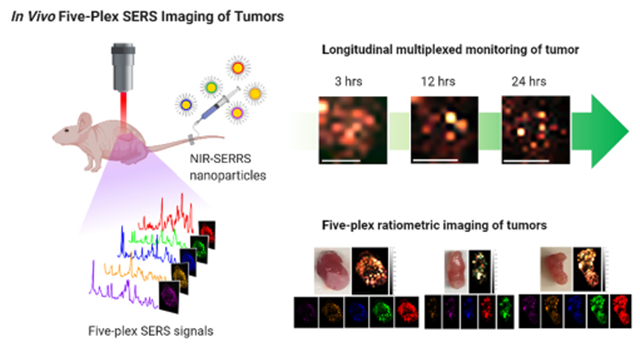
Abstract
In vivo multiplexed imaging aims for noninvasive monitoring of tumors with multiple channels without excision of the tissue. While most of the preclinical imaging has provided a number of multiplexing channels up to three, Raman imaging with surface-enhanced Raman scattering (SERS) nanoparticles was suggested to offer higher multiplexing capability originating from their narrow spectral width. However, in vivo multiplexed SERS imaging is still in its infancy for multichannel visualization of tumors, which require both sufficient multiplicity and high sensitivity concurrently. Here we create multispectral palettes of gold multicore-near-infrared (NIR) resonant Raman dyes-silica shell SERS (NIRSERRS) nanoparticle oligomers and demonstrate noninvasive and five-plex SERS imaging of the nanoparticle accumulation in tumors of living mice. We perform the five-plex ratiometric imaging of tumors by varying the administered ratio of the nanoparticles, which simulates the detection of multiple biomarkers with different expression levels in the tumor environment. Furthermore, since this method does not require the excision of tumor tissues at the imaging condition, we perform noninvasive and longitudinal imaging of the five-color nanoparticles in the tumors, which is not feasible with current ex vivo multiplexed tissue analysis platforms. Our work surpasses the multiplicity limit of previous preclinical tumor imaging methods while keeping enough sensitivity for tumor-targeted in vivo imaging and could enable the noninvasive assessment of multiple biological targets within the tumor microenvironment in living subjects.
Keywords: surface-enhanced Raman spectroscopy, surface-enhanced resonant Raman scattering, multiplexed imaging, in vivo imaging, cancer imaging
Graphical Abstract
Cancer is a heterogeneous disease in which the constituent cancer cells diversify to various molecular phenotypes.1 Multiplexed profiling of heterogeneous tumors is critical for classifying molecular subtypes of the tumors because it can reveal pathological variations among patients and guide the consequent patient-specific therapies.2–4 In translational and clinical stages, ex vivo multiplexed biopsy platforms have been developed to analyze tumor tissues with more than 20 biomarkers simultaneously.5–13 However, since this procedure is inevitably accompanied by excision of the tissues, it can only provide an instantaneous snapshot rather than longitudinal monitoring of multiple biomarkers at the same tumor site. Thus, it is necessary to develop a multiplexed imaging approach for noninvasive analysis of multiple biomarkers in vivo.14–16 Among the potential preclinical multiplexed imaging methods, optical imaging has inherent multiplexing capability through spectral deconvolution. For example, fluorescence imaging can provide up to five multiplexing channels across the visible wavelength region.17 Meanwhile, for noninvasive in vivo deep tissue imaging of living subjects, it is necessary to perform the imaging with both excitation and emission in the near-infrared (NIR) wavelength region, in which the broad fluorescence spectral bandwidth (~1500 cm−1) in the narrow NIR window limits the multiplicity up to three.18,19
Near-infrared Raman spectral imaging, on the other hand, offers higher multiplicity than the other optical imaging methods since it utilizes inelastic scattering emission with narrower spectral width (~10 cm−1), which corresponds to a characteristic “vibrational spectroscopic fingerprint”.20 While the intrinsic Raman intensities of molecules are weak, the sensitivity of Raman imaging can be greatly improved through the development of surface-enhanced Raman scattering (SERS) nanoparticles as imaging agents, in which plasmonic gold nanoparticles amplify the Raman signals of the adjacent Raman-active molecules (i.e., Raman reporters).21–30 The SERS nanoparticles provide picomolar-level sensitivity along with high multiplicity for the NIR Raman imaging. The multispectral SERS nanoparticles have enabled highly multiplexed in vivo deep tissue imaging of topically administered nanoparticles or nanoparticles uptake in the liver, in which nearly 100% of the administered nanoparticles are accumulated and imaged.31–34 However, the success of the multiplexed liver imaging has not been demonstrated in the in vivo multiplexed tumor imaging. The picomolar sensitivity of the nanoparticles was not sufficient for the noninvasive and multiplexed imaging of tumors, since only a limited amount (~1%) of the intravenously administered nanoparticles are expected to be accumulated and visualized in the tumors.35
Recent advances in the SERS nanoparticle synthesis, however, have further improved the imaging sensitivity by developing NIR surface-enhanced resonant Raman scattering (NIR-SERRS) nanoparticles. The NIR-SERRS nanoparticles utilize NIR dyes as resonant Raman reporters,36–40 which provide a higher Raman scattering cross-section (~10−25 cm2/photon) than nonresonant Raman reporters (~10−29 cm2/photon) by allowing Raman transition via electronically allowed states instead of virtual states.41 However, thus far, the NIR-SERRS nanoparticles have been utilized for other applications such as Raman-guided surgery,39,40 while the in vivo multiplexing capability of the NIR-SERRS nanoparticles has been limited to three—the same as other preclinical imaging modalities.42 This is mainly because the NIR-SERRS nanoparticles generate complex spectral fingerprints of the NIR-resonant Raman reporters with high spectral overlapping, which challenges their spectral unmixing to deliver a higher multiplicity than four.43 The complex spectral fingerprints of the NIR-SERRS nanoparticles also require further improvement of the sensitivity to be clearly resolved at the in vivo multiplexed imaging condition for accurate spectral unmixing.
In this paper, we utilize gold multicore-NIR Raman dyes-silica shell-structured multispectral NIR-SERRS nanoparticles to achieve fluorescence-like sensitivity via multicore nanoparticle geometry and provide both high multiplexing capability and enough sensitivity for in vivo imaging at the same time. Then, we demonstrate noninvasive and highly multiplexed imaging of NIR-SERRS nanoparticles-targeting tumors in living mice. While most in vivo preclinical imaging has provided up to three multiplexing channels, we successfully perform the five-plex ratiometric imaging of tumors by varying the administered ratio of the nanoparticles, which simulates variations of multiple biomarker expressions within the tumor microenvironment. In particular, since our multiplexed imaging method does not require the excision of tumor tissues at the imaging condition, it allows for noninvasive and longitudinal monitoring of the five-color NIR-SERRS nanoparticles retention in the tumors. Thereby it could enable the noninvasive assessment of multiple biological targets within tumors.
RESULTS AND DISCUSSION
Synthesis of Multispectral NIR-SERRS Nanoparticles.
We synthesized nine spectrally different NIR-SERRS nanoparticles by using heptamethine cyanine NIR dyes as Raman reporters.43 These NIR dyes have similar molecular structures to that of the clinically approved indocyanine green NIR dye and provide high NIR-resonant Raman cross-section that is several orders of magnitude brighter than nonresonant Raman reporter molecules (Figure 1).44 The multispectral NIR-SERRS nanoparticles were synthesized by the simultaneous addition of hydrophobic and cationic heptamethine cyanine dyes and tetraethylorthosilicate (TEOS) silica precursor to a 60 nm gold nanoparticle colloidal solution (Figure 2). The syntheses yielded NIR-resonant Raman dye-coated gold nanoparticles that are encapsulated in the 40 nm-thick silica shell (Figure 2a,b). The NIR-SERRS nanoparticles synthesized using a heptamethine NIR dye, 3,3’-diethylthiatricarbocyanine (DTTC), exhibited 110(±10)-fold higher brightness than the same structured SERS nanoparticles of nonresonant Raman dye, 1,2-bis(4-pyridyl)ethylene (BPE, S440, Oxonica Materials Inc., Figure 2c and Supporting Information Figure S1) at 10 pM. While the gold nanoparticle surface plasmon amplifies Raman scattering of the adsorbed Raman dyes, the silica shell stabilizes Raman signals by preventing desorption of the Raman dyes and adsorption of molecules from the outer environment onto the gold nanoparticle surface. We did not observe any influence of the silica shell thickness on the Raman intensity. However, the silication temperature was critical for the long-term duration of the Raman signal in the biological media.45,46 While the room-temperature grown silica shell allowed semipersistent protection of the Raman signal in serum, which was slowly decreased to 50% in 6 days due to the silica erosion, the silica shell grown at the elevated temperature of 60 °C allowed long-term duration of the Raman intensity over 20 days (Supporting Information Figure S2).46
Figure 1.
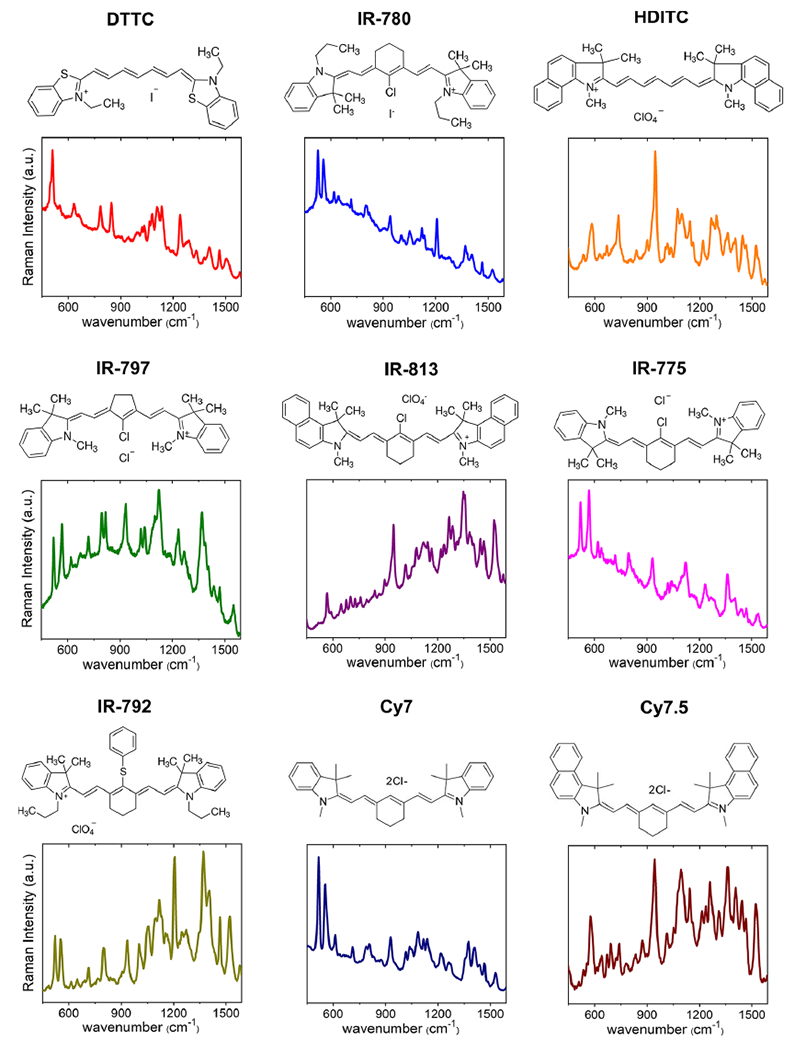
Nine spectrally different NIR-resonant Raman spectra. Molecular structures and the corresponding Raman spectra of heptamethine cyanine NIR-resonant Raman reporters that were embedded in the NIR-SERRS nanoparticles.
Figure 2.
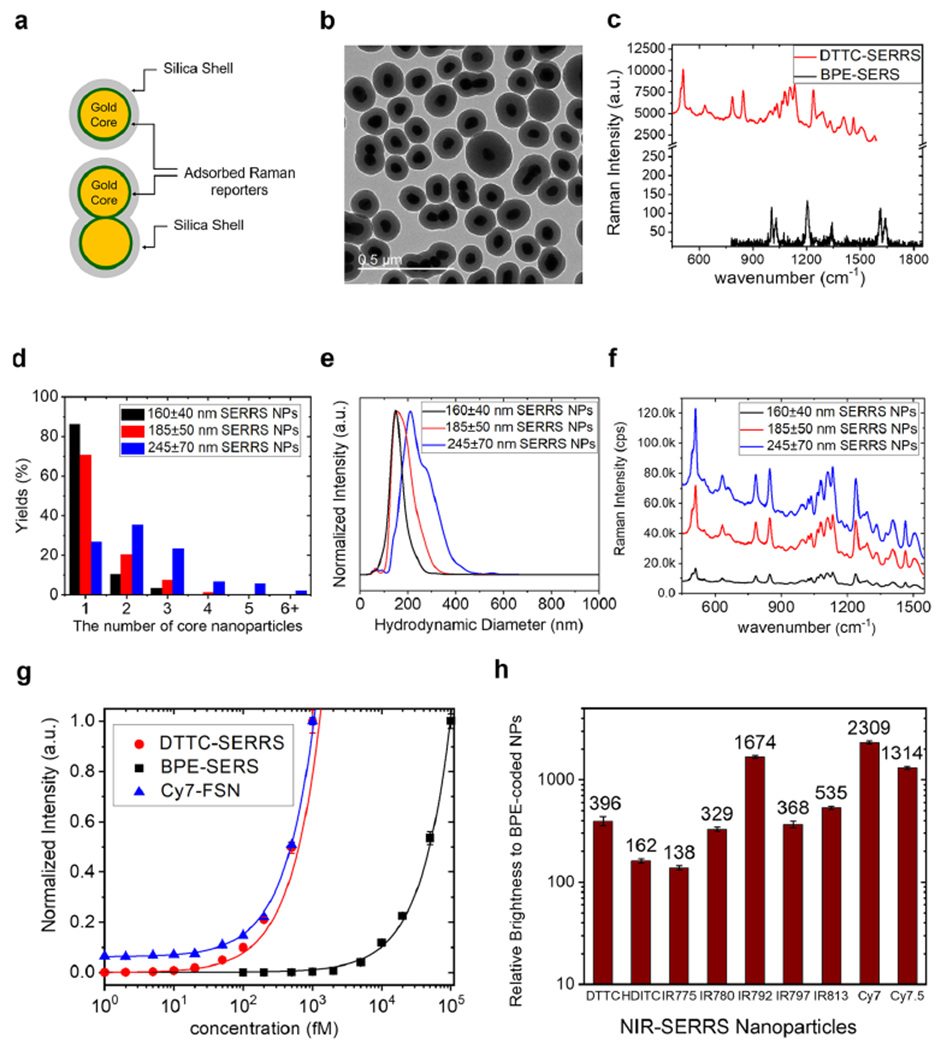
Synthesis and characterization of multispectral NIR-SERRS nanoparticles. (a) A schematic of gold nanoparticle core-Raman reporters-silica shell-structured NIR-SERRS nanoparticles. (b) A TEM image of the NIR-SERRS nanoparticles. The average size of the gold nanoparticle core is 60 nm in diameter, which is coated with a 40 nm-thick silica shell. The nanoparticles are mixtures of monomeric (single gold core in a silica shell), dimeric (double gold core in a silica shell), and oligomeric (multiple gold core in a silica shell) structures. Scale bar: 500 nm. (c) Raman spectra of the NIR-SERRS nanoparticles of 3,3’-diethylthiatricarbocyanine iodide (red, DTTC-SERRS, 160 ± 40 nm) and the nonresonant SERS nanoparticles of 1,2-bis(4-pyridyl)ethylene (black, BPE-SERS, S440, Oxonica Materials Inc., 180 ± 50 nm, see Supporting Information Figure S1) at the same nanoparticle concentration of 10 pM. (d–f) Controlled oligomerization of the nanoparticles upon the increase of the added DTTC dyes amount during the DTTC-coded SERRS nanoparticles syntheses: 10 nmol (160 ± 40 nm, black), 15 nmol (185 ± 50 nm, red), to 20 nmol (245 ± 70 nm, blue). The dose-dependent change in the number of gold nanoparticle cores in a single NIR-SERRS nanoparticle (d, see Supporting Information Figure S3), hydrodynamic diameter (e), and the Raman intensity (f) of the DTTC-coded SERRS nanoparticles. (g) Normalized standard curves of Raman intensities of the DTTC-coded SERRS nanoparticles (red, DTTC-SERRS, 185 ± 50 nm) and the BPE-coded SERS nanoparticles (black, BPE-SERS, 180 ± 50 nm), and the NIR fluorescence intensity of the Cy7 dye-doped silica nanoparticles (blue, Cy7-FSN, 215 ± 20 nm, see Supporting Information Figure S5). The LOD calculated from the curves were 3.8 ± 0.38 fM (red), 21.2 ± 6.6 fM (blue), and 650 ± 29 fM (black) concentrations of the nanoparticles, respectively (see Supporting Information Figure S6). (h) The Raman scattering brightness of the 9 NIR-SERRS nanoparticles with respect to the BPE-coded SERS nanoparticles (S440, Oxonica Materials), measured at 10 pM. The error bars (g and h) represent the standard deviations of the Raman intensities collected from multiple points (n = 300) scanning per measurement.
A transmission electron microscope image showed that the as-synthesized NIR-SERRS nanoparticles were a mixture of the silica-encapsulated single gold nanoparticles and the multicore gold nanoparticle oligomer structures such as dimers, trimers, and tetramers (Figure 2b). Such multicore oligomer structures are known to exhibit a large enhancement factor (EF) of 108 to amplify the Raman signals from the Raman reporters, which significantly outperform single-core monomeric nanopar-ticles.26,27 Therefore, we intentionally synthesized the multicore NIR-SERRS nanoparticle oligomers to achieve sufficient sensitivity for in vivo multiplexed imaging.
We increased the yield of the nanoparticle oligomers by increasing the amount of the added DTTC dyes during the synthesis (Figure 2d–f and Supporting Information Figure S3). The increment of the DTTC dyes induced more coating of the hydrophobic NIR dyes on the gold nanoparticles and more van der Waals interaction among the dyes-coated nanoparticles, which results in the controlled aggregation of gold nanoparticles into oligomers (Figure 2d and Supporting Information Figure S3).47 The increasing proportions of oligomers enlarged the average hydrodynamic diameter of the NIR-SERRS nanoparticles from 160 ± 40 nm (black) to 245 ± 70 nm (blue), and their cutoff sizes from 300 to 500 nm, respectively (Figure 2e), and further boosted up the Raman brightness from 110(±11) to 680(±66)-fold in comparison to the BPE-coded SERS nanoparticles (Figure 2f). It should be noted that such control of the oligomerization is also accompanied by the simultaneous control of the silica encapsulation. While the addition of the hydrophobic Raman dyes facilitates the oligomerization, the addition of ammonium hydroxide and silica precursor (TEOS) suppresses it by increasing the rate of the silica encapsulation process. Through this control, the degree of oligomerization, which is reflected on the average size and the SERS intensity of the synthesized NIR-SERRS nanoparticles, was highly reproducible as long as the same batch of gold nanoparticle solution was used (Supporting Information Figure S4).
To evaluate the enhanced Raman signal brightness of the NIR-SERRS nanoparticles in the context of in vivo NIR fluorescence imaging,48 we separately synthesized similarly sized Cy7-doped NIR-fluorescent silica nanobeads (215 ± 18 nm, and Supporting Information Figure S5). Then, we compared their fluorescence emission sensitivity to that of the Raman scattering emission from the DTTC-coded SERRS nanoparticles (186 ± 48 nm) in solution (Figure 2g and Supporting Information Figure S6). While the BPE-coded nonresonant SERS nanoparticles showed subpicomolar sensitivity for detection (650 ± 29 fM) under 785 nm excitation (black cubes in Figure 2g), both the DTTC-coded SERRS nanoparticles (3.8 ± 0.4 fM, red circles in Figure 2g) and the Cy7-doped NIR-fluorescent silica nanobeads (21.2 ± 6.6 fM, blue triangles in Figure 2g) exhibited femtomolar scale sensitivity. Note that the spectrally integrated intensity of the Raman spectrum of the DTTC-coded SERRS nanoparticles was 2.8(±0.1) times higher than that of the fluorescence spectra of the silica nanobeads, both of which were measured at the same concentration of 1 pM (Supporting Information Figure S7).
We extended the synthetic chemistry of the DTTC-coded NIR-SERRS nanoparticles for all the nine heptamethine cyanine Raman reporters described in Figure 1. For all the NIR-SERRS nanoparticles, we controlled the amount of Raman dyes during the nanoparticle synthesis to adjust the average size and the cutoff sizes to ~200 nm and ~400 nm, respectively (Supporting Information Figure S8). The produced NIR-SERRS nanoparticles exhibited 2–3 orders of magnitude higher Raman brightness than the BPE-coated nonresonant SERS nanoparticles (Figure 2h). All the experimental data discussed so far indicate that the multispectral NIR- SERRS nanoparticles will provide sufficient Raman scattering brightness for in vivo imaging in addition to their spectral diversity, that is, “multiplicity”.
Multiplicity Evaluation of the NIR-SERRS Nanoparticles.
For quantitative multiplexed imaging, it is essential to differentiate each NIR-SERRS nanoparticle reference spectrum from mixed spectra. As the number of the reference spectra increases, the probability for spectral overlap among the spectra also increases, which makes the unmixing of the multiplexed spectra more difficult. In particular, the bulkier NIR-resonant Raman reporters of the NIR-SERRS nanoparticles generate more complex Raman spectral fingerprints, which are more difficult to be unmixed (Figure 1). In this regard, we conducted the condition number analysis of the NIR-SERRS spectra to estimate the number of multiplexing channels that can be utilized for imaging (Figure 3a). A condition number (κ) of a function indicates how much deviation in the input will be amplified to deviation in the output with a factor of κ. Here, the condition number of the NIR-SERRS spectra is the amplification factor in the spectral unmixing error due to noise and spectral overlapping of multiple spectra. We derived the condition numbers for each combination of a given number of the normalized reference NIR-SERRS spectra. We then identified the combination that provides the lowest condition number for each number of the reference spectra (Table 1). We also performed the same analysis for the nonresonant SERS spectra for comparison (Figure 3a and Supporting Information Figure S9).
Figure 3.
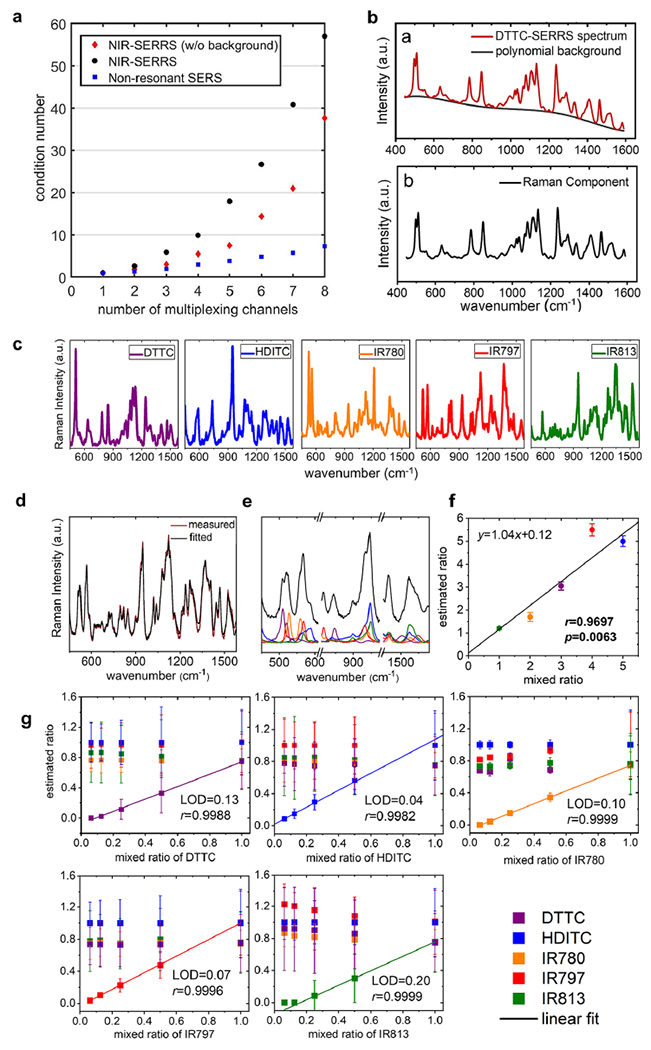
Multiplexing of the NIR-SERRS nanoparticles. (a) The plot of the lowest condition number versus the number of multiplex channels for the nonresonant SERS spectra (blue squares) and the NIR-SERRS spectra with (red diamonds) and without (black circles) polynomial background removals. (b) A measured spectrum of DTTC-coded SERRS nanoparticles (red in upper panel), in which the fluorescence background was fitted with a fifth-order polynomial (black in upper panel). The pure Raman component of the DTTC-coded SERRS spectrum after the polynomial-fitted background removal (lower panel). (c) The five NIR-SERRS reference spectra selected from the condition number analysis (a). (d–f) Spectral unmixing of a five-color NIR-SERRS nanoparticle mixture. A comparison between the measured Raman spectrum of the five-color NIR-SERRS nanoparticle mixture (red) and the best-fitted Raman spectrum (black) by an LMF least-squares method (d). The fitted Raman spectrum of the NIR-SERRS nanoparticle mixture in the selected wavenumber range (black), which shows the linear combination of the five NIR-SERRS spectra (e). The estimated ratios of the five-color NIR-SERRS nanoparticles from the spectral unmixing (d and e), which were normalized to the average concentration of the HDITC-coded nanoparticles that were set as 5. The five-color NIR-SERRS nanoparticles were mixed with 1:2:3:4:5 ratio of IR813, IR780, DTTC, IR797, and HDITC-coded SERRS nanoparticles, respectively. (g) The sensitivity of the five-plex Raman spectroscopy. The estimated ratio of the five-color NIR-SERRS nanoparticle mixtures, in which the concentrations of the four NIR-SERRS nanoparticles were kept constant as 1.0, and the fifth nanoparticle concentration was serially diluted. The ratios of all the nanoparticles were normalized to the average concentration of the IR797-coded nanoparticles (for the HDITC-coded nanoparticle variation) or that of HDITC-coded nanoparticles (for the other nanoparticles variations). The estimated ratio of the one nanoparticle variate was linearly fitted with the serially diluted ratio. The error bars (f and g) represent standard deviations of the nanoparticle concentrations, which were derived from the spectral unmixing of the multiple points spectra (n = 300) per single mixture measurement. Color codes of the five-color NIR-SERRS nanoparticles: DTTC, purple; HDITC, blue; IR780, orange; IR797, red; and IR813, green for (c), (e), (f), and (g).
Table 1.
Lowest Condition Numbers and the Corresponding NIR-SERRS Reference Spectra after the Polynomial Background Removal for Each Number of the Reference Spectra
| number of reference spectra | lowest condition number | choice of the NIR-SERRS spectra |
|---|---|---|
| 1 | 1 | Cy7 |
| 2 | 1.87 | DTTC, IR780 |
| 3 | 2.99 | DTTC, HDITC, IR780 |
| 4 | 5.44 | DTTC, HDITC, IR780, IR775 |
| 5 | 7.48 | DTTC, HDITC, IR780, lIR797, IR813 |
| 6 | 14.34 | Cy7, Cy7.5, DTTC, IR792, IR797, IR813 |
| 7 | 21.03 | Cy7, Cy7.5, DTTC, IR775, IR792, IR797, IR813 |
| 8 | 37.62 | Cy7, Cy7.5, DTTC, HDITC, IR775, IR792, IR797, IR813 |
Figure 3a shows the plot of the lowest condition number with respect to the number of Raman spectra. In all cases, the condition numbers increased as the number of the spectra increased, which reflects the spectral fitting errors increase when more spectra are taken into consideration. In particular, the condition numbers for the NIR-SERRS spectra exhibit a sharper increase with the number of spectra (black circles and red diamonds in Figure 3a) compared to those for the nonresonant SERS spectra (blue squares in Figure 3a). The analysis indicates that increasing the number of multiplexing channels for the NIR-SERRS nanoparticles is more complicated than the nonresonant SERS nanoparticles. This complexity stems from the fact that the NIR-resonant Raman reporters exhibit more complex spectral fingerprints than the nonresonant Raman reporters due to their bulkier molecular structures (compare Figure 1 with Supporting Information Figure S9). These complex spectra are also prone to more spectral overlapping. To limit the error in spectral deconvolution, we set a maximal threshold for the condition number to 10, which indicates a 10-fold increase in the spectral unmixing error upon multiplexing. Our target condition number allowed us to multiplex either 8 for the nonresonant SERS spectra (blue squares in Figure 3a) or 4 for the NIR-SERRS spectra (black circles in Figure 3a).
The NIR-SERRS nanoparticle spectrum is composed of a pure Raman spectral component and a broad background signal, which is mainly the fluorescence of the resonant Raman dyes (upper panel in Figure 3b). To further reduce the error and increase the number of multiplexing channels, the broad background signals were subtracted by fitting them with fifth-order polynomial, and only Raman spectral components were taken into account (lower panel in Figure 3b).49 The sole consideration of the pure Raman spectral components allowed us to reduce the condition number further and utilize five multiplexing channels (red diamonds in Figure 3a). Therefore, we chose five-color NIR-SERRS nanoparticles coded with DTTC, hexa-dibenzo-indotricarbocyanine (HDITC), IR780, IR797, and IR813 for further in vitro and in vivo multiplexed imaging, since this combination provides the lowest condition number and the smallest spectral unmixing error among the five-color nanoparticle combination from the nine NIR-SERRS spectra (Figure 3c and Table 1). It should be noted that the choice of condition number, 10, as a maximum threshold was arbitrary. It means that we can further exploit higher-order multiplexed imaging channels with higher condition numbers, as long as the higher error in the unmixed signals is experimentally acceptable or the spectral unmixing algorithm is advanced to reduce the unmixing error.
Next, we performed quantitative multiplexed assays of the five-color NIR-SERRS nanoparticles in solution.43 First, we measured Raman spectra of each NIR-SERRS nanoparticle solution, and the pure Raman components obtained by the fluorescence background subtractions were used as reference spectra. Then, we measured the Raman spectra of the five-color NIR-SERRS nanoparticle mixtures, in which the molar ratios of the nanoparticles were varied concurrently to 1:2:3:4:5 or 1:2:4:8:10 (Figure 3d–f and Supporting Information Figures S10 and S11). The spectra of the nanoparticle mixtures were unmixed based on the reference spectra using a non-negatively constrained Levenberg–Marquardt-Fletcher (LMF) least-squares algorithm (Figure 3d,e).
The five-plex spectra of the mixture solutions were successfully fitted with the spectral fitting error of 14.1(±2.1)%, defined as the average difference between the measured spectrum and the fitted spectrum over the entire wavenumber range (n = 5, Figure 3d and Supporting Information Figures S10 and S11). The mixture spectra were fitted to the linear combination of the five NIR-SERRS reference spectra (Figure 3e). Thereby we could estimate the ratio of the reference nanoparticles in the mixture from the fitting. We could also linearly correlate the nanoparticles mixed ratios with the estimated ratios with a spectral deconvolution error of 21.8(±15.2)%, defined as the average difference between the mixed ratio and the estimated ratio of the nanoparticles (n = 5, Pearson’s correlation coefficient r = 0.9588(±0.023), Figure 3f and Supporting Information Figure S11).
To evaluate the sensitivity of the five-plex spectroscopy for detection of the low-abundance nanoparticle from the five-color NIR-SERRS nanoparticle mixtures, the molar concentrations of four NIR-SERRS nanoparticles were kept constant, and the fifth nanoparticle concentration was serially diluted (Figure 3g). For all combinations of the five-color NIR-SERRS nanoparticles with a serial dilution of one of the five nanoparticles, we were able to detect the change in the concentrations of each NIR-SERRS nanoparticle variate within the mixtures and linearly correlate it with the input concentration (Pearson’s correlation coefficient r ≥ 0.998 in all cases). These results show sensitive multiplex detection of the low-abundance nanoparticle variate with the limit of detection (LOD) of 10.6(±6.2)% with respect to the other four nanoparticles.
In Vivo Five-Plex Raman Imaging of Tumors Using the NIR-SERRS Nanoparticles.
We performed noninvasive five-plex Raman imaging of subcutaneously implanted tumors in live nude mice using the NIR-SERRS nanoparticles. We simultaneously injected a total 600 fmol of the five-color NIR-SERRS nanoparticle mixture into the tail vein of each nude mouse bearing a subcutaneous LS174T tumor xenograft (n = 3). Then, we monitored the five-color nanoparticles accumulated at the tumor sites through multiplexed Raman imaging under the 785 nm NIR laser excitation (Figures 4 and 5) at 3, 12, and 24 h postinjection. In vivo Raman imaging detects signals through readout of the distinctive SERS spectra from nanoparticles, which can be separated from background fluorescence signals or weak intrinsic Raman signals of animals. Therefore, our NIR-SERRS nanoparticles that exhibit high sensitivity as NIR fluorescence in solution (Figure 2g) would also provide high sensitivity in vivo. It is important to note that while the SERS spectra inevitably have spectral overlap with the background Raman spectra from the living mice, the ratio of the SERS signal to the background Raman signal was large enough so that there was no significant contribution of the background Raman signal to increase the spectral unmixing error at the in vivo imaging condition (Supporting Information Figure S12). Benefiting from the high sensitivity of the NIR-SERRS nanoparticles (Figure 2), the Raman signals from the nanoparticles at the tumor site were clearly detected for over 24 h after injection (Figure 4a). During this period, the Raman signals at the tumor site peaked at 12 h postinjection and then decreased over time. Meanwhile, Raman signals from the nontumor sites gradually reduced except for the abdomen, where the reticular endothelial organs of the liver and spleen are located (Figure 4b). 244 h after injection, we excised the tumors, muscle, liver, spleen, and kidneys and imaged them under the Raman microscope (Supporting Information Figure S13). The tumor to muscle ratio of the accumulated nanoparticles calculated from the Raman intensities was 10.14(±7.39), which shows passive targeting of the NIR-SERRS nanoparticles to the tumors (Figure 4c and Supporting Information Figure S13).33 The tumor-targeting specificity of the NIR-SERRS nanoparticles, which was defined as the ratio of Raman intensity in tumors to that in the liver and spleen, was 2.60(±2.56)%. It should be noted that the femtomolar sensitivity of the NIR-SERRS nanoparticles enabled the detection of their Raman spectra in the tumors despite their low tumor-targeting specificity.
Figure 4.
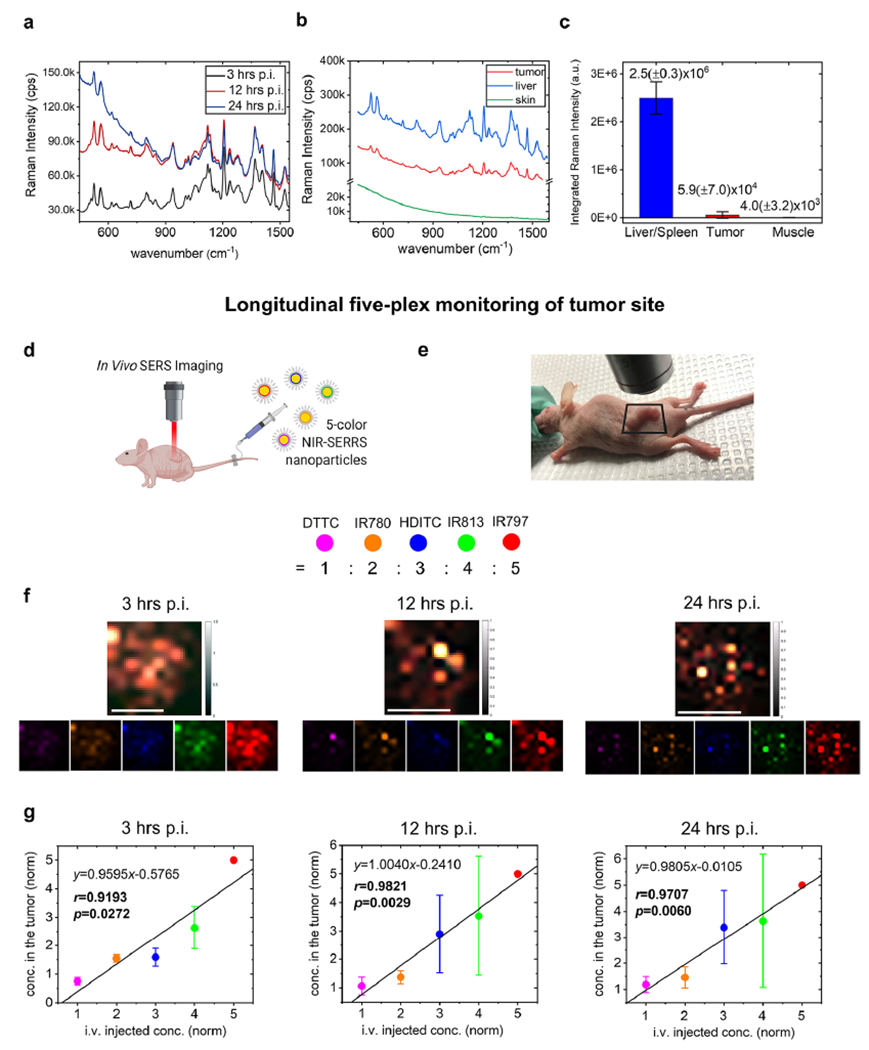
In Vivo five-plex imaging of tumor-targeted NIR-SERRS nanoparticles in a living mouse. (a) In vivo Raman spectra from the subcutaneous tumor at 3 h (black), 12 h (red), and 24 h (blue) postinjection (p.i.) of the five-color NIR-SERRS nanoparticles. (b) In vivo Raman spectra from the tumor (red), the liver (blue), and the noncancerous skin (green) of the tumor xenograft nude mouse after 24 h postinjection of the five-color NIR-SERRS nanoparticles. (c) Integrated Raman intensities from the excised tissues of the liver and spleen (blue), the tumor (red), and the muscle (green) after 24 h postinjection of the mixture of the five-color nanoparticles with different ratios. The error bars represent the standard deviations of the Raman intensities (n = 3, see Supporting Information Figure S13). (d) A schematic of Raman imaging of a subcutaneous tumor in a live nude mouse, to which the five-color NIR-SERRS nanoparticles were injected via tailvein. Image created with BioRender.com. (e–g) Noninvasive and multiplexed monitoring of the five-color NIR-SERRS nanoparticles at the tumor site on a live nude mouse. The ROI for the imaging was defined as a square (e). Five-plex Raman images of the subcutaneous tumor site at 3, 12, and 24 h postinjection of the five-color nanoparticles. The images were generated through the spectral unmixing of Raman spectra in each pixel and color-coded (lower panel, f) and merged (upper panel, f). Scale bar: 10 mm. The normalized concentrations (ratios) of the five-color nanoparticles within the tumor (g), obtained from the spectral unmixing of the Raman spectra in each pixel and averaged over the entire images (f). In each pixel, the five-color nanoparticle concentrations were normalized to the concentration of the IR797-coded NIR-SERRS nanoparticles that were set as 5. The error bars: the standard deviations of the spectrally unmixed concentrations calculated over the entire pixelated spectra. Color codes of the NIR-SERRS nanoparticles: DTTC, magenta-purple; HDITC, blue; IR780, orange; IR797, red; and IR813, green for (f and g). For all the experiments except (c), the NIR-SERRS nanoparticles were mixed with 1:2:3:4:5 molar ratio of DTTC, IR780, HDITC, IR813, and IR797-coded SERRS nanoparticles, respectively, and were concurrently administered.
Figure 5.
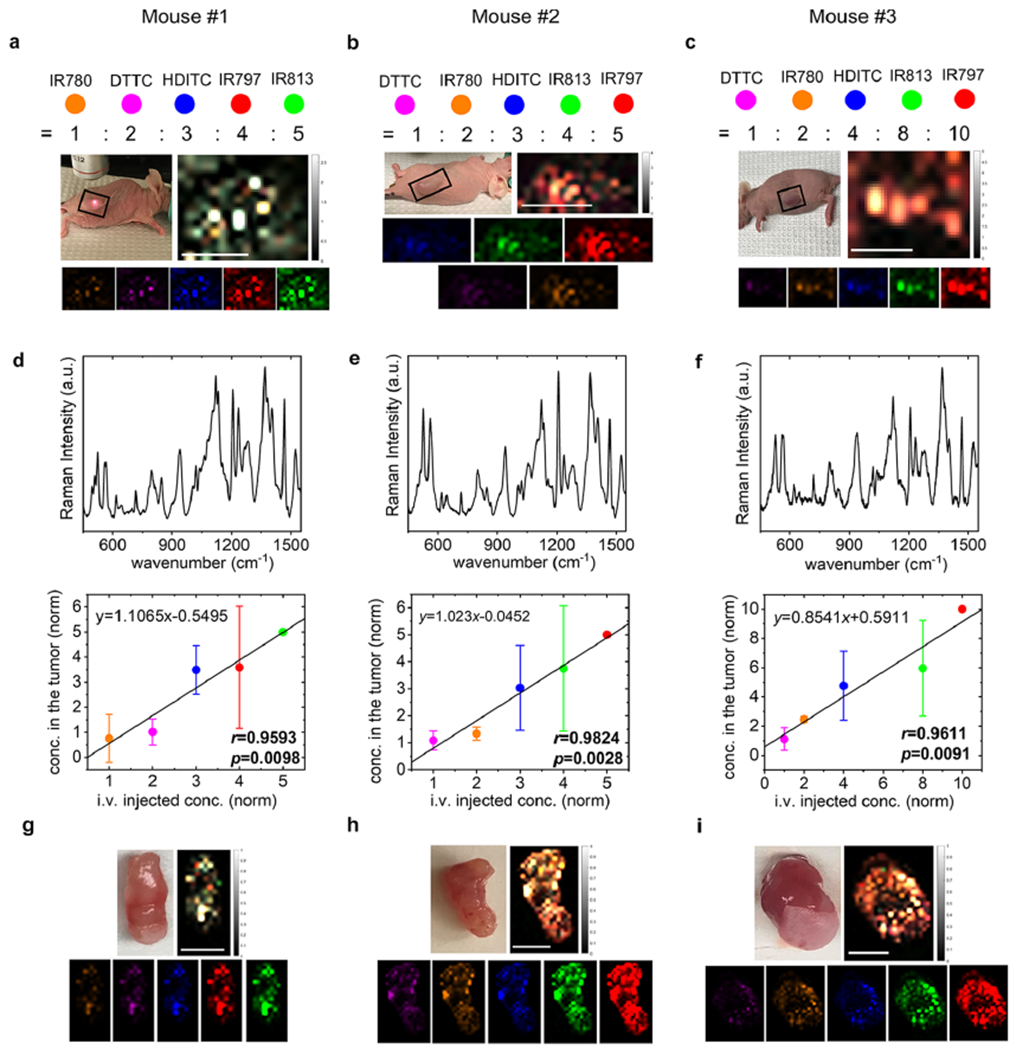
Correlation of the five-color NIR-SERRS nanoparticles injection ratio with the multiplexed Raman images of the tumors. (a–c) Tumor-xenograft nude mice (left, upper panel), and the five-color multiplexed Raman images of their tumors (right, upper panel), for which the ROI was defined as rectangles in the photograph. The multiplexed images were generated through the spectral unmixing of Raman spectra in each pixel, followed by color-coding (lower panels) and merge (right, upper panel). (d–f) Representative Raman spectra (upper panel) and the normalized concentrations (ratios) of the five-color NIR-SERRS nanoparticles (lower panel) in the tumors, obtained from the spectral unmixing of the Raman spectra in each pixel and averaged over the entire images of (a) (d), (b) (e), and (c) (f). In each pixel, the five-color nanoparticle concentrations were normalized to the concentration of the IR813-coded (d) or IR797-coded NIR-SERRS nanoparticles (e and f) that were set as 5 (d and e) or 10 (f). The error bars: the standard deviations of the spectrally unmixed concentrations calculated from all the pixelated Raman spectra in the ROIs. (g–i) Ex vivo tumor tissues (left, upper panel) and their respective five-color multiplexed Raman images (right, upper panel). The multiplexed images were generated through the spectral unmixing of Raman spectra in each pixel, followed by color-coding (lower panels) and merge (right, upper panel). The color-coded Raman images were organized with the order of increasing brightness from the left to right (lower panel, a–c, and g–i). The mixture of 1:2:3:4:5 molar ratio of IR780, DTTC, HDITC, IR797, and IR813-coded nanoparticles (mouse #1, (a), (d), and (g)), respectively, or 1:2:3:4:5 molar ratio of DTTC, IR780, HDITC, IR813, IR797 (mouse #2, (b), (e), and (h)), or 1:2:4:8:10 molar ratio of DTTC, IR780, HDITC, IR813, IR797 (mouse #3, (c), (f), and (i)), respectively, was prepared and concurrently injected via tail-vein. Color codes of the five-color NIR-SERRS nanoparticles: DTTC, magenta-purple; HDITC, blue; IR780, orange; IR797, red; and IR813, green. Scale bar: 10 mm.
Most importantly, the NIR-SERRS nanoparticles provided high multiplexing capability for imaging the nanoparticle-targeting tumors. Previously, the in vivo five-plex SERS imaging was successfully performed only in the deep imaging of the liver after systematic administration of five-color-coded nonresonant SERS nanoparticles.31,32 To demonstrate five-plex ratiometric Raman imaging of tumors, we intravenously (i.v.) injected a 5:4:3:2:1 molar ratio mixture of IR797, IR813, HDITC, IR780, and DTTC-coded SERRS nanoparticles, respectively, into a tumor-bearing mouse (Figure 4d), and the Raman signals at the tumor site were spectrally unmixed to derive the ratio of the five-color nanoparticles per each pixel (Figure 4e–g). In particular, the five-color NIR-SERRS nanoparticle mixture at the tumor site was noninvasively and longitudinally monitored at different time points, which cannot be performed with the currently available ex vivo multiplexing platforms.5–13 We observed that the distribution of the five-color nanoparticles in the tumor dynamically changed over 24 h, while it maintained their administered ratio with a spectral deconvolution error of 20.8(±12.4)%, that is, the average difference between the administered ratio and the estimated ratio of the five-color nanoparticles (Pearson’s correlation coefficient r = 0.9574(±0.0335), Figure 4f,g). The maintenance of the administered ratio of the nanoparticles in the tumor would have originated from the equivalent physiological characteristics of all the five-color NIR-SERRS nanoparticles through the adjustment of their average and cutoff sizes to ~200 nm and ~400 nm (Supporting Information Figure S8). The larger extravasation cutoff size of 500 nm for the LS174T subcutaneous tumor models would have also contributed to the access of the NIR-SERRS nanoparticles to the tumors without restriction for the cutoff sizes of the nanoparticles.50 The ratio of the administered five-color nanoparticles was also retained in the noninvasive five-plex Raman image of the abdomen (Supporting Information Figure S14), where a liver and a spleen are located. It is noteworthy that the development of the brighter NIR-SERRS nanoparticle palettes also enabled in vivo multiplexed imaging of the liver with a lower administration dose than the previously used nonresonant SERS nanoparticles (Supporting Information Figure S15). Such reduction of the administered dose is beneficial when considering the potential dose-dependent long-term toxicity and translation of the nanoparticle-based imaging method.
We also varied the five-color NIR-SERRS nanoparticle injection ratios to simulate various in vivo multiplexed imaging conditions, in which multiple biomarker expressions may vary simultaneously within the tumor microenvironment. We kept the total amount of the injected NIR-SERRS nanoparticle constant (600 fmol) and changed the molar ratios of the intravenously injected nanoparticles to 4:5:3:1:2 (mouse #1), 5:4:3:2:1 (mouse #2), and 10:8:4:2:1 (mouse #3) for IR797, IR813, HDITC, IR780, and DTTC-coded SERRS nanoparticles, respectively (Figure 5). The changes in the injection ratios of the five-color NIR-SERRS nanoparticles in tumorbearing mice were semiquantitatively reflected on the five-plex Raman spectral images of the tumors (Figure 5a–f, spectral deconvolution error, 22.5(±12.2)%) and were linearly correlated with the ratios of the five-color nanoparticle in the tumors (Pearson’s correlation coefficient r = 0.9676(±0.0118), Figure 5d–f). When we changed the injection ratio of the nanoparticles to 10:8:4:2:1 for IR797, IR813, HDITC, IR780, and DTTC-coded SERRS nanoparticles, respectively, we could detect the lowest abundant nanoparticle variate, the DTTC-coded SERRS nanoparticles from the tumor-targeted Raman image, of which the injection amount was 10% of the highest amount nanoparticle variate of the IR797-coded SERRS nanoparticles and only 4% of all the administered NIR-SERRS nanoparticles (mouse #3, Figure 5c,f). The ratiometric changes of the five-color NIR-SERRS nanoparticles distribution in the tumors were also reflected in the high-resolution ex vivo images of the excised tumor tissues, which were obtained after the in vivo imaging (Pearson’s correlation coefficient r = 0.9315(±0.0156), Figure 5g–i, Supporting Information Figure S16).
We also performed a histopathology examination of the tumor implanted mice after the five-plex imaging at 24 h postinjection of the NIR-SERRS nanoparticles (Supporting Information Figure S17). The examination of the reticuloendothelial organs through the hematoxylin and eosin (H&E) staining revealed no sign of acute toxicity, such as inflammation or necrosis in the liver, spleen, and kidneys. In particular, the liver image showed dark brown granules accumulating in the Kupffer cells, which would have been caused by the nanoparticle uptake. Still, there was no sign of damages and abnormal morphological changes. This suggests that the NIR-SERRS nanoparticles are biocompatible at the injection dose for multiplexed tumor-targeted imaging.51–53 The biocompatibility of the nanoparticles was further supported by in vitro cytotoxicity test. We performed cell viability (CCK-8 assay) and apoptosis assays of the NIR-SERRS nanoparticle incubated cells. The assays showed no significant effect of the nanoparticles on the viability or apoptosis of the cells (Supporting Information Figure S18).
CONCLUSION
In conclusion, we created gold multicore-NIR Raman dyes-silica shell-structured multispectral NIR-SERRS nanoparticles and demonstrated five-plex ratiometric and longitudinal SERS imaging of the nanoparticle accumulation in tumors of living mice. The current work demonstrates that highly multiplexed SERS imaging of tumors is possible without compromising either multiplexing capability or imaging sensitivity. The five-plex SERS imaging also surpasses the multiplicity limit of the previous preclinical tumor imaging methods while keeping enough sensitivity for tumor-targeted in vivo imaging.
Although there is no consensus for the number of multiplexing channels that will meet all the clinical needs, increasing the number of multiplexing channels from three to five is of particular clinical significance. For an example of breast cancer, analysis of three biomarkers only allows the instant therapeutic decision, while that of five biomarkers additionally enables the prognostic prediction, which makes it an important milestone for breast cancer diagnosis.54 While the currently used multiplexed biomarkers analysis is performed after the resection of the tumor tissue, we envision that noninvasive multiplexed SERS imaging will be particularly useful for longitudinal monitoring of therapeutic response or the expression of biomarkers with the cancer progression that needs to be performed without surgical resection. For this purpose, we also envision that this work will lay the foundation for noninvasive and higher-order multiplexed molecular imaging of tumors through the conjugation of the multispectral NIR-SERRS nanoparticles with multiple biomarkers.
MATERIALS AND METHODS
Synthesis of Near-Infrared-Resonant Surface-Enhanced Raman Scattering Nanoparticles.
For the synthesis of near-infrared-resonant surface-enhanced Raman scattering (NIR-SERRS) nanoparticles, a 60 nm gold nanoparticle was synthesized via a seed-mediated growth from 15 nm gold nanoparticles. First, the 15 nm gold nanoparticle seed solution was prepared by a Turkevich–Frens method. To a 500 mL boiling solution of 1 mM gold chloride trihydrate (Sigma-Aldrich) in water, 50 mL of 38.8 mM tribasic sodium citrate (Sigma-Aldrich) aqueous solution was added while vigorously stirring and kept at the boiling temperature for 10 min. The gold nanoparticle solution was cooled down to room temperature and filtered with a 0.8 μm surfactant-free cellulose acetate membrane syringe filter (Corning Inc.) for further use. Then, 100 mg of gold chloride was dissolved in 1 L of water and heated to 90 °C. 3.8 mL of the previously synthesized gold nanoparticle seed solution was mixed with 50 mg of tribasic sodium citrate and then rapidly added to the gold chloride solution. The solution was kept at the temperature for 30 min and then naturally cooled down. The produced gold nanoparticle solution was centrifuged at 2500g for 15 min, and the supernatant was discarded. The softly pelletized gold nanoparticles at the bottom of the falcon tube were collected and dialyzed for 2 days with a 7 kDa molecular weight cutoff dialysis cassette (Thermo Fisher).
Multispectral NIR-SERRS nanoparticles were synthesized by the simultaneous addition of hydrophobic and cationic heptamethine cyanine NIR-resonant Raman dyes and tetraethylorthosilicate (TEOS, 99.999% Sigma-Aldrich) silica precursor to the 60 nm gold nanoparticles colloidal solution. To 2 mL of 60 nm gold nanoparticle solution (2 nM), 300 μL of 28% NH4OH, 2 mL of water, and 30 mL of isopropyl alcohol were gently added while stirring at 500 rpm. Then, 80 μL of TEOS and 1 mg/mL of Raman dyes solution in N-dimethylformamide [75 μL for 3,3’-diethylthiatricarbocyanine iodide (DTTC, Sigma-Aldrich), 550 μL for IR780 perchlorate (HDITC, Sigma-Aldrich), 175 μL for IR780 iodide (Sigma-Aldrich), 85 μL for IR797 chloride (Sigma-Aldrich), and 60 μL for IR813 perchlorate (Santa Cruz Biotechnology), respectively] were rapidly added to the nanoparticle solution and reacted for 1.5 h at room temperature. Then the reacting solution was heated up to 60 °C and held at the temperature for 3 h. The produced NIR-SERRS nanoparticles were purified by five times washing with ethanol and pelletizing them through the centrifuge.
The NIR-SERRS nanoparticle surface was modified with thiol for further functionalization. To 1 mL of 1 nM NIR-SERRS nanoparticle solution in ethanol, 150 μL of NH4OH in ethanol (2 M, Sigma-Aldrich), 20 μL of TEOS, and 100 μL of (3-mercaptopropyl)-trimethoxysilane (MPTMS, Sigma-Aldrich) was added and reacted at 70 °C for 3 h. The nanoparticle was pelletized by centrifuge and washed with ethanol for 5 times. The purified NIR-SERRS nanoparticle solutions were stored in ethanol at 4 °C for further use. For in vivo imaging, the thiol-functionalized NIR-SERRS nanoparticle was PEGylated by adding 100 mg of methoxy polyethylene glycol maleimide (molecular weight 5 kDa, Sigma-Aldrich) to 1 mL of 1 nM thiolated SERRS nanoparticle in 10 mM 3-(N-morpholino) propanesulfonic acid buffer solution (pH 7.4). The PEGylated NIR-SERRS nanoparticles were purified by eluting the reacting solution through the PD-10 desalting column (GE Healthcare) and stored in 5% glucose in water solution at 4 °C.
Characterization of the NIR-SERRS Nanoparticles.
The size and morphology of the NIR-SERRS nanoparticles, the robustness and thickness of the silica shells, and the degree of gold nanoparticles oligomerization were characterized using transmission electron microscopy (TEM, JEM 1400, JEOL, 120 kV with La²6 emitter). The number of gold cores in Figure 2d was calculated by counting the 300 NIR-SERRS nanoparticles from the TEM images. The concentration and hydrodynamic size distribution of nanoparticles were measured with Nanoparticle Tracking Analysis (NanoSight, Malvern). The Raman spectra of the NIR-SERRS nanoparticles were obtained using a confocal Raman microscope with 785 nm NIR-diode laser excitation (InVia, Renishaw). For all the characterizations, surface-enhanced Raman scattering (SERS) nanoparticles of S440 (Oxonica Materials, Inc.) was used as a control, in which trans-1,2-bis(4-pyridyl)-ethylene (BPE) was coated onto 60 nm gold nanoparticles and encapsulated with a 35 nm-thick silica shell (BPE-SERS nanoparticles).
Synthesis of NIR-Fluorescent Cy7-Doped Silica Nanoparticles.
0.5 μmol of Cy7-NHS ester (lumiprobe) was reacted overnight with 2.5 μmol (3-aminopropyl)triethoxysilane (Sigma-Aldrich) in the solvent mixture of 900 μL of ethanol and 100 μL of DMSO at room temperature. Then, the solution was transferred to the mixture of 1 mL of TEOS, 16 mL of ethanol, and 2 mL of H2O. Finally, 500 μL of 28% NH4OH was added to initiate Cy7-doped silica synthesis. The colloidal solution was held at room temperature with gentle stirring (500 rpm) for 2 h. After the reaction, the Cy7-doped silica nanoparticles were pelletized by centrifuge and washed with ethanol for 5 times.
Raman Spectroscopy of the NIR-SERRS Nanoparticles.
Raman spectroscopy was conducted with a Renishaw InVia confocal Raman microscope, which utilized spontaneous Raman scattering from the NIR-SERRS nanoparticles and was further customized for in vivo mouse imaging experiments. To obtain Raman spectra of the NIR-SERRS nanoparticles, 200 μL of the NIR-SERRS nanoparticle solution was placed into a 96-well plate. The solution inside of a well was excited with a 785 nm diode near-infrared laser, equipped in Renishaw InVia confocal Raman microscope, of which the incident power was 20 mW from an objective lens (×5, NA = 0.15). We obtained Raman spectra with an integration time of 1 s from the CCD spectrometer with a grating of 1200 groove/mm, which provides a spectral resolution of 1.07 cm−1 . While monitoring Raman spectra, we adjusted the stage height to maximize the collection of Raman scattering signals. To obtain multiple numbers of spectra within the same solution for statistical analysis, the wells filled with the NIR-SERRS nanoparticles were scanned with the microscope with a step size from 100 to 200 μm to obtain 300 points per each well.
Quantitative Analysis of Raman Spectra.
For the quantitative analysis and spectral unmixing of Raman spectra, we used a nonnegative Levenberg–Marquardt–Fletcher (LMF) least-squares algorithm. First, the fluorescence background of the measured Raman spectrum was fitted with a fifth-order polynomial and removed so that the pure Raman component of the spectrum was taken into account for the quantitative analysis. Then, we used a non-negatively constrained LMF algorithm to match the Raman spectrum of interest with the linear combination of the pure Raman components of the NIR-SERRS nanoparticles to calculate their relative ratios. This algorithm minimizes the following objective function:
where is a vector of the estimated concentrations, A is a matrix containing all the pure Raman spectra, b is the vector of the measured spectra of mixtures, H(·) is the Heaviside step function, and α is a scalar controlling the non-negativity penalty. To improve the performance of the LMF algorithm, each of the reference spectra as well as the measured spectra were normalized to have a unitary norm. The weights used to normalize the reference spectra were reapplied after the LMF algorithm to undo the effect of normalization.
To present the spectrally unmixed ratio of the nanoparticles in solution, where there is a little deviation of the Raman intensities among multiple points within the same solution, the estimated concentrations of the nanoparticles were normalized to the average estimated concentration of the nanoparticle of the highest amount in solution. On the other hand, to present the ratio of the nanoparticle in vivo, where there is a considerable deviation of the Raman intensities among multiple points at the region of interest (ROI), the estimated concentrations of the nanoparticles were normalized to the concentration of the nanoparticle with the highest concentration per each point and averaged over the entire ROI.
The spectral fitting error was defined as the difference between the measured and fitted spectra, which was divided by the measured Raman intensity. The mean difference (eSF) was calculated by averaging the spectral fitting error over the entire wavenumber range as the following equation:
where k is the wavenumber, Imeas(k) is measured Raman intensity, and Ifit(k) is estimated Raman intensity.
The spectral deconvolution errors (eSM) were derived from the difference between the mixed ratios or the administered ratios of the five-color NIR-SERRS nanoparticles and their estimated ratios from the spectral unmixing:
where xadm is administered, or mixed ratios of the nanoparticles, and xest is their estimated ratio from the spectral unmixing. They were averaged over the multiplexing number of nanoparticles (n).
The sensitivity and the limits of detection for the concentration of NIR-SERRS nanoparticles (Figures 2 and 3) were calculated from the extrapolation of the sensitivity plots to the points, which intersect four times the standard deviation of the background signals.
Cytotoxicity Test of the NIR-SERRS Nanoparticles.
For the cytotoxicity test of the gold multicore-silica shell NIR-SERRS nanoparticles, cell viability assays and apoptosis assays were performed at 48 h after the incubation of the PEGylated NIR-SERRS nanoparticles (500 pM) with the MDA-MB-231 cells. For the cell viability assays, the cell plate was incubated with Cell Counting Kit-8 (CCK-8) solution (Sigma-Aldrich) for 2 h. Then, the optical density (OD) was measured at 450 nm by the UV–vis cell plate reader and was normalized with that of the control cells without the nanoparticle incubation. For the apoptosis assays, the nanoparticle-incubated cells were treated with annexin-V staining kit (ab176749, Abcam) and analyzed by fluorescence imaging using an EVOS FL Cell Imaging System.
Animal Experiments.
All procedures performed on the animals were approved by the Institutional Animal Care and Use Committee at Stanford University (APLAC #14465) and were conducted within the NIH guidelines for the humane care of laboratory animals. Female 8-week-old nude mice (Charles River Laboratories) were used for all in vivo studies. Subcutaneous tumor xenograft models were prepared by inoculating LS174T human colon cancer cells (2 × 106 cells/mouse) into the hind leg of the nude mice (n = 4). We performed in vivo Raman imaging when the tumor size was grown to ~1 cm3, as determined by caliper measurements.
In Vivo Multiplexed Raman Imaging of Tumor Xenograft Nude Mice.
For multiplexed imaging of subcutaneous tumors, the five spectrally different NIR-SERRS nanoparticles were mixed in the molar ratio of 1:2:3:4:5 or 1:2:4:8:10. Then, 200 μL of the 3 nM NIR-SERRS nanoparticles mixture solution in 5% glucose was intravenously injected through tail veins of the tumor xenograft nude mice (n = 4). Three to 24 h after the injection, the tumor-bearing mice were placed on the Raman microscope stage and put under anesthesia with 2–3% isoflurane delivered in 100% oxygen for noninvasive Raman imaging (Figures 4 and 5). The subcutaneous tumor was scanned with a 785 nm diode laser equipped in the confocal Raman microscope, of which the incident power was 20 mW from an objective lens (×5, NA = 0.15), for 10–20 s duration time per 1–2 mm step. For in vivo liver imaging, the abdomens of the nude mice were scanned for 10 s duration time per 1 mm step in the imaging plane. Twenty-4 h after the injection, the mice were sacrificed, and the tumor tissues, thigh muscles of the hind limb, liver, spleen, and kidneys were excised. After washing them with PBS several times, they were mounted onto a quartz slide and transferred to the microscope stage. The entire organs were scanned with the Raman microscope for 10 s duration time per 1 mm step.
Histological Analysis.
Histological examination of the organs was performed based on the tissue sections of the kidneys, liver, and spleen, which were collected from the nude mice 24 h after the administration of 600 pmol of the NIR-SERRS nanoparticles and stained with hematoxylin and eosin (H&E). A veterinary pathologist conducted the blind test for the H&E stained tissues.
Statistical Analysis.
The Raman data were obtained from multiple points scanning measurements, and the mean and the standard deviation were derived from the multiple points data. The error bars in the plots of the Raman intensities or the estimated ratios are the standard deviations. The reproducibility of the NIR-SERRS nanoparticles syntheses was assessed by the intraclass correlation from a nested random-effects model, which was calculated using Stata 16.1 (StataCorp LP, College Station, TX). Pearson’s correlation value, r, and p-value were calculated using linear regression through ORIGIN software without weighted selection on any data points. Biodistribution of the NIR-SERRS nanoparticles (Figures 4c) was derived from the analysis of ex vivo multiplexed Raman images of the tissues (Supporting Information Figure S11, n = 3) with ImageJ software (NIH).
Supplementary Material
ACKNOWLEDGMENTS
The transmission electron microscopy was conducted through the JEOL TEM 1400, which was provided by the Stanford Microscopy Facility, NIH grant SIG number 1S10RR02678001. We thank Prof. Kerriann Casey for the blind examination of the histology slides. The H&E slides of the mice organs were prepared through Animal Histology Services at the Department of Comparative Medicine at Stanford University. We thank Dr. Carmel T. Chan for the critical proofreading of the manuscript. We are sad to report that Prof. Sanjiv S. Gambhir passed away on July 18, 2020.
Funding
This work was supported by the Center for Cancer Nanotechnology Excellence for Translational Diagnostics (CCNE-TD) at Stanford University through an award number U54 CA199075 from the National Cancer Institute.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.1c07470.
Supporting Figures S1–S18. Detailed characterizations of the nanoparticles, supporting data on the spectral unmixing and the in vivo SERS imaging, in vitro and in vivo toxicity assessment (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsnano.1c07470
The authors declare no competing financial interest.
DEDICATION
This paper is dedicated to Prof. Sanjiv S. Gambhir, whom we lost during the preparation of this manuscript.
Contributor Information
Jung Ho Yu, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Idan Steinberg, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Ryan M. Davis, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Andrey V. Malkovskiy, Department of Plant Biology, Carnegie Institute for Science, Stanford, California 94305, United States
Aimen Zlitni, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Rochelle Karina Radzyminski, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Department of Applied Physics, Stanford University, Stanford, California 94305, United States.
Kyung Oh Jung, Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States; Department of Radiation Oncology, Stanford University School of Medicine, Stanford, California 94305, United States.
Daniel Tan Chung, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Luis Dan Curet, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Aloma L. D’Souza, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States
Edwin Chang, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Jarrett Rosenberg, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States.
Jos Campbell, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Hadas Frostig, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Seung-min Park, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Guillem Pratx, Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States; Department of Radiation Oncology, Stanford University School of Medicine, Stanford, California 94305, United States.
Craig Levin, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
Sanjiv S. Gambhir, Department of Radiology, Stanford University School of Medicine, Stanford, California 94305, United States; Molecular Imaging Program at Stanford (MIPS) and Bio-X Program, Stanford University, Stanford, California 94305, United States.
REFERENCES
- (1).Dagogo-Jack I; Shaw AT Tumour Heterogeneity and Resistance to Cancer Therapies. Nat. Rev. Clin. Oncol 2018, 15, 81–94. [DOI] [PubMed] [Google Scholar]
- (2).Heinzmann K; Carter LM; Lewis JS; Aboagye EO Multiplexed Imaging for Diagnosis and Therapy. Nat. Biomed. Eng 2017, 1, 697–713. [DOI] [PubMed] [Google Scholar]
- (3).Kobayashi H; Longmire MR; Ogawa M; Choyke PL; Kawamoto S Multiplexed Imaging in Cancer Diagnosis: Applications and Future Advances. Lancet Oncol. 2010, 11, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Wang YW; Reder NP; Kang S; Glaser AK; Liu JTC Multiplexed Optical Imaging of Tumor-Directed Nanoparticles: A Review of Imaging Systems and Approaches. Nanotheranostics 2017, 1, 369–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Angelo M; Bendall SC; Finck R; Hale MB; Hitzman C; Borowsky A; Levenson RM; Lowe JB; Liu SD; Zhao S; Natkunam Y; Nolan GP Multiplexed Ion Beam Imaging of Human Breast Tumors. Nat. Med 2014, 20, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Gerdes MJ; Sevinsky CJ; Sood A; Adak S; Bello MO; Bordwell A; Can A; Corwin A; Dinn S; Filkins R; Hollman D; Kamath V; Kaanumalle S; Kenny K; Larsen M; Lazare M; Li Q; Lowes C; McCulloch CC; McDonough E; et al. Highly Multiplexed Single-Cell Analysis of Formalin-Fixed, Paraffin-Embedded Cancer Tissue. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 11982–11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Liu J; Lau SK; Varma VA; Moffitt RA; Caldwell M; Liu T; Young AN; Petros JA; Osunkoya O; Krogstad T; Leyland-Jones B; Wang MD; Nie S Molecular Mapping of Tumor Heterogeneity on Clinical Tissue Specimens with Multiplexed Quantum Dots. ACS Nano 2010, 4, 2755–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Liu Z; Tabakman S; Sherlock S; Li X; Chen Z; Jiang K; Fan S; Dai H Multiplexed Five-Color Molecular Imaging of Cancer Cells and Tumor Tissues with Carbon Nanotube Raman Tags in the Near-Infrared. Nano Res. 2010, 3, 222–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Wang Y; Kang S; Khan A; Ruttner G; Leigh SY; Murray M; Abeytunge S; Peterson G; Abeytunge S; Peterson G; Rajadhyaksha M; Dintzis S; Javid S; Liu JTC Quantitative Molecular Phenotyping with Topically Applied SERS Nanoparticles for Intraoperative Guidance of Breast Cancer Lumpectomy. Sci. Rep 2016, 6, 21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wang YW; Reder NP; Kang S; Glaser AK; Yang Q; Wall MA; Javid SH; Dintzis SM; Liu JTC Raman-Encoded Molecular Imaging with Topically Applied SERS Nanoparticles for Intraoperative Guidance of Lumpectomy. Cancer Res. 2017, 77, 4506–4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wang YW; Yang Q; Kang S; Wall MA; Liu JTC High-Speed Raman-Encoded Molecular Imaging of Freshly Excised Tissue Surfaces with Topically Applied SERRS Nanoparticles. J. Biomed. Opt 2018, 23, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Davis RM; Kiss B; Trivedi DR; Metzner TJ; Liao JC; Gambhir SS Surface-Enhanced Raman Scattering Nanoparticles for Multiplexed Imaging of Bladder Cancer Tissue Permeability and Molecular Phenotype. ACS Nano 2018, 12, 9669–9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Nicolson F; Jamieson LE; Mabbott S; Plakas K; Shand NC; Detty MR; Graham D; Faulds K Multiplexed Imaging of Live Breast Cancer Tumour Models through Tissue Using Handheld Surface-Enhanced Spatially Offset Resonance Raman Spectroscopy. Chem. Commun 2018, 54, 8530–8533. [DOI] [PubMed] [Google Scholar]
- (14).Yuan E; Si P; Winetraub Y; Shevidi S; de la Zerda A A Spectral De-Mixing Model for Triplex in Vivo Imaging of Optical Coherence Tomography Contrast Agents. ACS Photonics 2020, 7, 893–900. [Google Scholar]
- (15).Zhong Y; Ma Z; Wang F; Wang X; Yang Y; Liu Y; Zhao X ; Li J; Du H; Zhang M; Cui Q; Zhu S; Sun Q; Wan H; Tian Y; Liu Q; Wang W; Garcia KC; Dai H In Vivo Molecular Imaging for Immunotherapy Using Ultra-Bright Near-Infrared-IIb Rare-Earth Nanoparticles. Nat. Biotechnol 2019, 37, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Fan Y; Wang P; Lu Y; Wang R; Zhou L; Zheng X; Li X; Piper JA; Zhang F Lifetime-Engineered NIR-II Nanoparticles Unlock Multiplexed in Vivo Imaging. Nat. Nanotechnol 2018, 13, 941–946. [DOI] [PubMed] [Google Scholar]
- (17).Kobayashi H; Hama Y; Koyama Y; Barrett T; Regino CAS; Urano Y; Choyke PL Simultaneous Multi-Color Imaging of Five Different Lymphatic Basins Using Quantum Dots. Nano Lett. 2007, 7, 1711–1716. [DOI] [PubMed] [Google Scholar]
- (18).Zhu S; Yang Q; Antaris AL; Yue J; Ma Z; Wang H; Huang W; Wan H; Wang J; Diao S; Zhang B; Li X; Zhong Y; Yu K; Hong G; Luo J; Liang Y; Dai H Molecular Imaging of Biological System with a Clickable Dye in the Broad 800- to 1,800-nm Near-Infrared Window. Proc. Natl. Acad. Sci. U. S. A 2017, 114, 962–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Cosco ED; Spearman AL; Ramakrishnan S; Lingg JGP; Saccomano M; Pengshung M; Arus BA; Wong KCY; Glasl S; Ntziachristos V; Warmer M; McLaughlin RR; Bruns OT; Sletten EM Shortwave Infrared Polymethine Fluorophores Matched to Excitation Lasers Enable Non-Invasive Multicolour in Vivo Imaging in Real Time. Nat. Chem 2020, 12, 1123–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wei L; Chen Z; Shi L; Long R; Anzalone AV; Zhang L; Hu F; Yuste R; Cornish VW; Min W Super-Multiplex Vibrational Imaging. Nature 2017, 544, 465–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Henry A-I; Sharma B; Cardinal MF; Kurouski D; Van Duyne RP Surface-Enhanced Raman Spectroscopy Biosensing: In Vivo Diagnostics and Multimodal Imaging. Anal. Chem 2016, 88, 6638–6647. [DOI] [PubMed] [Google Scholar]
- (22).Laing S; Jamieson LE; Faulds K; Graham D Surface-Enhanced Raman Spectroscopy for in Vivo Biosensing. Nat. Rev. Chem 2017, 1, 0060. [Google Scholar]
- (23).Lane LA; Qian X; Nie S SERS Nanoparticles in Medicine: From Label-Free Detection to Spectroscopic Tagging. Chem. Rev 2015, 115, 10489–10529. [DOI] [PubMed] [Google Scholar]
- (24).Kneipp J Interrogating Cells, Tissues, and Live Animals with New Generations of Surface-Enhanced Raman Scattering Probes and Labels. ACS Nano 2017, 11, 1136–1141. [DOI] [PubMed] [Google Scholar]
- (25).Langer J; de Aberasturi DJ; Aizpurua J; Alvarez-Puebla RA; Auguié B; Baumberg JJ; Bazan GC; Bell SEJ; Boisen A; Brolo AG; Choo J; Cialla-May D; Deckert V; Fabris L; Faulds K; de Abajo JG; Goodacre R; Graham D; Haes AJ; Haynes CL; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Wustholz KL; Henry A-I; McMahon JM; Freeman RG; Valley N; Piotti ME; Natan MJ; Schatz GC; Van Duyne RP Structure-Activity Relationships in Gold Nanoparticle Dimers and Trimers for Surface-Enhanced Raman Spectroscopy. J. Am. Chem. Soc 2010, 132, 10903–10910. [DOI] [PubMed] [Google Scholar]
- (27).Freitag I; Matthäus C; Csaki A; Clement JH; Cialla-May D; Weber K; Krafft C; Popp J Differentiation of MCF-7 Tumor Cells from Leukocytes and Fibroblast Cells Using Epithelial Cell Adhesion Molecule Targeted Multicore Surface-Enhanced Raman Spectroscopy Labels. J. Biomed. Opt 2015, 20, 055002. [DOI] [PubMed] [Google Scholar]
- (28).Nam J-M; Oh J-W; Lee H; Suh YD Plasmonic Nanogap-Enhanced Raman Scattering with Nanoparticles. Acc. Chem. Res 2016, 49, 2746–2755. [DOI] [PubMed] [Google Scholar]
- (29).Matschulat A; Drescher D; Kneipp J Surface-Enhanced Raman Scattering Hybrid Nanoprobe Multiplexing and Imaging in Biological Systems. ACS Nano 2010, 4, 3259–3269. [DOI] [PubMed] [Google Scholar]
- (30).de Aberasturi DJ; Serrano-Montes AB; Langer J; Henriksen-Lacey M; Parak WJ; Liz-Marzán LM Surface-Enhanced Raman Scattering Encoded Gold Nanostars for Multiplexed Cell Discrimination. Chem. Mater 2016, 28, 6779–6790. [Google Scholar]
- (31).Zavaleta CL; Smith BR; Walton I; Doering W; Davis G; Shojaei B; Natan MJ; Gambhir SS Multiplexed Imaging of Surface-Enhanced Raman Scattering Nanotags in Living Mice Using Noninvasive Raman Spectroscopy. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 13511–13516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Bohndiek SE; Wagadarikar A; Zavaleta CL; Van de Sompel D; Garai E; Jokerst J; Yazdanfar S; Gambhir SS A Small Animal Raman Instrument for Rapid, Wide-Area, Spectroscopic Imaging. Proc. Natl. Acad. Sci. U. S. A 2013, 110, 12408–12413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Kang H; Jeong S; Park Y; Yim J; Jun B-H; Kyeong S; Yang J-K; Kim G; Hong S; Lee LP; Kim J-H; Lee H-Y; Jeong DH; Lee Y-S Near-Infrared SERS Nanoprobes with Plasmonic Au/Ag Hollow-Shell Assemblies for in Vivo Multiplex Detection. Adv. Funct. Mater 2013, 23, 3719–3727. [Google Scholar]
- (34).Dinish US; Balasundaram G; Chang Y-T; Olivo M Actively Targeted in Vivo Multiplex Detection of Intrinsic Cancer Biomarkers Using Biocompatible SERS Nanotags. Sci. Rep 2015, 4, 4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW Analysis of Nanoparticle Delivery to Tumours. Nat. Rev. Mater 2016, 1, 16014. [Google Scholar]
- (36).Qian X; Peng X-H; Ansari DO; Yin-Goen Q; Chen GZ; Shin DM; Yang L; Young AN; Wang MD; Nie S in Vivo Tumor Targeting and Spectroscopic Detection with Surface-Enhanced Raman Nanoparticle Tags. Nat. Biotechnol 2008, 26, 83–90. [DOI] [PubMed] [Google Scholar]
- (37).Jokerst JV; Cole AJ; Van de Sompel D; Gambhir SS Gold Nanorods for Ovarian Cancer Detection with Photoacoustic Imaging and Resection Guidance via Raman Imaging in Living Mice. ACS Nano 2012, 6, 10366–10377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Harmsen S; Bedics MA; Wall MA; Huang R; Detty MR; Kircher MF Rational Design of a Chalcogenopyrylium-Based Surface-Enhanced Resonance Raman Scattering Nanoprobe with Attomolar Sensitivity. Nat. Commun 2015, 6, 6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Huang R; Harmsen S; Samii JM; Karabeber H; Pitter KL; Holland EC; Kircher MF High Precision Imaging of Microscopic Spread of Glioblastoma with a Targeted Ultrasensitive SERRS Molecular Imaging Probe. Theranostics 2016, 6, 1075–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Andreou C; Neuschmelting V; Tschaharganeh DF; Huang C-H; Oseledchyk A; Iacono P; Karabeber H; Colen RR; Mannelli L; Lowe SW; Kircher MF Imaging of Liver Tumors Using Surface-Enhanced Raman Scattering Nanoparticles. ACS Nano 2016, 10, 5015–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Silva WR; Keller EL; Frontiera RR Determination of Resonance Raman Cross-Sections for Use in Biological SERS Sensing with Femtosecond Stimulated Raman Spectroscopy. Anal. Chem 2014, 86, 7782–7787. [DOI] [PubMed] [Google Scholar]
- (42).Maiti KK; Dinish US; Samanta A; Vendrell M; Soh K-S; Park S-J; Olivo M; Chang Y-T Multiplex Targeted in Vivo Cancer Detection Using Sensitive Near-Infrared SERS Nanotags. Nano Today 2012, 7, 85–93. [Google Scholar]
- (43).Yuan H; Liu Y; Fales AM; Li YL; Liu J; Vo-Dinh T Quantitative Surface-Enhanced Resonant Raman Scattering Multiplexing of Biocompatible Gold Nanostars for in Vitro and ex Vivo detection. Anal. Chem 2013, 85, 208–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yang X; Shi C; Tong R; Qian W; Zhau HE; Wang R; Zhu G; Cheng J; Yang VW; Cheng T; Henary M; Strekowski L; Chung LWK Near IR Heptamethine Cyanine Dye-Mediated Cancer Imaging. Clin. Cancer Res 2010, 16, 2833–2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Wong YJ; Zhu L; Teo WS; Tan YW; Yang Y; Wang C; Chen H Revisiting the Stöber Method: Inhomogeneity in Silica Shells. J. Am. Chem. Soc 2011, 133 (30), 11422–11425. [DOI] [PubMed] [Google Scholar]
- (46).Yang S-A; Choi S; Jeon SM; Yu J Silica Nanoparticle Stability in Biological Media Revisited. Sci. Rep 2018, 8, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lim I-IS; Goroleski F; Mott D; Kariuki N; Ip W; Luo J; Zhong C-J. Adsorption of Cyanine Dyes on Gold Nanoparticles and Formation of J-Aggregates in the Nanoparticle Assembly. J. Phys. Chem. B 2006, 110, 6673–6682. [DOI] [PubMed] [Google Scholar]
- (48).Pallaoro A; Braun GB; Moskovits M Biotags Based On Surface-Enhanced Raman Can Be as Bright as Fluorescence Tags. Nano Lett. 2015, 15, 6745–6750. [DOI] [PubMed] [Google Scholar]
- (49).Lutz BR; Dentinger CE; Nguyen LN; Sun L; Zhang J; Allen AN; Chan S; Knudsen BS Spectral Analysis of Multiplex Raman Probe Signatures. ACS Nano 2008, 2, 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Yuan F; Dellian M; Fukumura D; Leunig M; Berk DA; Torchilin VP; Jain RK Vascular Permeability in a Human Tumor Xenograft: Molecular Size Dependence and Cutoff Size. Cancer Res. 1995, 55, 3752–3756. [PubMed] [Google Scholar]
- (51).Thakor AS; Luong R; Paulmurugan R; Lin FI; Kempen P; Zavaleta C; Chu P; Massoud TF; Sinclair R; Gambhir SS The Fate and Toxicity of Raman-Active Silica-Gold Nanoparticles in Mice. Sci. Transl Med 2011, 3, 79ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Falagan-Lotsch P; Grzincic EM; Murphy CJ One Low-Dose Exposure of Gold Nanoparticles Induces Long-Term Changes in Human Cells. Proc. Natl. Acad. Sci. U. S. A 2016, 113, 13318–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Park S.-m.; Aalipour A; Vermesh O; Yu JH; Gambhir SS Towards Clinically Translatable in Vivo Nanodiagnostics. Nat. Rev. Mater 2017, 2, 17014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Cheang MCU; Voduc D; Bajdik C; Leung S; McKinney S; Chia SK; Perou CM; Nielsen TO Basal-Like Breast Cancer Defined by Five Biomarkers Has Superior Prognostics Value than Triple-Negative Phenotype. Clin. Cancer Res 2008, 14, 1368–1376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


