.; Porro S.; Lewkowicz E. S.; Sinisterra J. V.; Iribarren A. M. Immobilized Escherichia Coli Bl21 as a Catalyst for the Synthesis of Adenine and Hypoxanthine Nucleosides. Chem. Biodivers. 2004, 1, 280–288. 10.1002/cbdv.200490024. [DOI] [PubMed] [Google Scholar]
- Schaffner M.; Ruhs P. A.; Coulter F.; Kilcher S.; Studart A. R. 3d Printing of Bacteria into Functional Complex Materials. Sci. Adv. 2017, 3, eaao6804 10.1126/sciadv.aao6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freyman M. C.; Kou T.; Wang S.; Li Y. 3d Printing of Living Bacteria Electrode. Nano Res. 2020, 13, 1318–1323. 10.1007/s12274-019-2534-1. [DOI] [Google Scholar]
- Ceballos-González C. F.; Bolívar-Monsalve E. J.; Quevedo-Moreno D. A.; Lam-Aguilar L. L.; Borrayo-Montaño K. I.; León J. F. Y.-d.; Zhang Y. S.; Alvarez M. M. s.; Santiago G. T.-d. High-Throughput and Continuous Chaotic Bioprinting of Spatially Controlled Bacterial Microcosms. ACS Biomater. Sci. Eng. 2021, 7, 2408–2419. 10.1021/acsbiomaterials.0c01646. [DOI] [PubMed] [Google Scholar]
- Dubbin K.; Dong Z.; Park D. M.; Alvarado J.; Su J.; Wasson E.; Robertson C.; Jackson J.; Bose A.; Moya M. L.; Jiao Y.; Hynes W. F. Projection Microstereolithographic Microbial Bioprinting for Engineered Biofilms. Nano Lett. 2021, 21, 1352–1359. 10.1021/acs.nanolett.0c04100. [DOI] [PubMed] [Google Scholar]
- Connell J. L.; Ritschdorff E. T.; Whiteley M.; Shear J. B. 3d Printing of Microscopic Bacterial Communities. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 18380–18385. 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Yuk H.; Lin S.; Parada G. A.; Tang T. C.; Tham E.; de la Fuente-Nunez C.; Lu T. K.; Zhao X. 3d Printing of Living Responsive Materials and Devices. Adv. Mater. 2018, 30, 1704821 10.1002/adma.201704821. [DOI] [PubMed] [Google Scholar]
- Schmieden D. T.; Basalo Vazquez S. J.; Sanguesa H.; van der Does M.; Idema T.; Meyer A. S. Printing of Patterned, Engineered E. coli Biofilms with a Low-Cost 3d Printer. ACS Synth. Biol. 2018, 7, 1328–1337. 10.1021/acssynbio.7b00424. [DOI] [PubMed] [Google Scholar]
- Lehner B. A. E.; Schmieden D. T.; Meyer A. S. A Straightforward Approach for 3d Bacterial Printing. ACS Synth. Biol. 2017, 6, 1124–1130. 10.1021/acssynbio.6b00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Madero C.; Len-Derby M. D. d.; Samandari M.; Ceballos-Gonzlez C. F.; Bolvar-Monsalve E. J.; Mendoza-Buenrostro C.; Holmberg S.; Garza-Flores N. A.; Almajhadi M. A.; Gonzlez-Gamboa I.; Len J. F. Y.-d.; Martnez-Chapa S. O.; Rodrguez C. A.; Wickramasinghe H. K.; Madou M.; Dean D.; Khademhosseini A.; Zhang Y. S.; Alvarez M. M.; Santiago G. T.-d. Using Chaotic Advection for Facile High-Throughput Fabrication of Ordered Multilayer Micro- and Nanostructures: Continuous Chaotic Printing. Biofabrication 2020, 12, 035023 10.1088/1758-5090/ab84cc. [DOI] [PubMed] [Google Scholar]
- Ning E.; Turnbull G.; Clarke J.; Picard F.; Riches P.; Vendrell M.; Graham D.; Wark A. W.; Faulds K.; Shu W. 3d Bioprinting of Mature Bacterial Biofilms for Antimicrobial Resistance Drug Testing. Biofabrication 2019, 11, 045018 10.1088/1758-5090/ab37a0. [DOI] [PubMed] [Google Scholar]
- Oh S. H.; Ward C. L.; Atala A.; Yoo J. J.; Harrison B. S. Oxygen Generating Scaffolds for Enhancing Engineered Tissue Survival. Biomaterials 2009, 30, 757–762. 10.1016/j.biomaterials.2008.09.065. [DOI] [PubMed] [Google Scholar]
- Pedraza E.; Coronel M. M.; Fraker C. A.; Ricordi C.; Stabler C. L. Preventing Hypoxia-Induced Cell Death in Beta Cells and Islets Via Hydrolytically Activated, Oxygen-Generating Biomaterials. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 4245–4250. 10.1073/pnas.1113560109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemdar N.; Leijten J.; Camci-Unal G.; Hjortnaes J.; Ribas J.; Paul A.; Mostafalu P.; Gaharwar A. K.; Qiu Y.; Sonkusale S.; Liao R.; Khademhosseini A. Oxygen-Generating Photo-Cross-Linkable Hydrogels Support Cardiac Progenitor Cell Survival by Reducing Hypoxia-Induced Necrosis. ACS Biomater. Sci. Eng. 2017, 3, 1964–1971. 10.1021/acsbiomaterials.6b00109. [DOI] [PubMed] [Google Scholar]
- Tabor J. J.; Salis H. M.; Simpson Z. B.; Chevalier A. A.; Levskaya A.; Marcotte E. M.; Voigt C. A.; Ellington A. D. A Synthetic Genetic Edge Detection Program. Cell 2009, 137, 1272–1281. 10.1016/j.cell.2009.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamsir A.; Tabor J. J.; Voigt C. A. Robust Multicellular Computing Using Genetically Encoded nor Gates and Chemical ’Wires’. Nature 2011, 469, 212–215. 10.1038/nature09565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant P. K.; Dalchau N.; Brown J. R.; Federici F.; Rudge T. J.; Yordanov B.; Patange O.; Phillips A.; Haseloff J. Orthogonal Intercellular Signaling for Programmed Spatial Behavior. Mol. Syst. Biol. 2016, 12, 849–13. 10.15252/msb.20156590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaerli Y.; Munteanu A.; Gili M.; Cotterell J.; Sharpe J.; Isalan M. A Unified Design Space of Synthetic Stripe-Forming Networks. Nat. Commun. 2014, 5, 4905 10.1038/ncomms5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Moreno J.; Tasiudi E.; Stelling J.; Schaerli Y. Multistable and Dynamic Crispri-Based Synthetic Circuits. Nat. Commun. 2020, 11, 2746 10.1038/s41467-020-16574-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J.; Jäkel A. C.; Schwarz D.; Aufinger L.; Simmel F. C. Programming Diffusion and Localization of DNA Signals in 3d-Printed DNA-Functionalized Hydrogels. Small 2020, 16, 2001815 10.1002/smll.202001815. [DOI] [PubMed] [Google Scholar]
- Bronikowski A. M.; Bennett A. F.; Lenski R. E. Evolutionary Adaptation to Temperature. Viii. Effects of Temperature on Growth Rate in Natural Isolates of Escherichia Coli and Salmonella Enterica from Different Thermal Environments. Evolution 2001, 55, 33–40. 10.1111/j.0014-3820.2001.tb01270.x. [DOI] [PubMed] [Google Scholar]
- Cooper V. S.; Bennett A. F.; Lenski R. E. Evolution of Thermal Dependence of Growth Rate of Escherichia Coli Populations During 20,000 Generations in a Constant Environment. Evolution 2001, 55, 889–96. 10.1554/0014-3820(2001)055[0889:EOTDOG]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Rudolph B.; Gebendorfer K. M.; Buchner J.; Winter J. Evolution of Escherichia Coli for Growth at High Temperatures. J. Biol. Chem. 2010, 285, 19029–19034. 10.1074/jbc.M110.103374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Marcial G. R.; Zeng A. Y.; Osuna C.; Dennis J.; García J. M.; O’Connell G. D. Agarose-Based Hydrogels as Suitable Bioprinting Materials for Tissue Engineering. ACS Biomater. Sci. Eng. 2018, 4, 3610–3616. 10.1021/acsbiomaterials.8b00903. [DOI] [PubMed] [Google Scholar]
- Strack R. L.; Strongin D. E.; Mets L.; Glick B. S.; Keenan R. J. Chromophore Formation in Dsred Occurs by a Branched Pathway. J. Am. Chem. Soc. 2010, 132, 8496–505. 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien R. Y. The Green Fluorescent Protein. Annu. Rev. Biochem. 1998, 67, 509–44. 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- Mitchell A. J.; Wimpenny J. W. T. The Effects of Agar Concentration on the Growth and Morphology of Submerged Colonies of Motile and Non-Motile Bacteria. J. Appl. Microbiol. 1997, 83, 76–84. 10.1046/j.1365-2672.1997.00192.x. [DOI] [PubMed] [Google Scholar]
- Pernodet N.; Maaloum M.; Tinland B. Pore Size of Agarose Gels by Atomic Force Microscopy. Electrophoresis 1997, 18, 55–58. 10.1002/elps.1150180111. [DOI] [PubMed] [Google Scholar]
- Ramalho T.; Meyer A.; Muckl A.; Kapsner K.; Gerland U.; Simmel F. C. Single Cell Analysis of a Bacterial Sender-Receiver System. PLoS One 2016, 11, e0145829 10.1371/journal.pone.0145829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler J. Chemotaxis in Bacteria. J. Supramol. Str. Cell 1976, 4, 305–317. 10.1002/jss.400040302. [DOI] [PubMed] [Google Scholar]
- Croze O. A.; Ferguson G. P.; Cates M. E.; Poon W. C. Migration of Chemotactic Bacteria in Soft Agar: Role of Gel Concentration. Biophys. J. 2011, 101, 525–34. 10.1016/j.bpj.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J.; Berg H. C. Migration of Bacteria in Semisolid Agar. Proc. Natl. Acad. Sci. U.S.A. 1989, 86, 6973–6977. 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. E.; Endy D. Engineered Cell-Cell Communication Via DNA Messaging. J. Biol. Eng. 2012, 6, 16 10.1186/1754-1611-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuñez I. N.; Matute T. F.; Valle I. D. D.; Kan A.; Choksi A.; Endy D.; Haseloff J.; Rudge T. J.; Federici F. Artificial Symmetry-Breaking for Morphogenetic Engineering Bacterial Colonies. ACS Synth. Biol. 2017, 6, 256–265. 10.1021/acssynbio.6b00149. [DOI] [PubMed] [Google Scholar]
- Gardener S.; Jones J. G. A New Solidifying Agent for Culture Media Which Liquefies on Cooling. J. Gen. Microbiol. 1984, 130, 731–733. 10.1099/00221287-130-3-731. [DOI] [Google Scholar]
- Jayaraman S.; Gunasekaran M. Use of a Synthetic Polymer (Pluronic F-127) as an Agar Substitute in Culture Medium to Detect and Measure Microbial Enzyme Production. Biotechnol. Technol. 1991, 5, 483–484. 10.1007/BF00155499. [DOI] [Google Scholar]
- Portolés M.; et al. Poloxarner 407 as a Bacterial Adhesive for Hydrogel Contact Lenses. J. Biomed. Mater. Res. 1994, 28, 303–309. 10.1002/jbm.820280304. [DOI] [PubMed] [Google Scholar]
- Khattak S. F.; Bhatia S. R.; Roberts S. C. Pluronic F127 as a Cell Encapsulation Material: Utilization of Membrane-Stabilizing Agents. Tissue Eng. 2005, 11, 974–983. 10.1089/ten.2005.11.974. [DOI] [PubMed] [Google Scholar]
- Balleza E.; Kim J. M.; Cluzel P. Systematic Characterization of Maturation Time of Fluorescent Proteins in Living Cells. Nat. Methods 2018, 15, 47–51. 10.1038/nmeth.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leicht U.; Volkmer E.; Wiese H.; Gutbrod M.; Schieker M.. Hydrogels as Cell Carriers for Tissue Engineering Calcium Peroxide Used as an Oxygen Releasing Additive, Presents Application Note, 2011.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.