Abstract
The mRNA SARS-CoV-2 vaccines were brought to market in response to the public health crises of Covid-19. The utilization of mRNA vaccines in the context of infectious disease has no precedent. The many alterations in the vaccine mRNA hide the mRNA from cellular defenses and promote a longer biological half-life and high production of spike protein. However, the immune response to the vaccine is very different from that to a SARS-CoV-2 infection. In this paper, we present evidence that vaccination induces a profound impairment in type I interferon signaling, which has diverse adverse consequences to human health. Immune cells that have taken up the vaccine nanoparticles release into circulation large numbers of exosomes containing spike protein along with critical microRNAs that induce a signaling response in recipient cells at distant sites. We also identify potential profound disturbances in regulatory control of protein synthesis and cancer surveillance. These disturbances potentially have a causal link to neurodegenerative disease, myocarditis, immune thrombocytopenia, Bell's palsy, liver disease, impaired adaptive immunity, impaired DNA damage response and tumorigenesis. We show evidence from the VAERS database supporting our hypothesis. We believe a comprehensive risk/benefit assessment of the mRNA vaccines questions them as positive contributors to public health.
Keywords: SARS-CoV-2 mRNA vaccines, Type I interferon Response, Exosomes, G-quadruplexes, microRNAs, Cancer
Graphical abstract
1. Introduction
Vaccination is an endeavor to utilize non-pathogenic material to mimic the immunological response of a natural infection, thereby conferring immunity in the event of pathogen exposure. This goal has been primarily pursued through the use of both whole organism and attenuated virus vaccines. Use of fragments of virus or their protein products, referred to as “subunit vaccines,” has been more technically challenging (Bhurani et al., 2018). In any event, an implicit assumption behind the deployment of any vaccination campaign is that the vaccine confers the effects of a ‘benign infection,’ activating the immune system against future exposure, while avoiding the health impacts of actual infection.
Much of the literature on this related to COVID-19 suggests that the immune response to mRNA-based vaccination is similar to natural infection. A preprint study found “high immunogenicity of BNT162b2 vaccine in comparison with natural infection.” The authors found there to be many qualitative similarities though quantitative differences (Psichogiou et al., 2021a). Jhaveri (2021) suggests that mRNA vaccines do what infection with the virus does: “The protein is produced and presented in the same way as natural infection.” The U.S. Centers for Disease Control and Prevention (CDC) makes the case based upon antibody titers generated by prior infection vs. vaccination, in addition to production of memory B cells, to argue that the immune response to vaccination is analogous to the response to natural infection (Centers for Disease Control and Prevention, 2021a). It is this similarity in the humoral immune response to vaccination vs natural infection, paired with both trial and observational data demonstrating reduced risk of infection following vaccination, that stands as the justification for the mass vaccination campaign.
Our paper summarizes the current literature on mRNA and its effects on the molecular biology within human cells. We recognize that there is a wide range of opinions in this nascent phase of mRNA technology. Given its widespread deployment ahead of basic work on so many of the mechanisms we discuss here, we believe that our work is important for providing a broad understanding of present and future reviews that relate to the burgeoning preclinical molecular work being done in this area.
In this paper, we explore the scientific literature suggesting that vaccination with an mRNA vaccine initiates a set of biological events that are not only different from that induced by infection but are in several ways demonstrably counterproductive to both short- and long-term immune competence and normal cellular function. These vaccinations have now been shown to downregulate critical pathways related to cancer surveillance, infection control, and cellular homeostasis. They introduce into the body highly modified genetic material. A preprint has revealed a remarkable difference between the characteristics of the immune response to an infection with SARS-CoV-2 as compared with the immune response to an mRNA vaccine against COVID-19 (Ivanova et al., 2021). Differential gene expression analysis of peripheral dendritic cells revealed a dramatic upregulation of both type I and type II interferons (IFNs) in COVID-19 patients, but not in vaccinees. One remarkable observation they made was that there was an expansion of circulating hematopoietic stem and progenitor cells (HSPCs) in COVID-19 patients, but this expansion was notably absent following vaccination. A striking expansion in circulating plasmablasts observed in COVID-19 patients was also not seen in the vaccinees. All of these observations are consistent with the idea that the anti-COVID-19 vaccines actively suppress type I IFN signaling, as we will discuss below. In this paper we will be focusing extensively, though not exclusively, on vaccination-induced type I IFN suppression and the myriad downstream effects this has on the related signaling cascade.
Since long-term pre-clinical and Phase I safety trials were combined with Phase II trials, then phase II and III trials were combined (Kwok, 2021); and since even those were terminated early and placebo arms given the injections, we look to the pharmacosurveillance system and published reports for safety signals. In doing so, we find that that evidence is not encouraging. The biological response to mRNA vaccination as it is currently employed is demonstrably not similar to natural infection. In this paper we will illustrate those differences, and we will describe the immunological and pathological processes we expect are being initiated by mRNA vaccination. We will connect these underlying physiological effects with both realized and yet-to-be-observed morbidities. We anticipate that implementation of booster vaccinations on a wide scale will amplify all of these problems.
The mRNA vaccines manufactured by Pfizer/BioNTech and Moderna have been viewed as an essential aspect of our efforts to control the spread of COVID-19. Countries around the globe have been aggressively promoting massive vaccination programs with the hope that such efforts might finally curtail the ongoing pandemic and restore normalcy. Governments are reticent to consider the possibility that these injections might cause harm in unexpected ways, and especially that such harm might even surpass the benefits achieved in protection from severe disease. It is now clear that the antibodies induced by the vaccines fade in as little as 3–10 weeks after the second dose (Shrotri et al., 2021), such that people are being advised to seek booster shots at regular intervals (Centers for Disease Control and Prevention, 2021b). It has also become apparent that rapidly emerging variants such as the Delta and now the Omicron strain are showing resistance to the antibodies induced by the vaccines, through mutations in the spike protein (Yahi et al., 2021). Furthermore, it has become clear that the vaccines do not prevent transmission of the disease, but can only be claimed to reduce symptom severity (Kampf, 2021a). A study comparing vaccination rates with COVID-19 infection rates across 68 countries and 2947 counties in the United States in early September 2021, found no correlation between the two, suggesting that these vaccines do not protect from spread of the disease (Subramanian and Kumar, 2947). Regarding symptom severity, even this aspect is beginning to be in doubt, as demonstrated by an outbreak in an Israeli hospital that led to the death of five fully vaccinated hospital patients (Shitrit et al., 2021). Similarly, Brosh-Nissimov et al. (2021) reported that 34/152 (22%) of fully vaccinated patients among 17 Israeli hospitals died of COVID-19.
The increasing evidence that the vaccines do little to control disease spread and that their effectiveness wanes over time make it even more imperative to assess the degree to which the vaccines might cause harm. That SARS-CoV-2 modified spike protein mRNA vaccinations have biological impacts is without question. Here we attempt to distinguish those impacts from natural infection, and establish a mechanistic framework linking those unique biological impacts to pathologies now associated with vaccination. We recognize that the causal links between biological effects initiated by mRNA vaccination and adverse outcomes have not been established in the large majority of cases.
2. Interferons: an overview with attention to cancer surveillance
Discovered in 1957, interferon (IFN) earned its name with the recognition that cells challenged by attenuated influenza A virus created a substance that “interfered with” a subsequent infection by a live virus (Lindenmann, 1982). IFN is now understood to represent a very large family of immune-modulating proteins, divided into three types, designated as type I, II, and III based upon the receptors each IFN interacts with. Type I IFN includes both IFN-α and IFN-β, and this type is the most diverse, being further divided into seventeen subtypes. IFN-α alone has thirteen subtypes currently identified, and each of those is further divided into multiple categories (Wang et al., 2017a). Type I IFNs play a powerful role in the immune response to multiple stressors. In fact, they have enjoyed clinical therapeutic value as a treatment option for a variety of diseases and conditions, including viral infections, solid tumors, myeloproliferative disorders, hematopoietic neoplasms and autoimmune diseases such as multiple sclerosis (Passegu and Ernst, 2009).
As a group, IFNs play exceedingly complicated and pleiotropic roles that are coordinated and regulated through the activity of the family of IFN regulatory factors, or IRFs (Kaur and Fang, 2020). IRF9 is most directly involved in anti-viral as well as anti-tumor immunity and genetic regulation (Alsamman and El-Masry, 2018; Huang et al., 2019; Zitvogel et al., 2015).
Closely related to this are plasmacytoid dendritic cells (pDCs), a rare type of immune cell that circulate in the blood but migrate to peripheral lymphoid organs during a viral infection. They respond to a viral infection by sharply upregulating production of type I IFNs. The IFN-α released in the lymph nodes induces B cells to differentiate into plasmablasts. Subsequently, interleukin-6 (Il-6) induces plasmablasts to evolve into antibody-secreting plasma cells (Jego et al., 2003). Thus, IFNs play a critical role in both controlling viral proliferation and inducing antibody production. Central to both antiviral and anticancer immunity, IFN-α is produced by macrophages and lymphocytes when either is challenged with viral or bacterial infection or encounters tumor cells (De Andrea et al., 2002). Its role as a potent antiviral therapy has been recognized in the treatment of hepatitis C virus complications (Feng et al., 2012), Cytomegalovirus infection (Delannoy et al., 1999), chronic active ebola virus infection (Sakai et al., 1998), inflammatory bowel disease associated with herpes virus infection (Ruther et al., 1998), and others.
Impaired type I IFN signaling is linked to many disease risks, most notably cancer, as type I IFN signaling suppresses proliferation of both viruses and cancer cells by arresting the cell cycle, in part through upregulation of p53, a tumor suppressor gene, and various cyclin-dependent kinase inhibitors (Musella et al., 2017; Matsuoka et al., 1998). IFN-α also induces major histocompatibility (MHC) class 1 antigen presentation by tumor cells, causing them to be more readily recognized by the cancer surveillance system (Heise et al., 2016; Sundstedt et al., 2008). The range of anticancer effects initiated by IFN-α expression is astounding and occurs through both direct and indirect mechanisms. Direct effects include cell cycle arrest, induction of cell differentiation, initiation of apoptosis, activation of natural killer and CD8+ T cells, and others (Schneider et al., 2014).
The indirect anticancer effects are predominantly carried out through gene transcription activation of the Janus kinase signal transducer and activator of transcription (JAK/STAT) pathway. IFN-α binding on the cell surface initiates JAK, a tyrosine kinase, to phosphorylate STAT1 and STAT2 (Asmana Ningrum, 2014). Once phosphorylated, these STATs form a complex with IRF9, one of a family of IRFs that play a wide range of roles in oncogene regulation and other cell functions (Takaoka et al., 2008). It is this complex, named IFN-stimulated gene factor 3 (ISGF3), that translocates to the cell nucleus to enhance the expression of at least 150 genes (Schneider et al., 2014). IRF9 has been suggested to be the primary member of the IRF family of proteins responsible for activation of the IFN-α antiproliferative effects, and that appears to be through its binding to the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) receptor 1 and 2 (TRAIL-R1/2) (Tsuno et al., 2009). IRF7 is another crucial member of the IRF family of proteins involved early in the response to a viral infection. It is normally expressed in low amounts but is strongly induced by ISGF3. IRF7 also undergoes serine phosphorylation and nuclear translocation to further activate the immune response. IRF7 has a very short half-life, so its gene-induction process is transient, perhaps to avoid overexpression of IFNs (Honda et al., 2006).
Once TRAIL is bound by IRF9, it is then able to act as a ligand for Death Receptor 4 (DR4) or DR5, initiating a cascade of events involving production of caspase 8 and caspase 3, and ultimately triggering apoptosis (Sayers, 2011). Dysregulation of this pathway, through suppression of either IFN-α or IRF9 and the resulting failure to bind TRAIL-R, has been associated with several hematologic malignancies (Testa, 2010) and has been shown to increase the metastatic potential in animal models of melanoma, colorectal cancer, and lymphoma (Finnberg and El-Deiry, 2008).
IFN-α both initiates and orchestrates a wide range of cancer suppressing roles. Dunn et al. (2005) showed that IFN-α plays an active role in cancer immunoediting, its locus of action being hematopoietic cells that are “programmed” via IFN-α binding for tumor surveillance. It is via the exceedingly complex interactions between type I IFNs and IRF7 and IRF9 in particular that a great deal of antiproliferative effects are carried out. This is evidenced by the large number of studies showing increased tumor growth and/or metastases associated with a wide number of cancer types.
For example, Bidwell et al. (2012) found that, among over 800 breast cancer patients, those with high expression of IRF7-regulated genes had significantly fewer bone metastases, and they propose assessment of these IRF7-related gene signatures as a way to predict those at greatest risk. Use of microRNA to target IRF7 expression has also been shown to enhance breast cancer cell proliferation and invasion in vitro (Li et al., 2015). Zhao et al. (2017) found a similar role for IRF7 in relation to bone metastases in a mouse model of prostate cancer. Regarding the anti-cancer mechanism behind IRF7 expression, Solis et al. (2006) found that IRF7 induces transcription of multiple genes and translation of their downstream protein products including TRAIL, IL-15, ISG-56 and CD80, with the noted therapeutic implications.
IRF9, too, has a central role to play in cancer surveillance and prevention. Erb et al. (2013) demonstrated that IRF9 is the mediator through which IL-6 augments the anti-proliferation effects of IFN-α against prostate cancer cells. Tian et al. (2018) found IRF9 to be a key negative regulator of acute myeloid leukaemia cell proliferation and evasion of apoptosis. It does so, at least in part, through acetylation of the master regulatory protein p53.
Both IFN-α and IRF9 are also apparently necessary for the cancer-preventative properties of a fully functional BRCA2 gene. In a study presented as an abstract at the First AACR International Conference on Frontiers in Basic Cancer Research, Mittal and Chaudhuri (2009) describe a set of experiments which show for the first time that BRCA2 expression leads to increased IFN-α production and augments the signal transduction pathway resulting in the complexing of IRF9, STAT1 and STAT2 described previously. Two years prior, Buckley et al. (2007) had established that BRCA1 in combination with IFN-γ promotes type I IFNs and subsequent production of IRF7, STAT1, and STAT2. Thus, the exceedingly important cancer regulatory genes BRCA1 and BRCA2 rely on IRF7 and IRF9, respectively, to carry out their protective effects. Rasmussen et al. (2021) reviewed compelling evidence that deficiencies of either IRF7 or IRF9 lead to significantly greater risk of severe COVID-19 illness. Importantly, they also note that evidence suggests type I IFNs play a singularly important role in protective immunity against COVID-19 illness, a role that is shared by multiple cytokines in most other viral illnesses including influenza.
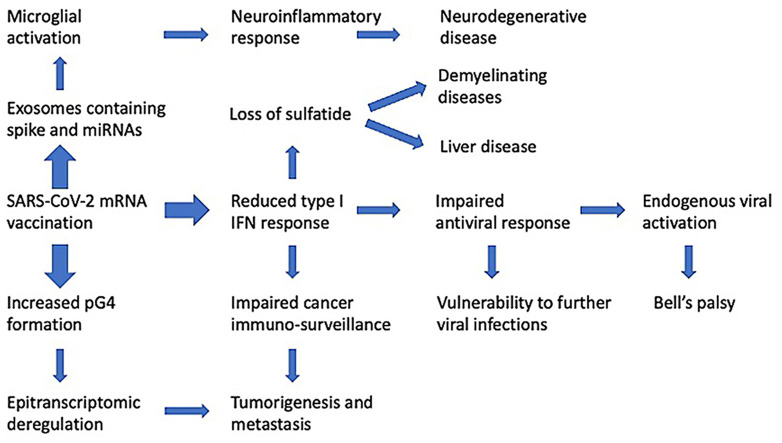
As will be discussed in more detail below, the SARS-CoV-2 spike glycoprotein modifies host cell exosome production. Transfection of cells with the spike protein's gene and subsequent SARS-CoV-2 spike protein production results in those cells generating exosomes containing microRNAs that suppress IRF9 production while activating a range of pro-inflammatory gene transcripts (Mishra and Banerjea, 2021). Since these vaccines are specifically designed to induce high and ongoing production of SARS-CoV-2 spike glycoproteins, the implications are ominous. As described above, inhibition of IRF9 will suppress TRAIL and all its regulatory and downstream apoptosis-inducing effects. IRF9 suppression via exosomal microRNA should also be expected to impair the cancer-protective effects of BRCA2 gene activity, which depends on that molecule for its activity as described above. BRCA2-associated cancers include breast, fallopian tube, and ovarian cancer for women, prostate and breast cancer for men, acute myeloid leukaemia in children, and others (National Cancer Institute, 2021).
Vaccination has also been demonstrated to suppress both IRF7 and STAT2 (Liu et al., 2021). This can be expected to interfere with the cancer-protective effects of BRCA1 as described above. Cancers associated with impaired BRCA1 activity include breast, uterine, and ovarian cancer in women; prostate and breast cancer in men; and a modest increase in pancreatic cancer for both men and women (Cancer risk and BRCA1 gene, 2021).
Reduced BRCA1 expression is linked to both cancer and neurodegeneration. BRCA1 is a well-known breast cancer susceptibility gene. BRCA1 inhibits breast cancer cell proliferation through activation of SIRT1 and subsequent suppression of the androgen receptor (Zhang et al., 2016). In a study conducted by Suberbielle et al. (2015), reduced levels of BRCA1 were found in the brains of Alzheimer's patients. Furthermore, experiments with knocking down neuronal BRCA1 in the dentate gyrus of mice showed that DNA double-strand breaks were increased, along with neuronal shrinkage and impairments in synaptic plasticity, learning and memory.
Analysis detailed in a recent case study on a patient diagnosed with a rare form of lymphoma called angioimmunoblastic T cell lymphoma provided strong evidence for unexpected rapid progression of lymphomatous lesions after administration of the BNT162b2 mRNA booster shot (Goldman et al., 2021). Comparisons of detailed metrics for hypermetabolic lesions conducted immediately before and 21 days after the vaccine booster revealed a five-fold increase after the vaccine, with the post-booster test revealing a 2-fold higher activity level in the right armpit compared to the left one. The vaccine had been injected on the right side. It is worth pointing out in this regard that lymphoid malignancies have been associated with suppression of TRAIL-R1 (MacFarlane et al., 2005).
Given the universally recognized importance of optimally functioning BRCA1/2 for cancer prevention and given the central role of the TRAIL signal transduction pathway for additional cancer surveillance, the suppression of IRF7 and IRF9 through vaccination and subsequent SARS-CoV-2 spike glycoprotein production is extremely concerning for long-term cancer control in SARS-CoV-2 mRNA genetic vaccine injected populations.
3. Considerations in the design of mRNA vaccines
Over the last three decades, the mRNA technological platform aimed to develop effective and safe nucleic acid therapeutic tools is said to have overcome serious obstacles on the coded product instability, the overwhelming innate immunogenicity, and on the delivery methodologies (Pardi et al., 2018). One of the major success stories of mRNA use as a genetic vaccination tool is on the introduction of robust immunity against cancer (Van Lint et al., 2015). In addition, the potential of mRNAs to restore or replace various types of proteins in cases of rare genetic metabolic disorders like Fabry disease has offered great potential therapeutic alternatives where no other medication has proved to be successful (Martini and Guey, 2019). However, in the case of mRNA use as genetic vaccines against infectious diseases, the preliminary safety investigations seemed to be premature for a world-wide use in the general population (Pardi et al., 2018; Doulberis et al., 2021).
Although there are essential epitopes on other SARS-CoV-2 proteins where an antibody response could have provided essential immunogenicity, well known from SARS-CoV-1 (Gordon et al., 2020), the primary goal of the developers of the SARS-CoV-2 mRNA vaccines was to design a vaccine that could induce a robust antibody response exclusively to the spike glycoprotein. Such antibodies, especially IgA in the nasopharynx, should cause the invading viruses to be quickly cleared before they could invade host cells, thus arresting the disease process early on. As stated succinctly by Kaczmarek et al. (2021):
“The rationale behind vaccination is to provide every vaccinated person with protection against the SARS‐CoV‐2 virus. This protection is achieved by stimulating the immune system to produce antibodies against the virus and to develop lymphocytes that will retain memory and the ability to fight off the virus for a long time.” However, since vaccination is given parenterally, IgG is the principal antibody class that is raised against the SARS-CoV-2 spike glycoprotein, not IgA (Wisnewski et al., 2021).
Vaccines generally depend upon adjuvants such as aluminum and squalene to provoke immune cells to migrate to the injection site immediately after vaccination. In the history of mRNA vaccine development, it was initially hoped that the mRNA itself could serve as its own adjuvant. This is because human cells recognize viral RNA as foreign, and this leads to upregulation of type I IFNs, mediated via toll like receptors such as TLR3, TLR7 and TLR8 (Karik ó et al., 2005).
However, with time it became clear that there were problems with this approach, both because the intense reaction could cause flu-like symptoms and because IFN-α could launch a cascade response that would lead to the breakdown of the mRNA before it could produce adequate amounts of SARS-CoV-2 spike glycoprotein to induce an immune response (de Beuckelaer et al., 2016). A breakthrough came when it was discovered experimentally that the mRNA coding for the spike protein could be modified in specific ways that would essentially fool the human cells into recognizing it as harmless human RNA. A seminal paper by Karikó et al. (2005) demonstrated through a series of in vitro experiments that a simple modification to the mRNA such that all uridines were replaced with pseudouridine could dramatically reduce innate immune activation against exogenous mRNA. Andries et al. (2015) later discovered that 1-methylpseudouridine as a replacement for uridine was even more effective than pseudouridine and could essentially abolish the TLR response to the mRNA, preventing the activation of blood-derived dendritic cells. This modification is applied in both the mRNA vaccines on the market (Park et al., 2021).
Rather prophetically, the extensive review by Forni and Mantovani (2021) has raised serious questions about the development of innate immunity by the mRNA SARS-CoV-2 genetic vaccinations. As the authors declared: “Due to the short development time and the novelty of the technologies adopted, these vaccines will be deployed with several unresolved issues that only the passage of time will permit to clarify.” Subsequently, the authors recommended including certain molecules such as the long pentraxin PTX3 as representative humoral immunity markers to assess the early activation of innate immune mechanisms and the underlying reactogenicity under the BIOVACSAFE consortium protocols (Forni and Mantovani, 2021; Weiner et al., 2019). However, to the best of our knowledge these safety protocols have not been included in the assessment of induced innate immunity by the SARS-CoV-2 mRNA genetic vaccines (Mulligan et al., 2020).
In this regard, in the case of SARS-CoV-2 BNT162b2 mRNA vaccine, unlike the immune response induced by natural SARS-CoV-2 infection, where a robust interferon response is observed, those vaccinated with BNT162b2 mRNA vaccines developed a robust adaptive immune response which was restricted only to memory cells, i.e., an alternative route of immune response that bypassed the IFN mediated pathways (Mulligan et al., 2020). Furthermore, due to subsequent mutations in the SARS-CoV-2 spike protein, there is a substantial loss of neutralizing antibodies induced by the BNT162b2 mRNA vaccine compared to those conferred by the SARS-CoV-2 mutants alone (Collier et al., 2021). In that respect, as vaccine developers admit: “Vaccine RNA can be modified by incorporating 1-methylpseudouridine, which dampens innate immune sensing and increases mRNA translation in vivo.” (Mulligan et al., 2020; Katalin Karikó et al., 2008). Bearing in mind the multiple mutations that SARS-CoV-2 develops, as for example in the Brazil outbreaks (Timmers et al., 2021), an effective immune response that prevents the spread of SARS-CoV2 mutants necessarily involves the development of a robust IFN-I response as a part of the innate immune system. This response also requires the involvement of a functional NF-κB response. Unfortunately, spike glycoprotein overexpression dismantles the NF-κB pathway responses, and this molecular event can be augmented by spike-protein-coding mRNAs (Kyriakopoulos and McCullough, 2021; Jiang and Mei, 2021).
For successful mRNA vaccine design, the mRNA needs to be encapsulated in carefully constructed particles that can protect the RNA from degradation by RNA depolymerases. The mRNA vaccines are formulated as lipid nanoparticles containing cholesterol and phospholipids, with the modified mRNA complexed with a highly modified polyethylene glycol (PEG) lipid backbone to promote its early release from the endosome and to further protect it from degradation (Hou et al., 2021). The host cell's existing biological machinery is co-opted to facilitate the natural production of protein from the mRNA through endosomal uptake of a lipid particle (Hou et al., 2021). A synthetic cationic lipid is added as well, since it has been shown experimentally to work as an adjuvant to draw immune cells to the injection site and to facilitate endosomal escape. de Beuckelaer et al. (2016) observed that “condensing mRNA into cationic lipoplexes increases the potency of the mRNA vaccine evoked T cell response by several orders of magnitude.” Another important modification is that they replaced the code for two adjacent amino acids in the genome with codes for proline, which causes the spike glycoprotein to stay in a prefusion stabilized form (Wrapp et al., 2020).
The SARS-CoV-2 spike glycoprotein mRNA is further “humanized” with the addition of a guanine-methylated cap, 3′ and 5′ untranslated regions (UTRs) copied from those of human proteins, and finally a long poly(A) tail to further stabilize the RNA (Kyriakopoulos and McCullough, 2021). In particular, researchers have cleverly selected the 3′UTR taken from globins which are produced in large quantities by erythrocytes, because it is very effective at protecting the mRNA from degradation and maintaining sustained protein production (Orlandini von Niessen et al., 2019). This is to be expected, since erythrocytes have no nucleus, so they are unable to replace the mRNAs once they are destroyed. Both the Moderna and the Pfizer vaccines adopted a 3′UTR from globins, and the Pfizer vaccine also uses a slightly modified globin 5′UTR (Xia, 2021). de Beuckelaer et al. (2016) aptly summed up the consequences of such modifications as follows: “Over the past years, technical improvements in the way IVT [in vitro transcribed] mRNAs are prepared (5′ Cap modifications, optimized GC content, improved polyA tails, stabilizing UTRs) have increased the stability of IVT mRNAs to such extent protein expression can now be achieved for days after direct in vivo administration of the mRNA.”
However, the optimized analogue cap formation of synthetic mRNAs inevitably forces the recipient cells to undergo a cap-dependent prolonged translation, ignoring homeostatic demands of cellular physiology (Kyriakopoulos and McCullough, 2021). The cap 2'-O methylation carried out by cap 2′-O methyltransferase (CMTR1) serves as a motif that marks the mRNA as “self,” to prevent recognition by IFN-induced RNA binding proteins (Williams et al., 2020). Thus, the mRNA in the vaccines, equipped with the cap 2'-O methylation motif, evades detection as a viral invasion. Furthermore, the overwhelming impetus for cells to perform a single and artificial approach to translation according to the robust capping and synthetic methylations of mRNAs in vaccines is fundamentally associated with disease progression due to differential rather than normal signaling of pattern recognition receptors (PRRs) (Leung and Amarasinghe, 2016).
The regulatory process controlling mRNA translation is extremely complex, and it is highly disturbed in the context of mRNA vaccines (Kyriakopoulos and McCullough, 2021; Leung and Amarasinghe, 2016). Briefly, the idea is for mRNA vaccines to achieve the intended goal (i.e., production of the modified spike protein) through a stealth strategy that bypasses the natural immunological response to RNA-type viral infection. Injected lipid nanoparticles containing mRNA are brought to the cell interior via endocytosis. The mRNA escapes its lipid carrier and migrates to the ribosome, where it is abundantly translated into its final protein product, following an optimized program for producing large quantities of a specific protein over an extended period of time. These modified SARS-CoV-2 spike glycoproteins then follow one of three primary pathways. Some are proteolytically degraded and fragments are bound by MHC class I molecules for surface presentation to cytotoxic T-cells. A second pathway has those same spike glycoprotein fragments bind MHC class II molecules, move to the cell surface, and activate T-helper cells. A final pathway has soluble spike glycoproteins extruded from the cell in exosomes, where they can be recognized by B-cell-activated spike-glycoprotein-specific antibodies (Chaudhary et al., 2021).
A recent early-release study has found that the mRNA in the COVID-19 vaccines is present in germinal centers in secondary lymphoid tissue long after the vaccine is administered, and that it continues to synthesize spike glycoprotein up to at least sixty days post-vaccination (Röltgen et al., 2022). This suggests that immune cells taking up the mRNA in the arm muscle migrate into the lymph system to the lymph nodes, presumably in order to expose B-cells and T-cells to the toxic antigen. The persistence of the mRNA in the lymph nodes and its sustained synthesis of SARS-CoV-2 spike glycoprotein reflect the clever engineering involved in the mRNA technology, as described above.
In the end, it is through utilization of nanolipids and sophisticated mRNA technology that the normal immune response to exogenous RNA is evaded in order to produce a strong antibody response against an exogenous RNA virus.
4. GC enrichment and potential G4 (pG4) structures in vaccine mRNAs
Recently, members of our team investigated possible alterations in secondary structure of mRNAs in SARS-CoV-2 vaccines due to codon optimization of synthetic mRNA transcripts (McKernan et al., 2021). This study has shown that there is a significant enrichment of GC content in mRNAs in vaccines (53% in BNT162b2 and 61% in Moderna mRNA-1273) as compared to the native SARS-CoV-2 mRNA (36%). The enriched GC content of mRNAs is the result of codon optimization performed during the development of the mRNAs used in SARS-CoV-2 vaccines, apparently without determining the effect on secondary structures, particularly the Guanine quadruplex (G quadruplex) formation (McKernan et al., 2021).
Codon optimization describes the production of synthetic, codon-optimized polypeptides and proteins used in biotechnology therapeutics (such as the synthetic mRNAs used for SARS-CoV-2 vaccination). The altered codon assignments within the mRNA template dramatically increase the quantity of polypeptides and/or proteins produced (Mauro and Chappell, 2014). Synonymous codon replacement also results in a change in the multifunctional regulatory and structural roles of resulting proteins (Shabalina et al., 2013). For this reason, codon optimization has been cautioned against due to its consequent changes causing perturbation in the secondary conformation of protein products with potentially devastating effects on their resulting immunogenicity, efficacy and function (Zhou et al., 2013; Agashe et al., 2013). Notably, various human diseases are the result of synonymous nucleotide polymorphisms (McCarthy et al., 2017).
In an experiment where GC-rich and GC-poor versions of mRNA transcripts for heat shock protein 70 were configured in the context of identical promoters and UTR sequences, it was found that GC-rich genes were expressed several-fold to over a hundred-fold more efficiently than their GC-poor counterparts (Kudla et al., 2006). This is partly because all of the preferred mammalian codons have G or C nucleotides in the third position. It is also well documented that AU-rich elements in the 3’ UTRs can destabilize mRNA (Otsuka et al., 2019). What may be of particular concern is the fact that GC enrichment content in vaccine mRNAs results in an enhanced ability for potential G-quadruplex (pG4) formations in these structures, and this could cause onset of neurological disease (Wang et al., 2021). Remarkably, the human prion protein (PrP) genetic sequence contains multiple G4 forming motifs, and their presence may form the missing link in the initial conversion of PrP to the misfolded form, PrPsc (Olsthoorn, 2014). PrP binding to its own mRNA may be the seed that causes the protein to misfold. This observation is particularly concerning in light of the fact that the SARS-CoV-2 spike glycoprotein has prion-like characteristics (Tetz and Tetz, 2022).
On the one hand, the GC content has a key role in the modulation of translation efficiency and control of mRNA expression in mammals (Babendure et al., 2006). Especially during translation initiation, the GC content operating as a cis-acting mRNA element orchestrates the 43S ribosomal pre-initiation complex attachment and thereafter the assembly of the eukaryotic translation initiation factor 4F (eIF4F) complex. One representative example of this system in action is the regulation of α and β globin mRNA expression through their 5′ untranslated regions (5′UTRs) (Babendure et al., 2006).
On the other hand, the presence of pG4s in RNAs is implicated in cancer biology as key determinants of the regulation of G4 RNA binding proteins such as helicase (Herdy et al., 2018). Generally, the G-quadruplexes in RNAs have essential roles in a) the regulation of gene expression, b) the localization of ribonuclear proteins, c) the mRNA localization and d) the regulation of proto-oncogene expression (Fay et al., 2017).
Regarding SARS-CoV-2, relevant studies reveal overwhelming similarities between SARS-CoV-2 pG4s, including in RNA coding for SARS-CoV-2 spike glycoprotein, and those sequenced in the human transcriptome (Zhang et al., 2020). Thus, it can be inferred that synthetic mRNAs in vaccines carrying more pG4 structures in their coding sequence for SARS-CoV-2 spike glycoprotein will amplify and compound the potential post-transcriptional disorganization due to G4-enriched RNA during natural SARS-CoV-2 infection. Moreover, the cellular nucleic acid binding protein (CNBP), which is the main cellular protein that binds to the SARS-CoV-2 RNA genome in human-infected cells (Schmidt et al., 2021), binds to and promotes the unfolding of SARS-CoV-2 G4s formed by both positive and negative sense template strands of the SARS-CoV-2 RNA genome. A similar modulation of CNBP on vaccine mRNA G4s and promotion of G4 equilibrium towards unfolded conformations create favorable conditions for miRNA binding, and this will have a direct impact on miRNA-dependent regulation of gene expression (Rouleau et al., 2017).
The negative-sense RNAs are intermediate molecules produced by the replicase transcriptase complex (RTC) formed by the nonstructural proteins of coronaviruses (including SARS-CoV-2) to provide efficiency in replication and transcription (Bezzi et al., 2021; Sola et al., 2015). This, however, introduces another potentially serious complication associated with vaccination. Co-infection with other negative sense RNA viruses such as hepatitis C (Jaubert et al., 2018) or infection by other coronaviruses contemporaneous with vaccination periods would provide the necessary machinery of RTC to reproduce negative sense intermediates from synthetic mRNAs and therefore amplify the presence of pG4s by negative sense templates. This would result in further epitranscriptomic dysregulation (Spiegel et al., 2020).
Summarizing the topic to this point, the enrichment of GC content in vaccine mRNA will inevitably lead to an increase in the pG4 content of the vaccines. This, in turn, will lead to dysregulation of the G4-RNA-protein binding system and a wide range of potential disease-associated cellular pathologies including suppression of innate immunity, neurodegeneration, and malignant transformation (Herdy et al., 2018).
Concerning the post translational dysregulation due to emergence of new G4 structures introduced by vaccination, one other important issue related to miRNA regulation and pG4s arises. In miRNA structures, hundreds of pG4 sequences are identified (Rouleau et al., 2018). In their unfolded conformation, as during binding to their respective targets in 3′ to 5′ sequences of mRNAs, miRNAs switch off the translation of their respective target mRNA. Alternatively, when in the presence of a G4 ligand, the translation of their target mRNAs is promoted (Chan et al., 2018). Moreover, a vast number of putative miRNA binding sites overlap with G4s in 3’ UTRs of mRNAs as there are at least 521 specific miRNAs that are predicted to bind to at least one of these G4s. Overall, 44,294 potential G4-miRNA binding sites have been traced to possess putative overlapping G4s in humans (Rouleau et al., 2017).
As described elsewhere, during the cellular translation of vaccine mRNAs, an increased assembly of a number of RNA binding protein helicases, such as eIF4A bound to eIF4G, will occur (Kyriakopoulos and McCullough, 2021). The presence of increased pG4s in synthetic mRNAs can potentially amplify binding of RNA binding proteins and miRNAs. This form of molecular crowding of protein components (helicases) with great affinity for G4 binding (Rouleau et al., 2017) will decrease the number of RNA binding proteins binding G4s normally available for miRNA regulation. This loss of RNA binding proteins as well as miRNA availability for regulation by binding to G4s can dramatically alter the translational regulation of miRNAs present in cells and thereby disrupt essential regulation of oncogene expression. An example is the p16-dependent regulation of the p53 tumor suppressor protein (Rouleau et al., 2017; Al-Khalaf and Aboussekhra, 2018).
This process is exceedingly complicated yet tantamount to cellular homeostasis. So, again, it merits summarizing. If pG4s accumulate, as would be expected with an increased amount of GC content in the vaccine mRNA, this would have an effect of increasing potential G4 structures available during translation events and this can affect miRNA post-transcriptional regulation. This, in turn, would either favor greater expression of the oncogenes related to a range of cancers, or drive cells towards apoptosis and cell death (Weldon et al., 2018). The case study described earlier in this paper strongly supports the hypothesis that these injections induce accelerated lymphoma progression in follicular B-cells (Goldman et al., 2021).
miRNA binding recognition patterns are imperfectly complementary to their target regions, and for this reason they are referred to as “master regulators,” since one miRNA affects a plethora of different targets (Rouleau et al., 2018). The multitude of pG4s in the mRNA of the vaccine would predictably act as decoys, distracting miRNAs from their normal function in regulating human protein expression. The increase in G4 targets due to the vaccine would decrease the availability of miRNAs to target human-expressed G4s for regulation of gene expression. This can result in downregulation of miRNA expression which is implicated in cardiovascular pathology (Small and Olson, 2011), onset of neurodegeneration (Abe and Bonini, 2013), and/or cancer progression (Farazi et al., 2013).
In most respects within epitranscriptomic machinery, miRNAs are involved in translation repression. One example, vital for cellular normal housekeeping, is that of Mouse double minute 2 homolog (MDM2), a physical negative regulatory protein of p53. P53 itself is considered the master regulator of the cellular tumor suppression network of genes. P16 controls the expression of many miRNAs, and, via miR-141 and mIR-146b-5p binding to MDM2 mRNA, it induces the negative regulation of MDM2, thus enabling p53 ubiquitination and promotion of cell survival upon DNA damage events (Al-Khalaf and Aboussekhra, 2018). Dysregulation of miRNAs that control MDM2 suppression of p53 would predictably lead to an increased risk to a range of cancers (Ozaki and Nakagawara, 2011).
5. Type I IFNs and COVID-19
Type I IFNs play an essential role in fighting viral infections, and deficiencies in type I IFN signaling have been associated with poor outcomes from COVID-19 in multiple studies. These cases are often associated with autoantibodies to type I IFNs. As reviewed below, type I IFNs have been used with some success in treating severe COVID-19, particularly if administered very early in the disease process. If, as argued above, the mRNA vaccines interfere with type I IFN signaling, this could lead to increased susceptibility to COVID-19 in the two weeks following the first vaccine, before an antibody response has been initiated.
Cells infected with a virus detect the presence of virus replication through a number of pattern recognition receptors (PRRs), which serve as sentinels sensing aberrant RNA structures that often form during viral replication. These receptors respond by oligomerizing and subsequently inducing type I IFNs, ultimately upregulating a large number of proteins involved in suppressing viral proliferation (Janeway and Medzhitov, 2002).
A multi-author study by researchers in Paris, France, involving a cohort of 50 COVID-19 patients with varying degrees of disease severity, revealed that patients with severe disease were characterized by a highly impaired type I IFN response (Hadjadj et al., 2020). These patients had essentially no IFN-β and low IFN-α production and activity. This was associated with a persistent blood viral load and an exacerbated inflammatory response, characterized by high levels of tumor necrosis factor α (TNF-α) and Il-6. The authors proposed type I IFN therapy as a potential treatment option. A paper by several researchers in the United States also identified a unique and inappropriate inflammatory response in severe COVID-19 patients, characterized by low levels of both type I and type III IFNs along with elevated chemokines and elevated expression of Il-6 (Blanco-Melo et al., 2020).
Type I IFNs have even been proposed as a treatment option for severe COVID-19. In a hamster model, researchers exposed hamsters to SARS-CoV-2 and induced an inflammatory response in the lungs and systemic inflammation in distal tissues. They found that intranasal administration of recombinant IFN-α resulted in a reduced viral load and alleviation of symptoms (Hoagland et al., 2021). A retrospective cohort study of 446 COVID-19 patients determined that early administration of IFN-α2b was associated with reduced in-hospital mortality. However, late IFN therapy increased mortality and delayed recovery, revealing that early administration of interferon therapy is essential for a favorable response (Wang et al., 2020a).
A surprising number of people have neutralizing autoantibodies against type I IFNs, although the underlying etiology of this phenomenon is not understood. A study using longitudinal profiling of over 600,000 peripheral blood mononuclear cells and transcriptome sequencing from 54 patients with COVID-19 and 26 controls found a notable lack of type I IFN-stimulated gene responses in myeloid cells from patients with critical disease (van der Wijst et al., 2021). Neutralizing autoantibodies against type I IFNs were found in 19% of patients with critical disease, 6% of patients with severe disease, and 0% of patients with moderate disease. Another study based in Madrid, Spain revealed that 10% of patients with severe COVID-19 disease had autoimmune antibodies to type I IFNs (Troya et al., 2021). A multi-author study based in France found that COVID-19 mortality was significantly more frequent in patients with neutralizing autoantibodies against type I interferon than those without neutralizing antibodies (55% vs. 23%) (Chauvineau ‐ Grenier et al., 2022). Finally, Stertz and Hale (2021) note that, whether due to autoantibodies or perhaps loss-of-function polymorphisms associated with interferon system genes, deficiencies in interferon production are associated with as many as 15% of all life-threatening COVID-19 cases.
6. Are the methylation strategies for cellular housekeeping generally omitted by vaccine mRNAs?
Methylation of mRNAs has been evolutionarily devised to control translation of transcripts and therefore expression of genes by a complex cascade of methylator (writers), de-methylator (eraser) and reader proteins. Adenosine methylation is the most abundant epitranscriptomic mRNA modification, and it occurs at multiple sites across the mRNA molecule (Zaccara et al., 2019). A key methylation of adenosine “N6-methyladenosine (m6A)” specifically in the 5′ UTR of mRNAs regulates normal cell physiology, the inflammatory response and cancer progression. The role and mechanisms of m6A in human disease is extensive, and it is excellently covered in other comprehensive reviews (Yang et al., 2020; Knuckles and Bühler, 2018). Foremost among these, the SARS-CoV-2 molecular vaccination induces cell stress conditions, as is described by the elevated NF-κB signaling after vaccination (Liu et al., 2021; Koo et al., 2010).
Under conditions of cellular stress, which can be induced by a viral infection or disease states such as cancer, m6A mediates mRNAs to undergo translation preferentially in a cap-independent way (Meyer et al., 2015). As discussed previously, this is opposite to the impact of mRNA SARS-CoV-2 vaccination, which drives cells toward a cap-dependent translation. Furthermore, under diversified conditions of cellular stress, there is an overwhelming induction of transcriptome-wide addition of m6A that causes an increased number of mRNAs to possess 5′UTRs enriched with m6A (Meyer et al., 2015).
Eukaryotic translation initiation factor 4E (eIF4E) is the initial mRNA cap-binding protein that directs ribosomes to the cap structure of mRNAs, in order to initiate translation into protein. The dependence on cap-dependent translation of vaccine mRNAs will consume a surplus of eIF4E availability needed to translate an unnaturally high number of synthetic mRNAs. However, cap-independent translation takes place without requiring eIF4E to be bound to eIF4F. The competition for ribosomes will shift towards the cap-independent translation of transcripts, since the mRNAs undergoing cap-independent translation are equipped, apart from internal ribosome entry sites (IRES), with special binding motifs that bind to factors that actively recruit mRNAs to the ribosome cap-independent translational enhancers (CITEs) (Shatsky et al., 2018).
Furthermore, this also means that eIF4E, which is a powerful oncogene regulator and cell proliferation modulator, will sustain its activities by this competition for an unnaturally prolonged period of time, trying to counterbalance the competition between robustly-capped mRNAs in vaccines and IRES-containing mRNAs (Kyriakopoulos and McCullough, 2021; Svitkin et al., 2005). This type of condition results in dysregulation of co-transcriptional m6A mRNA modifications and seriously links to molecular progressions of various cancers (Han and Choe, 2020), as well as creating predisposing conditions for subsequent viral infections (Svitkin et al., 2005).
We next consider the impact of mRNA-vaccination-derived SARS-CoV-2 spike glycoprotein on the cellular IFN system via massive exosome production.
7. Exosomes and MicroRNAs
An important communication network among cells consists of extracellular vesicles (EVs) that are constantly released by one cell and later taken up by another cell, which could be in a distant organ. Small vesicles known as exosomes, formed inside endosomes, are similar in size to viruses, and are released through exocytosis into the extracellular space to subsequently circulate throughout the body (Yoshikawa et al., 2019). Exosomes can deliver a diverse collection of biologically active molecules, including mRNA, microRNAs (miRNAs), proteins, and lipids (Ratajczak and Ratajczak, 2016). During a viral infection, infected cells secrete large quantities of exosomes that act as a communication network among the cells to orchestrate the response to the infection (Chahar et al., 2015).
In a collaborative effort by a team of researchers from Arizona and Connecticut, it was found that people who were vaccinated with the mRNA vaccines acquired circulating exosomes containing the SARS-CoV-2 spike glycoprotein by day 14 following vaccination (Bansal et al., 2021). They also found that there were no circulating antibodies to the spike glycoprotein fourteen days after the first vaccine. After the second vaccine, however, the number of circulating spike-glycoprotein-containing exosomes increased by up to a factor of 12. Furthermore, antibodies first appeared on day 14. The exosomes presented spike glycoprotein on their surface, which, the authors argued, facilitated antibody production. When mice were exposed to exosomes derived from vaccinated people, they developed antibodies to the spike glycoprotein. Interestingly, following peak expression, the number of circulating spike-glycoprotein-containing exosomes decreased over time, in step with the decrease in the level of antibodies to the spike glycoprotein.
Exosomes exist as a part of the mRNA decay mechanism in close association under stress conditions with stress granules (SGs) and P-bodies (PBs) (Decker and Parker, 2012; Kothandan et al., 2020). Under conditions of vaccine-mRNA-induced translation, which could be called “excessive dependence on cap-dependent translation,” there is an obvious resistance to promotion and assembly of the large decapping complex (Kyriakopoulos and McCullough, 2021), and therefore resistance against physiological mRNA decay processes (Decker and Parker, 2012). This would mean that the fate of particular synthetic mRNAs that otherwise would be determined by the common cellular strategy for mRNA turnover involving messenger ribonucleinproteins (mRNPs) is being omitted (Borbolis and Syntichaki, 2015).
Furthermore, under conditions of over-reliance on cap-dependent translation by the synthetic mRNAs in SARS-CoV-2 vaccines (Kyriakopoulos and McCullough, 2021), many native mRNAs holding considerable IRES and specific methylations (m6A) in their structure will favorably choose cap-independent translation, which is strongly linked to mRNA decay quality control mechanisms (Han and Choe, 2020). In this sense, considerable deadenylated mRNA products as well as products derived from mRNA metabolism (decay) are directly linked to exosome cargoes (Borbolis and Syntichaki, 2015).
An example of dependence on cap-dependent translation is described in T-cell acute lymphoblastic leukaemia (T-ALL). Due to mechanistic target of rapamycin C (mTORC)-1 over-functioning in T-ALL, the cells are driven completely towards cap-dependent translation (Girardi and De Keersmaecker, 2015). An analogous condition is described by Kyriakopoulos and McCullough (2021). Even in this highly aggressive cancerous state, during inhibition of cap-dependent translation in T-ALL cells, there is a rapid reversion to cap-independent translation (Girardi and De Keersmaecker, 2015). Similarly, a picornavirus infection (Jang et al., 1990) drives cells towards cap-independent translation due to inhibition of components of eIF4F complex and pluralism of IRES in viral RNA.
In humans, there is an abundance of mostly asymptomatic picornavirus infections like the Safford Virus with an over 90% seroprevalence in young children and adults (Zoll et al., 2009). In either case, whether an apoptotic event due to a stress-like condition (Rusk, 2008) or an mRNA-cap-driven-like carcinomatous effect (De Paolis et al., 2021), the miRNA levels will be increased due to the increased epitranscriptomic functioning and enhanced mRNA decay. Because of the high demand for gene expression, high levels of certain miRNAs will be expected to be contained in exosomes via P bodies (Yu et al., 2016).
Also, under conditions of overwhelming production of SARS-CoV-2 spike glycoprotein due to SARS-CoV-2 molecular vaccination, it would of course be expected that a significant proportion of over-abundant intracellular spike glycoproteins would also be exported via exosome cargoes (Wei et al., 2021).
Mishra and Banerjea (2021) investigated the role of exosomes in the cellular response of SARS-CoV-2 spike-transfected cells. They wrote in the abstract:
“We propose that SARS-CoV-2 gene product, Spike, is able to modify the host exosomal cargo, which gets transported to distant uninfected tissues and organs and can initiate a catastrophic immune cascade within Central Nervous System (CNS).”
Their experiments involved growing human HEK293T cells in culture and exposing them to SARS-CoV-2 spike gene plasmids, which induced synthesis of spike glycoprotein within the cells. They found experimentally that these cells released abundant exosomes housing spike glycoprotein along with specific microRNAs. They then harvested the exosomes and transferred them to a cell culture of human microglia (the immune cells that are resident in the brain). They showed that the microglia readily took up the exosomes and responded to the microRNAs by initiating an acute inflammatory response. The role of microglia in causing neuroinflammation in various viral diseases, such as Human Immunodeficiency Virus (HIV), Japanese Encephalitis Virus (JEV), and Dengue, is well established. They proposed that long-distance cell-cell communication via exosomes could be the mechanism by which neurological symptoms become manifest in severe cases of COVID-19.
In further exploration, the authors identified two microRNAs that were present in high concentrations in the exosomes: miR-148a and miR-590. They proposed a specific mechanism by which these two microRNAs would specifically disrupt type I interferon signaling, through suppression of two critical proteins that control the pathway: ubiquitin specific peptidase 33 (USP33) and IRF9. Phosphorylated STAT1 and STAT2 heterodimers require IRF9 in order to bind IFN-stimulated response elements, and therefore IRF9 plays an essential role in the signaling response. The authors showed experimentally that microglia exposed to the exosomes extracted from the HEK293 culture had a 50% decrease in cellular expression of USP33 and a 60% decrease in IRF9. They further found that miR-148a specifically blocks USP33 and miR-590 specifically blocks IRF9. USP33 removes ubiquitin from IRF9, and in so doing it protects it from degradation. Thus, the two microRNAs together conspire to interfere with IRF9, thus blocking receptor response to type I interferons.
A study by de Gonzalo-Calvo et al. (2021) looked at the microRNA profile in the blood of COVID-19 patients and their quantitative variance based upon disease severity. Multiple miRNAs were found to be up- and down-regulated. Among these was miR-148a-3p, the guide strand precursor to miR-148a. However, miR-148a itself was not among the microRNAs catalogued as excessive or deficient in their study, nor was miR-590. It appears from these findings that miR148a and miR-590 and their inflammatory effects are unique to vaccination-induced SARS-CoV-2 spike glycoprotein production.
Tracer studies have shown that, following injection into the arm muscle, the mRNA in mRNA vaccines is carried into the lymph system by immune cells and ultimately accumulates in the spleen in high concentrations (Bahl et al., 2017). Other studies have shown that stressed immune cells in germinal centers in the spleen release large quantities of exosomes that travel to the brain stem nuclei along the vagus nerve (as reviewed in Seneff and Nigh (2021)). The vagus nerve is the 10th cranial nerve and it enters the brainstem near the larynx. The superior and recurrent laryngeal nerves are branches of the vagus that innervate structures involved in swallowing and speaking. Lesions in these nerves cause vocal cord paralysis associated with difficulty swallowing (dysphagia) difficulty speaking (dysphonia) and/or shortness of breath (dyspnea) (Gould et al., 2019; Erman et al., 2009). We will return to these specific pathologies in our review of VAERS data below.
HEK293 cells were originally derived from cultures taken from the kidney of a human fetus several decades ago and immortalized through infection with adenovirus DNA. While they were extracted from the kidney, the cells show through their protein expression profile that they are likely to be of neuronal origin (Shaw et al., 2002). This suggests that neurons in the vagus nerve would respond similarly to the SARS-CoV-2 spike glycoprotein. Thus, the available evidence strongly suggests that endogenously produced SARS-CoV-2 spike glycoprotein creates a different microRNA profile than does natural infection with SARS-CoV-2, and those differences entail a potentially wide range of deleterious effects.
A central point of our analysis below is the important distinction between the impact of vaccination versus natural infection on type I IFN. While vaccination actively suppresses its production, natural infection promotes type I IFN production very early in the disease cycle. Those with preexisting conditions often exhibit impaired type I IFN signaling, which leads to more severe, critical, and even fatal COVID-19. If the impairment induced by the vaccine is maintained as antibody levels wane over time, this could lead to a situation where the vaccine causes a more severe disease expression than would have been the case in the absence of the vaccine.
Another expected consequence of suppressing type I IFN would be reactivation of preexisting, chronic viral infections, as described in Section 9.
8. Impaired DNA repair and adaptive immunity
The immune system and the DNA repair system are the two primary systems that higher organisms rely on for defense against diverse threats, and they share common elements. Loss of function of key DNA repair proteins leads to defects in repair that inhibit the production of functional B- and T-cells, resulting in immunodeficiency. Non-homologous end joining (NHEJ) repair plays a critical role in lymphocyte-specific V(D)J recombination, which is essential for producing the highly diverse repertoire of B-cell antibodies in response to antigen exposure (Jiang and Mei, 2021). Impaired DNA repair is also a direct pathway towards cancer.
A paper published by Liu et al., in 2021 monitored several parameters associated with immune function in a cohort of patients by conducting single-cell mRNA sequencing of peripheral blood mononuclear cells (PBMCs) harvested from the patients before and 28 days after the first injection of a COVID-19 vaccine based on a weakened version of the virus (Liu et al., 2021). While these vaccines are different from the mRNA vaccines, they also work by injecting the contents of the vaccine into the deltoid muscle, bypassing the mucosal and vascular barriers. The authors found consistent alteration of gene expression following vaccination in many different immune cell types. Observed increases in NF-κB signaling and reduced type I IFN responses were further confirmed by biological assays. Consistent with other studies, they found that STAT2 and IRF7 were significantly downregulated 28 days after vaccination, indicative of impaired type I IFN responses. They wrote: “Together, these data suggested that after vaccination, at least by day 28, other than generation of neutralizing antibodies, people's immune systems, including those of lymphocytes and monocytes, were perhaps in a more vulnerable state.” (Liu et al., 2021).
These authors also identified disturbing changes in gene expression that would imply impaired ability to repair DNA. Up to 60% of the total transcriptional activity in growing cells involves the transcription of ribosomal DNA (rDNA) to produce ribosomal RNA (rRNA). The enzyme that transcribes ribosomal DNA into RNA is RNA polymerase I (Pol I). Pol I also monitors rDNA integrity and influences cell survival (Kakarougkas et al., 2013). During transcription, RNA polymerases (RNAPs) actively scan DNA to find bulky lesions (double-strand breaks) and trigger their repair. In growing eukaryotic cells, most transcription involves synthesis of ribosomal RNA by Pol I. Thus, Pol I promotes survival following DNA damage (Kakarougkas et al., 2013). Many of the downregulated genes identified by Liu et al. (2021) were linked to the cell cycle, telomere maintenance, and both promoter opening and transcription of POL I, indicative of impaired DNA repair processes.
One of the gene sets that were suppressed was due to “deposition of new CENPA [centromere protein A] containing nucleosomes at the centromere.” Newly synthesized CENPA is deposited in nucleosomes at the centromere during late telophase/early G1 phase of the cell cycle. This points to arrest of the cell cycle in G1 phase as a characteristic feature of the response to the inactivated SARS-CoV-2 vaccine. Arrest of pluripotent embryonic stem cells in the G1 phase (prior to replication initiation) would result in impaired self-renewal and maintenance of pluripotency (Choi et al., 2013).
Two checkpoint proteins crucially involved in DNA repair and adaptive immunity are BRCA1 and 53BP1, which facilitate both homologous recombination (HR) and NHEJ, the two primary repair processes (Zhang and Powell, 2005; Panier and Boulton, 2014). In an in vitro experiment on human cells, the SARS-CoV-2 full-length spike glycoprotein was specifically shown to enter the nucleus and hinder the recruitment of these two repair proteins to the site of a double-strand break (Jiang and Mei, 2021). The authors summarized their findings by saying, “Mechanistically, we found that the spike protein localizes in the nucleus and inhibits DNA damage repair by impeding key DNA repair protein BRCA1 and 53BP1 recruitment to the damage site.”
Another mechanism by which the mRNA vaccines could interfere with DNA repair is through miR-148. This microRNA has been shown to downregulate HR in the G1 phase of the cell cycle (Choi et al., 2014). As was mentioned earlier in this paper, this was one of the two microRNAs found in exosomes released by human cells following SARS-CoV-2 spike glycoprotein synthesis in the experiments by Mishra and Banerjea (2021).
9. Reactivation of varicella-zoster
Type I IFN receptor signaling in CD8+ T cells is critical for the generation of effector and memory cells in response to a viral infection (Kolumam et al., 2005). CD8+ T cells can block reactivation of latent herpes infection in sensory neurons (Liu et al., 2000). If type I IFN signaling is impaired, as happens following vaccination but not following natural infection with SARS-CoV-2, CD8+ T cells’ ability to keep herpes in check would also be impaired. Might this be the mechanism at work in response to the vaccines?
Shingles is an increasingly common condition caused by reactivation of latent herpes zoster viruses (HZV), which also causes chicken pox in childhood. In a systematic review, Katsikas Triantafyllidis et al. (2021) identified 91 cases of herpes zoster occurring an average of 5.8 days following mRNA vaccination. While acknowledging that causality is not yet confirmed, “Herpes zoster is possibly a condition physicians and other healthcare professionals may expect to see in patients receiving COVID-19 vaccines” (Katsikas Triantafyllidis et al., 2021). In a letter to the editor published in September 2, 2021, Fathy et al. (2022) reported on 672 cases of skin reactions that were presumably vaccine-related, including 40 cases of herpes zoster and/or herpes simplex reactivation. These cases had been reported to the American Academy of Dermatology and the International League of Dermatologic Societies’ COVID-19 Dermatology Registry, established specifically to track dermatological sequalae from the vaccines. There are multiple additional case reports of herpes zoster reactivation following COVID-19 vaccination in the literature (Psichogiou et al., 2021b; Iwanaga et al., 2021). Lladó et al. (2021) noted that 51 of 52 reports of reactivated herpes zoster infections happened following mRNA vaccination. Herpes zoster itself also interferes with IFN-α signaling in infected cells both through interfering with STAT2 phosphorylation and through facilitating IRF9 degradation (Verweij et al., 2015).
An additional case of viral reactivation is noteworthy as well. It involved an 82-year-old woman who had acquired a hepatitis C viral (HCV) infection in 2007. A strong increase in HCV load occurred a few days after vaccination with an mRNA Pfizer/BioNTech vaccine, along with an appearance of jaundice. She died three weeks after vaccination from liver failure (Lensen et al., 2021).
10. Immune thrombocytopenia
Immune thrombocytopenia is an autoimmune disorder, where the immune system attacks circulating platelets. Immune thrombocytopenic purpura (ITP) has been associated with several vaccinations, including measles, mumps, rubella (MMR), hepatitis A, varicella, diphtheria, tetanus, pertussis (DPT), oral polio and influenza (Perricone et al., 2014). While there is broad awareness that the adenovirus DNA-based vaccines can cause vaccine-induced immune thrombotic thrombocytopenia (VITT) (Kelton et al., 2021), the mRNA vaccines are not without risk to VITT, as case studies have been published documenting such occurrences, including life threatening and fatal cerebral venous sinus thrombosis (Lee et al., 2021; Akiyama et al., 2021; Atoui et al., 2022; Zakaria et al., 2021). The mechanism is believed to involve VITT antibodies binding to platelet factor 4 (PF4) and forming immune complexes that induce platelet activation. Subsequent clotting cascades cause the formation of diffuse microclots in the brain, lungs, liver, legs and elsewhere, associated with a dramatic drop in platelet count (Kelton et al., 2021). The reaction to the vaccine has been described as being very similar to heparin-induced thrombocytopenia (HIT), except that heparin administration is notably not involved (Cines and Bussel, 2021).
It has been shown that the mRNA vaccines elicit primarily an immunoglobulin G (IgG) immune response, with lesser amounts of IgA induced (Wisnewski et al., 2021), and even less IgM production (Danese et al., 2021). The amount of IgG antibodies produced is comparable to the response seen in severe cases of COVID-19. It is IgG antibodies in complex with heparin that induce HIT. One can hypothesize that IgG complexed with the SARS-CoV-2 spike glycoprotein and PF4 is the complex that induces VITT in response to mRNA vaccines. It has in fact been shown experimentally that the receptor binding domain (RBD) of the spike protein binds to PF4 (Passariello et al., 2021).
The underlying mechanism behind HIT has been well studied, including through the use of humanized mouse models. Interestingly, human platelets, but not mouse platelets, express the FcγRIIA receptor, which responds to PF4/heparin/IgG complexes through a tyrosine phosphorylation cascade to induce platelet activation. Upon activation, platelets release granules and generate procoagulant microparticles. They also take up calcium, activate protein kinase C, clump together into microthrombi, and launch a cell death cascade via calpain activation. These activated platelets release PF4 into the extracellular space, supporting a vicious cycle, as this additional PF4 also binds to heparin and IgG antibody to further promote platelet activation. Thus, FcγRIIA is central to the disease process (Nevzorova et al., 2019).
Studies on mice engineered to express the human FcγRIIA receptor have shown that these transgenic mice are far more susceptible to thrombocytopenia than their wild type counterparts (McKenzie et al., 1999). It has been proposed that platelets may serve an important role in the clearance of antibody-antigen complexes by trapping the antigen in thrombi and/or carrying them into the spleen for removal by immune cells. Platelets are obviously rapidly consumed in the process, which then results in low platelet counts (thrombocytopenia).
Platelets normally circulate with an average lifespan of only five to nine days, so they are constantly synthesized in the bone marrow and cleared in the spleen. Antibody-bound platelets, subsequent to platelet activation via Fcγ receptors, migrate to the spleen where they are trapped and removed through phagocytosis by macrophages (Crow and Lazarus, 2003). Fully one third of the body's total platelets are found in the spleen. Since the mRNA vaccines are carried into the spleen by immune cells initially attracted to the injection site in the arm muscle, there is tremendous opportunity for the release of spike-glycoprotein-containing exosomes by dendritic cells in the spleen synthesizing spike protein. One can speculate that platelet activation following the formation of a P4F/IgG/spike protein complex in the spleen is part of the mechanism that attempts to clear the toxic spike glycoprotein.
We mentioned earlier that one of the two microRNAs highly expressed in exosomes released by human cells exposed to the SARS-CoV-2 spike glycoprotein was miR-148a. miR-148a has been shown experimentally to suppress expression of a protein that plays a central role in regulating FcγRIIA expression on platelets. This protein, called T-cell ubiquitin ligand-2 (TULA-2), specifically inhibits activity of the platelet Fcγ receptor. miR-148a targets TULA-2 mRNA and downregulates its expression. Thus, miR-148a, present in exosomes released by macrophages that are compelled by the vaccine to synthesize SARS-CoV-2 spike glycoprotein, acts to increase the risk of thrombocytopenia in response to immune complexes formed by spike glycoprotein antigen and IgG antibodies produced against the spike glycoprotein.
11. PPAR-α, sulfatide and liver disease
As we have already stated, an experiment by Mishra and Banerjea (2021) demonstrated that the SARS-CoV-2 spike glycoprotein induces the release of exosomes containing microRNAs that specifically interfere with IRF9 synthesis. In this section we will show that one of the consequences of suppression of IRF9 would be reduced synthesis of sulfatide in the liver, mediated by the nuclear receptor peroxisome proliferator-activated receptor α (PPAR-α).
Sulfatides are major mammalian serum sphingoglycolipids which are synthesized and secreted mainly from the liver (Lu et al., 2019). They are the only sulfonated sphingolipids in the body. Sulfatides are formed by a two-step process involving the conversion of ceramide to galactocerebroside and its subsequent sulfation. Sulfatide is expressed on the surface of platelets, erythrocytes and lymphocytes. Serum sulfatides exert both anti-coagulative and anti-platelet-activation functions. The enzyme in the liver that synthesizes sulfatide, cerebroside sulfotransferase, has specifically been found to be induced by activation of PPAR-α in mice (Kimura et al., 2012). Therefore, reduced expression of PPAR-α leads to sulfatide deficiency.
PPAR-α ligands exhibit anti-inflammatory and anti-fibrotic effects, whereas PPAR-α deficiency leads to hepatic steatosis, steatohepatitis, steatofibrosis, and liver cancer (Wang et al., 2020b). In 2019, an experiment was conducted by a team of researchers in Japan on mice with a defective gene for PPAR-α (Lu et al., 2019). These mice, when fed a high cholesterol diet, were susceptible to excess triglyceride accumulation and exacerbated inflammation and oxidative stress in the liver, along with increased levels of coagulation factors. The mice also manifested with decreased levels of sulfatides in both the liver and the serum. The authors hypothesized that cholesterol overload exerts its toxic effects in part by enhancing thrombosis, following abnormal hepatic lipid metabolism and oxidative stress. They showed that PPAR-α can attenuate these toxic effects through transcriptional regulation of coagulation factors and upregulation of sulfatide synthesis, in addition to its effects in ameliorating liver disease. They proposed that therapies such as fibrates aimed at activating PPAR-α might prevent high-cholesterol-diet-induced cardiovascular disease.
Tracer studies have shown that the mRNA from mRNA vaccines migrates preferentially to the liver and spleen, reaching higher concentration there than in any other organs (Bahl et al., 2017). Thus, there is potential for suppression of IRF9 in the liver by the vaccine. IRF9 is highly expressed in hepatocytes, where it interacts with PPAR-α, activating PPAR-α target genes. A study on IRF9 knockout mice showed that these mice developed steatosis and hepatic insulin resistance when exposed to a high-fat diet. In contrast, adenoviral-mediated hepatic IRF9 overexpression in obese mice improved insulin sensitivity and ameliorated steatosis and inflammation (Wang et al., 2013).
Multiple case reports in the research literature describe liver damage following mRNA vaccines (Zin Tun et al., 2021; Dumortiera, 2022; Mann et al., 2021). A plausible factor leading to these outcomes is the suppression of PPAR-α through downregulation of IRF9, and subsequently decreased sulfatide synthesis in the liver.
12. Guillain Barré syndrome and neurologic injury syndromes
GBS is an acute inflammatory demyelinating neuropathy associated with long-lasting morbidity and a significant risk of mortality (Cr é ange, 2000). The disease involves an autoimmune attack on the nerves associated with the release of pro-inflammatory cytokines.
GBS is often associated with autoantibodies to sulfatide and other sphingolipids (Ilyas et al., 1991). Activated T-cells produce cytokines in response to antigen presentation by macrophages, and these cytokines can induce autoantibody production through epitope spreading (Vanderlugt and Miller, 2002). The antibodies, in turn, induce complement activation, which causes demyelination and axonal damage, leading to severe injury to peripheral neurons (Kuwahara and Kusunoki, 2018). The SARS-CoV-2 spike glycoprotein has been shown to bind to heparan sulfate, which is a sulfated amino-sugar complex resembling the sulfated galactose in sulfatide (Kalra and Kandimalla, 2021). Thus, it is conceivable that the spike glycoprotein also binds to sulfatide, and this might trigger an immune reaction to the spike-glycoprotein-sulfatide complex.
As described in the previous section, impaired sulfatide synthesis in the liver due to suppression of IRF9 will lead to systemic sulfatide deficiency over time. Sulfatide deficiency can have major impact in the brain and nervous system. Twenty percent of the galactolipids found in the myelin sheath are sulfatides. Sulfatide is a major component of the nervous system, found in especially high concentrations in the myelin sheath in both the peripheral and the central nervous system. Deficiencies in sulfatide can lead to muscle weakness, tremors, and ataxia (Honke, 2013), which are common symptoms of GBS. Chronic neuroinflammation mediated by microglia and astrocytes in the brain leads to dramatic losses of brain sulfatide, and brain deficiencies in sulfatide are a major feature of Alzheimer's disease (Qiu et al., 2021). Mice with a defect in the ability to synthesize sulfatide from ceramide show an impaired ability to maintain the health of axons as they age. Over time, they develop redundant, uncompacted and degenerating myelin sheaths as well as deteriorating structure at the nodes of Ranvier in the axons, causing the loss of a functionally competent axoglial junction (Marcus et al., 2006).
Angiotensin II (Ang II), in addition to its profound effects on cardiovascular disease, also plays a role in inflammation in the brain leading to neurodegenerative disease (Lanz. et al., 2010). The SARS-CoV-2 spike glycoprotein contains a unique furin cleavage site not found in SARS-CoV, which allows the extracellular enzyme furin to detach the S1 segment of the spike glycoprotein and release it into circulation (Letarov et al., 2021). S1 has been shown to cross the blood-brain barrier in mice (Rhea et al., 2021). S1 contains the receptor binding domain that binds to ACE2 receptors, disabling them. When ACE2 receptor signaling is reduced, Ang II synthesis is increased. Neurons in the brain possess ACE2 receptors that would be susceptible to disruption by S1 released from spike-glycoprotein-containing exosomes or spike-glycoprotein-producing cells that had taken up the nanoparticles in the vaccines. Ang II enhances TLR4-mediated signaling in microglia, inducing microglial activation and increasing the production of reactive oxygen species leading to tissue damage, within the paraventricular nucleus in the brain (Rodriguez-Perez et al., 2015).
Elevated levels of Ang II is a causal factor in neurodegeneration of the optic nerve, causing optic neuritis, which can result in severe irreversible visual loss (Guo et al., 2017). Multiple case reports have described cases of optic neuropathy appearing shortly after mRNA vaccination for COVID-19 (Maleki, 2021; Barone et al., 2021). Other debilitating neurological conditions are also appearing shortly after vaccination, where a causal relationship is suspected. A case study based in Europe tracking neurological symptoms following COVID-19 vaccination identified 21 cases developing within a median of 11 days post-vaccination. The cases had diverse diagnoses including cerebral venous sinus thrombosis, nervous system demyelinating diseases, inflammatory peripheral neuropathies, myositis, myasthenia, limbic encephalitis, and giant cell arteritis (Kaulen et al., 2021). Khayat-Khoei et al. (2021) describe a case series of 7 patients, ages ranging from 24 to 64, presenting with demyelinating disease within 21 days of a first or second mRNA vaccination. Four had a prior history of (controlled) MS, while three were previously healthy.
Hearing loss and tinnitus are also well-known side effects of COVID-19. A case study involved a series of ten COVID-19 patients who suffered from audiovestibular symptoms such as hearing loss, vestibular dysfunction and tinnitus (Jeong et al., 2021). The authors demonstrated that human inner ear tissue expresses ACE2, furin and the transmembrane protease serine 2 (TMPRSS2), which facilitates viral entry. They also showed that SARS-CoV-2 can infect specific human inner ear cell types.
Another study evaluating the potential for the SARS-CoV-2 virus to infect the ear specifically examined expression of the receptor ACE2 and the enzymes furin and TM-PRSS2 various types of cells in the middle and inner ears of mice. They found that ACE2 and furin were “diffusely present in the eustachian tube, middle ear spaces, and cochlea, suggesting that these tissues are susceptible to SARS-CoV-2 infection.” (Uranaka et al., 2021). Tinnitus is positively associated with hypertension, which is induced by elevated levels of Ang II (Rodrigues Figueiredo et al., 2016).
Headache is a very common adverse reaction to the COVID-19 mRNA vaccines, particularly for people who are already susceptible to headaches. In a study based on a questionnaire involving 171 participants, the incidence of headaches was found to be 20.5% after the first vaccine, rising to 45.6% after the second shot (Sekiguchi et al., 2021). A case study described a 37-year-old woman suffering from a debilitating migraine attack lasting for 11 days following the second Pfizer/BioNtech mRNA vaccine (Consoli et al., 2021).
Steroids are often used as adjunct therapy to treat migraine (Huang et al., 2013). Dexamethasone and other steroids stimulate PPAR-α receptors in the liver through the steroid receptor, thus offsetting the effects of IRF9 suppression (Lemberger et al., 1994). A theory for the origins of migraine involves altered processing of sensory input in the brainstem, primarily trigeminal neurons (Dodick and Silberstein, 2006). The trigeminal nerve is in close proximity to the vagus nerve in the brainstem, so spike-glycoprotein-carrying exosomes could easily reach it along the vagal route. Magnetic resonance imaging has revealed that structural changes in the trigeminal nerve reflecting aberrant microstructure and demyelination are a characteristic feature of people who suffer from frequent migraine headaches (Mungoven et al., 2020). A potential factor linked to either SARS-CoV-2 infection or mRNA vaccination is an excessive level of Ang II in the brainstem due to SARS-CoV-2 spike glycoprotein inhibition of ACE2 receptors. ACE inhibitors and Ang II receptor antagonists have become popular drugs to treat migraine headaches off-label (Tronvik et al., 2003; Nandha and Singh, 2012). Migraine headache could thus arise from both the spike glycoprotein's disruption of ACE2 receptors and the destruction of the myelin sheath covering critical facial nerves through a microglial inflammatory response and loss of sulfatide. The source of that spike glycoprotein could be either exogenous or endogenous.
13. Bell's palsy
Bell's palsy is a common cranial neuropathy causing unilateral facial paralysis. Even in the Phase III clinical trials, Bell's palsy stood out, with seven cases appearing in the treatment arm as compared to only one in the placebo group (FDA, 2021a; FDA, 2021b). A case study reported in the literature involved a 36-year-old man who developed weakness in the left arm one day after vaccination, progressing to numbness and tingling in the arm and subsequent symptoms of Bell's palsy over the next few days. A common cause of Bell's palsy is reactivation of herpes simplex virus infection centered around the geniculate ganglion (Eviston et al., 2015). This, in turn, can be caused by disruption of type I IFN signaling.
14. Myocarditis
There has been considerable media attention devoted to the fact that COVID-19 vaccines cause myocarditis and pericarditis, with an increased risk in particular for men below the age of 50 (Simone et al., 2021; Jain et al., 2021). The SARS-CoV-2 spike glycoprotein has been demonstrated to injure cardiac pericytes, which support the capillaries and the cardiomyocytes (Avolio et al., 2020). Myocarditis is associated with platelet activation, so this could be one factor at play in the response to the vaccines (Weikert. et al., 2002). However, another factor could be related to exosomes released by macrophages that have taken up the mRNA nanoparticles, and the specific microRNAs found in those exosomes.
A study involving patients suffering from severe COVID-19 disease looked specifically at the expression of circulating microRNAs compared to patients suffering from influenza and to healthy controls. One microRNA that was consistently upregulated in association with COVID-19 was miR-155, and the authors suggested that it might be a predictor of chronic myocardial damage and inflammation. By contrast, influenza infection was not associated with increased miR-155 expression. They concluded: “Our study identified significantly altered levels of cardiac-associated miRs [microRNAs] in COVID-19 patients indicating a strong association of COVID-19 with cardiovascular ailments and respective biomarkers” (Garg et al., 2021).
A study comparing 300 patients with cardiovascular disease to healthy controls showed a statistically significant increase in circulating levels of miR-155 in the patients compared to controls. Furthermore, those with more highly constricted arteries (according to a Gensini score) had higher levels than those with lesser disease (Qiu and Ma, 2018).
Importantly, exosomes play a role in inflammation in association with heart disease. During myocardial infarction, miR-155 is sharply upregulated in macrophages in the heart muscle and released into the extracellular milieu within exosomes. These exosomes are delivered to fibroblasts, and miR-155 downregulates proteins in the fibroblasts that protect from inflammation and promote fibroblast proliferation. The resulting impairment leads to cardiac rupture (Wang et al., 2017b).
We have already discussed how the S1 segment of the SARS-CoV-2 spike glycoprotein can be cleaved by furin and released into circulation. It binds to ACE2 receptors through its receptor binding domain (RBD), and this inhibits their function. Because ACE2 degrades Ang II, disabling ACE2 leads directly to overexpression of Ang II, further enhancing risk to cardiovascular disease. AngII-induced vasoconstriction is an independent mechanism to induce permanent myocardial injury even when coronary obstruction is not present. Repeated episodes of sudden constriction of a cardiac artery due to Ang II can eventually lead to heart failure or sudden death (Gavras and Gavras, 2002). Fatal cases of COVID-19 vaccination have been described (Choi et al., 2021; Verma et al., 2021).
ACE2 suppression had already been seen in studies on the original SARS-CoV virus. An autopsy study on patients succumbing to SARS-CoV revealed an important role for ACE2 inhibition in promoting heart damage. SARS-CoV viral RNA was detected in 35% of 20 autopsied human heart samples taken from patients who died. There was a marked increase in macrophage infiltration associated with myocardial damage in the patients whose hearts were infected with SARS-CoV. Importantly, the presence of SARS-CoV in the heart was associated with marked reduction in ACE2 protein expression (Oudit et al., 2009).
15. Considerations regarding the Vaccine Adverse Event Reporting System (VAERS)
The Food and Drug Administration's Vaccine Adverse Event Reporting System (VAERS) is an imperfect but valuable resource for identifying potential adverse reactions to vaccines. Established through collaboration between the CDC and FDA, VAERS is “a national early warning system to detect possible safety problems in U.S.-licensed vaccines.” According to the CDC it is “especially useful for detecting unusual or unexpected patterns of adverse event reporting that might indicate a possible safety problem with a vaccine.” (https://vaers.hhs.gov/about.html) Even the CDC recognizes that adverse events reported to VAERS represent “only a small fraction of actual adverse events” (Vaers Home, 2021). A widely cited report noted that fewer than 1% of all vaccine-related adverse events are reported to VAERS (Lazarus et al., 2010). That assertion, though, has no citation so the basis for the claim is unclear. Rose (2021) published a much more sophisticated analysis of VAERS data to offer an estimate of underreporting by a factor of 31 (Rose, 2021). While it is impossible to determine underreporting with precision, the available evidence is that underreporting very strongly characterizes the VAERS data. The information presented below should be understood in that light.
In mining VAERS for ‘signals’ that might indicate adverse reactions (AEs) to mRNA vaccinations, we acknowledge that no report to VAERS establishes a causal link with the vaccination. That said, the possibility of a causal relationship is strengthened through both the causal pathways we have described in this paper, and the strong temporal association between injections and reported AEs. Nearly 60% of all mRNA-injection-related -AEs have happened within 48 h of injection (https://medalerts.org/vaersdb/findfield.php?TABLE=ON&GROUP1=ONS&EVENTS=ON&VAX=COVID19&VAXTYPES=COVID-19&STATE=NOTFR).
Two important cautions regarding analysis of VAERS data should be noted. The first is that, in addition to health care professionals submitting reports, VAERS is open for public submissions as well. Members of the public may lack the skills necessary to evaluate a symptom appropriately to determine if it merits a VAERS entry. A second caution is that public access also allows for the possibility of anti-vaccination activists to populate VAERS with false reports to exaggerate the appearance of AE risk.
An interim analysis of deaths cited previously found that health service employees were the VAERS reporter in 67% of reports analyzed (Nandha and Singh, 2012), suggesting a large portion of VAERS reports are submitted by medical professionals and not the public. This finding also belies the notion that anti-vaccination activists are filing an excessive number of egregious reports of vaccine injury.
All of the data reported in this section were obtained by querying the online resource, http://wonder.cdc.gov/vaers.html. Over the 31-year history of VAERS, up to February 3, 2022, there were a total of 10,321 deaths reported as a “symptom” in association with any vaccine, and 8,241 (80%) of those deaths were linked to COVID-19 vaccines. Importantly, only 14% of COVID-19 VAERS-reported deaths as of June 2021 could have vaccination ruled out as a cause (McLachlan et al., 2021). This strongly suggests that these unprecedented vaccines exhibit unusual mechanisms of toxicity that go well beyond what is seen with more traditional vaccines.
We decided that a reasonable way to characterize the significance of adverse events linked to COVID-19 vaccines was to focus on events received in the year 2021, and to compare the counts in the “SYMPTOM” field for the events associated with COVID-19 vaccines to the total counts for that same symptom for all vaccines over that same year. In total, there were 737,689 events reported in VAERS for COVID-19 vaccines in 2021, representing a shocking 93% of the total cases reported for any vaccine that same year. While we recognize that some of the COVID-19 vaccines are based on DNA vector technology rather than mRNA technology, this class (i.e., the Johnson & Johnson vaccine) represents less than 9% of the COVID-19 reports, and its reaction profile is surely much more similar to that of the mRNA vaccines than to that of all the other vaccines.
The total number of adverse event reports for COVID-19 injections is far greater than the cumulative number of annual vaccine adverse event reports combined in all prior years, as shown by Rose (2021). The influenza vaccine is a good one to compare against. Given that the protocol for the mRNA vaccines requires two doses, and that many were persuaded to receive a booster shot as well, it is clear that the sheer number of COVID-19 vaccines administered is large compared to other vaccines. We can actually estimate what percent of the adverse reactions in 2021 would be expected to be associated with COVID-19 vaccines if the likelihood of an adverse reaction were similar to that of the influenza vaccine. The CDC tells us that 52% of the US population received a flu shot in 2021. The USAFacts web site provides percentages of the US population that received one, two or three doses of COVID-19 vaccines as a function of time (see: https://usafacts.org/visualizations/covid-vaccine-tracker-states/). The numbers they report for December 30, 2021 are 73% single dose, 62% fully vaccinated, and 21% boosted. This tallies up to 156% of the population as the total number of COVID-19 vaccines administered. This is exactly three times as many COVID vaccines as flu shots.
From VAERS, one can easily obtain the total number of adverse reactions associated with COVID-19 vaccines, the total number associated with flu vaccines, and the total number associated with all vaccines, for the US-restricted VAERS data from 2021. These come out as: COVID-19: 737,587, FLU: 9,124, and ALL: 792,935. First, we can observe that 93% of all the events reported were linked to COVID-19 vaccines. If we remove the counts for COVID-19 and replace them with three times the counts for flu (since COVID-19 vaccines were administered three times as often), we find that COVID-19 should have accounted for 32.6% of all the events, which can be compared with the actual result, which is 93%. We can also conclude that any event that shows up more than 93% as often for COVID-19 vaccines as for all other vaccines is especially significant as a potential toxic effect of these vaccines. Finally, we find that there are 27 times as many reports for COVID-19 vaccines as would be expected if its adverse reactions were comparable to those from the flu vaccine.
15.1. VAERS data indicative of nerve damage and vagus nerve involvement
Table 1 lists a number of symptoms in VAERS that can be associated with inflammation of or damage to various major nerves of the body, particularly those in the head. Strikingly, COVID-19 vaccines represented from 96 to 98% of the reports in the year 2021 related to each of these debilitating conditions. There were nearly 100,000 cases of nausea or vomiting, which are common symptoms of vagus nerve stimulation or damage (Babic and Browning, 2014). 14,701 cases of syncope linked to COVID-19 vaccines represented 96.3% of all cases of syncope, a well-established feature of vagus nerve dysfunction (Fenton et al., 2000). There were 3,657 cases of anosmia (loss of smell), clearly demonstrating that the SARS-CoV-2 spike glycoprotein from the injection in the arm was reaching the olfactory nerve. Dyspnea (shortness of breath) is related to vagus nerve impairment in the lungs, and there were 39,551 cases of dyspnea connected to COVID-19 vaccines in 2021.
Table 1.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various adverse effects that could be caused by inflammation in associated major nerves, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Inflamed Nerve(s) | Covid-19 Vaccines | All Vaccines | Percent COVID-19 |
|---|---|---|---|---|
| Anosmia | olfactory nerve | 3,657 | 3,677 | 99.5 |
| Tinnitus | vestibulo-cochlear nerve | 13,275 | 13,522 | 98.2 |
| Deafness | cochlea | 2,895 | 3,033 | 95.5 |
| Bell's Palsy/facial palsy | facial nerve | 5,881 | 6,129 | 96.0 |
| Vertigo | vestibular nerve | 7,638 | 7,819 | 97.7 |
| Migraine headache | trigeminal nerve | 8,872 | 9,059 | 97.9 |
| Dysphonia | glossopharyngeal nerve | 1,692 | 1,751 | 96.6 |
| Dysphagia | several lower cranial nerves | 4,711 | 4,835 | 97.4 |
| Nausea | vagus nerve | 69,121 | 71,275 | 97.0 |
| Vomiting | vagus nerve | 27,885 | 28,955 | 96.3 |
| Dyspnea | vagus nerve | 39,551 | 40,387 | 97.9 |
| Syncope | vagus nerve | 14,701 | 15,268 | 96.3 |
| Bradycardia | vagus nerve | 673 | 699 | 96.3 |
| TOTAL | -- | 200,552 | 206,409 | 97.2 |
Altogether, these events add up to a total of over 200,000 events, representing 97.2% of all the entries related to any vaccine in 2021. This is also a substantial 27.2% of all the events listed for 2021 in association with COVID-19 vaccines.
15.2. VAERS data on the heart and liver
In this paper, we have identified both the heart and the liver as organs that can be expected to be affected by the mRNA vaccines. The VAERS database shows a strong signal for both organs. Table 2 shows the statistics for 2021 on major disorders of the heart, including myocarditis, arrest (cardiac, cardiorespiratory and sinus arrest), arrhythmia (including supraventricular, nodal, sinus, tachyarrhythmia and ventricular arrhythmia), myocardial infarction (including acute and silent), and cardiac failure (including acute, chronic and congestive). Altogether, there were a total of 8,090 COVID-19 events related to these heart conditions, representing nearly 98% of all the events for all the vaccines for these symptoms in 2021.
Table 2.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various disorders of the heart, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Covid-19 Vaccines | All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Myocarditis | 2,322 | 2,361 | 98.3 |
| Arrest | 1,319 | 1,371 | 96.2 |
| Arrhythmia | 1,069 | 1,087 | 98.3 |
| Myocardial infarction | 2,224 | 2,272 | 97.9 |
| Cardiac failure | 1,156 | 1,190 | 97.1 |
| TOTAL | 8,090 | 8,281 | 97.7 |
It is difficult to find all of the symptoms associated with liver damage in VAERS, but we selected a number that had high enough counts to be of interest and that clearly represent serious liver problems. Altogether there were 731 events in these categories for COVID-19 vaccines, as shown in Table 3 , representing over 97% of all the cases connecting these conditions to any vaccine in 2021.
Table 3.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various indicators of liver disease, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Covid-19 Vaccines | All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Liver disorder | 83 | 87 | 95.4 |
| [Drug-induced] liver injury | 65 | 65 | 100 |
| [Acute] hepatic failure | 86 | 88 | 97.7 |
| Hepatic cancer [metastatic] | 12 | 12 | 100 |
| Hepatic cirrhosis | 67 | 69 | 97.1 |
| Hepatic cyst | 33 | 34 | 97.0 |
| Liver function test increased | 238 | 245 | 97.1 |
| Liver function test abnormal | 90 | 94 | 95.7 |
| Hepatic function abnormal | 34 | 34 | 100 |
| Haemangioma of liver | 10 | 10 | 100 |
| Liver abscess | 7 | 7 | 100 |
| Liver transplant | 6 | 6 | 100 |
| TOTAL | 731 | 751 | 97.3 |
15.3. VAERS data related to thrombosis
There were 78 unique symptoms in VAERS involving thrombosis, specifying different arteries and veins. Table 4 shows nine symptoms with the highest counts, totaling 7,356 events. We investigated the time interval for the three dominant ones (thrombosis, deep vein thrombosis and pulmonary thrombosis), and found that these all have a sharp peak in the 15-30-day range for onset interval (time after vaccination). This coincides with a sharp peak in pulmonary embolism, a life-threatening condition, also in the 15-30-day time interval. Overall, for these nine thrombotic symptoms, a random sampling from the year 2021 would yield a COVID vaccine as opposed to any other vaccine 98.7% of the time. Pulmonary embolism, a life-threatening condition that can be caused by a blood clot that travels to the lungs, has a slightly higher probability of 98.8%, with 3,100 cases listed for COVID-19.
Table 4.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various specific types of thrombosis, showing total counts for COVID-19 vaccines and for all vaccines. Pulmonary embolism, a highly related symptom, is also shown.
| Symptom | Covid-19 Vaccines | All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Thrombosis | 3,899 | 3,951 | 98.7 |
| Deep vein thrombosis | 2,275 | 2,297 | 99.0 |
| Pulmonary thrombosis | 631 | 646 | 97.7 |
| Cerebral thrombosis | 211 | 215 | 98.1 |
| Portal vein thrombosis | 89 | 90 | 98.9 |
| Superficial vein thrombosis | 81 | 81 | 100 |
| Peripheral artery thrombosis | 74 | 74 | 100 |
| Mesenteric vein thrombosis | 55 | 56 | 98.2 |
| Venous thrombosis | 41 | 41 | 100 |
| TOTAL | 7,356 | 7,451 | 98.7 |
| Pulmonary embolism | 3,100 | 3,137 | 98.8 |
15.4. VAERS data related to neurodegenerative disease
Table 5 lists results for several conditions that are linked to neurodegenerative disease. Decreased mobility can be caused by Parkinson's disease, and there were a striking 8,975 cases listed for 2021 and COVID-19 vaccines. Alzheimer's and Parkinson's are diseases that normally take decades to develop, and ordinarily one would assume that a vaccine has nothing to do with it. While the numbers are small, most of the cases in VAERS were linked to COVID-19 vaccines. Anosmia, also included in the table on the vagus nerve, is especially interesting, because it is a well-known early sign of Parkinson's disease, and it is also a well-identified feature of SARS-CoV-2 infection. 99.5% of the cases with anosmia as a symptom were linked to COVID-19 vaccines. Overall, the symptoms in this table were linked to COVID-19 vaccines nearly 95% of the time.
Table 5.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various disorders linked to neurodegenerative disease, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Covid-19 Vaccines | All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Alzheimer's dementia | 37 | 39 | 94.9 |
| Parkinsonian symptoms | 83 | 89 | 93.3 |
| Memory impairment | 1,681 | 1,720 | 97.7 |
| Anosmia | 3,657 | 3,677 | 99.5 |
| Mobility decreased | 8,975 | 9,743 | 92.1 |
| Cognitive disorder | 779 | 815 | 92.1 |
| TOTAL | 15,212 | 16,083 | 94.6 |
15.5. VAERS signal for cancer
Cancer is a disease generally understood to take months or, more commonly, years to progress from an initial malignant transformation in a cell to development of a clinically recognized condition. Since VAERS reports of adverse events are happening primarily within the first month or even the first few days after vaccination (Rose, 2021), it seems likely that the acceleration of cancer progression following vaccines would be a difficult signal to recognize. Furthermore, most people do not expect cancer to be an adverse event that could be caused by a vaccine, and hence they fail to enter a report when cancer develops shortly after vaccination. However, as we have outlined in our paper, if the mRNA vaccinations are leading to widespread dysregulation of oncogene controls, cell cycle regulation, and apoptosis, then VAERS reports should reflect an increase in reports of cancer, relative to the other vaccines, even if the numbers are small. The experiment demonstrating impairment of DNA repair mechanisms by SARS-CoV-2 spike protein in an in vitro study provides compelling evidence that the vaccines could accelerate the rate of DNA mutations, increasing cancer risk (Jiang and Mei, 2021).
For our analysis of evidence of increased cancer risk in VAERS, we focused on two somewhat distinct approaches. One, represented by the results in Table 6 , was to gather the counts for any terms that contained keywords clearly linked to cancer, namely, “cancer,” “lymphoma,” “leukaemia,” “metastasis,” “carcinoma,” and “neoplasm.” Overall, we found 1,474 entries linking these terms to COVID-19 vaccines, representing 96% of all the entries for any of these terms for any vaccine in that year.
Table 6.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for various cancer-related terms, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Counts COVID-19 vaccines | Counts All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Cancer | 396 | 403 | 98.3 |
| Lymphoma | 144 | 153 | 94.1 |
| Leukaemia | 155 | 161 | 96.3 |
| Metastatic/metastasis | 175 | 179 | 97.8 |
| Carcinoma | 176 | 187 | 94.1 |
| Neoplasm | 428 | 452 | 94.7 |
| TOTAL | 1,474 | 1,535 | 96.0 |
The complementary approach was to find terms involving cancer in specific organs, namely, breasts, prostate, bladder, colon, brain, lungs, pancreas and ovaries, as shown in Table 7 . Although all the numbers are small, the highest by far was for breast cancer (246 cases), with nearly four times as many hits as for lung cancer, the second most common type. All of the cases for pancreatic, ovarian and bladder cancer were linked to COVID-19 vaccines, with zero cases for any other vaccine. Altogether, we tabulated 534 cases of cancer of specific organs linked to COVID-19 vaccines, representing 97.3% of all the cases for any vaccine in 2021.
Table 7.
Number of symptoms reported in VAERS, restricted to the US population, for the year 2021, for cancer of specific organs, showing total counts for COVID-19 vaccines and for all vaccines.
| Symptom | Counts COVID-19 vaccines | Counts All Vaccines | Percent COVID-19 |
|---|---|---|---|
| Breast cancer | 246 | 254 | 96.8 |
| Prostate cancer | 50 | 52 | 96.2 |
| Bladder cancer | 30 | 30 | 100 |
| Colon cancer | 40 | 41 | 97.6 |
| Brain neoplasm | 53 | 55 | 96.4 |
| Lung cancer | 64 | 66 | 97.0 |
| Pancreatic cancer | 24 | 24 | 100 |
| Ovarian cancer | 27 | 27 | 100 |
| Total | 534 | 549 | 97.3 |
16. Conclusions
There has been an unwavering message about the safety and efficacy of mRNA vaccinations against SARS-CoV-2 from the public health apparatus in the US and around the globe. The efficacy is increasingly in doubt, as shown in a recent letter to the Lancet Regional Health by Günter Kampf (2021b). Kampf provided data showing that the vaccinated are now as likely as the unvaccinated to spread disease. He concluded: “It appears to be grossly negligent to ignore the vaccinated population as a possible and relevant source of transmission when deciding about public health control measures.” Moreover, the inadequacy of phase I, II, and III trials to evaluate mid-term and long-term side effects from mRNA genetic vaccines may have been misleading on their suppressive impact on the innate immunity of the vaccinees.
In this paper, we call attention to three very important aspects of the safety profile of these vaccinations. First is the extensively documented subversion of innate immunity, primarily via suppression of IFN-α and its associated signaling cascade. This suppression will have a wide range of consequences, not the least of which include the reactivation of latent viral infections and the reduced ability to effectively combat future infections. Second is the dysregulation of the system for both preventing and detecting genetically driven malignant transformation within cells and the consequent potential for vaccination to promote those transformations. Third, mRNA vaccination potentially disrupts intracellular communication carried out by exosomes, and induces cells taking up spike glycoprotein mRNA to produce high levels of spike-glycoprotein-carrying exosomes, with potentially serious inflammatory consequences. Should any of these potentials be fully realized, the impact on billions of people around the world could be enormous and could contribute to both the short-term and long-term disease burden our health care system faces.
Given the current rapidly expanding awareness of the multiple roles of G4s in regulation of mRNA translation and clearance through stress granules, the increase in pG4s due to enrichment of GC content as a consequence of codon optimization has unknown but likely far-reaching consequences. Specific analytical evaluation of the safety of these constructs in vaccines is urgently needed, including mass spectrometry for identification of cryptic expression and immunoprecipitation studies to evaluate the potential for disturbance of or interference with the essential activities of RNA and DNA binding proteins.
It is essential that further studies be conducted to determine the extent of the potential pathological consequences outlined in this paper. It is not practical for these vaccinations to be considered part of a public health campaign without a detailed analysis of the human impact of the potential collateral damage. VAERS and other monitoring systems should be optimized to detect signals related to the health consequences of mRNA vaccination we have outlined. We believe the upgraded VAERS monitoring system described in the Harvard Pilgrim Health Care, Inc. study, but unfortunately not supported by the CDC, would be a valuable start in this regard (Lazarus et al., 2010).
In the end, billions of lives are potentially at risk, given the large number of individuals injected with the SARS-CoV-2 mRNA vaccines and the broad range of adverse outcomes we have described. We call on the public health institutions to demonstrate, with evidence, why the issues discussed in this paper are not relevant to public health, or to acknowledge that they are and to act accordingly. Furthermore, we encourage all individuals to make their own health care decisions with this information as a contributing factor in those decisions.
Author contributions
S.S., G.N and A.K. all contributed substantially to the writing of the original draft. P.M. participated in the process of editorial revisions.
Funding
This research was funded in part by Quanta Computers, Inc., Taipei, Taiwan, under the auspices of the Qmulus project.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling Editor: Dr. Jose Luis Domingo
References
- Abe M., Bonini N.M. MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol. 2013;23(1):30–36. doi: 10.1016/j.tcb.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agashe D., Martinez-Gomez N.C., Drummond D.A., Marx C.J. Good codons, bad transcript: large reductions in gene expression and fitness arising from synonymous mutations in a key enzyme. Mol. Biol. Evol. 2013;30:549–560. doi: 10.1093/molbev/mss273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama H., Kakiuchi S., Rikitake J., Matsuba H., Sekinada D., Kozuki Y., Iwata N. Immune thrombocytopenia associated with Pfizer-BioNTech’s BNT162b2 mRNA COVID-19 vaccine. IDCases. 2021;25 doi: 10.1016/j.idcr.2021.e01245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khalaf H.H., Aboussekhra A. p16 controls p53 protein expression through miR-dependent destabilization of MDM2. Mol. Cancer Res. 2018;16(8):1299–1308. doi: 10.1158/1541-7786.MCR-18-0017. [DOI] [PubMed] [Google Scholar]
- Alsamman K., El-Masry O.S. Interferon regulatory factor 1 inactivation in human cancer. Biosci. Rep. 2018;38(3) doi: 10.1042/BSR20171672. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Contr. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- Asmana Ningrum R. Human interferon α-2b: a therapeutic protein for cancer treatment. Sci. Tech. Rep. 2014 doi: 10.1155/2014/970315. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atoui A., Jarrah K., Al Mahmasani L., Bou‐Fakhredin R., Taher A.T. Deep venous thrombosis and pulmonary embolism after COVID‐19 mRNA vaccination. Ann. Hematol. 2022:1–3. doi: 10.1007/s00277-021-04743-1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avolio E., Gamez M., Gupta K., Foster R., Berger I., Caputo M., Davidson A., Hill B., Madeddu P. The SARS-CoV-2 spike protein disrupts the cooperative function of human cardiac pericytes - endothelial cells through CD147 receptor-mediated signalling: a potential non-infective mechanism of COVID-19 microvascular disease. bioRxiv preprint. 2020 doi: 10.1101/2020.12.21.423721. December 21. [DOI] [Google Scholar]
- Babendure J.R., Babendure J.L., Ding J.H., Tsien R.Y. Control of mammalian translation by mRNA structure near caps. RNA. 2006;12(5):851–861. doi: 10.1261/rna.2309906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babic T., Browning K.N. The role of vagal neurocircuits in the regulation of nausea and vomiting. Eur. J. Pharmacol. 2014;722:38–47. doi: 10.1016/j.ejphar.2013.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl K., Senn J.J., Yuzhakov O., Bulychev A., Brito L.A., Hassett K.J., Laska M.E., Smith M., Almarsson Ö., Thompson J., et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol. Ther. 2017;25(6):1316–1327. doi: 10.1016/j.ymthe.2017.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal S., Perincheri S., Fleming T., Poulson C., Tiffany B., Bremner R.M., Mohanakumar T. Cutting edge: circulating exosomes with COVID spike protein are induced by BNT162b2 (PfizerBioN-Tech) vaccination prior to development of antibodies: a novel mechanism for immune activation by mRNA vaccines. J. Immunol. 2021;207(10):2405–2410. doi: 10.4049/jimmunol.2100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barone V., Camilli F., Crisci M., Scandellari C., Barboni P., Lugaresia A. Inflammatory optic neuropathy following SARS-CoV-2 mRNA vaccine: description of two cases. J. Neurol. Sci. 2021;429:118186. doi: 10.1016/j.jns.2021.118186. [DOI] [Google Scholar]
- Bezzi G., Piga E.J., Binolfi A., Armas P. CNBP binds and unfolds in vitro G-quadruplexes formed in the SARS-CoV-2 positive and negative genome strands. Int. J. Mol. Sci. 2021;22(5):2614. doi: 10.3390/ijms22052614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhurani V., Mohankrishnan A., Morrot A., Dalai S.K. Developing effective vaccines: cues from natural infection. Int. Rev. Immunol. 2018;37(5):249–265. doi: 10.1080/08830185.2018.1471479. [DOI] [PubMed] [Google Scholar]
- Bidwell B.N., Slaney C.Y., Withana N.P., Forster S., Cao Y., Loi S., Andrews D., Mikeska T., Mangan N.E., Samarajiwa S.A., et al. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat. Med. 2012;18(8):1224–1231. doi: 10.1038/nm.2830. [DOI] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181(5):1036–1045 e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbolis F., Syntichaki P. Cytoplasmic mRNA turnover and ageing. Mech. Ageing Dev. 2015;152:32–42. doi: 10.1016/j.mad.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh-Nissimov T., Orenbuch-Harroch E., Chowers M., Elbaz M., Nesher L., Stein M., Maor Y., Cohen R., Hussein K., Weinberger M., et al. BNT162b2 vaccine breakthrough: clinical characteristics of 152 fully vaccinated hospitalized COVID-19 patients in Israel. Clin. Microbiol. Infect. 2021;27(11):1652–1657. doi: 10.1016/j.cmi.2021.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley N.E., Hosey A.M., Gorski J.J., Purcell J.W., Mulligan J.M., Harkin D.P., Mullan P.B. BRCA1 regulates IFN-γ signaling through a mechanism involving the type I IFNs. Mol. Cancer Res. 2007;5(3):261–270. doi: 10.1158/1541-7786.MCR-06-0250. [DOI] [PubMed] [Google Scholar]
- Cancer risk and BRCA1 gene mutations. 2021. https://www.facingourrisk.org/info/hereditary-cancer-and-genetic-testing/hereditary-cancer-genes-and-risk/genes-by-name/brca1/cancer-risk Available at:
- Centers for Disease Control and Prevention . 2021. Coronavirus Disease 2019 (COVID-19)https://www.cdc.gov/coronavirus/2019-ncov/science/science-briefs/vaccine-induced-immunity.html#anchor_1635540449320 [online] Available at: [Google Scholar]
- Centers for Disease Control and Prevention . 2021. COVID-19 Booster Shot.https://www.cdc.gov/coronavirus/2019-ncov/vaccines/booster-shot.html [online] Available at: [Google Scholar]
- Chahar H.S., Bao X., Casola A. Exosomes and their role in the life cycle and pathogenesis of RNA viruses. Viruses. 2015;7:3204–3225. doi: 10.3390/v7062770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.L., Peng B., Umar M.I., Chan C.Y., Sahakyan A.B., Le M.T.N., Kwok C.K. Structural analysis reveals the formation and role of RNA G-quadruplex structures in human mature microRNAs. Chem. Commun. 2018;54(77):10878–10881. doi: 10.1039/c8cc04635b. [DOI] [PubMed] [Google Scholar]
- Chaudhary N., Weissman D., Whitehead K.A. mRNA vaccines for infectious diseases: principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021;20:817–838. doi: 10.1038/s41573-021-00283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvineau‐Grenier A., Bastard P., Servajeank A., Gervais A., Rosain J., Jouanguy E., Cobat A., Casanova J.-L., Rossi B. Autoantibodies neutralizing type I interferons in 20% of COVID‐19 deaths in a French hospital. J. Clin. Immunol. 2022;27 doi: 10.1007/s10875-021-01203-3. January. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.S., Lee H.M., Jang Y.-J., Kim C.-H., Ryua C.J. Heterogeneous nuclear ribonucleoprotein A2/B1 regulates the self-renewal and pluripotency of human embryonic stem cells via the control of the G1/S transition. Stem Cell. 2013;31:2647–2658. doi: 10.1002/stem.1366. [DOI] [PubMed] [Google Scholar]
- Choi Y.E., Pan Y., Park E., Konstantinopoulos P., De S., D'Andrea A., Chowdhury D. MicroRNAs downregulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife. 2014;3 doi: 10.7554/eLife.02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Lee S., Seo J.-W., Kim M.-J., Jeon Y.H., Park J.H., Lee J.K., Yeo N.S. Myocarditis-induced sudden death after BNT162b2 mRNA COVID-19 vaccination in korea: case report focusing on histopathological findings. J. Kor. Med. Sci. 2021;36(40):e286. doi: 10.3346/jkms.2021.36.e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cines D.B., Bussel J.B. SARS-CoV-2 vaccine-induced immune thrombotic thrombocytopenia. N. Engl. J. Med. 2021;384:2254–2256. doi: 10.1056/NEJMe2106315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consoli S., Dono F., Evangelista G., D'Apolito M., Travaglini D., Onofrj M., Bonanni L. Status migrainosus: a potential adverse reaction to Comirnaty (BNT162b2, BioNtech/Pfizer) COVID-19 vaccinea case report. Neurol. Sci. 2021;22:1–4. doi: 10.1007/s10072-021-05741-x. [Epub ahead of print] Nov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Créange A. A role for interferon-beta in Guillain-Barré Syndrome? BioDrugs. 2000;14(1):1–11. doi: 10.2165/00063030-200014010-00001. [DOI] [PubMed] [Google Scholar]
- Crow A.R., Lazarus A.H. Role of Fcgamma receptors in the pathogenesis and treatment of idiopathic thrombocytopenic purpura. J. Pediatr. Hematol. Oncol. 2003;25(Suppl. 1):S14S18. doi: 10.1097/00043426-200312001-00004. [DOI] [PubMed] [Google Scholar]
- Danese E., Montagnana M., Salvagno G.L., Peserico D., Pighi L., De Nitto S., Henry B.M., Porru S., Lippi G. Comprehensive assessment of humoral response after Pfizer BNT162b2 mRNA Covid-19 vaccination: a three-case series. Clin. Chem. Lab. Med. 2021;59(9):1585–1591. doi: 10.1515/cclm-2021-0339. [DOI] [PubMed] [Google Scholar]
- De Andrea M., Ravera R., Gioia D., Gariglio M., Landolfo S. The interferon system: an overview. Eur. J. Paediatr. Neurol. 2002;6:A41–A46. doi: 10.1053/ejpn.2002.0573. [DOI] [PubMed] [Google Scholar]
- de Beuckelaer A., Pollard C., Van Lint S., Roose K., Van Hoecke L.V., Naessens T., Udhayakumar V.K., Smet M., Sanders N., Lienenklaus S., et al. Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol. Ther. 2016;24(11):2012–2020. doi: 10.1038/mt.2016.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gonzalo-Calvo D., Benítez I.D., Pinilla L., Carratalá A., Moncusí-Moix A., Gort-Paniello C., Molinero M., González J., Torres G., Bernal M., et al. Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients. Transl. Res. 2021;236:147–159. doi: 10.1016/j.trsl.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Paolis V., Lorefice E., Orecchini E., Carissimi C., Laudadio I., Fulci V. Epitranscriptomics: a new layer of microRNA regulation in cancer. Cancers. 2021;13(13):3372. doi: 10.3390/cancers13133372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harbor Perspect. Biol. 2012;4(9):a012286. doi: 10.1101/cshperspect.a012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy A.S., Hober D., Bouzidi A., Wattre P. Role of interferon alpha (IFN‐α) and interferon gamma (IFN‐γ) in the control of the infection of monocyte‐like cells with Human Cytomegalovirus (HCMV) Microbiol. Immunol. 1999;43(12):1087–1096. doi: 10.1111/j.1348-0421.1999.tb03365.x. [DOI] [PubMed] [Google Scholar]
- Dodick D., Silberstein S. Central sensitization theory of migraine: clinical implications. Headache. 2006;46(Suppl. 4):S18291. doi: 10.1111/j.1526-4610.2006.00602.x. [DOI] [PubMed] [Google Scholar]
- Doulberis M., Papaefthymiou A., Kotronis G., Gialamprinou D., Soteriades E.S., Kyriakopoulos A., et al. Does COVID-19 vaccination warrant the classical principle "ofelein i mi vlaptin. Medicina (Kaunas). 2021;57(3):253. doi: 10.3390/medicina57030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortiera J. Liver injury after mRNA-based SARS-CoV-2 vaccination in a liver transplant recipient. Clin. Res. Hepatol. Gastroenterol. 2022;46 doi: 10.1016/j.clinre.2021.101743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn G.P., Bruce A.T., Sheehan K.C.F., Shankaran V., Uppaluri R., Bui J.D., Diamond M.S., Koebel C.M., Arthur C., White J.M., et al. A critical function for type I interferons in cancer immunoediting. Nat. Immunol. 2005;6(7):722–729. doi: 10.1038/ni1213. [DOI] [PubMed] [Google Scholar]
- Erb H.H., Langlechner R.V., Moser P.L., Handle F., Casneuf T., Verstraeten K., Schlick B., Schäfer G., Hall B., Sasser K., Culig Z., Santer F.R., et al. IL6 sensitizes prostate cancer to the antiproliferative effect of IFNα2 through IRF9. Endocr. Relat. Cancer. 2013;20(5):677. doi: 10.1530/ERC-13-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erman A.B., Kejner A.E., Norman B.S., Hogikyan D., Feldman E.L. Disorders of cranial nerves IX and X. Semin. Neurol. 2009;29(1):8592. doi: 10.1055/s-0028-1124027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eviston T., Croxson G.R., Kennedy P.G.E., Hadlock T., Krishnan A.V. Bell's palsy: aetiology, clinical features and multidisciplinary care. J. Neurol. Neurosurg. Psychiatry. 2015;86 doi: 10.1136/jnnp-2014-309563. [DOI] [PubMed] [Google Scholar]
- Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathy R.A., McMahon D.E., Lee C., Chamberlin G.C., Rosenbach M., Lipoff J.B., Tyagi A., Desai S.R., French L.E., Lim H.W., et al. Varicella-zoster and herpes simplex virus reactivation post-COVID-19 vaccination: a review of 40 cases in an International Dermatology Registry. JEADV. 2022;36(1):e6–e9. doi: 10.1111/jdv.17646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay M.M., Lyons S.M., Ivanov P. RNA G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017;429(14):2127–2147. doi: 10.1016/j.jmb.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA . Vaccines and Related Biological Products Advisory Committee December 10, 2020 Meeting Announcement. 2021. https://www.fda.gov/advisory-committees/advisory-committee-calendar/vaccines-and-related-biological-products-advisory-committee-december-10-2020-meeting- announcement [Google Scholar]
- FDA . Vaccines and Related Biological Products Advisory Committee December 17, 2020 Meeting Announcement. 2021. https://www.fda.gov/advisory-committees/advisory-committee-calendar/vaccines-and-related-biological-products-advisory-committee-december-17-2020-meeting-announcement [Google Scholar]
- Feng B., Eknoyan G., Guo Z.S., Jadoul M., Rao H.Y., Zhang W., Wei L. Effect of interferon- alpha-based antiviral therapy on hepatitis C virus-associated glomerulonephritis: a meta-analysis. Nephrol. Dial. Transplant. 2012;27(2):640–646. doi: 10.1093/ndt/gfr236. [DOI] [PubMed] [Google Scholar]
- Fenton A.M., Hammill S.C., Rea R.F., Low P.A., Shen W.-K. Vasovagal syncope. Ann. Intern. Med. 2000;133(9):714–725. doi: 10.7326/0003-4819-133-9-200011070-00014. [DOI] [PubMed] [Google Scholar]
- Finnberg N.K., El-Deiry W.S. TRAIL death receptors as tumor suppressors and drug targets. Cell Cycle. 2008;7(11):1525–1528. doi: 10.4161/cc.7.11.5975. [DOI] [PubMed] [Google Scholar]
- Forni G., Mantovani A. COVID-19 Commission of Accademia Nazionale dei Lincei, Rome. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28(2):626–639. doi: 10.1038/s41418-020-00720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Seeliger B., Derda A.A., Xiao K., Gietz A., Scherf K., Sonnenschein K., Pink I., Hoeper M.M., Welte T., et al. Circulating cardiovascular microRNAs in critically ill COVID-19 patients. Eur. J. Heart Fail. 2021;23(3):468–475. doi: 10.1002/ejhf.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavras I., Gavras H. Angiotensin II as a cardiovascular risk factor. J. Hum. Hypertens. 2002;16(Suppl. 2):S2–S6. doi: 10.1038/sj.jhh.1001392. [DOI] [PubMed] [Google Scholar]
- Girardi T., De Keersmaecker K. T-ALL: ALL a matter of translation? Haematologica. 2015;100(3):293–295. doi: 10.3324/haematol.2014.118562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S., Bron D., Tousseyn T., Vierasu I., Dewispelaere L., Heimann P., Cogan E., Goldman M. Rapid progression of angioimmunoblastic T cell lymphoma following BNT162b2 mRNA vaccine booster shot: a case report. Front. Med. 2021;8 doi: 10.3389/fmed.2021.798095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370(6521) doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould F.D.H., Lammers A.R., Mayer C.J., German R.Z. Specific vagus nerve lesion have distinctive physiologic mechanisms of dysphagia. Front. Neurol. 2019;10:1301. doi: 10.3389/fneur.2019.01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Namekata K., Kimura A., Harada C., Harada T. The renin-angiotensin system regulates neurodegeneration in a mouse model of optic neuritis. Am. J. Pathol. 2017;187(12):2876–2885. doi: 10.1016/j.ajpath.2017.08.012. [DOI] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369(6504):718–724. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.H., Choe J. Diverse molecular functions of m6A mRNA modification in cancer. Exp. Mol. Med. 2020;52(5):738–749. doi: 10.1038/s12276-020-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise R., Amann P.M., Ensslen S., Marquardt Y., Czaja K., Joussen S., Beer D., Abele R., Plewnia G., Tampé R., et al. Interferon alpha signaling and its relevance for the upregulatory effect of transporter proteins associated with antigen processing (TAP) in patients with malignant melanoma. PLoS One. 2016;11(1) doi: 10.1371/journal.pone.0146325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B., Mayer C., Varshney D., Marsico G., Murat P., Taylor C., D'Santos C., Tannahill D., Balasubramanian S. Analysis of NRAS RNA G-quadruplex binding proteins reveals DDX3X as a novel interactor of cellular G-quadruplex containing transcripts. Nucleic Acids Res. 2018;46(21):11592–11604. doi: 10.1093/nar/gky861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland D.A., Møller R., Uhl S.A., Oishi K., Frere J., Golynker T., Horiuchi S., Panis M., Blanco-Melo D., Sachs D., et al. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity. 2021;54 doi: 10.1016/j.immuni.2021.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Takaoka A., Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25(3):349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Honke K. Biosynthesis and biological function of sulfoglycolipids. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2013;89(4) doi: 10.2183/pjab.89.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X., Zaks T., Langer R., Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 2021;6:1078–1094. doi: 10.1038/s41578-021-00358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Cai X., Song X., Tang H., Huang Y., Xie S., Hu Y. Steroids for preventing recurrence of acute severe migraine headaches: a meta-analysis. Eur. J. Neurol. 2013;20(8):1184–1190. doi: 10.1111/ene.12155. [DOI] [PubMed] [Google Scholar]
- Huang F.T., Sun J., Zhang L., He X., Zhu Y.H., Dong H.J., Wang H.-Y., Zhu L., Zou Huang J.W., et al. Role of SIRT1 in hematologic malignancies. J. Zhejiang Univ. - Sci. B. 2019;20(5):391–398. doi: 10.1631/jzus.B1900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyas A.A., Mithen F.A., Dalakas M.C., Wargo M., Chen Z.W., Bielory L., Cook S.D. Antibodies to sulfated glycolipids in Guillain-Barr syndrome. J. Neurol. Sci. 1991;105(1):108–117. doi: 10.1016/0022-510x(91)90126-r. [DOI] [PubMed] [Google Scholar]
- Ivanova E.N., Devlin J.C., Buus T.B., Koide A., Cornelius A., Samanovic M.I., Herrera A., Zhang C., Desvignes L., Odum N., Ulrich R., Mulligan M.J., Koide S., Ruggles K.V., Herati R.S., Koralov S.B. Discrete immune response signature to SARS-CoV-2 mRNA vaccination versus infection. medRxiv preprint. 2021 doi: 10.1101/2021.04.20.21255677. April 21. [DOI] [Google Scholar]
- Iwanaga J., Fukuoka H., Fukuoka N., Yutori H., Ibaragi S., Tubbs R.S. A narrative review and clinical anatomy of Herpes zoster infection following COVID‐19 vaccination. Clin. Anat. 2021;35(1):45–51. doi: 10.1002/ca.23790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S.S., Steele J.M., Fonseca B., Huang S., Shah S., Maskatia S.A., Buddhe S., Misra N., Ramachandran P., Gaur L., et al. COVID-19 vaccination–associated myocarditis in adolescents. Pediatrics. 2021;148(5) doi: 10.1542/peds.2021-053427. [DOI] [PubMed] [Google Scholar]
- Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jang S.K., Pestova T.V., Hellen C.U.T., Witherell G.W., Wimmer E. Cap-independent translation of picornavirus RNAs: structure and function of the internal ribosomal entry site. Enzyme. 1990;44:292–309. doi: 10.1159/000468766. [DOI] [PubMed] [Google Scholar]
- Jaubert C., Bedrat A., Bartolucci L., Di Primo C., Ventura M., Mergny J.-L., Amrane S., Andreola M.-L. RNA synthesis is modulated by G-quadruplex formation in Hepatitis C virus negative RNA strand. Sci. Rep. 2018;8:8120. doi: 10.1038/s41598-018-26582-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jego G.A., Palucka K., Blanck J.-P., Chalouni C., Pascual V., Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity. 2003;19 doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- Jeong M., Ocwieja K.E., Han D., Wackym P.A., Zhang Y., Brown A., Moncada C., Vambutas A., Kanne T., Crain R., et al. Direct SARS-CoV-2 infection of the human inner ear may underlie COVID-19-associated audiovestibular dysfunction. Commun. Med. 2021;1:44. doi: 10.1038/s43856-021-00044-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhaveri R. The COVID-19 mRNA vaccines and the pandemic: do they represent the beginning of the end or the end of the beginning? Clin. Therapeut. 2021;43(3):549–556. doi: 10.1016/j.clinthera.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Mei Y.-F. SARS-CoV-2 spike impairs DNA damage repair and inhibits V(D)J recombination in vitro. Viruses. 2021;13(2056) doi: 10.3390/v13102056. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kaczmarek R., El Ekiaby M., Hart D.P., Hermans C., Makris M., Noone D., O'Mahony B., Page D., Peyvandi F., Pipe S.W., et al. Vaccination against COVID‐19: rationale, modalities and precautions for patients with haemophilia and other inherited bleeding disorders. Haemophilia. 2021;27(4):515–518. doi: 10.1111/hae.14271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakarougkas A., Ismail A., Klement K., Goodarzi A.A., Conrad S., Freire R., Shibata A., Lobrich M., Jeggo P.A. Opposing roles for 53BP1 during homologous recombination. Nucleic Acids Res. 2013;41(21):9719–9731. doi: 10.1093/nar/gkt729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalra R.S., Kandimalla R. Engaging the spikes: heparan sulfate facilitates SARS-CoV-2 spike protein binding to ACE2 and potentiates viral infection. Signal Transduct. Targeted Ther. 2021;6:39. doi: 10.1038/s41392-021-00470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G. The epidemiological relevance of the COVID-19-vaccinated population is increasing. Lancet. Reg. Health – Europ. 2021;11 doi: 10.1016/j.lanepe.2021.100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf G. The epidemiological relevance of the COVID-19-vaccinated population is increasing. Lancet Reg. Health - Europ. 2021;11 doi: 10.1016/j.lanepe.2021.100272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23 doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Katalin Karikó K., Muramatsu H., Welsh F.A., Ludwig J., Kato H., Akira S., Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol. Ther. 2008;16:1833–1840. doi: 10.1038/mt.2008.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsikas Triantafyllidis K., Giannos P., Mian I.T., Kyrtsonis G., Kechagias K.S. Varicella zoster virus reactivation following COVID-19 vaccination: a systematic review of case reports. Vaccines. 2021;9(9):1013. doi: 10.3390/vaccines9091013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulen L.D., Doubrovinskaia S., Mooshage C., Jordan B., Purrucker J., Haubner C., Seliger C., Lorenz H.-M., Nagel S., Wildemann B., Bendszus M., Wick W., Schnenberger S. Neurological autoimmune diseases following vaccinations against SARS-CoV-2: a case series. Eur. J. Neurol. 2021:1–9. doi: 10.1111/ene.15147. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur A., Fang C.M. An overview of the human immune system and the role of interferon regulatory factors (IRFs) Prog. Microb. Mol. Biol. 2020;3(1) doi: 10.36877/pmmb.a0000129. 2020. [DOI] [Google Scholar]
- Kelton J.G., Arnold D.M., Nazy I. Lessons from vaccine-induced immune thrombotic thrombocytopenia. Nat. Rev. Immunol. 2021;21(12):753–755. doi: 10.1038/s41577-021-00642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat-Khoei M., Bhattacharyya S., Katz J., Harrison D., Tauhid S., Bruso P., Houtchens M.K., Edwards K.R., Bakshi R. COVID-19 mRNA vaccination leading to CNS inflammation: a case series. J. Neurol. 2021 Sep 4:1–14. doi: 10.1007/s00415-021-10780-7. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Nakajima T., Kamijo Y., Tanaka N., Wang L., Hara A., Sugiyama E., Tanaka E., Gonzalez F.J., Aoyama T. Hepatic cerebroside sulfotransferase is induced by PPAR activation in mice. PPAR Res. 2012 doi: 10.1155/2012/174932. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Bühler M. Adenosine methylation as a molecular imprint defining the fate of RNA. FEBS Lett. 2018;592(17):2845–2859. doi: 10.1002/1873-3468.13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolumam G.A., Thomas S., Thompson L.J., Sprent J., Murali-Krishna K. Type I interferons act directly on CD8 T cells to allow clonal expansion and memory formation in response to viral infection. J. Exp. Med. 2005;202(5) doi: 10.1084/jem.20050821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo J.W., Russo S.J., Ferguson D., Nestler E.J., Duman R.S. Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc. Natl. Acad. Sci. U. S. A. 2010;107(6):2669–2674. doi: 10.1073/pnas.0910658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothandan V.K., Kothandan S., Kim D.H., Byun Y., Lee Y.-K., Park I.-K., Hwang S.R. Crosstalk between stress granules, exosomes, tumour antigens, and immune cells: significance for cancer immunity. Vaccines. 2020;8(2):172. doi: 10.3390/vaccines8020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G., Lipinski L., Caffin F., Helwak A., Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4(6) doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara M., Kusunoki S. Mechanism and spectrum of anti-glycolipid antibody-mediated chronic inflammatory demyelinating polyneuropathy. Clin. Exper. Neuroimmunol. 2018;9(1):65–74. doi: 10.1111/cen3.12452. [DOI] [Google Scholar]
- Kwok H.F. Review of COVID-19 vaccine clinical trials -- A puzzle with missing pieces. Int. J. Biol. Sci. 2021;7(6):1461. doi: 10.7150/ijbs.59170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulos A.M., McCullough P.A. Synthetic mRNAs; their analogue caps and contribution to disease. Diseases. 2021;9(3):57. doi: 10.3390/diseases9030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz T.V., Ding Z., Ho P.P., Luo J., Agrawal A.N., Srinagesh H., Axtell R., Zhang H., Platten M., Wyss-Coray T., Steinman L. Angiotensin II sustains brain inflammation in mice via TGF-beta. J. Clin. Invest. 2010;120(8):2782–2794. doi: 10.1172/JCI41709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarus R., Klompas M., Bernstein S. 2010. Electronic Support for Public Health–Vaccine Adverse Event Reporting System (ESP: VAERS). Grant. Final Report, Grant ID: R18 HS; p. 17045. [Google Scholar]
- Lee E.-J., Cines D.B., Gernsheimer T., Kessler C., Michel M., Tarantino M.D., Semple J.W., Arnold D.M., Godeau B., Lambert M.P., Bussel J.B. Thrombocytopenia following pfizer and Moderna SARS-CoV-2 vaccination. Am. J. Hematol. 2021;96(5):534–537. doi: 10.1002/ajh.26132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T., Staels B., Saladin R., Desvergne B., Auwerx J., Wahli W. Regulation of the peroxisome proliferator-activated receptor alpha gene by glucocorticoids. J. Biol. Chem. 1994;269(40):24527–24530. doi: 10.1093/toxsci/kfn260. [DOI] [PubMed] [Google Scholar]
- Lensen R., Netea M.G., Rosendaal F.R. Hepatitis C virus reactivation following COVID-19 vaccination -- A case report. Int. Med. Case Rep. J. 2021;14:573–575. doi: 10.2147/IMCRJ.S328482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letarov A.V., Babenko V.V., Kulikov E.E. Free SARS-CoV-2 spike protein S1 particles may play a role in the pathogenesis of COVID-19 infection. Biochemistry (Mosc.) 2021;86(3):257–261. doi: 10.1134/S0006297921030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Amarasinghe G.K. When your cap matters: structural insights into self vs non-self recognition of 5' RNA by immunomodulatory host proteins. Curr. Opin. Struct. Biol. 2016;36:133–141. doi: 10.1016/j.sbi.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang R., Wang L., Hao J., Zhang Q., Ling R., Yun J. Micro RNA‐762 promotes breast cancer cell proliferation and invasion by targeting IRF7 expression. Cell Prolif. 2015;48(6):643–649. doi: 10.1111/cpr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmann J. From interference to interferon: a brief historical introduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1982;299(1094):3–6. doi: 10.1098/rstb.1982.0101. [DOI] [PubMed] [Google Scholar]
- Liu T., Khanna K.M., Chen X., Fink D.J., Hendricks R.L. CD8(+) T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. J. Exp. Med. 2000;191(9):1459–1466. doi: 10.1084/jem.191.9.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Wang J., Xu J., Xia H., Wang Y., Zhang C., Chen W., Zhang H., Liu Q., Zhu R., et al. Comprehensive investigations revealed consistent pathophysiological alterations after vaccination with COVID-19 vaccines. Cell Discov. 2021;7(1):99. doi: 10.1038/s41421-021-00329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lladó I., Fernández-Bernáldez A., Rodríguez-Jiménez P. Varicella zoster virus reactivation and mRNA vaccines as a trigger. JAAD. Case Rep. 2021;15:62–63. doi: 10.1016/j.jdcr.2021.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Harada M., Kamijo Y., Nakajima T., Tanaka N., Sugiyama E., Kyogashima M., Gonzalez F.J., Aoyama T. Peroxisome proliferator-activated receptor attenuates high-cholesterol diet-induced toxicity and pro-thrombotic effects in mice. Arch. Toxicol. 2019;93(1) doi: 10.1007/s00204-018-2335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane M., Kohlhaas S.L., Sutcliffe M.J., Dyer M.J., Cohen G.M. TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res. 2005;65(24):11265–11270. doi: 10.1158/0008-5472.CAN-05-2801. [DOI] [PubMed] [Google Scholar]
- Maleki A. COVID-19 recombinant mRNA vaccines and serious ocular inflammatory side effects: real or coincidence? J. Ophthalmic Vis. Res. 2021;16(3) doi: 10.18502/jovr.v16i3.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Sekhon S., Sekhon S. Drug-induced liver injury after COVID-19 vaccine. Cureus. 2021;13(7) doi: 10.7759/cureus.16491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus J., Honigbaum S., Shroff S., Honke K., Rosenbluth J., Dupree J.L. Sulfatide is essential for the maintenance of CNS myelin and axon structure. Glia. 2006;53(4):372–381. doi: 10.1002/glia.20292. [DOI] [PubMed] [Google Scholar]
- Martini P.G.V., Guey L.T. A new era for rare genetic diseases: messenger RNA therapy. Hum. Gene Ther. 2019;30(10):1180–1189. doi: 10.1089/hum.2019.090. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Tani K., Asano S. Interferon-alpha-induced G1 phase arrest through upregulated expression of CDK inhibitors, p19Ink4D and p21Cip1 in mouse macrophages. Oncogene. 1998;16:2075–2086. doi: 10.1038/sj.onc.1201745. [DOI] [PubMed] [Google Scholar]
- Mauro V.P., Chappell S.A. A critical analysis of codon optimization in human therapeutics. Trends Mol. Med. 2014;20(11):604–613. doi: 10.1016/j.molmed.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy C., Carrea A., Diambra L. Bicodon bias can determine the role of synonymous SNPs in human diseases. BMC Genom. 2017;18(1):227. doi: 10.1186/s12864-017-3609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie S.E., Taylor S.M., Malladi P., Yuhan H., Cassel D.L., Chien P., Schwartz E., Schreiber A.D., Surrey S., Reilly M.P. The role of the human Fc receptor FcRIIA in the immune clearance of platelets: a transgenic mouse model. J. Immunol. 1999;162:4311–4318. http://www.jimmunol.org/content/162/7/4311 [PubMed] [Google Scholar]
- McKernan K., Kyriakopoulos A.M., McCullough P.A. Differences in vaccine and SARS-CoV-2 replication derived mRNA: implications for cell biology and future disease. OSF Prepr. 2021 doi: 10.31219/osf.io/bcsa6. November 26. [DOI] [Google Scholar]
- McLachlan S., Osman M., Dube K., Chiketero P., Choi Y., Fenton N. Analysis of COVID-19 vaccine death reports from the vaccine adverse events reporting system (VAERS) database. Preprint. 2021 doi: 10.13140/RG.2.2.26987.26402. [DOI] [Google Scholar]
- Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. 5' UTR m(6)A promotes cap-independent translation. Cell. 2015;163(4):999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Banerjea A.C. SARS-CoV-2 Spike targets USP33-IRF9 axis via exosomal miR-148a to activate human microglia. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.656700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal M.K., Chaudhuri G. Abstracts: First AACR International Conference on Frontiers in Basic Cancer Research--Oct 8–11, 2009. Boston, MA. 2009. https://cancerres.aacrjournals.org/content/69/23_Supplement/A16.short [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Raabe V., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Walsh E.E., Frenck R., Falsey A.R., Dormitzer P.R., Gruber W.C., Şahin U., Jansen K.U. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Mungoven T.J., Meylakh N., Marciszewski K.K., Macefield V.G., Macey P.M., Henderson L.A. Microstructural changes in the trigeminal nerve of patients with episodic migraine assessed using magnetic resonance imaging. J. Headache Pain. 2020;21:59. doi: 10.1186/s10194-020-01126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musella M., Manic G., de Maria R., Vitale I., Sistigue A. Type-I-interferons in infection and cancer: unanticipated dynamics with therapeutic implications. OncoImmunology. 2017;6(5) doi: 10.1080/2162402X.2017.1314424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandha R., Singh H. Renin angiotensin system: a novel target for migraine prophylaxis. Indian J. Pharmacol. 2012;44(2) doi: 10.4103/0253-7613.93840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute BRCA gene mutations: cancer risk and genetic testing fact sheet. 2021. https://www.cancer.gov/about-cancer/causes-prevention/genetics/brca-fact-sheet#what-other-cancers-are-linked-to-harmful-variants-in-brca1-and-brca2 [online] Available at:
- Nevzorova T.A., Mordakhanova E.R., Daminova A.G., Ponomareva A.A., Andrianova I.A., Minh G.L., Rauova L., Litvinov R.L., Weisel J.W. Platelet factor 4-containing immune complexes induce platelet activation followed by calpain-dependent platelet death. Cell Death Dis. 2019;5:106. doi: 10.1038/s41420-019-0188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn R.C. G-quadruplexes within prion mRNA: the missing link in prion disease? Nucleic Acids Res. 2014;42:9327–9333. doi: 10.1093/nar/gku559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandini von Niessen A.G., Poleganov M.A., Rechner C., Plaschke A., Kranz L.M., Fesser S., Diken M., Löwer M., Vallazza B., Beissert T., et al. Improving mRNA-based therapeutic gene delivery by expression-augmenting 3’ UTRs identified by cellular library screening. Mol. Ther. 2019;27(4):824–836. doi: 10.1016/j.ymthe.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka H., Fukao A., Funakami Y., Duncan K.E., Fujiwara T. Emerging evidence of translational control by AU-rich element-binding proteins. Front. Genet. 2019;10:332. doi: 10.3389/fgene.2019.00332.g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudit G.Y., Kassiri Z., Jiang C., Liu P.P., Poutanen S.M., Penninger J.M., Butany J. SARS coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur. J. Clin. Invest. 2009;39(7) doi: 10.1111/j.1365-2362.2009.02153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki T., Nakagawara A. Role of p53 in cell death and human cancers. Cancers. 2011;3(1):994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. 2014;15:9. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17(4):261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.W., Lagniton P., Liu Y., Xu R.H. mRNA vaccines for COVID-19: what, why and how. Int. J. Biol. Sci. 2021;17(6):1446–1460. doi: 10.7150/ijbs.59233. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passariello M., Vetrei C., Amato F., De Lorenzo C. Interactions of spike-RBD of SARS-CoV-2 and platelet factor 4: new insights in the etiopathogenesis of thrombosis. Int. J. Mol. Sci. 2021;22:8562. doi: 10.3390/ijms22168562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegu E., Ernst P.A. IFN-alpha wakes up sleeping hematopoietic stem cells. Nat. Med. 2009;15(6) doi: 10.1038/nm0609-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perricone C., Ceccarelli F., Nesher G., Borella E., Odeh Q., Conti F., Shoenfeld Y., Valesini G. Immune thrombocytopenic purpura (ITP) associated with vaccinations: a review of reported cases. Immunol. Res. 2014;60:226–235. doi: 10.1007/s12026-014-8597-x. [DOI] [PubMed] [Google Scholar]
- Psichogiou M., Karabinis A., Poulakou G., Antoniadou A., Kotanidou A., Degiannis D., Pavlopoulou I.D., Chaidaroglou A., Roussos S., Mastrogianni E., et al. Comparative immunogenicity of BNT162b2 mRNA vaccine with natural COVID-19 infection. Vaccines (Basel) 2021;9(9):1017. doi: 10.3390/vaccines9091017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psichogiou M., Samarkos M., Mikos N., Hatzakis A. Reactivation of Varicella zoster virus after vaccination for SARS-CoV-2. Vaccines. 2021;9:572. doi: 10.3390/vaccines9060572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.-K., Ma J. Alteration in microRNA-155 level correspond to severity of coronary heart disease. Scand. J. Clin. Lab. Invest. 2018;78(3):219–223. doi: 10.1080/00365513.2018.1435904. [DOI] [PubMed] [Google Scholar]
- Qiu S., Palavicini J.P., Wang J., Gonzalez N.S., He S., Dustin E., Zou C., Ding L., Bhattacharjee A., Van Skike C.E., et al. Adult-onset CNS myelin sulfatide deficiency is sufficient to cause Alzheimer's disease-like neuroinflammation and cognitive impairment. Mol. Neurodegener. 2021;16:64. doi: 10.1186/s13024-021-00488-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röltgen K., Nielsen S.C.A., Silva O., Younes S.F., Zaslavsky M., Costales C., Yang F., Wirz O.F., Solis D., Hoh R.A. Immune imprinting, breadth of variant recognition and germinal center response in human SARS-CoV-2 infection and vaccination. Cell. 2022 doi: 10.1016/j.cell.2022.01.018. Jan 25; S0092-8674(22)00076-9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S.A., Abul-Husn N.S., Casanova J.L., Daly M.J., Rehm H.L., Murray M.F. The intersection of genetics and COVID-19 in 2021: preview of the 2021 Rodney Howell Symposium. Genet. Med. 2021;23(6):1001–1003. doi: 10.1038/s41436-021-01113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak M.Z., Ratajczak J. Horizontal transfer of RNA and proteins between cells by extracellular microvesicles: 14 years later. Clin. Transl. Med. 2016;5:7. doi: 10.1186/s40169-016-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhea E.M., Logsdon A.F., Hanse K.M., Williams L.M., Reed M.J., Baumann K.K., Holden S.J., Raber J., Banks W.A., Erickson M.A. The S1 protein of SARS-CoV-2 crosses the blood-brain barrier in mice. Nat. Neurosci. 2021;24:368–378. doi: 10.1038/s41593-020-00771-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues Figueiredo R., Aparecida Azevedo A., De Oliveira Penido N. Positive association between tinnitus and arterial hypertension. Front. Neurol. 2016;7:171. doi: 10.3389/fneur.2016.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Perez A.I., Borrajo A., Rodriguez-Pallares J., Guerra M.J., Labandeira-Garcia J.L. Interaction between NADPH-oxidase and Rho-kinase in angiotensin II-induced microglial activation. Glia. 2015;63:466e482. doi: 10.1002/glia.22765. [DOI] [PubMed] [Google Scholar]
- Rose J. Critical appraisal of VAERS pharmacovigilance: is the U.S. vaccine adverse events reporting system (VAERS) a Functioning pharmacovigilance system? Sci. Publ. Health Pol. the Law. 2021;3:100–129. [Google Scholar]
- Rouleau S., Glouzon J.S., Brumwell A., Bisaillon M., Perreault J.P. 3' UTR G-quadruplexes regulate miRNA binding. RNA. 2017;23(8):1172–1179. doi: 10.1261/rna.060962.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau S.G., Garant J.-M., Balduc F., Bisaillon M., Perreault J.-P. G-Quadruplexes influence pri-microRNA processing. RNA Biol. 2018;15(2):198–206. doi: 10.1080/15476286.2017.1405211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusk N. When microRNAs activate translation. Nat. Methods. 2008;5:122–123. doi: 10.1038/nmeth0208-122a. [DOI] [Google Scholar]
- Ruther U., Nunnensiek C., Muller H.A., Bader H., May U., Jipp P. Interferon alpha (IFN alpha 2a) therapy for herpes virus-associated inflammatory bowel disease (ulcerative colitis and Crohn's disease) Hepato-Gastroenterology. 1998;45(21):691–699. doi: 10.1111/j.1348-0421.1999.tb03365.x. [DOI] [PubMed] [Google Scholar]
- Sakai Y., Ohga S., Tonegawa Y., Takada H., Nakao F., Nakayama H., Aoki T., Yamamori S., Hara T. Interferon-alpha therapy for chronic active Epstein-Barr virus infection: potential effect on the development of T- lymphoproliferative disease. J. Pediatr. Hematol. Oncol. 1998;20(4):342–346. doi: 10.1097/00043426-199807000-00013. [DOI] [PubMed] [Google Scholar]
- Sayers T.J. Targeting the extrinsic apoptosis signaling pathway for cancer therapy. Cancer Immunol. Immunother. 2011;60(8):1173–1180. doi: 10.1007/s00262-011-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., Melanson R., Werner S., Wei Y., Zimmer M., et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021;6(3):339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Chevillotte M.D., Rice C.M. Interferon-stimulated genes: a complex web of host defenses. Anni. Rev. Immunol. 2014;32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi K., Watanabe N., Miyazaki N., Ishizuchi K., Iba C., Tagashira Y., Uno S., Shibata M., Hasegawa N., Takemura R., et al. Incidence of headache after COVID-19 vaccination in patients with history of headache: a cross-sectional study. Cephalalgia. 2021 doi: 10.1177/03331024211038654. [Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneff S., Nigh G. Worse than the disease? Reviewing some possible unintended consequences of the mRNA vaccines against COVID-19. IJVTPR. 2021;2(1):38–79. [Google Scholar]
- Shabalina S.A., Spiridonov N.A., Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41(4):2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatsky I.N., Terenin I.M., Smirnova V.V., Andreev D.E. Cap-independent translation: what's in a name? Trends Biochem. Sci. 2018;43(11):882–895. doi: 10.1016/j.tibs.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Shaw G., Morse S., Ararat M., Graham F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb. J. 2002;16(8):869–871. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- Shitrit P., Zuckerman N.S., Mor O., Gottesman B.-S., Chowers M. Nosocomial outbreak caused by the SARS-CoV-2 Delta variant in a highly vaccinated population, Israel, July 2021. Euro Surveill. 2021;26(39) doi: 10.2807/1560-7917.ES.2021.26.39.2100822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrotri M., Navaratnam A.M., Nguyen V., Byrne T., Geismar C., Fragaszy E., Beale S., Fong W.L.E., Patel P., Kovar J., et al. Spike-antibody waning after second dose of BNT162b2 or ChAdOx1. Lancet. 2021;398(10298):385–387. doi: 10.1016/S0140-6736(21)01642-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone A., Herald J., Chen A. Acute myocarditis following COVID-19 mRNA vaccination in adults aged 18 years or older. JAMA Intern. Med. 2021;181(12):1668–1670. doi: 10.1001/jamainternmed.2021.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small E.M., Olson E.N. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469(7330):336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sola I., Almazán F., Zúñiga S., Enjuanes L. Continuous and discontinuous RNA synthesis in coronaviruses. Ann. Rev. Virol. 2015;2(1):265–288. doi: 10.1146/annurev-virology-100114-055218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis M., Goubau D., Romieu-Mourez R., Genin P., Civas A., Hiscott J. Distinct functions of IRF-3 and IRF-7 in IFN-alpha gene regulation and control of anti-tumor activity in primary macrophages. Biochem. Pharmacol. 2006;72(11):1469–1476. doi: 10.1016/j.bcp.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Spiegel J., Adhikari S., Balasubramanian S. The structure and function of DNA G-quadruplexes. Trend. Chem. 2020;2(2):123–136. doi: 10.1016/j.trechm.2019.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S., Hale B.G. Interferon system deficiencies exacerbating severe pandemic virus infections. Trends Microbiol. 2021;29(11):973–982. doi: 10.1016/j.tim.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suberbielle E., Djukic B., Evans M., Kim D.H., Taneja P., Wang X., Finucane M., Knox J., Ho K., Devidze N., et al. DNA repair factor BRCA1 depletion occurs in Alzheimer brains and impairs cognitive function in mice. Nat. Commun. 2015;6:8897. doi: 10.1038/ncomms9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, S.V.; Kumar, A. Increases in COVID-19 are unrelated to levels of vaccination across 68 countries and 2947 counties in the United States. Eur. J. Epidemiol. 2021, 1-4. doi: 10.1007/s10654-021-00808-7. [DOI] [PMC free article] [PubMed]
- Sundstedt A., Celander M., Hedlund G. Combining tumor-targeted superantigens with interferon-alpha results in synergistic anti-tumor effects. Int. Immunopharm. 2008;8(3):442–452. doi: 10.1016/j.intimp.2007.11.006. 2008. [DOI] [PubMed] [Google Scholar]
- Svitkin U.V., Herdy B., Costa-Mattioli M., Gingras A.-C., Raught B., Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell Biol. 2005;25(23):10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A., Tamura T., Taniguchi T. Interferon regulatory factor family of transcription factors and regulation of oncogenesis. Cancer Sci. 2008;99(3):467–478. doi: 10.1111/j.1349-7006.2007.00720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa U. TRAIL/TRAIL‐R in hematologic malignancies. J. Cell. Biochem. 2010;110(1):21–34. doi: 10.1002/jcb.22549. [DOI] [PubMed] [Google Scholar]
- Tetz G., Tetz V. Prion-like domains in spike protein of SARS-CoV-2 differ across its variants and enable changes in affinity to ACE2. Microorganisms. 2022;10:280. doi: 10.3390/microorganisms10020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W.-L., Guo R., Wang F., Jiang Z.-X., Tang P., Huang Y.-M., Sun L. The IRF9-SIRT1-P53 axis is involved in the growth of human acute myeloid leukemia. Exp. Cell Res. 2018;365:185–193. doi: 10.1016/j.yexcr.2018.02.036. [DOI] [PubMed] [Google Scholar]
- Timmers L.F.S.M., Peixoto J.V., Ducati R.G., Bachega J.F.R., de Mattos Pereira L., Caceres R.A., Majolo F., da Silva G.L., Anton D.B., Dellagostin O.A., Henriques J.A.P., Xavier L.L., Goettert M.I., Laufer S. SARS-CoV-2 mutations in Brazil: from genomics to putative clinicalconditions. Sci. Rep. 2021;11:11998. doi: 10.1038/s41598-021-91585-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronvik E., Stovner L.J., Helde G., Sand T., Bovim G. Prophylactic treatment of migraine with an angiotensin II receptor-blocker: a randomized controlled trial. JAMA. 2003;289(1):65–69. doi: 10.1001/jama.289.1.65. [DOI] [PubMed] [Google Scholar]
- Troya J., Bastard P., Planas-Serra L., Ryan P., Ruiz M., de Carranza M., Torres J., Martnez A., Abel L., Casanova J.-L., Pujol A. Neutralizing autoantibodies to type I IFNs in >10% of patients with severe COVID-19 pneumonia hospitalized in Madrid, Spain. J. Clin. Immunol. 2021;41 doi: 10.1007/s10875-021-01036-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuno T., Mejido J., Zhao T., Morrow A., Zoon K.C. IRF9 is a key factor for eliciting the antiproliferative activity of IFN-α. J. Immunother. 2009;32(8):803. doi: 10.1097/CJI.0b013e3181ad4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uranaka T., Kashio A., Ueha R., Sato T., Bing H., Ying G., Kinoshita M., Kondo K., Yamasoba T. Expression of ACE2, TMPRSS2, and furin in mouse ear tissue, and the implications for SARS-CoV-2 infection. Laryngoscope. 2021;131(6):E2013–E2017. doi: 10.1002/lary.29324. [DOI] [PubMed] [Google Scholar]
- Vaers Home VAERS. https://vaers.hhs.gov/data/dataguide.html n.d.). Retrieved December 5, 2021, from.
- van der Wijst M.G.P., Vazquez S.E., Hartoularos G.C., Bastard P., Grant T., Bueno R.>, Lee D.S., Greenland J.R., Sun Y., Perez R., et al. Type I interferon autoantibodies are associated with systemic immune alterations in patients with COVID-19. Sci. Transl. Med. 2021;13(612) doi: 10.1126/scitranslmed.abh2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint S., Renmans D., Broos K., Dewitte H., Lentacker I., Heirman C., Breckpot K., Thielemans K. The ReNAissanCe of mRNA-based cancer therapy. Expert Rev. Vaccines. 2015;14(2):235–251. doi: 10.1586/14760584.2015.957685. [DOI] [PubMed] [Google Scholar]
- Vanderlugt C.L., Miller S.D. Epitope spreading in immune-mediated diseases: implications for immunotherapy. Nat. Rev. Immunol. 2002;2:85–95. doi: 10.1038/nri724. [DOI] [PubMed] [Google Scholar]
- Verma A.K., Lavine K.J., Lin C.-Y. Myocarditis after covid-19 mRNA vaccination. NEJM. 2021;385:1332–1334. doi: 10.1056/NEJMc2109975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verweij M.C., Wellish M., Whitmer T., Malouli D., Lapel M., Jonjić S., Haas J.G., DeFilippis V.R., Mahalingam R., Früh K. Varicella viruses inhibit interferon-stimulated JAK-STAT signaling through multiple mechanisms. PLoS Pathog. 2015;11(5) doi: 10.1371/journal.ppat.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-A., Zhang R., Jiang D., Deng W., Zhang S., Deng S., Zhong J., Wang T., Zhu L.-H., Yang L., et al. Interferon regulatory factor 9 protects against hepatic insulin resistance and steatosis in male mice. Hepatology. 2013;58(2):603–616. doi: 10.1002/hep.26368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hu H., Zhang K. Overview of interferon: characteristics, signaling and anti-cancer effect. Arch. Biotechnol. Biomed. 2017;1:1–16. [Google Scholar]
- Wang C., Zhang C., Liu L., A X., Chen B., Li Y., Du J. Macrophage-derived mir-155-containing exosomes suppress fibroblast proliferation and promote fibroblast inflammation during cardiac injury. Mol. Ther. 2017;25(1):192–204. doi: 10.1016/j.ymthe.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Zhan Y., Zhu L., Hou Z., Liu F., Song P., Qiu F., Wang X., Zou X., Wan D., et al. Retrospective multicenter cohort study shows early interferon therapy is associated with favorable clinical responses in COVID-19 patients. Cell Host Microbe. 2020;28(3):455–464. doi: 10.1016/j.chom.2020.07.005. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Nakajima T., Gonzalez F.J., Tanaka N. PPARs as metabolic regulators in the liver: lessons from liver-specific PPAR-null mice. Int. J. Mol. Sci. 2020;21:2061. doi: 10.3390/ijms21062061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E., Thombre R., Shah Y., Latanich R., Wang J. G-Quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res. 2021;49(9):4816–4830. doi: 10.1093/nar/gkab164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H., Chen Q., Lin L., Sha C., Li T., Liu Y., Yin X., Xu Y., Chen L., Gao W., Li Y., Zhu X. Regulation of exosome production and cargo sorting. Int. J. Biol. Sci. 2021;17(1):163–177. doi: 10.7150/ijbs.53671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weikert U., Kühl U., Schultheiss H.-P., Rauch U. Platelet activation is increased in patients with cardiomyopathy: myocardial inflammation and platelet reactivity. Platelets. 2002;13(8):487–491. doi: 10.1080/0953710021000057857. [DOI] [PubMed] [Google Scholar]
- Weiner J., Lewis D., Maertzdorf J., Mollenkopf H., Bodinham C., Pizzoferro K., Linley C., Greenwood A., Mantovani A., Bottazzi B., Denoel P., Leroux-Roels G., Kester K., Jónsdóttir I., van den Berg R.A., Kaufmann S., Del Giudice G. Characterization of potential biomarkers of reactogenicity of licensed antiviral vaccines: randomized controlled clinical trials conducted by the BIOVACSAFE consortium. Sci. Rep. 2019;9(1):20362. doi: 10.1038/s41598-019-56994-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weldon C., Dacanay J.G., Gokhale V., Boddupally P.V.L., Behm-Ansmant I., Burley G.A., Branlant C., Hurley L.M., Dominguez C., Eperon I.C. Specific G-quadruplex ligands modulate the alternative splicing of Bcl-X. Nucleic Acids Res. 2018;46(2):886–896. doi: 10.1093/nar/gkx1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G.D., Gokhale N.S., Snider D.L., Horner S.M. The mRNA cap 2'-O-methyltransferase CMTR1 regulates the expression of certain interferon-stimulated genes. mSphere. 2020;5(3) doi: 10.1128/mSphere.00202-20. e00202-e00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisnewski A.V., Campillo Luna J., Redlich C.A. Human IgG and IgA responses to COVID-19 mRNA vaccines. PLoS One. 2021;16(6) doi: 10.1371/journal.pone.0249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X. Detailed dissection and critical evaluation of the Pfizer/BioNTech and Moderna mRNA vaccines. Vaccines. 2021;9:734. doi: 10.3390/vaccines9070734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahi N., Chahinian H., Fantini J. Infection-enhancing anti-SARS-CoV-2 antibodies recognize both the original Wuhan/D614G strain and Delta variants. A potential risk for mass vaccination? J. Infect. 2021;83(5):607–635. doi: 10.1016/j.jinf.2021.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Hu Y., Zhou B., Bao Y., Li Z., Gong C., Yang H., Wang S., Xiao Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020;11:960. doi: 10.1038/s41419-020-03143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa F.S., Teixeira F.M., Sato M.N., Oliveira L.M. Delivery of microRNAs by extracellular vesicles in viral infections: could the news be packaged? Cells. 2019;8(6):611. doi: 10.3390/cells8060611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Odenthal M., Fries J.W.U. Exosomes as miRNA carriers: formation–function–future. Int. J. Mol. Sci. 2016;17:2028. doi: 10.3390/ijms17122028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S., Ries R.J., Jaffrey S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. 2019;20:608–624. doi: 10.1038/s41580-019-0168-5. [DOI] [PubMed] [Google Scholar]
- Zakaria Z., Sapiai N.A., Izaini Ghani A.R. Cerebral venous sinus thrombosis 2 weeks after the first dose of mRNA SARS‐CoV‐2 vaccine. Acta Neurochir. 2021;163(8):2359–2362. doi: 10.1007/s00701-021-04860-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Powell S.N. The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol. Cancer Res. 2005;3(10):531–539. doi: 10.1158/1541-7786.MCR-05-0192. [DOI] [PubMed] [Google Scholar]
- Zhang W., Luo J., Yang F., Wang Y., Yin Y., Strom A., Gustafsson J.Å., Guan X. BRCA1 inhibits AR-mediated proliferation of breast cancer cells through the activation of SIRT1. Sci. Rep. 2016;6:22034. doi: 10.1038/srep22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Xiao K., Gu Y., Liu H., Sun X. Whole genome identification of potential G-quadruplexes and analysis of the G-quadruplex binding domain for SARS-CoV-2. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.587829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen W., Zhu W., Meng H., Chen J., Zhang J. Overexpression of interferon regulatory factor 7 (IRF7) reduces bone metastasis of prostate cancer cells in mice. Oncol. Res. 2017;25(4):511. doi: 10.3727/096504016X14756226781802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Guo J., Cha J., Chae M., Chen S., Barral J.M., Sachs M.S., Liu Y. Non-optimal codon usage affects expression, structure and function of clock protein FRQ. Nature. 2013;495(7439):111–115. doi: 10.1038/nature11833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zin Tun G.S., Gleeson D., Al-Joudeh A., Dube A. Immune-mediated hepatitis with the Moderna vaccine, no longer a coincidence but confirmed. J. Hepatol. 2021 doi: 10.1016/j.jhep.2021.09.031. Oct 5. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel L., Galluzzi L., Kepp O., Smyth M.J., Kroemer G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015;15(7):405–414. doi: 10.1038/nri3845. [DOI] [PubMed] [Google Scholar]
- Zoll J., Erkens Hulshof S., Lanke K., Verduyn Lunel F., Melchers W.J., Schoondermark-van de Ven E., Roivainen M., Galama J.M., van Kuppeveld F.J. Saffold virus, a human Theiler's-like cardiovirus, is ubiquitous and causes infection early in life. PLoS Pathog. 2009;5(5) doi: 10.1371/journal.ppat.1000416. [DOI] [PMC free article] [PubMed] [Google Scholar]