Abstract
Genetic CLN5 variants are associated with childhood neurodegeneration and Alzheimer’s disease; however, the molecular function of ceroid lipofuscinosis neuronal protein 5 (Cln5) is unknown. We solved the Cln5 crystal structure and identified a region homologous to the catalytic domain of members of the N1pC/P60 superfamily of papain-like enzymes. However, we observed no protease activity for Cln5; and instead, we discovered that Cln5 and structurally related PPPDE1 and PPPDE2 have efficient cysteine palmitoyl thioesterase (S-depalmitoylation) activity using fluorescent substrates. Mutational analysis revealed that the predicted catalytic residues histidine-166 and cysteine-280 are critical for Cln5 thioesterase activity, uncovering a new cysteine-based catalytic mechanism for S-depalmitoylation enzymes. Last, we found that Cln5-deficient neuronal progenitor cells showed reduced thioesterase activity, confirming live cell function of Cln5 in setting S-depalmitoylation levels. Our results provide new insight into the function of Cln5, emphasize the importance of S-depalmitoylation in neuronal homeostasis, and disclose a new, unexpected enzymatic function for the N1pC/P60 superfamily of proteins.
Cln5 structure reveals thioesterase activity among N1pC/P60 proteins, stressing the role of S-depalmitoylation in neurodegeneration.
INTRODUCTION
The neuronal ceroid lipofuscinoses (NCLs) are the most common neurodegenerative disorders of childhood and share the accumulation of intracellular autofluorescent storage material and progressive loss of neurons. Characteristic clinical symptoms include developmental regression, ataxia, gradual loss of motor function and vision, epileptic seizures, and premature death (1). Various types of NCL diseases have been linked to at least 13 different gene loci (2). Mutations in the ceroid lipofuscinosis neuronal protein 5 (Cln5), first described in Finland, cause late infantile NCL (OMIM, 256731) and have later been identified in various ethnic populations as well as in juvenile and adult patients. More than 50 sequence variations and mutations in the CLN5 gene have been reported to be associated with variable types of dementias ranging from late infantile epileptic encephalopathy to Alzheimer’s disease (3–6).
The precursor Cln5 protein is a 407–amino acid–long membrane-located protein that is cleaved by signal peptide peptidase (SPP) and its homologs the SPP-like proteases (SPPL), resulting in the soluble Cln5 protein (7). Several studies have demonstrated Cln5 localized to lysosomes (8, 9) and associated with endolysosomal dysfunction (10–12). However, the structure and molecular function of the Cln5 protein remains unknown (13), precluding a mechanistic understanding and potential treatment development for Cln5-dependent NCL.
We crystallized the glycosylated human Cln5 protein, determined the structure by single-wavelength anomalous diffraction (SAD), and found a minor but substantial structural similarity in its core domain to the PPPDE (permuted papain fold peptidases of double-stranded RNA viruses and eukaryotes) proteins. A thorough analysis revealed a highly conserved triad of histidine-tyrosine-cysteine within the catalytic centers of Cln5 and PPPDE. The PPPDE proteins belong to N1pC/P60 superfamily of papain-like enzymes, which comprise an extremely diverse group of proteins originating from archaea, bacteria, bacteriophages, viruses, and eukaryotes with predicted protease, amidase, transglutaminases, or acyltransferase activity but often uncharacterized function (14, 15). Here, we present evidence that this evolutionarily conserved structural motif can also function as a palmitoyl thioesterase, which differs substantially from the currently known palmitoyl thioesterases, which all belong to the serine hydrolase superfamily (16–20).
Protein lipidation is an important co- or posttranslational modification that results in membrane attachment of about 10% of cellular proteins by a lipid anchor. Palmitoylation via cysteine residues is a unique form of protein lipidation because it is readily reversible through the action of cellular thioesterases (21). The dynamic interplay between palmitoylation and S-depalmitoylation is a universal mechanism to control protein function and localization in cells, and defects in either process may result in endocrine syndromes, malignant diseases, or neurological disorders (22). The identification of Cln5 as a new member of the palmitoyl thioesterase family of proteins that regulate cellular protein S-palmitoylation opens a new avenue for the study of neurodegeneration and stimulates the search for other cysteine-based S-depalmitoylases.
RESULTS
Cln5 structure
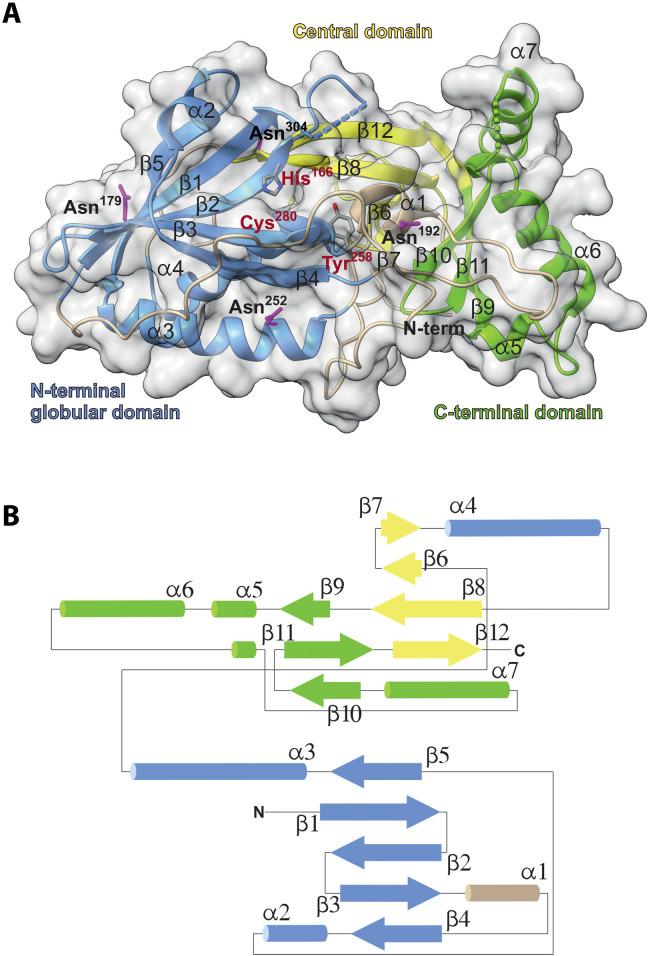
We present a 2.7-Å-resolution crystal structure of human Cln5 [Protein Data Bank (PDB), 6R99], revealing a globular protein with three tightly interconnected domains and one molecule of Cln5 in the asymmetric unit (Fig. 1A). A topological diagram of the secondary structure displays two centered antiparallel β sheets sandwiched by α-helical elements (Fig. 1B).
Fig. 1. The Cln5 overall structure.
(A) The three-dimensional (3D) structure of Cln5 colored by domain. The N-terminal (blue), central (yellow), and C-terminal (green) domains are tightly interconnected. The catalytic residues Cys280, His166, and Tyr258 are labeled dark red. The side chains of the four glycosylated asparagine residues Asn179, Asn192, Asn252, and Asn304 are marked in purple. Dashed lines represent the missing loops where model building was not possible. (B) Schematic representation of the Cln5 structure indicating the relative positions of the six α helices and the 12 β strands with corresponding domain coloring such as in (A), as well as the N terminus (N) and the C terminus (C).
Residues 99 to 399 could be modeled in one chain with two gaps of 10 and 6 amino acids at positions 151 and 347, respectively. Electron densities of four glycosylation sites with up to three sugar residues were identified at Asn179, Asn192, Asn252, and Asn304. At the N terminus, the first 40 amino acids form an extended strand covering nearly one complete flank of the molecule. Two disulfide bridges, from Cys119 to Cys208 and from Cys126 to Cys214 (fig. S1, A and B) keep the N terminus in place between two sugar modifications at Asn192 and Asn252. The N-terminal strand proceeds into a globular domain, colored blue (Fig. 1A), consisting of an antiparallel β sheet (strands β1 to β5) and four α helices (α1 to α4). The central domain, colored yellow, is a short antiparallel β sheet (strands β6 to β8 and β12) bridging the N- and C-terminal folds (Fig. 1, A and B). The C-terminal domain, colored green, consists of a small antiparallel β sheet (strands β9 to β11) and three α helices (α5 to α7).
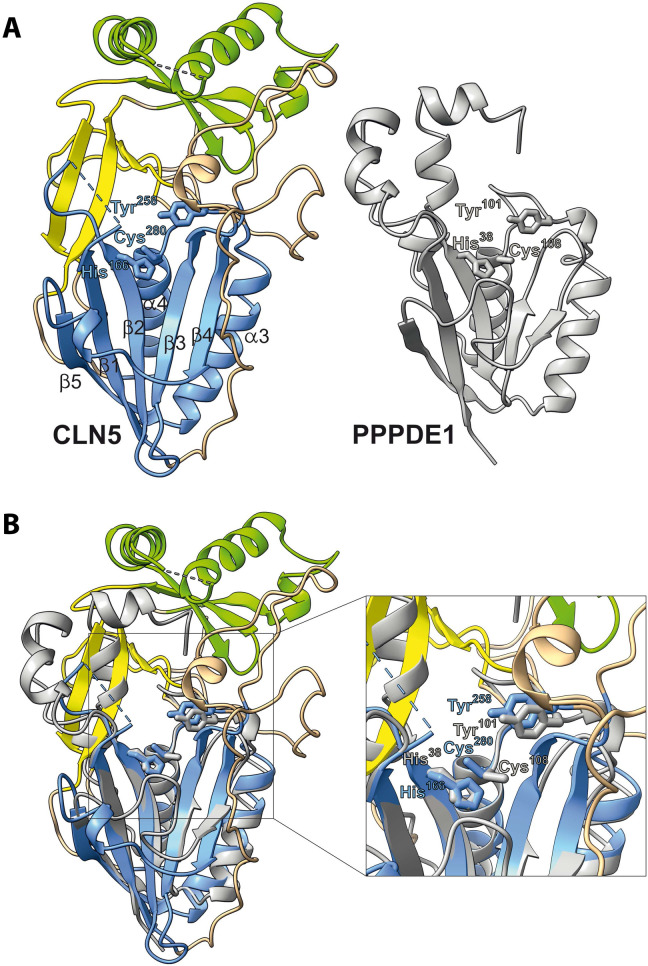
A bioinformatic search for Cln5 homologs, combining both the primary amino acid sequence and the three-dimensional (3D) structural data, revealed the PPPDE1 and PPPDE2 (PDB, 3ebq and 2wq7) as homologous proteins. These proteins belong to the N1pC/P60 superfamily of papain-like cysteine peptidases and are members of a subgroup of the permuted papain-like N1pC/P60 enzymes displaying a circularly permuted catalytic domain consisting of a conserved catalytic Cys-His dyad and a polar third residue.
By protein structure comparison using distance matrix alignment (DALI) (23, 24), a weak but significant structural similarity between Cln5, PPPDE1, and PPPDE2 was identified (Table 1). Despite a sequence identity of only 11%, the structure overlay of Cln5 and PPPDE1 using 3D alignment based on secondary structure matching with PDBeFold (25) revealed that the complete structural fold of the circularly permuted papain-like domain around the catalytic center of PPPDE1 is preserved in the structure of Cln5 (Fig. 2). The catalytic center in PPPDE proteins comprises a His-Tyr-Cys triad (15), and, for both Cln5 and PPPDE1, these three residues perfectly superimpose with each other (Fig. 2B). In addition, both proteins share five β strands (β1 to β5) and two α helices (α3 and α4) as structural elements. Furthermore, the catalytic Cys-His in PPPDE1 corresponds to the reactive center of the protein’s assumed deubiquitinating or deSUMOylation activity (26–29), indicating that the residues His166 and Cys280 of Cln5 may also have a catalytic function.
Table 1. Structure similarity matches using DALI web service (23, 24).
RMSD, root mean square deviation; lali, length of the alignment.
| PDB ID | Chain | z score | RMSD (Å) | lali | N res | %ID |
Protein
description |
| 3ebq | A | 6.8 | 3.2 | 109 | 144 | 11 | PPPDE1 |
| 2wq7 | A | 6.7 | 3.3 | 111 | 158 | 12 | PPPDE2 |
Fig. 2. The structural homology between Cln5 and PPPDE1.
(A) The ribbon models of CLN5 (left) and PPPDE1 (right) reveal homologous secondary structure elements in the region of the N-terminal domain comprising β strands 1 to 5 and α helices 3 and 4 (numbering as in Fig. 1) (B) Overlay of permuted papain-like fold of Cln5 (coloring as in Fig. 1) and PPPDE1 (gray). Only the globular N-terminal domain shows significant structural homology. The enlarged box around the conserved residues indicates the catalytic center that is shared by PPPDE1 and Cln5. In PPPDE1, the catalytic center consists of the residues His38, Tyr101, and Cys108 (colored light gray). In Cln5, the corresponding residues are His166, Tyr258, and Cys280 (colored blue).
Elucidation of Cln5 thioesterase activity
Although first predicted to be deubiquitinating and deSUMOylating peptidases, PPPDE1 and PPPDE2 were later found to have very low endopeptidase activity toward precursor forms of SUMO1 and SUMO2, and PPPDE2 did not show any deubiquitinating activity (30). We investigated purified Cln5 for possible deubiquitinase or deSUMOylation activity, but Cln5 did neither cleave off SUMO1 and SUMO2 residues from synthesized probes nor cut ubiquitin chains (fig. S2, A and B).
Because several members of the N1pC/P60 family were described to be associated with peptidase functions and lytic activity on bacterial peptidoglycan (14), we tested Cln5 for a proteolytic function. We carried out screening assays with protein substrates such as resorufin-labeled casein and performed multiplex quantitative proteomics analyses by terminal amine isotopic labeling of substrates (TAILS) to identify possible protease substrates and cleavage sites in biological samples. However, no conclusive results could be obtained.
In eukaryotes, two members of the N1pC/P60 family are known to be involved in the biochemical transformation of lipids. First, lecithin retinol acyltransferase catalyzes the transfer of the sn-1 acyl group of phosphatidylcholine to all-trans retinol to form a retinyl ester (31). Second, H-ras revertant protein 107 was described as a Ca2+-independent phospholipase A1/2 that also acts as a class II tumor suppressor (32). Therefore, we assayed purified Cln5 for possible lipase activity using commercially available phospholipase A1 and A2 assay kits. Last, we were able to detect a minor but reproducible enzymatic activity for Cln5 when applying a phospholipase A2 kit containing the fluorescent compound BODIPY PC-A2 as substrate.
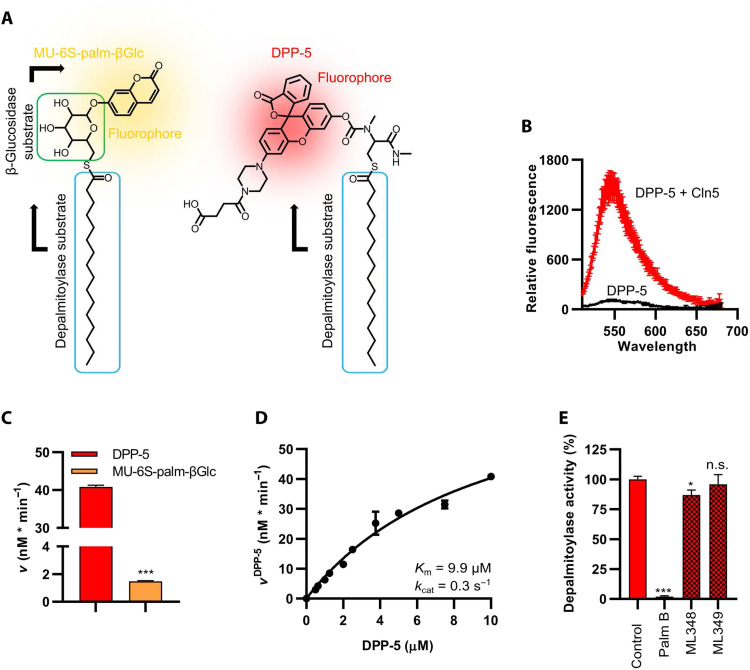
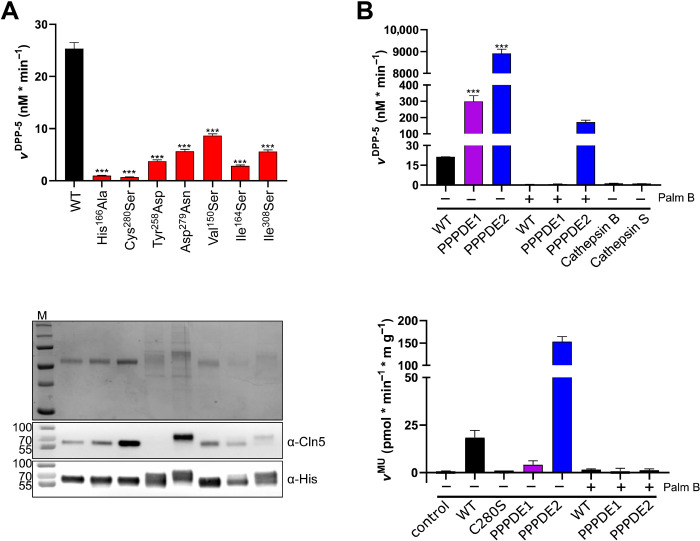
We further tested several acyl esters, acyl acid amides, and acyl thioesters as possible substrates. Unexpectedly, Cln5 processed only palmitoyl thioester substrates with a significant enzymatic conversion rate. Cln5 thioesterase activity was further characterized with the two fluorogenic substrates of S-depalmitoylation, 4-methylumbelliferyl 6-thio-palmitate-β-d-glucopyranoside (MU-6S-palm-βGlc) and S-depalmitoylation probe 5 (DPP-5) (Fig. 3A) that has recently been developed (33). While both substrates were processed by Cln5, DPP-5 showed a more than 20-fold-higher turnover rate when compared with MU-6S-palm-βGlc (Fig. 3, B and C). Incubation of Cln5 with increasing DPP-5 concentrations indicated a DPP-5 concentration–dependent first-order reaction kinetic with a Michaelis constant Km = 9.9 μM DPP-5 and a catalytic efficacy with a turnover number of the Michaelis complex of kcat = 0.3 s−1 (Fig. 3D). Probing Cln5 activity with 10 μM DPP-5 at varying pH values revealed two activity optima at pH 6.5 and pH 8.5 (fig. S3). The thioesterase activity of Cln5 was completely blocked by the inhibitor palmostatin B, but not through the addition of ML348 or ML349, which are known to specifically inhibit thioesterase activity of the acyl protein thioesterases APT1 and APT2, respectively (Fig. 3E). The two key residues within the active center of Cln5, His166 and Cys280; two residues reported in patient mutations (Tyr258Asp and Asp279Asn); and three amino acids (Val150Ser, Ile164Ser, and Ile308Ser) within the putative palmitate binding pocket were replaced, and the mutant proteins were produced as stable, soluble proteins and purified to homogeneity (Fig. 4A and figure in Discussion). All mutants showed severely decreased thioesterase activity (Fig. 4B), confirming the S-depalmitoylase activity of Cln5, substantiating the identical active site composition as indicated for PPPDE1 and PPPDE2 (see Fig. 2, A and B), and demonstrating the relevance of the putative palmitate binding pocket for substrate binding and cleavage.
Fig. 3. Thioesterase activity of Cln5 using fluorescent probes.
(A) Structural models of two different fluorescent S-depalmitoylase substrates: MU-6S-palm-βGlc (left) and the DPP-5 (right). (B) Fluorescent emission spectrum of 5 μM DPP-5 excited at λex = 485 nm either in the absence (black) or in the presence of 1 μg (5.3 nM) wild-type (WT) Cln5 (red) for 30 min. (C) Comparison of 1 μg (5.3 nM) WT Cln5 activity using either 10 μM DPP-5 or 10 μM MU-6S-palm-βGlc. Both enzymatic reactions were run at 37°C in reaction buffer as described in Materials and Methods. For the MU-6S-palm-βGlc reaction, 20 μg of β-glucosidase was present to allow fluorophore release and detection. (D) DPP-5–dependent kinetic measurement using 5.3 nM WT Cln5 hyperbolically fitted using PRISM 8. (E) Detection of remaining Cln5 S-depalmitoylase activity in the presence of different inhibitors. Reaction of 1 μg of WT Cln5 with 5 μM DPP-5 was time-dependently recorded at λem = 540 nm for 30 min in the absence (control) or in the presence of 2.5 μM palmostatin B (Palm B), 10 μM ML349, or 10 μM ML349. Values represent means ± SD (*P ≤ 0.05 and ***P ≤ 0.001). n.s., not significant.
Fig. 4. Thioesterase activity of Cln5 variants, PPPDE1 and PPPDE2.
(A) Top: S-Depalmitoylase activity of Cln5 variants His166Ala, Cys280Ser, Tyr258Asp, Asp279Asn, Val150Ser, Ile164Ser, and Ile308Ser in comparison to WT Cln5. Reaction of 1 μg of each purified enzyme with 5 μM DPP-5 was recorded for 60 min. Values represent means ± SD (***P ≤ 0.001). Similarly, reduced activities were detected with 10 μM MU-6S-palm-βGlc plus 20 μg of β-glucosidase (fig. S4). Middle: Coomassie brilliant blue staining of a 12% SDS gel loaded with 1 μg of purified human Cln5 variants. Bottom: Corresponding Western blot loaded with 30 ng of purified proteins and decorated with either primary monoclonal antibodies raised against human Cln5 (α-Cln5) or against polyhistidine tag (α-His). (B) Top: S-Depalmitoylase activity of WT Cln5, PPPDE1, and PPPDE2. Reaction of each 1 μg of purified enzyme with 5 μM DPP-5 was recorded for 30 min in the absence or presence of 2.5 μM Palm B. No activity was detected in the presence of 1 μg of cathepsin L or cathepsin S (controls). Bottom: S-Depalmitoylase activity of WT Cln5, Cln5 Cys280Ser, PPPDE1, and PPPDE2. Reaction of each 1 μg of purified enzyme or blank (control) with 10 μM MU-6S-palm-βGlc plus 20 μg β-glucosidase (MU) for 30 min at 37°C in the absence or presence of 2.5 μM Palm B. Values represent means ± SD (***P ≤ 0.001).
Furthermore, we tested the two structurally related proteins PPPDE1 and PPPDE2 for thioesterase activity. Unexpectedly, both showed high S-depalmitoylation activity by assaying DPP-5 or MU-6S-palm-βGlc that was inhibited by palmostatin B (Fig. 4B). Cln5, PPPDE1, and PPPDE2 showed differences in the relative cleavage activity for both probes, suggesting that they have distinct substrate preferences. While both probe compounds use the cleavage of a palmitic acid thioester to generate response, DPP-5 displays the thioester in a more native peptide substrate context, whereas MU-6S-palm-βGlc displays the palmitate thioester in the context of β-d-glucopyranoside. This preference not only for S-palmitoylation activity but also for peptide S-depalmitoylation activity suggests a role for Cln5 in protein S-depalmitoylation.
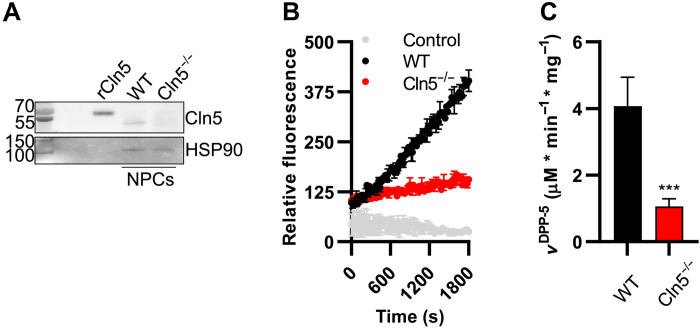
Last, we compared the thioesterase activity of cell lysates of neural progenitor cells (NPCs) derived from either wild-type or Cln5-deficient (Cln5−/−) mice. The Cln5−/− cells showed greatly diminished thioesterase activity when compared to wild-type cells (Fig. 5, A to C). This indicates that Cln5 represents one of the major palmitoyl thioesterases in neuronal cells.
Fig. 5. Thioesterase activity in Cln5−/− and WT NPC cells.
(A) Western blot analysis of Cln5 in 2 μg of total protein from WT and Cln5 knockout human NPCs (WT and Cln5−/− NPCs) in comparison to 10 ng of purified recombinantly expressed WT Cln5 (rCln5). Heat shock protein 90 (HSP90) was used as loading control. (B) Time-dependent traces of DPP-5 fluorescent signal at 540 nm in the presence of 1 μg of total protein of WT and Cln5−/− NPCs. (C) Evaluation of DPP-5 activity of NPC total extracts. Values represent means ± SD (***P ≤ 0.001).
DISCUSSION
Cln5 shows no significant homology to other proteins by primary amino acid sequence, and the molecular functions of Cln5 have not been previously demonstrated. Our study, which has determined the protein structure of Cln5, indicated its enzymatic function and demonstrated an unexpected thioesterase activity of Cln5. This finding is an important step toward understanding the neurodegenerative disease mechanisms associated with CLN5 mutations and developing causative therapies against this fatal disorder. In addition, we found thioesterase activity as a completely new function within the N1pC/P60 protein family of enzymes. While the catalytic triad of Cys-His-X represents an evolutionarily conserved structural motif of almost all cysteine peptidases (34), the circularly permuted triad His-Tyr-Cys is shared by several subfamilies of the N1pC/P60 superfamily of largely uncharacterized enzymes including the PPPDEs (15). Thus, the nucleophile Cys and the imidazolium ring of the catalytic His are shared but the additional polar residue (Tyr) and the permuted orientation of these residues may determine the functional specificity of these enzymes.
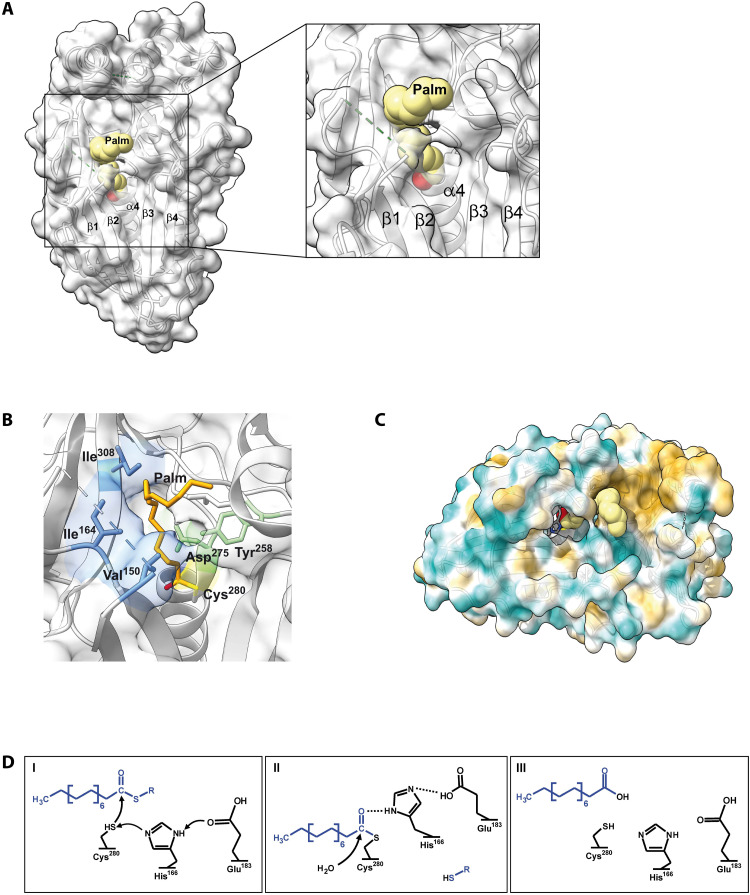
Cln5 is remotely related to PPPDE1 and PPPDE2 and contains the same conserved residues in its active center. PPPDE1 and PPPDE2 were previously shown to have weak deubiquinase or desumoylase activity. However, our results demonstrate high S-depalmitoylation activity for both enzymes, suggesting a similar thioesterase function for other members of the PPPDE family. We modeled the palmitate substrate binding site of Cln5 (Fig. 6, A and C) and found that mutations affecting either the residues of the palmitate binding pocket or the active site center resulted in loss of thioesterase activity, underlining the functional importance of this region (Fig. 6B). On the basis of our structural and biochemical data, we propose a catalytic mechanism for Cln5 and possibly for other PPPDE proteins that relates to the one described for the DHHC (Asp-His-His-Cys) family of S-palmitoyl transferases (Fig. 6D) (35).
Fig. 6. Model of Cln5 with its substrate and proposed active site mechanism.
(A) Modeling of the palmitoyl group (Palm) associated with the active center of Cln5. The secondary structural elements are labeled according to Fig. 1. The hydrophilic head group of the palmitoyl molecule binds into the active center, while the acyl part is located to a lipophilic groove on the surface of Cln5. (B) Localization of residues that were mutated to elucidate their role in CLN5 thioesterase activity. Ile308, Ile164, and Val150 (colored blue) form parts of the proposed hydrophobic binding pocket of palmitate (Palm; marked light brown) and were mutated to serine residues. The positions of the clinically described mutations Tyr258Asp and Asp279Gln are colored green; the catalytic Cys280 is marked yellow. (C) Surface representation of Cln5 including the palmitoyl group (yellow) and the two catalytic residues His166 and Cys280. Cyan color indicates hydrophilic regions, and sand-colored regions are lipophilic. (D) Proposed catalytic mechanism of Cln5 thioesterase activity. (I) In the first step, the nucleophilic sulfur atom of Cys280 attacks the carbonyl carbon of the Cln5-bound thiopalmitoyl substrate (blue). “R” denotes unknown proteins that are palmitoylated. The reacting SH group of Cys280 is stabilized by a free electron pair from nitrogen in the imidazole ring of His166 that functions also as a proton acceptor. The position of the His166 side chain, in turn, is stabilized by Glu183. SH-R is the leaving group of the nucleophilic substitution reaction. (II) His166 forms a hydrogen bridge to the carbonyl oxygen of the acyl protein intermediate, accomplishing the hydrolysis reaction with water. HS-R has been eliminated. (III) Palmitate is the reaction product beside SH-R; the SH group of Cys280 is in the reduced and reactive state again.
All other known palmitoyl thioesterases such as the acyl protein thioesterases APT1 and APT2 (16, 36), the ABHD17 (α/β-hydrolase domain) family thioesterases (17), the ABHD10 S-depalmitoylase (37), and the lysosomal palmitoyl thioesterases PPT1 and PPT2 (20, 38) belong to the family of α/β-serine hydrolases. Note that mutations in the PPT1 gene are associated with a similar childhood-onset neurodegenerative disorder referred to as infantile NCL (39). It is intriguing that disturbed S-depalmitoylation may compromise neuronal functioning because the reversible attachment of a palmitic acid onto a cysteine residue via thioester linkage (S-palmitoylation) determines the assembly and compartmentalization of many neuronal proteins at specific subcellular domains and is crucial for neuronal development and synaptic plasticity (40). Considering the fundamental importance of reversible protein palmitoylation in establishing the spatial localization of many well-studied intracellular signaling pathways, it is quite likely that there will be a battery of thioesterases that are required to regulate the reversible membrane attachment of a multitude of proteins and that Cln5 might not be the only member of the N1pC/P60 protein family of enzymes with thioesterase activity. Hence, Cln5, PPPDE1, PPPDE2, and related proteins may be members of a new subfamily of cysteine-based S-depalmitoylase belonging to the N1pC/P60 protein family of enzymes. The neutral pH optimum of Cln5 (fig. S3) points to a possible relation to other nonlyosomal thioesterases and indicates that it may also function as depalmitoylase outside the lysosomal compartment. Neutral pH optima were also found for the lysosomal palmitoyl thioesterases PPT1 and PPT2 (38). Furthermore, our results emphasize the relevance of lysosomal S-depalmitoylation for neuronal functioning. The fact that neither Cln5 nor PPT1 can compensate for each other’s functional loss and both cause an early-onset neurodegenerative disorder may be explained either by distinct substrate specificity or by functional interactions. PPT1 itself is palmitoylated and partially inhibited by its palmitoylation (41). It is currently not known that thioesterase catalyzes its S-depalmitoylation. However, we hypothesize that palmitoylation may regulate the enzymatic activity of various enzymes that are associated with neurodegeneration.
Because we detected the depalmitoylase activity of Cln5, PPPDE1, and PPPDE2 by the cleavage of the two artificial substrates DPP-5 and MU-6S-palm-βGlc, we cannot assess the physiological importance of their activities. Additional experiments are needed to identify natural Cln5 substrates and to delineate the substrate specificity among the cysteine-based S-depalmitoylases of the PPPDE proteins.
Proteases that are involved in the formation of the Alzheimer β-amyloid plaques are palmitoylated. The transmembrane aspartyl protease BACE1 (β-site β-amyloid precursor protein cleaving enzyme 1) contains several palmitoylation sites that determine the extent of its shedding (42). Lack of BACE1 palmitoylation reduced cerebral amyloid burden and ameliorated memory deficits in transgenic mouse models of Alzheimer’s disease (43). We speculate that impaired S-depalmitoylation may increase the amount of palmitoylated BACE1 and, thus, promote the formation of the β-amyloid plaques. Further experiments are required to clarify whether the Cln5 variant Asn320Ser is associated with increased BACE1 palmitoylation, explaining the segregation of this allele with multiplex Alzheimer’s disease families (3). In the end, our results highlight the pivotal role of S-depalmitoylation in the pathology of early- and late-onset neurodegeneration and will stimulate further investigations to elucidate the mechanism of neuronal loss in the state of unbalanced protein lipidation.
MATERIALS AND METHODS
Materials
Dulbecco’s modified Eagle’s medium was purchased from Invitrogen. Oligonucleotides for the introduction of the C-terminal tag were synthesized by MWG-Biotech (Munich, Germany). Endoglycosidase H (Endo H) (cleaves within the core of high mannose oligosaccharides from N-linked glycoproteins leaving one N-acetyl-d-glucosamine residue bound after cleavage) was supplied by New England Biolabs (Frankfurt, Germany). The resorufin-labeled casein was purchased from Sigma-Aldrich (Munich, Germany) as the universal protease substrate, Roche. PPPDE1 expressed in human embryonic kidney (HEK) 293T was purchased from OriGene Technologies GmbH (Herford, Germany), and PPPDE2 expressed in Escherichia coli was obtained from Novus Biologicals (Centennial, USA). Human cathepsin L and cathepsin S expressed in mouse myeloma cell lines were purchased from R&D Systems (Minneapolis, USA). All other reagents were purchased from Merck Sigma-Aldrich (Munich, Germany).
Expression and purification of Cln5
The amino acid sequence “RSHHHHHH” was introduced at the C terminus of the CLN5 cDNA to facilitate the purification of Cln5. Active-site Cln5 variants were generated using primers 5′-tgaatccaatggcatcagccataattttcaagtgtcccaggag-3′ and 5′-ctcctgggacacttgaaaattatggctgatgccattggattca-3′ for substitution of histidine-166 to alanine, primers 5′-gtccttaacacaaatttggaagagtcgtaggaatcaaaccatg-3′ and 5′-catggtttgattcctacgactcttccaaatttgtgttaaggac-3′ for substitution of cysteine-280 to serine, 5′-ttcgattacaagccccatcatgggaatttaaatatg-3′ and 5′-catatttaaattcccatgatggggcttgtaatcgaa-3′ for substitution of valine-150 to serine, 5′-ctcctgggacacttgaaaagtatgcatgatgccattgga-3′ and 5′-tccaatggcatcatgcatacttttcaagtgtcccaggag-3′ for substitution of leucine-164 to serine, 5′-accaactatacaagaagttttctttacagtggag-3′ and 5′-ctccactgtaaagaaaacttcttgtatagttggt-3′ for substitution of isoleucine-308 to serine, 5′-gaaacaggaatttatgatgagacatggaatgtaa-3′ and 5′-ttacattccatgtctcatcataaattcctgtttc-3′ for substitution of tyrosine-258 to aspartate, and 5′-ggtttgattcctacaactgttccaaatttg-3′ and 5′-caaatttggaacagttgtaggaatcaaacc-3′ for substitution of aspartate-279 to asparagine. Transfection of the modified CLN5 cDNA and selection of HEK293 cells were performed as described previously (44).
Both the native and selenomethionine derivative recombinant protein samples of Cln5 were purified from the cell culture supernatant. The medium was cleared by centrifugation at 3000g and 4°C for 60 min and then filtered with a 0.2-μm pore membrane. After addition of 20 mM K2HPO4 (pH 7.5), 0.5 M NaCl, and 40 mM imidazole, the crude solution was loaded on a HisTrap HP column (GE Healthcare). Bound Cln5 was eluted with an imidazole gradient and was typically released at 70 to 90 mM imidazole. For crystallization, the purified Cln5 fractions were pooled and concentrated using a centrifugal filter (Millipore, Schwalbach, Germany). The final solution contained Cln5 (10 mg/ml) and was adapted to 20 mM KH2PO4 (pH 4.6) and 50 mM NaCl. Deglycosylation of Cln5 Endo H digestion was performed at nondenaturing conditions according to the supplier’s protocol.
Crystallization
Crystals were obtained using the sitting-drop vapor diffusion method at 293 K. Robotic crystallization screening trials were set up in 96-well AxyGem plates (Axygen Scientific, Union City, USA) using Tecan Genesis and TTP Labtech Mosquito robots. The reservoir and drop volumes were 100 μl and 200 nl, respectively. The drop was prepared by mixing protein solution with reservoir solution in a 1:1 ratio.
Diamond-shaped crystals were grown from precipitate that had formed under conditions of 22.3 to 25.9% polyethylene glycol 3350 (w/v) (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) and 179 to 193 mM magnesium nitrate (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) after 12 weeks. For cryoprotection, the crystals were soaked in reservoir solution with addition of 5% glycerol (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) or with an increased concentration of precipitant of 35% polyethylene glycol 3350 (w/v). The crystals were mounted on the MiTeGen MicroMounts (MiTeGen LLC, Ithaca, USA) and flash-cooled in liquid nitrogen.
Data collection and processing
Single-crystal x-ray diffraction data were collected on the PXII-X10SA beamline at the Swiss Light Source using the monochromatic radiation of selenium inflection wavelength of 0.979 Å with a Pilatus 6M detector, an oscillation range of 0.1° or 0.2°, and an exposure time of 0.1 s. A total of 900 to 3600 images were collected per scan with multiple scans for each crystal. The space group was determined to be P3221 with unit cell parameters of a = 58.406 Å and c = 178.235 Å. Data integration and merging were performed using XDS (X-ray Detector Software) and XSCALE (45) (see table S1). Resolution limits were set per run from 2.65 to 3.27 Å on the basis of the self-correlation coefficients and Rmeas of 65 and 80%, respectively. Data were selected and merged by optimizing the strength of the anomalous signal characterized by maximizing Combined figure of merit (CFOM) in SHELXD (46) and the averaged anomalous density in ANODE (47).
Structure solution and refinement
The crystal structure of Cln5 was solved by selenium SAD from the intrinsic selenomethionine using SHELXD (48). The initial auto trace was accomplished with SHELXE and extended using ARP/wARP (49). Iterative steps of manual model building using COOT (50) and refinement by REFMAC 5.8.0238 (51) and Phenix_refine (52) were used. Final model validation was achieved using PDB-REDO (53) and MolProbity (54) and by the validation of the sugar moieties with Privateer (55). The occupancy of the selenium sites was refined using SHELXL (56) after conversion of the files into the appropriate format using PDB2INS (57). The structural refinement of human CLN5 protein converged to an Rfree of 0.247 and an R value of 0.221. The overall geometry shows no abnormalities, with an all-atom clash score of 4.30 and a score of 1.58 as given by MolProbity (54). The structure had 0.72% outliers, and 94.62% of all residues were in favored regions of the Ramachandran plot. The PDB ID of the structure is 6R99. Refinement statistics are shown in table S2.
Synthesis of activity-based probes
To synthesize the activity-based probes [Ubiquitin propargylamide (Ub-PA) and SUMO1-PA], both proteins were recombinantly expressed in E. coli with a C-terminal intein tag. The proteins were purified in PA buffer [20 mM Hepes, 50 mM sodium acetate (pH 6.5), and 75 mM NaCl) via affinity chromatography using chitin resin (New England Biolabs). In a 24-hour reaction, the proteins were cleaved off the chitin beads by addition of 100 mM MesNA (sodium 2-mercaptoethanesulfonate) to the PA buffer, leading to the formation of Ub-MesNA/SUMO1-MesNA. The eluted fractions were subjected to size exclusion chromatography (HiLoad 26/600 Superdex 75) with PA buffer. Subsequently, 300 μM Ub-MesNA/SUMO1-MesNa were incubated with 600 μM propargylamine hydrochloride (Sigma-Aldrich) in PA buffer containing 150 mM NaOH overnight at 4°C. Unreacted propargylamine was removed by size exclusion chromatography, and Ub-PA/SUMO1-PA was collected.
Ubiquitin chain generation
M1-linked di-ubiquitin (2M1) was expressed as a linear fusion protein and purified by ion exchange chromatography and size exclusion chromatography. K48-linked ubiquitin chains were enzymatically assembled using CDC34 (K48) as previously described. Briefly, ubiquitin chains were generated by incubation of 1 μM E1, 25 μM of the respective E2, and 2 mM ubiquitin in reaction buffer [10 mM adenosine triphosphate, 40 mM tris (pH 7.5), 10 mM MgCl2, and 1 mM dithiothreitol (DTT)] for 18 hours at room temperature. The reaction was stopped by 20-fold dilution in 50 mM sodium acetate (pH 4.5), and chains of different lengths were separated by cation exchange using a RESOURCE S column (GE Healthcare). Elution of different chain lengths was achieved with a gradient from 0 to 600 mM NaCl.
SUMO2 chains were synthesized by incubation of 1 μM E1, 25 μM E2 [glutathione S-transferase (GST)–tagged Ubc9], and 2 mM SUMO2 in reaction buffer (pH 7.5) for 24 hours at 4°C (58). The synthesized chains were separated by size exclusion chromatography (HiLoad 16/600 Superdex 75). The high molecular SUMO2 chains used for this study were conjugated to the GST tag of Ubc9.
TAILS analysis
To examine a potential role of Cln5 in proteolysis of target proteins, we performed TAILS analysis on Cln5−/− proteome from mouse neural progenitor cells (NPCs) incubated with recombinant human Cln5. Cln5−/− mouse NPCs were provided by K. Kanninen (A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland) and previously characterized (59). NPCs were grown overnight in serum-free medium, and cells were then washed three times with phosphate-buffered saline before protein extraction using radioimmunoprecipitation assay (RIPA) buffer. Cells were incubated in RIPA buffer for 15 min at 4°C, vortexed, and then centrifuged at 14,500 rpm for 15 min at 4°C. Bicinchoninic acid (BCA) assay was performed according to the manufacturer’s directions, and 500 μg of protein was then taken for buffer exchange to 0.1 M MES (pH 7). Buffer exchange was achieved using ReadyLyzer 3, 3.5-kDa molecular weight cutoff dialysis tubes according to the manufacturer’s directions. Protein concentration following buffer exchange was determined using BCA assay.
Purified recombinant Cln5 (1 μg) or buffer alone was added to 100 μg of proteome in 0.1 M MES (pH 7) and incubated at 37°C for 4 hours. Protein quality before and after incubation with Cln5 was determined by gel electorophoresis using a 10% Bio-Rad stain-free gel and visualized after activation with ultraviolet using a Bio-Rad ChemiDoc XR imager.
TAILS processing and analysis was performed by the Functional Genomics Centre Zürich. Samples were precipitated by acetone/methanol precipitation (100 μg for each sample). The samples were reduced, alkylated, and labeled with TMT (TMTduplex Isobaric Mass Tagging Kit by Thermo Fisher Scientific). After combining the samples, unreacted reagents were cleaned up by acetone/methanol precipitation. The combined samples were digested with trypsin (6 hours at 42°C), and after digestion, an aliquot was taken (pre-TAILS sample). The HPG-ALDII (hyperbranched polyglycerol polyaldehyde-derivatized type II) polymer was added, and the reaction was performed overnight at 37°C. After quenching, the labeled peptides were recovered by spinning through a 30-kDa centrifugal filter unit (Microcon, Merck Millipore Ltd.; TAILS sample).
Both the pre-TAILS and the TAILS sample were desalted by C18 and injected into a liquid chromatography–tandem mass spectrometry system. The protein identification and quantification were performed using Proteome Discoverer 2.2 against the Mus musculus protein database.
Phospholipase A2 activity assay
To check for a rudimentary phospholipase A2-like activity, 25 μg of purified Cln5 was analyzed using the Invitrogen EnzCheck Phospholipase A2 Assay Kit (Thermo Fisher Scientific, Eugene, USA) as described by the manufacturer. Briefly, a lipid mix containing 10 mM dioleoylphosphatidylcholine, 10 mM dioleoylphosphatidylglycerol, and 1 mM of an intrinsically quenched fluorescencent phospholipase substrate (Red/Green BODIPY PC-A) was prepared. Using a narrow-bore pipette tip, the lipid-mix was introduced into the reaction buffer placed in a beaker with a magnetic stir bar to form a liposome emulsion. Purified Cln5 was placed in a 96-well plate (Greiner Bio-One, Frickenhausen, Germany), and the assay liposome emulsion was added. The reaction was monitored continuously in terms of fluorescence units (excitation at 460 nm, emission at 515 nm) using a SynergyMx microplate reader (BioTek, Winooski, USA), and an assay blank was subtracted.
Thioesterase activity assay
Thioesterase activity of Cln5 as well as PPPDE1 and PPPDE2 was tested using two different substrates with C16-palmitoyl moieties, MU-6S-palm-βGlc and DPP-5. In either case, the reaction buffer was prewarmed to 37°C and composed of 50 mM NaP (pH 5.0), 150 mM NaCl, and 0.1% Triton X-100. In general, reactions took place in 200 μl using 1 μg of purified Cln5 protein (unless otherwise stated) in a 96-well plate–based format (black, polystyrene) in a Cytation 5 instrument (BioTek). When using MU-6S-palm-βGlc as a Cln5 substrate, 20 μg of β-glucosidase was in the mixture. The reaction of Cln5 was initiated by adding 100 μl of substrate solution of different concentrations. The fluorescence change of MU-6S-palm-βGlc was recorded with λex = 360 nm and λem = 450 nm. The fluorescence change of the catalytic reaction was normalized to a free methylumbelliferyl standard regression to calculate the enzymatic rate. Similarly, the fluorescence change of DPP-5 was recorded with λex = 485 nm and λem = 540 nm, and the resulting slopes of the catalytic reaction were normalized to the regression of free DPP-5 fluorophore to calculate the enzymatic rate. For standard regression generation, DPP-5 was reduced by addition of 50 mM DTT for 4 hours at 37°C to chemically release the fluorophore.
Western blot analysis
For Western blot analysis of crude protein extracts, 2 μg of total protein was loaded onto a 10% SDS gel. Separated proteins were electroblotted using the semidry method onto a polyvinylidene difluoride membrane. After membrane blocking with 5% bovine serum albumin (BSA) in TBS-T (tris buffered saline with tween 20)solution, blotted proteins were immunodecorated with antibodies raised against human Cln5 (rabbit, monoclonal, EPR12197; 1:1000 in 5% BSA TBS-T) and heat shock protein 90 (rabbit, monoclonal, EPR16621; 1:1000 in 5% TBS-T). Horseradish peroxidase bioluminescence from secondary antibody decorations was detected using a Bio-Rad ChemiDoc detection system.
Supplementary Material
Acknowledgments
We are grateful to K. Schreiber, T. Wilke, and M. Ziegenbein (University Medical Center Göttingen, Germany) for excellent technical assistance. We thank K. Kanninen (A.I. Virtanen Institute for Molecular Sciences, University of Eastern Finland; katja.kanninen@uef.fi] for providing the Cln5−/− mouse neural progenitor cells. Proteomics analysis and TAILS analysis were performed at the Functional Genomics Center Zurich (FGCZ), and we would like to thank P. Nanni, T. Kockmann, and L. Kunz for technical expertise and support. We would like to acknowledge L. Neuenroth and C. Lenz at the UMG Core Facility Proteomics for analytical support and discussions. This study was supported by the Max Planck Society. We would like to thank the staff at beamline X10SA of Swiss light source for support with data collection.
Funding: This work was supported by Swiss National Science Foundation grant 310030_185298/1 (to R.S.), National Institutes of Health R35 GM119840 (to B.C.D.), Deutsche Forschungsgemeinschaft (DFG) GA 354/14-1, and the DFG under Germany’s Excellence Strategy (EXC 2067/1) (to J.G.).
Author contributions: Conceptualization: R.S. and R.K. Methodology: G.M.S., R.K., A.V.L., S.B., and R.S. Experimental work: A.V.L., D.B., S.B., L.M.C., H.K., I.E., T.Q., R.S.K., C.B., and A.S. Funding acquisition: R.S., J.G., and B.C.D. Supervision: G.M.S., R.S., R.K., S.B., J.S., K.H., and J.G. Writing—original draft: R.S., A.V.L., S.B., and L.M.C. Writing—review and editing: R.S., R.K., B.C.D., J.S., J.G., H.K., G.M.S., and S.B.
Competing interests: B.C.D. and R.S.K. are inventors on a patent related to this work filed by the University of Chicago (no. 10,413,583; filed 30 November 2017, published 31 May 2018). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Correction (12 May 2022): Due to a production error, the x-axis labels in Figure 4B (bottom) were erroneously removed in an earlier version. The x-axis labels have now been restored.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Tables S1 and S2
REFERENCES AND NOTES
- 1.Haltia M., The neuronal ceroid-lipofuscinoses. J. Neuropathol. Exp. Neurol. 62, 1–13 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Mole S. E., Cotman S. L., Genetics of the neuronal ceroid lipofuscinoses (Batten disease). Biochim. Biophys. Acta 1852, 2237–2241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qureshi Y. H., Patel V. M., Berman D. E., Kothiya M. J., Neufeld J. L., Vardarajan B., Tang M., Reyes-Dumeyer D., Lantigua R., Medrano M., Jimenez-Velazquez I. J., Small S. A., Reitz C., An Alzheimer’s disease-linked loss-of-function CLN5 variant impairs cathepsin D maturation, consistent with a retromer trafficking defect. Mol. Cell. Biol. 38, e00011–e00018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xin W., Mullen T. E., Kiely R., Min J., Feng X., Cao Y., O’Malley L., Shen Y., Chu-Shore C., Mole S. E., Goebel H. H., Sims K., CLN5 mutations are frequent in juvenile and late-onset non-finnish patients with NCL. Neurology 74, 565–571 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Kousi M., Lehesjoki A. E., Mole S. E., Update of the mutation spectrum and clinical correlations of over 360 mutations in eight genes that underlie the neuronal ceroid lipofuscinoses. Hum. Mutat. 33, 42–63 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Savukoski M., Klockars T., Holmberg V., Santavuori P., Lander E. S., Peltonen L., CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat. Genet. 19, 286–288 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Jules F., Sauvageau E., Dumaresq-Doiron K., Mazzaferri J., Haug-Kroper M., Fluhrer R., Costantino S., Lefrancois S., CLN5 is cleaved by members of the SPP/SPPL family to produce a mature soluble protein. Exp. Cell Res. 357, 40–50 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Isosomppi J., Vesa J., Jalanko A., Peltonen L., Lysosomal localization of the neuronal ceroid lipofuscinosis CLN5 protein. Hum. Mol. Genet. 11, 885–891 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Schmiedt M. L., Bessa C., Heine C., Ribeiro M. G., Jalanko A., Kyttala A., The neuronal ceroid lipofuscinosis protein CLN5: New insights into cellular maturation, transport, and consequences of mutations. Hum. Mutat. 31, 356–365 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Basak I., Hansen R. A., Ward M. E., Hughes S. M., Deficiency of the lysosomal protein CLN5 alters lysosomal function and movement. Biomolecules 11, 1412 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmiedt M. L., Blom T., Blom T., Kopra O., Wong A., von Schantz-Fant C., Ikonen E., Kuronen M., Jauhiainen M., Cooper J. D., Jalanko A., Cln5-deficiency in mice leads to microglial activation, defective myelination and changes in lipid metabolism. Neurobiol. Dis. 46, 19–29 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Yasa S., Sauvageau E., Modica G., Lefrancois S., CLN5 and CLN3 function as a complex to regulate endolysosome function. Biochem. J. 478, 2339–2357 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Basak I., Wicky H. E., McDonald K. O., Xu J. B., Palmer J. E., Best H. L., Lefrancois S., Lee S. Y., Schoderboeck L., Hughes S. M., A lysosomal enigma CLN5 and its significance in understanding neuronal ceroid lipofuscinosis. Cell. Mol. Life Sci. 78, 4735–4763 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anantharaman V., Aravind L., Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4, R11 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q., Rawlings N. D., Chiu H. J., Jaroszewski L., Klock H. E., Knuth M. W., Miller M. D., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A., Structural analysis of papain-like NlpC/P60 superfamily enzymes with a circularly permuted topology reveals potential lipid binding sites. PLoS One 6, e22013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan J. A., Gilman A. G., A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J. Biol. Chem. 273, 15830–15837 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Lin D. T., Conibear E., ABHD17 proteins are novel protein depalmitoylases that regulate N-Ras palmitate turnover and subcellular localization. eLife 4, e11306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long J. Z., Cravatt B. F., The metabolic serine hydrolases and their functions in mammalian physiology and disease. Chem. Rev. 111, 6022–6063 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yokoi N., Fukata Y., Sekiya A., Murakami T., Kobayashi K., Fukata M., Identification of PSD-95 depalmitoylating enzymes. J. Neurosci. 36, 6431–6444 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camp L. A., Hofmann S. L., Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J. Biol. Chem. 268, 22566–22574 (1993). [PubMed] [Google Scholar]
- 21.Chamberlain L. H., Shipston M. J., The physiology of protein S-acylation. Physiol. Rev. 95, 341–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornemann T., Palmitoylation and depalmitoylation defects. J. Inherit. Metab. Dis. 38, 179–186 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Holm L., Laakso L. M., Dali server update. Nucleic Acids Res. 44, W351–W355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L., Kaariainen S., Rosenstrom P., Schenkel A., Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E., Henrick K., Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Shin E. J., Shin H. M., Nam E., Kim W. S., Kim J. H., Oh B. H., Yun Y., DeSUMOylating isopeptidase: A second class of SUMO protease. EMBO Rep. 13, 339–346 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gillies J., Hochstrasser M., A new class of SUMO proteases. EMBO Rep. 13, 284–285 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie X., Wang X., Jiang D., Wang J., Fei R., Cong X., Wei L., Wang Y., Chen H., PPPDE1 is a novel deubiquitinase belonging to a cysteine isopeptidase family. Biochem. Biophys. Res. Commun. 488, 291–296 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Iyer L. M., Koonin E. V., Aravind L., Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle 3, 1440–1450 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Suh H. Y., Kim J. H., Woo J. S., Ku B., Shin E. J., Yun Y., Oh B. H., Crystal structure of DeSI-1, a novel deSUMOylase belonging to a putative isopeptidase superfamily. Proteins 80, 2099–2104 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Saari J. C., Bredberg D. L., Lecithin:retinol acyltransferase in retinal pigment epithelial microsomes. J. Biol. Chem. 264, 8636–8640 (1989). [PubMed] [Google Scholar]
- 32.Ren X., Lin J., Jin C., Xia B., Solution structure of the N-terminal catalytic domain of human H-REV107—A novel circular permutated NlpC/P60 domain. FEBS Lett. 584, 4222–4226 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Qiu T., Kathayat R. S., Cao Y., Beck M. W., Dickinson B. C., A fluorescent probe with improved water solubility permits the analysis of protein S-depalmitoylation activity in live cells. Biochemistry 57, 221–225 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barrett A. J., Rawlings N. D., Evolutionary lines of cysteine peptidases. Biol. Chem. 382, 727–733 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Rana M. S., Kumar P., Lee C. J., Verardi R., Rajashankar K. R., Banerjee A., Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 359, eaao6326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomatis V. M., Trenchi A., Gomez G. A., Daniotti J. L., Acyl-protein thioesterase 2 catalyzes the deacylation of peripheral membrane-associated GAP-43. PLoS One 5, e15045 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao Y., Qiu T., Kathayat R. S., Azizi S. A., Thorne A. K., Ahn D., Fukata Y., Fukata M., Rice P. A., Dickinson B. C., ABHD10 is an S-depalmitoylase affecting redox homeostasis through peroxiredoxin-5. Nat. Chem. Biol. 15, 1232–1240 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soyombo A. A., Hofmann S. L., Molecular cloning and expression of palmitoyl-protein thioesterase 2 (PPT2), a homolog of lysosomal palmitoyl-protein thioesterase with a distinct substrate specificity. J. Biol. Chem. 272, 27456–27463 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Vesa J., Hellsten E., Verkruyse L. A., Camp L. A., Rapola J., Santavuori P., Hofmann S. L., Peltonen L., Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature 376, 584–587 (1995). [DOI] [PubMed] [Google Scholar]
- 40.Fukata Y., Fukata M., Protein palmitoylation in neuronal development and synaptic plasticity. Nat. Rev. Neurosci. 11, 161–175 (2010). [DOI] [PubMed] [Google Scholar]
- 41.Segal-Salto M., Sapir T., Reiner O., Reversible cysteine acylation regulates the activity of human palmitoyl-protein thioesterase 1 (PPT1). PLoS One 11, e0146466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benjannet S., Elagoz A., Wickham L., Mamarbachi M., Munzer J. S., Basak A., Lazure C., Cromlish J. A., Sisodia S., Checler F., Chretien M., Seidah N. G., Post-translational processing of beta-secretase (beta-amyloid-converting enzyme) and its ectodomain shedding. The pro- and transmembrane/cytosolic domains affect its cellular activity and amyloid-beta production. J. Biol. Chem. 276, 10879–10887 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Andrew R. J., Fernandez C. G., Stanley M., Jiang H., Nguyen P., Rice R. C., Buggia-Prevot V., De Rossi P., Vetrivel K. S., Lamb R., Argemi A., Allaert E. S., Rathbun E. M., Krause S. V., Wagner S. L., Parent A. T., Holtzman D. M., Thinakaran G., Lack of BACE1 S-palmitoylation reduces amyloid burden and mitigates memory deficits in transgenic mouse models of Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, E9665–E9674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pal A., Kraetzner R., Gruene T., Grapp M., Schreiber K., Gronborg M., Urlaub H., Becker S., Asif A. R., Gartner J., Sheldrick G. M., Steinfeld R., Structure of tripeptidyl-peptidase I provides insight into the molecular basis of late infantile neuronal ceroid lipofuscinosis. J. Biol. Chem. 284, 3976–3984 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Kabsch W., XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider T. R., Sheldrick G. M., Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Thorn A., Sheldrick G. M., ANODE: Anomalous and heavy-atom density calculation. J. Appl. Cryst. 44, 1285–1287 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uson I., Sheldrick G. M., An introduction to experimental phasing of macromolecules illustrated by SHELX; new autotracing features. Acta Crystallogr D Struct Biol 74, 106–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langer G., Cohen S. X., Lamzin V. S., Perrakis A., Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov G. N., Skubak P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A., REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H., PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joosten R. P., Long F., Murshudov G. N., Perrakis A., The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen V. B., Arendall W. B. III, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C., MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agirre J., Iglesias-Fernandez J., Rovira C., Davies G. J., Wilson K. S., Cowtan K. D., Privateer: Software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Biol. 22, 833–834 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Sheldrick G. M., Crystal structure refinement with SHELXL. Acta Crystallogr C Struct Chem 71, 3–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lubben A. V., Sheldrick G. M., PDB2INS: Bridging the gap between small-molecule and macromolecular refinement. J. Appl. Cryst. 52, 669–673 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomanov K., Zeschmann A., Hermkes R., Eifler K., Ziba I., Grieco M., Novatchkova M., Hofmann K., Hesse H., Bachmair A., Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26, 4547–4560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savchenko E., Singh Y., Konttinen H., Lejavova K., Mediavilla Santos L., Grubman A., Karkkainen V., Keksa-Goldsteine V., Naumenko N., Tavi P., White A. R., Malm T., Koistinaho J., Kanninen K. M., Loss of Cln5 causes altered neurogenesis in a mouse model of a childhood neurodegenerative disorder. Dis. Model. Mech. 10, 1089–1100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2