Abstract
Spatiotemporal allostery is the source of complex but ordered biological phenomena. To identify the structural basis for allostery that drives the cyanobacterial circadian clock, we crystallized the clock protein KaiC in four distinct states, which cover a whole cycle of phosphor-transfer events at Ser431 and Thr432. The minimal set of allosteric events required for oscillatory nature is a bidirectional coupling between the coil-to-helix transition of the Ser431-dependent phospho-switch in the C-terminal domain of KaiC and adenosine 5′-diphosphate release from its N-terminal domain during adenosine triphosphatase cycle. An engineered KaiC protein oscillator consisting of a minimal set of the identified master allosteric events exhibited a monophosphorylation cycle of Ser431 with a temperature-compensated circadian period, providing design principles for simple posttranslational biochemical circadian oscillators.
Coupling between a phospho-switch and KaiC ATPase-dependent nucleotide exchange drives the cyanobacterial circadian clock.
INTRODUCTION
The cooperative nature of allosteric regulation is a source of nonlinearity that gives rise to oscillatory phenomena in diverse cellular functions (1, 2). Circadian clock systems are a typical example, wherein clock proteins are posttranslationally phosphorylated at multiple sites in a programmed (3–5) or pseudo-random manner (6, 7) as a means to allosterically regulate the stability of heteromultimeric complexes of clock proteins (8, 9), delays for feedback loops (6), and period length (10). Accordingly, a great deal of effort has been devoted to characterizing the phosphorylation-dependent allosteric structural changes in the clock proteins along the circadian reaction coordinate (8, 9, 11–15).
The cyanobacterium Synechococcus elongatus PCC7942 is one of the simplest prokaryotic model organisms (16). Only two adjacent amino acid residues, S431 and T432, in the core clock protein KaiC are phosphorylation targets (17, 18). In the presence of KaiA, KaiB, and adenosine 5′-triphosphate (ATP), KaiC exhibits a phosphorylation cycle in vitro (P-cycle): ST → SpT → pSpT → pST → ST, where S, T, pS, and pT indicate S431, T432, phosphorylated S431 (pS431), and phosphorylated T432 (pT432), respectively (17–19). Whereas the KaiC P-cycle has been investigated in vivo (20), in vitro (19, 21), and in silico (18, 22–24), little is known about the allosteric regulation of the KaiC P-cycle because all KaiC structures reported to date share nearly identical conformations at the phosphorylation sites, irrespective of the presence or absence of phosphoryl modifications (8, 9, 14, 15).
RESULTS
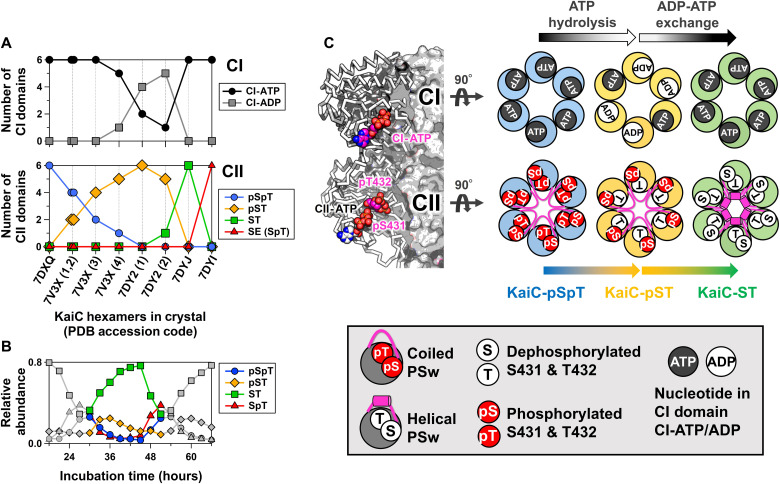
To visualize the structural basis for allosteric oscillatory regulation, we crystallized the KaiC hexamer in eight distinct states and sorted them from the fully phosphorylated KaiC-pSpT to the fully dephosphorylated KaiC-ST so that fractional changes in pS431 and pT432 per hexamer in the crystalline phase (Fig. 1A) reproduce those observed during the circadian cycle in solution (Fig. 1B). Each subunit of the KaiC hexamer consists of an N-terminal adenosine triphosphatase (ATPase) (CI) domain (11, 25) and a C-terminal autokinase/autophosphatase (CII) domain (14, 21) (Fig. 1C, left). Dephosphorylation of pT432 in the fully KaiC-pSpT hexamer occurred in a stepwise manner, but no specific order was observed in regard to which subunit was dephosphorylated at each step (fig. S1). However, systematic adenosine 5′-diphosphate (ADP) accumulation was observed for the CI domain (CI-ADP) of KaiC-pST (Fig. 1, A and C, and fig. S1). The accumulated CI-ADP molecules were replaced with CI-ATP during the transition from KaiC-pST to KaiC-ST (Fig. 1C). These results indicate that the autodephosphorylation events in the CII domain are linked to CI-ATP hydrolysis (ADP production) and CI-ADP exchange.
Fig. 1. KaiC crystal structures sorted by phosphorylation, ATP hydrolysis, and nucleotide exchange events.
(A) Fractional changes for ATP/ADP bound in the CI domain (top) and phosphorylation status of S431 and T432 in the CII domain (bottom) in the crystalline phase. Horizontal axis depicts the Protein Data Bank (PDB) accession codes for nine KaiC hexamers crystallized in eight distinct states. For 7V3X and 7DY2, four and two hexamers present in each asymmetric unit are distinguished by the number in parentheses. Vertical axes represent the number of CI and CII domains that are in the ATP/ADP-bound and phosphorylated/dephosphorylated states, respectively, in the hexamer of interest. (B) Phosphorylation cycle for KaiC in solution. (C) Schematic drawings for three representative hexamers for KaiC-pSpT (7DXQ), KaiC-pST (7DY2), and KaiC-ST (7DYJ). Arrows above and below the hexameric rings represent biochemical reactions and state transitions, respectively, that are suggested by the sorted structures. The nomenclatures used in the drawings are defined in the bottom panel.
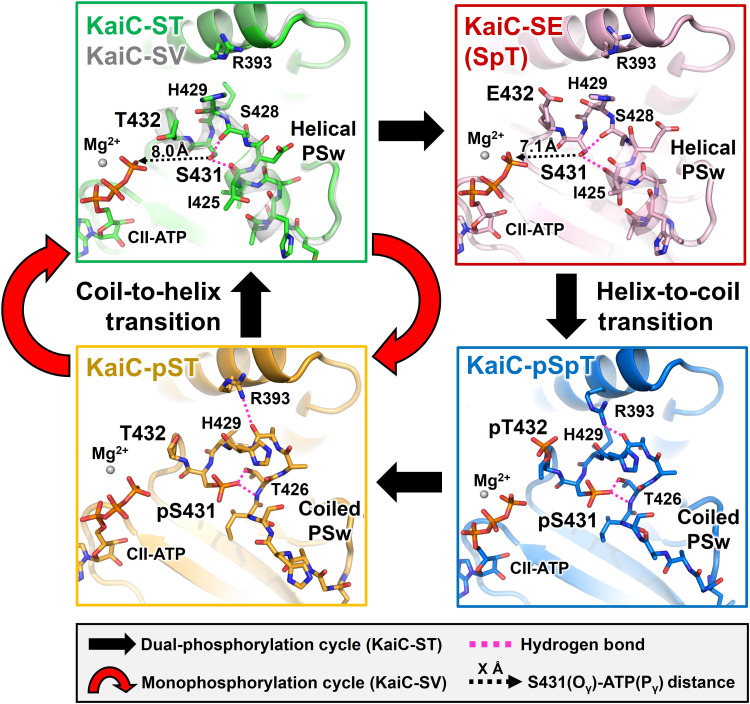
Our crystal structure library indicates that key allosteric communication occurs between the CI and CII domains during the transition from KaiC-pST to KaiC-ST. We observed that the region upstream of S431 (T416–H429), which adopts a coiled structure in KaiC-pST, folds into a previously unidentified helical structure in KaiC-ST (Fig. 2, left). A helix-to-coil reversal was observed in the transition from KaiC-SpT to KaiC-pSpT (Fig. 2, right). These results enable us to assign the upstream region of S431 as a local phospho-switch (PSw). As shown in the left panels of Fig. 2, removal of the phosphoryl group from pS431 disrupted hydrogen bond interactions between pS431 and T426 in KaiC-pST, which provided the space necessary for the PSw to fold into the compact helical structure in KaiC-ST.
Fig. 2. Cyclic structural changes in the PSw located upstream of S431 in KaiC-ST (green), KaiC-SV (white), KaiC-SE (pink), KaiC-pSpT (blue), and KaiC-pST (orange).
Black and red arrows represent the dual-phosphorylation and monophosphorylation cycles in KaiC-ST and KaiC-SV, respectively. Dashed magenta lines correspond to hydrogen bonds around S431/pS431. Dashed black arrows indicate the distances between the S431 Oγ atom and CII-ATP Pγ atom.
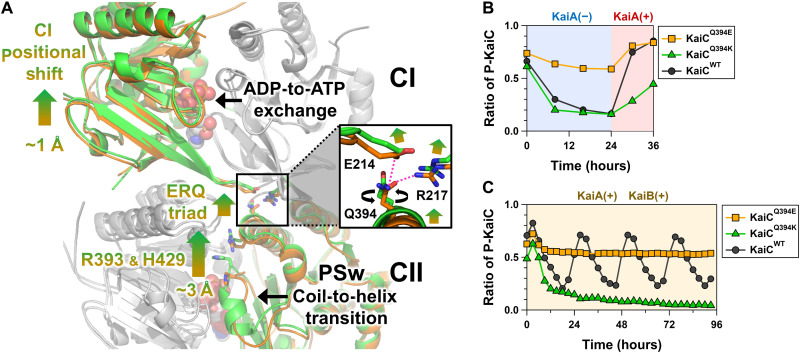
The PSw coil-to-helix transition in the CII domain was coupled with the ADP release from the CI domain. Indicated by arrows in Fig. 3A (gradient from orange to green), the ADP-to-ATP exchange at the CI-CI interface resulted in a slight but systematic positional shift of the neighboring CI domain. This rearrangement drove repositioning of the side chain of the basic residue R217 away from the neutral residue Q394 located in the CI-CII interface (box in Fig. 3A), eventually disrupting the hydrogen bonding interaction between them that was observed in KaiC-pST. Instead, the acidic residue E214 captured the released Q394 through a new hydrogen bond, thereby causing the CII domain to move closer to the CI domain in KaiC-ST. R393 and H429 are particular examples of this CI-directed movement by up to ~3 Å, assisting the coil-to-helix switching of the PSw through multiple hydrogen bond formation of S431 with I425 and S428 (Fig. 2, top left).
Fig. 3. Structural basis for master allostery between CI-ATPase and CII-PSw in KaiC.
(A) Tertiary and quaternary structural changes in the transition from KaiC-pST (orange) to KaiC-ST (green). The ADP-ATP exchange occurring at the CI-CI interfaces and the coil-to-helix transition for PSw are allosterically coupled via rearrangements of E214, R217, Q394, R393, and H429. The inserted box indicates the zoomed-in view of the ERQ triad mediating the communication between the CI and CII domains. (B) Effects of Q394E and Q394K mutations on the switching ability of CI-CII allostery. Proportion of phosphorylated KaiC (P-KaiC) was measured for 24 hours at 30°C and after KaiA addition at 24 hours. (C) Effects of Q394E and Q394K mutations on the P-cycle.
The neutral character of residue 394 was critical to this allosteric communication because a Q394E mutation (KaiCQ394E) stabilized its electrostatic interaction with R217 but destabilized with E214, which resulted in constitutive phosphorylation, even with KaiC alone (Fig. 3B), and arrhythmic accumulation of its phosphorylated forms even in the presence of KaiA and KaiB (Fig. 3C). Consistent with this, KaiCQ394K exhibited very slow phosphorylation rates in the presence of KaiA (Fig. 3B) and remained dephosphorylated even in the presence of KaiA and KaiB (Fig. 3C). Hence, most of the large-scale bidirectional CI-CII communication during the P-cycle takes place through an E214-R217-Q394 (ERQ) triad in the transition from KaiC-pST to KaiC-ST.
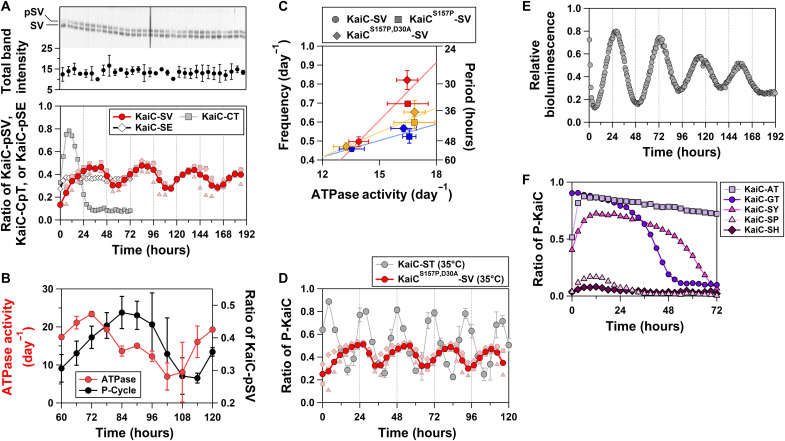
Our biochemical analysis demonstrated that the observed CI-CII communication is necessary and sufficient to allosterically drive the P-cycle. We designed a KaiC T432V mutant (KaiC-SV) to investigate the monophosphorylation/dephosphorylation of S431 with minimal structural perturbation around residue 432. Valine is closest to threonine in terms of topology and volume (fig. S2 and Supplementary Text), as evidenced by conservation of the helical PSw in KaiC-SV (Fig. 2, top left). Astonishingly, KaiC-SV exhibited an in vitro mono–P-cycle for S431 (Fig. 4A) (KaiC-SV ↔ KaiC-pSV; red arrows in Fig. 2), an in vitro ATPase cycle (Fig. 4B), and an in vitro assembly and disassembly cycle (fig. S3 and Supplementary Text) with a prolonged but temperature-compensated period [51.0 ± 2.2 hours (n = 5), Q10 = 1.08 ± 0.1 (n = 3)] (Fig. 4C). By contrast, KaiC-CT, which was designed to allow for monophosphorylation/dephosphorylation of T432 with minimal structural perturbation around residue 431, was arrhythmic at 30°C (Fig. 4A). Furthermore, KaiC-SE, which was generated to allow for monophosphorylation/dephosphorylation of S431 without going through the fully dephosphorylated KaiC-ST, was also arrhythmic (Fig. 4A). The minimal set of allostery that ensures oscillatory nature is the coupling between the PSw transition associated with monophosphorylation/dephosphorylation of S431 and nucleotide exchange in the CI domain (Fig. 2, left).
Fig. 4. Monophosphorylation rhythms for KaiC-SV.
(A) In vitro P-cycle at 30°C. The top panel represents a magnified view of the upper phosphorylated (pSV) and lower dephosphorylated (SV) bands in SDS-PAGE gels. Dark red–colored circles correspond to the mean from independent preparations and measurements (pale red–colored squares, triangles, and diamonds). (B) In vitro ATPase rhythm at 30°C. (C) Correlation plot between ATPase activity for KaiC alone and the frequency (24 per period) of the in vitro P-cycle in the presence of KaiA and KaiB. Blue, orange, and red markers correspond to data analyzed at 25°, 30°, and 35°C, respectively. (D) Near-circadian mono–P-cycle (29.2 ± 1.8 hours) for KaiCS157P,D30A-SV at 35°C. Dark red–colored circles correspond to the mean from independent preparations and measurements (pale red–colored squares, triangles, and diamonds). Data for KaiC-ST at 35°C were taken from the previous study (27). (E) In vivo bioluminescence rhythm of KaiCS157P,D30A-SV at 30°C. (F) Arrhythmicity of KaiC-AT, KaiC-GT, KaiC-SY, KaiC-SP, and KaiC-SH at 30°C.
The prolonged mono–P-cycle observed for KaiC-SV implies that pT432 plays a role in accelerating the KaiC-ST P-cycle. The Oγ atom of S431 was positioned 1 Å closer to CII-ATP in the KaiC-SpT–mimicking mutant KaiC-SE (Fig. 2A, top right) relative to the position in KaiC-ST, which likely facilitates the subsequent formation of KaiC-pSpT. To investigate whether other mutations can allosterically suppress the period-prolonging T432V substitution, we introduced various ATPase-activating mutations into KaiC-SV. A S157P substitution (KaiCS157P-SV), which is a known period-shortening and ATPase-activating mutation (11, 25, 26), resulted in a less-prolonged period [40.2 ± 2.1 hours (n = 4)] at 30°C (Fig. 4C). We also introduced the ATPase-activating mutation D30A (27). The resultant KaiCS157P,D30A-SV exhibited a near-circadian mono–P-cycle [29.2 ± 1.8 hours (n = 3); Fig. 4D] at 35°C with a slight impairment in temperature compensation [Q10 = 1.42 ± 0.1 (n = 3)]. However, note that the ATPase activities of KaiCS157P-SV and KaiCS157P,D30A-SV are temperature-compensated, as was observed for KaiC-SV (Fig. 4C). Furthermore, a cyanobacterial reporter strain with KaiCS157P,D30A-SV exhibited in vivo bioluminescence rhythm at 30°C (Fig. 4E), whose period [43.6 ± 2.0 hours (n = 20)] did not deviate significantly from that of in vitro mono–P-cycle [36.8 ± 3.5 hours (n = 3); Fig. 4C]. Combined with KaiA and KaiB, KaiCS157P,D30A-SV constitutes a simple biochemical posttranslational circadian oscillator with a sole phosphorylation site.
The crucial role of the observed allostery is further supported by evolutional conservation and divergence of the dual phosphorylation sites. Multiple sequence alignment of KaiC and its homologs from other organisms revealed a near-perfect conservation of S431 (28). Only alanine (Fibrella aestuarina BUZ 2) and glycine (Allochromatium vinosum DSM 180) were the observed exceptions, but the arrhythmicity of KaiC-AT and KaiC-GT in the presence of KaiA and KaiB supports our present interpretation (Fig. 4F). By contrast, amino acids other than threonine, such tyrosine (Pseudomonas oryzihabitans), histidine (Massilia sp. WG5), and proline (Pontibacter korlensis), are observed at position 432. We generated KaiC-SY, KaiC-SH, and KaiC-SP to investigate the effects of these substitutions and found that all of them exhibited arrhythmicity (Fig. 4F). These results suggest that different evolutionary selection pressures acted on the two sites; position 431 was conserved to allow for acquisition of master CI-CII allostery that gives rise to the temperature-compensated rhythmicity, and amino acid variances at position 432 fine-tune the period length into the circadian time scale by accelerating the CI-CII allostery mediated by S431.
DISCUSSION
The significance of allosteric regulation in KaiC is also pointed out by previous observations, such as the correlation between the CI-ATPase activity and CII–P-cycle frequency (11, 25, 26) and the dynamic stacking/unstacking between CI and CII rings (29). These phenomena are closely related to the nonlocal mutual regulation of the ATPase and kinase/phosphatase active sites through the CI-CII allostery demonstrated in this study. It is also important to note that this CI-CII allostery is linked on the CII domain side to a local regulatory mechanism reminiscent of positive feedback. This view is supported not only by previous studies showing that pT432 promotes the phosphorylation of S431 (17, 30–32) but also by the present biochemical result that the slowdown caused by the T432V mutation is recovered by the CI mutations via the CI-CII allostery (Fig. 4, C and D) and by the structural observation that the side chain of S431 is more proximal to ATP in KaiC-SE (Fig. 2). It remains possible that T426, which has been proposed to be temporarily and partially phosphorylated (33, 34), is also involved in this local regulation in some way. However, any electron densities that might suggest the phosphorylation of T426 have not been observed in our structural library. Furthermore, since KaiC-SV oscillated in a two-state manner (Fig. 4A) and a further S431C substitution of KaiC-SV resulted in a single-band state even in the presence of an excessive amount of KaiA (fig. S4), the contribution of T426 is considered to be very limited.
Mathematical modeling approaches have highlighted the importance of multisite phosphorylation in oscillatory phenomena. Simple enzymatic futile cycles in which a substrate shuttles between two states, e.g., monophosphorylated and dephosphorylated forms, via forward and backward reactions catalyzed by an opposing enzyme converge into an equilibrium (35). Unbiased systems targeting a substrate with two modification sites retain a limited (~0.1%) potential to exhibit self-sustained oscillation (36). This apparent inconsistency can be resolved by taking into consideration the fact that KaiC-SV can still be regarded as a two-site system because its single site for phosphorylation is influenced allosterically by another biochemically active site for ATP hydrolysis in the CI domain (Fig. 3A).
The cyanobacterial circadian clock is the simplest circadian clock system known to date in terms of the number of components, but mechanistically, it is a very complex system involving the ATPase cycle and a four-state P-cycle. The oscillators we designed using KaiC-SV and KaiCS157P,D30A-SV reconstruct the complex system into a minimal unit by extracting the core allostery from the complex cycle. This will serve as a research tool for further elucidation of mechanisms, such as period determination, temperature compensability, and entrainment, and will provide design principles for the simplest posttranslational biochemical oscillator that oscillates with a temperature-compensated circadian period.
MATERIALS AND METHODS
Expression and purification of KaiC
Plasmid vectors for wild-type KaiC and KaiC mutants were generated for glutathione S-transferase–tagged (pGEX-6P-1) or hexa-histidine (His)–tagged (pET-3a) proteins. Kai proteins were expressed in BL21(DE3) or BL21(DE3)pLysE Escherichia coli cells and purified (17, 27, 37).
In vivo and in vitro rhythm assays
In vivo bioluminescence assays were conducted as previously reported using a cooled charge-coupled device camera system (38). KaiC phosphorylation and ATPase cycles were reconstituted in vitro in the presence of KaiA, KaiB, and ATP (19). The relative abundances of the four phosphorylation states of KaiC were quantified using SDS–polyacrylamide gel electrophoresis (PAGE) and LOUPE (looking over undulatory phospho-patterns on electrophoresis) software (39). The ATPase activity of KaiC was measured as previously described (13, 27) and presented as the number of ATP molecules hydrolyzed into ADP molecules per KaiC monomer per unit time. Unless otherwise noted, all the rhythm assays were conducted at 30°C.
Crystallization of KaiC
All crystals were obtained using the vapor diffusion method. The purified samples were concentrated to 3.5 mg/ml and stored in a solution of 20 mM tris-HCl (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), and 1 mM ATP. To obtain crystals of dephosphorylated KaiC-ST, the sample was preincubated for 2 days at 40°C. The incubated solution was mixed with the crystallization solution containing 100 mM tris-HCl (pH 7.0), 1 M KCl, 0.7 to 1.0 M sodium/potassium tartrate, 0.3 to 1.2 M sodium acetate, and 1 to 5 mM adenylyl-imidodiphosphate. For the crystallization of KaiC-SE and KaiC-SV, the preincubation was not conducted. The sample solution kept under the on-ice condition was directly mixed with the same crystallization solution as KaiC-ST. The crystals of phosphorylated KaiC-pSpT and KaiC-pST were obtained in the solution containing 80 to 250 mM acetic acid and 1.0 to 1.5 M sodium acetate without the preincubation and frozen with 30% (w/v) glycerol or 25% (w/v) polyethylene glycol 8000. The crystals of KaiC-pSpT and KaiC-pST were picked up 2 and 15 days, respectively, after mixing the sample and crystallization solutions. While the crystals of KaiC-ST, KaiC-SE, and KaiC-SV were obtained in the P63 space group, KaiC-pSpT and KaiC-pST crystals belonged to P212121 and P21, respectively. All crystals were soaked into the liquid nitrogen and frozen for the radiation experiment conducted at the cryo-temperature.
Data collection and structure determination
X-ray diffraction data were collected on beamline BL44XU at SPring-8 (Harima, Japan). Crystals were mounted at 100 K under a cryostream, and diffraction images were recorded with an MX300-HE (Rayonix) or EIGER X 16M (DECTRIS) detector and processed using HKL2000 (40) and XDSGUI (41). Initial phases were obtained using molecular replacement with previously deposited structures [2GBL (42) or 7DYJ] and MOLREP (43). Refinement and modeling were conducted using Refmac5 (44) and COOT (45), respectively. The crystal structure of KaiC-ST was refined using reflections merged from two crystals that had only a 0.7% difference in lattice sizes (acrystal1 = 94.7 Å and ccrystal1 = 180.5 Å; acrystal2 = 94.6 Å and ccrystal2 = 181.8 Å) (table S1). The asymmetric unit for the P63 crystal contained a dimer of KaiC-ST and was arranged along a crystallographic threefold axis to form a hexamer. The KaiC-SE and KaiC-SV crystals also belonged to the P63 space group. KaiC-pSpT crystallized in the space group P212121 with a hexamer in the asymmetric unit, whereas KaiC-pST crystallized in the space group P21 with two hexamers in the asymmetric unit. Graphic representations were generated using PyMOL (Schrödinger). The statistics for data collection and refinement are listed in table S1.
KaiA-KaiB-KaiC interaction assay
KaiA (0.04 mg/ml) and KaiB (0.04 mg/ml) were incubated with KaiC-SV (0.2 mg/ml) at 30°C in a buffer containing 20 mM tris-HCl (pH 8.0), 150 mM NaCl, 0.5 mM EDTA, 1 mM ATP, 5 mM MgCl2, and 1 mM DTT. Every aliquot taken from the KaiA/KaiB/KaiC-SV mixture incubated at 30°C was used for size exclusion chromatography analysis at room temperature. A Superdex 200 Increase 10/300 GL (Cytiva) column was connected to a right-angle light scattering system (Viscotek TDA305, Malvern) to estimate the molecular masses of the eluted peaks.
Acknowledgments
Diffraction data were collected at BL44XU at the SPring-8 facility under the proposals 2017A6700, 2017B6700, 2018A6700, 2018B6700, 2019A6700, 2019B6700, 2020A6700, 2020A6500, 2017A6702, 2017B6702, 2018A6802, 2018B6802, 2019A6902, 2019B6902, and 2020A6502.
Funding: This research was partly supported by the Platform Project for Supporting Drug Discovery and Life Science Research (BINDS) from AMED under grant number JP20am0101072 (support number 0583). This study was supported by Grants-in-Aid for Scientific Research (17H06165 to S.A., 19K16061 to Y.F., and 18K06171 to A.M.).
Author contributions: Y.F., A.M., and S.A. designed the experiments. Y.F. collected the diffraction data and analyzed it with input from E.Y. Y.F., A.M., D.O., and D.S. performed the biochemical experiments. K.I.-M. and Y.F. performed in vivo experiments with input from T.K. S.A. and Y.F. drafted the manuscript with input from all authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The atomic coordinates and structure factors are deposited in the Protein Data Bank with accession codes 7DYK (KaiC-SV), 7DYJ (KaiC-ST), 7DY2 (KaiC-pST), 7DYI (KaiC-SE), 7DXQ (KaiC-pSpT), and 7V3X (KaiC-pSpT and pST).
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S4
Table S1
References
REFERENCES AND NOTES
- 1.Goldbeter A., Dupont G., Allosteric regulation, cooperativity, and biochemical oscillations. Biophys. Chem. 37, 341–353 (1990). [DOI] [PubMed] [Google Scholar]
- 2.Goodwin B. C., Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 3, 425–437 (1965). [DOI] [PubMed] [Google Scholar]
- 3.Narasimamurthy R., Hunt S. R., Lu Y. N., Fustin J. M., Okamura H., Partch C. L., Forger D. B., Kim J. K., Virshup D. M., CK1δ/ε protein kinase primes the PER2 circadian phosphoswitch. Proc. Natl. Acad. Sci. U.S.A. 115, 5986–5991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narasimamurthy R., Virshup D. M., The phosphorylation switch that regulates ticking of the circadian clock. Mol. Cell 81, 1133–1146 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Partch C. L., Orchestration of circadian timing by macromolecular protein assemblies. J. Mol. Biol. 432, 3426–3448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upadhyay A., Marzoll D., Diernfellner A., Brunner M., Herzel H., Multiple random phosphorylations in clock proteins provide long delays and switches. Sci. Rep. 10, 22224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diernfellner A. C. R., Brunner M., Phosphorylation timers in the neurospora crassa circadian clock. J. Mol. Biol. 432, 3449–3465 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Snijder J., Schuller J. M., Wiegard A., Lossl P., Schmelling N., Axmann I. M., Plitzko J. M., Forster F., Heck A. J. R., Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Science 355, 1181–1184 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Tseng R., Goularte N. F., Chavan A., Luu J., Cohen S. E., Chang Y. G., Heisler J., Li S., Michael A. K., Tripathi S., Golden S. S., LiWang A., Partch C. L., Structural basis of the day-night transition in a bacterial circadian clock. Science 355, 1174–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaguchi H. Q., Ode K. L., Ueda H. R., A design principle for posttranslational chaotic oscillators. Iscience 24, 101946 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abe J., Hiyama T. B., Mukaiyama A., Son S., Mori T., Saito S., Osako M., Wolanin J., Yamashita E., Kondo T., Akiyama S., Atomic-scale origins of slowness in the cyanobacterial circadian clock. Science 349, 312–316 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Aryal R. P., Kwak P. B., Tamayo A. G., Gebert M., Chiu P. L., Walz T., Weitz C. J., Macromolecular assemblies of the mammalian circadian clock. Mol. Cell 67, 770–782.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murayama Y., Mukaiyama A., Imai K., Onoue Y., Tsunoda A., Nohara A., Ishida T., Maeda Y., Terauchi K., Kondo T., Akiyama S., Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution. EMBO J. 30, 68–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattanayek R., Wang J. M., Mori T., Yao X., Johnson C. H., Egli M., Visualizing a circadian clock protein: Crystal structure of KaiC and functional insights. Mol. Cell 15, 375–388 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Pattanayek R., Xu Y., Lamichhane A., Johnson C. H., Egli M., An arginine tetrad as mediator of input-dependent and input-independent ATPases in the clock protein KaiC. Acta Crystallog. D Biol. Crystallogr. 70, 1375–1390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiura M., Kutsuna S., Aoki S., Iwasaki H., Andersson C. R., Tanabe A., Golden S. S., Johnson C. H., Kondo T., Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281, 1519–1523 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Nishiwaki T., Satomi Y., Kitayama Y., Terauchi K., Kiyohara R., Takao T., Kondo T., A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J. 26, 4029–4037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rust M. J., Markson J. S., Lane W. S., Fisher D. S., O’Shea E. K., Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318, 809–812 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyarna T., Kondo T., Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308, 414–415 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Tomita J., Nakajima M., Kondo T., Iwasaki H., No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307, 251–254 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Nishiwaki T., Satomi Y., Nakajima M., Lee C., Kiyohara R., Kageyama H., Kitayama Y., Temamoto M., Yamaguchi A., Hijikata A., Go M., Iwasaki H., Takao T., Kondo T., Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc. Natl. Acad. Sci. U.S.A. 101, 13927–13932 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hatakeyama T. S., Kaneko K., Generic temperature compensation of biological clocks by autonomous regulation of catalyst concentration. Proc. Natl. Acad. Sci. U.S.A. 109, 8109–8114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sasai M., Mechanism of autonomous synchronization of the circadian KaiABC rhythm. Sci. Rep. 11, 4713 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Zon J. S., Lubensky D. K., Altena P. R. H., ten Wolde P. R., An allosteric model of circadian KaiC phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 104, 7420–7425 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terauchi K., Kitayama Y., Nishiwaki T., Miwa K., Murayama Y., Oyama T., Kondo T., ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc. Natl. Acad. Sci. U.S.A. 104, 16377–16381 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito-Miwa K., Furuike Y., Akiyama S., Kondo T., Tuning the circadian period of cyanobacteria up to 6.6 days by the single amino acid substitutions in KaiC. Proc. Natl. Acad. Sci. U.S.A. 117, 20926–20931 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ouyang D. Y., Furuike Y., Mukaiyama A., Ito-Miwa K., Kondo T., Akiyama S., Development and optimization of expression, purification, and ATPase assay of KaiC for medium-throughput screening of circadian clock mutants in cyanobacteria. Int. J. Mol. Sci. 20, 2789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmelling N. M., Lehmann R., Chaudhury P., Beck C., Albers S. V., Axmann I. M., Wiegard A., Minimal tool set for a prokaryotic circadian clock. BMC Evol. Biol. 17, 169 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang Y. G., Tseng R., Kuo N. W., LiWang A., Rhythmic ring-ring stacking drives the circadian oscillator clockwise. Proc. Natl. Acad. Sci. U.S.A. 109, 16847–16851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tseng R., Chang Y. G., Bravo I., Latham R., Chaudhary A., Kuo N. W., LiWang A., Cooperative KaiA-KaiB-KaiC interactions affect KaiB/SasA competition in the circadian clock of cyanobacteria. J. Mol. Biol. 426, 389–402 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin J., Chew J., Chockanathan U., Rust M. J., Mixtures of opposing phosphorylations within hexamers precisely time feedback in the cyanobacterial circadian clock. Proc. Natl. Acad. Sci. U.S.A. 111, E3937–E3945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egli M., Pattanayek R., Sheehan J. H., Xu Y., Mori T., Smith J. A., Johnson C. H., Loop-loop interactions regulate KaiA-stimulated KaiC phosphorylation in the cyanobacterial KaiABC circadian clock. Biochemistry 52, 1208–1220 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pattanayek R., Mori T., Xu Y., Pattanayek S., Johnson C. H., Egli M., Structures of KaiC circadian clock mutant proteins: A new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. PLOS ONE 4, e7529 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y., Mori T., Qin X., Yan H., Egli M., Johnson C. H., Intramolecular regulation of phosphorylation status of the circadian clock protein KaiC. PLOS ONE 4, e7509 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angeli D., Sontag E. D., Translation-invariant monotone systems, and a global convergence result for enzymatic futile cycles. Nonlinear Anal. Real World Appl. 9, 128–140 (2008). [Google Scholar]
- 36.Jolley C. C., Ode K. L., Ueda H. R., A design principle for a posttranslational biochemical oscillator. Cell Rep. 2, 938–950 (2012). [DOI] [PubMed] [Google Scholar]
- 37.Mukaiyama A., Ouyang D. Y., Furuike Y., Akiyama S., KaiC from a cyanobacterium Gloeocapsa sp. PCC 7428 retains functional and structural properties required as the core of circadian clock system. Int. J. Biol. Macromol. 131, 67–73 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Kondo T., Tsinoremas N. F., Golden S. S., Johnson C. H., Kutsuna S., Ishiura M., Circadian clock mutants of cyanobacteria. Science 266, 1233–1236 (1994). [DOI] [PubMed] [Google Scholar]
- 39.Furuike Y., Abe J., Mukaiyama A., Akiyama S., Accelerating in vitro studies on circadian clock systems using an automated sampling device. Biophys. Physicobiol. 13, 235–241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otwinowski Z., Minor W., [20] Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997). [DOI] [PubMed] [Google Scholar]
- 41.Kabsch W., XDS. Acta Crystallogr. D 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattanayek R., Williams D. R., Pattanayek S., Xu Y., Mori T., Johnson C. H., Stewart P. L., Egli M., Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J. 25, 2017–2028 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vagin A., Teplyakov A., MOLREP: An automated program for molecular replacement. J. Appl. Cryst. 30, 1022–1025 (1997). [Google Scholar]
- 44.Murshudov G. N., Skubak P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oyama K., Azai C., Nakamura K., Tanaka S., Terauchi K., Conversion between two conformational states of KaiC is induced by ATP hydrolysis as a trigger for cyanobacterial circadian oscillation. Sci. Rep. 6, 32443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S4
Table S1
References