Abstract
The main objectives of this study were to determine whether the nitroreductase enzyme encoded by the rdxA gene of Helicobacter pylori was responsible for reductive activation of nitrofurantoin and whether a triple-therapy regimen with nitrofurantoin was able to eradicate metronidazole-sensitive and -resistant H. pylori infections from mice. The susceptibilities to nitrofurantoin of parent and isogenic rdxA mutant strains (three pairs), as well as a series of matched metronidazole-sensitive and -resistant strains isolated from mice (30) and patients (20), were assessed by agar dilution determination of the MIC. Groups of mice colonized with the metronidazole-sensitive H. pylori SS1 strain or a metronidazole-resistant rdxA SS1 mutant were treated with either metronidazole or nitrofurantoin as part of a triple-therapy regimen. One month after the completion of treatment the mice were sacrificed and their stomachs were cultured for H. pylori. The nitrofurantoin MICs for all strains tested were between 0.5 and 4.0 μg/ml. There was no significant difference between the susceptibility to nitrofurantoin of the parental strains and those of respective rdxA mutants or between those of matched metronidazole-sensitive and -resistant H. pylori isolates. The regimen with metronidazole eradicated infection from all eight SS1-infected mice and from one of eight mice inoculated with the rdxA mutant (P ≤ 0.001). The regimen with nitrofurantoin failed to eradicate infection from any of the six SS1-infected mice (P ≤ 0.001) and cleared infection from one of seven mice inoculated with the rdxA mutant. These results demonstrate that, despite the good in vitro activity of nitrofurantoin against H. pylori and the lack of cross-resistance between metronidazole and nitrofurantoin, eradication regimens involving nitrofurantoin are unable to eradicate either metronidazole-sensitive or -resistant H. pylori infections from mice.
Helicobacter pylori is a gram-negative, microaerobic, spiral bacterium that colonizes the stomachs of approximately one-half the world's population (54). Infection with H. pylori is associated with chronic gastritis and peptic ulceration, and the bacterium is also considered a risk factor for the development of gastric adenocarcinoma and mucosa-associated lymphoid tissue lymphoma (3, 41, 42). Although the 5-nitroimidazole metronidazole is an important component of many currently used H. pylori eradication regimens, resistance to this class of antibiotics is relatively common. It has been estimated that 10 to 30% of clinical strains isolated in western Europe and the United States are metronidazole resistant, and this prevalence is far higher in developing countries and in certain immigrant populations (11, 13). Although there have been conflicting reports concerning the clinical impact of metronidazole resistance in H. pylori, many studies have now demonstrated that resistance to the 5-nitroimidazoles reduces the efficacy of eradication regimens involving metronidazole and is therefore an important predictor of treatment failure (5, 23, 27, 43, 51). Several reports also suggest that the prevalence of metronidazole resistance is rising and is likely to become an increasingly important problem in the clinical management of H. pylori infection (34, 52). This, combined with the expense of currently used antimicrobial regimens, means that there is a need to evaluate alternative antibiotics for combination therapy of H. pylori infections. We have previously used the H. pylori SS1 mouse model to characterize the evolution of metronidazole resistance by H. pylori in vivo and to examine the contribution of underlying resistance mechanisms (26, 27). This model system may also be used to assess the efficacy of novel anti-H. pylori agents in vivo and to determine optimal regimens for the eradication of resistant strains.
Recently it was demonstrated that loss of oxygen-insensitive NADPH nitroreductase activity resulted in the development of resistance to metronidazole in H. pylori (18). It was proposed that this enzyme reduces the nitro group of metronidazole to active metabolites that are toxic to the bacterium and that resistance arose from mutational inactivation of the underlying gene, rdxA (HP0954 in the H. pylori genome database [49]). Subsequent studies have suggested that, while the development of metronidazole resistance in H. pylori is highly associated with mutational inactivation of the rdxA gene, other mechanisms of resistance are likely to exist in this bacterium (26, 48; D. H. Kwon, D. Y. Graham, and F. A. K. El-Zaatari, Gut 43(Suppl. 2):A6).
The nitrofuran group of compounds, which includes furazolidone and nitrofurantoin, appears a particularly promising source of alternative agents for metronidazole in H. pylori eradication regimens (7, 46, 57). Like that of the 5-nitroimidazoles, the biological activity of these nitroaromatic compounds is largely derived from reductive metabolism of the parent compound's nitro moiety by oxygen-insensitive nitroreductases (2, 36, 55). A number of recent clinical trials have demonstrated that short-term triple therapies with furazolidone are effective in the treatment of H. pylori infection (10, 33, 46, 57). Based on nitrofurantoin's in vitro activity, there is evidence that it might also be a suitable alternative agent in combination antimicrobial therapy, particularly against metronidazole-resistant H. pylori strains (7). Although nitrofurantoin has failed to eradicate H. pylori in a limited number of clinical trials, in all cases the agent was given either as a monotherapy or in combination with bismuth subsalicylate and not as part of a triple-agent regimen (4, 21, 24, 39, 44).
The aims of this study were to (i) determine whether the rdxA gene of H. pylori is responsible for reductive activation of nitrofurantoin, (ii) evaluate the in vitro activity of nitrofurantoin against a series of matched metronidazole-sensitive and -resistant H. pylori isolates, and (iii) determine the efficacy of a triple-therapy regimen with nitrofurantoin in eradicating established metronidazole-sensitive and -resistant H. pylori infections from mice.
MATERIALS AND METHODS
Bacteria and growth conditions.
Escherichia coli strain MC1061 (6) was used as the host for plasmid cloning experiments and was grown at 37°C in L broth (10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter, pH 7.0) or on L agar plates (1.5% agar) at 37°C. Antibiotics were used at the following final concentrations: 100 μg of spectinomycin (Upjohn Laboratories, Paris, France) and 25 μg of kanamycin (Serva, Frankfurt, Germany)/ml.
H. pylori strains (Table 1) were routinely cultured on a blood agar medium (blood agar base no. 2 [Oxoid, Lyon, France]) supplemented with 10% horse blood (bioMérieux, Marcy L'Etoile, France) and the following antibiotics: 10 μg of vancomycin (Dakota Pharmaceuticals, Creteil, France)/ml, 2.5 IU of polymyxin (Pfizer Laboratories, Orsay, France)/liter, 5 μg of trimethoprim (Sigma Chemicals, Saint-Quentin Fallavier, France)/ml, and 4 μg of amphotericin B (Bristol-Myers Squibb, Paris, France)/ml. The plates were incubated at 37°C under microaerobic conditions in an anaerobic jar (Oxoid) with a carbon dioxide generator (CampyGen; Oxoid) without a catalyst. For the selection of metronidazole- and nitrofurantoin-resistant colonies and their subsequent subculture, the medium was additionally supplemented with 8 μg of metronidazole (Sigma) or nitrofurantoin (Sigma)/ml, respectively. H. pylori cells that had undergone chromosomal allelic exchange were selected on medium supplemented with 25 μg of kanamycin.
TABLE 1.
H. pylori strains used in this study
| Strain(s) | Characteristics | Reference or source |
|---|---|---|
| SS1 | H. pylori wild-type strain; metronidazole sensitive; colonizes mice | 32 |
| G27 | H. pylori wild-type strain; metronidazole sensitive | 8 |
| HAS-141 | H. pylori wild-type strain; metronidazole sensitive | 25 |
| SS1-rdxA | H. pylori SS1 isogenic rdxA deletion mutant | This work |
| G27-rdxA | H. pylori G27 isogenic rdxA deletion mutant | This work |
| HAS-141-rdxA | H. pylori HAS-141 isogenic rdxA deletion mutant | This work |
| SS1-1 to SS1-10 | Mouse-derived metronidazole-sensitive H. pylori SS1 | 27 |
| SS1-11 to SS1-19 | Mouse-derived metronidazole-resistant H. pylori SS1 with defined mutations in rdxA | 27 |
| SS1-20 | Mouse-derived metronidazole-resistant H. pylori SS1 with no defined mutation in rdxA | 27 |
| SS1-21 to SS1-30 | Mouse-derived metronidazole-resistant H. pylori SS1 (rdxA gene not sequenced) | 27 |
| T1S/T1R to T10S/T10R | Paired metronidazole-sensitive and -resistant clinical H. pylori isolates | 48 |
To determine viable counts of H. pylori, samples to be tested were serially diluted in sterile saline and then plated in duplicate onto blood agar plates supplemented with either 10% horse blood or fetal calf serum (Gibco BRL, Cergy Pontoise, France) and 10 g of agar (bacteriological agar no. 1; Oxoid)/liter, 200 μg of bacitracin/ml, and 10 μg of nalidixic acid (Sigma)/ml. After 5 days of incubation, colonies with H. pylori morphology were identified using standard criteria (morphology on Gram staining and the presence of catalase, oxidase, and urease enzyme activities) and enumerated (14).
General molecular-biology techniques and construction of a defined mutation in the H. pylori rdxA gene.
The alkaline lysis procedure was used for small-scale plasmid preparation (45). MIDI columns (Qiagen, Courtaboeuf, France) were used for large-scale plasmid preparation. Genomic DNA from individual H. pylori strains was extracted using the QIAamp tissue kit (Qiagen) according to the manufacturer's instructions. Standard procedures for cloning and DNA analysis were used (45).
H. pylori strains with a defined mutation in the rdxA gene were generated by allelic exchange. For this purpose, a recombinant plasmid was constructed in E. coli MC1061 as follows. Oligonucleotide primers were designed to amplify a fragment of 510 bp from the 5′ end (HP0954-1 and HP0954-2) and 490 bp from the 3′ end (HP0954-3 and HP094-4) of the rdxA gene (Table 2). The two generated fragments were restricted with EcoRI and BamHI and with PstI and BamHI, respectively, and were cloned simultaneously into the plasmid vector pILL570-1 (30) linearized with EcoRI and PstI. The resulting plasmid was restricted with BamHI, and the BamHI-digested kanamycin cassette from pILL600 (31) was introduced to generate the final construct.
TABLE 2.
Oligonucleotide primers used for PCR and cloning
| Primer | Oligodeoxynucleotide sequence (5′–3′)a |
|---|---|
| HP0954-1 | ggaattcCTGATTGTGGTTTATGGTTTGGGG |
| HP0954-2 | gcggatccATAGAGATTTTGCATGTAGTGGCCG |
| HP0954-3 | gcggatccCTGTGGGGCAAATTTGCATGGGCG |
| HP0954-4 | aaaactgcagAATCCCTAAATATTTATTATTAACAGGG |
Underlining, EcoRI (GAATTC), BamHI (GGATCC), and PstI (CTGCAG) sites; lowercase letters, nucleotides that were added to the 5′ end to create a restriction site.
Resultant recombinant plasmids were introduced into H. pylori for allelic exchange by natural transformation. H. pylori strains were naturally transformed with circular plasmid DNA (∼2 μg per transformation) using a modification of the technique of Wang et al. (53). Briefly, bacteria were inoculated as 1-cm patches and grown for 5 h before the addition of 10 μl of supercoiled plasmid DNA. After further incubation for 18 h, the bacteria from each individual patch were harvested and plated directly onto a single plate of selective medium containing 25 μg of kanamycin/ml.
To determine whether natural transformation of H. pylori was followed by allelic replacement of the intact chromosomal gene by the mutated gene, chromosomal DNA was isolated from paired H. pylori parent and mutant strains. Genotypic analysis was performed by PCR using a single pair of oligonucleotide primers that flanked the point of insertion of the antibiotic resistance cassette (HP0954-1 and HP0954-4) (Table 2).
Susceptibility testing.
Susceptibility to metronidazole and nitrofurantoin was assessed by agar dilution determination of the MIC. Inoculates yielding 104 CFU/spot were inoculated onto plates of IsoSensitest agar (Oxoid) enriched with 10% horse blood containing doubling dilutions of metronidazole or nitrofurantoin. The MIC was defined as the lowest concentration of antibiotic inhibiting growth when the plates were read after 72 h of incubation under microaerobic conditions (generated as described above) at 37°C. Isolates were considered resistant to nitrofurantoin or metronidazole if the MIC of either was ≥8 μg/ml (56).
Infection of mice with H. pylori SS1.
Six-week-old specific-pathogen-free Swiss mice (Centre d'Elevage R. Janvier, Le-Genest-St-Isle, France) were housed in polycarbonate cages in isolators and fed a commercial pellet diet with water ad libitum. All animal experimentation was performed in accordance with institutional guidelines. Mice were inoculated intragastrically with a suspension of either H. pylori SS1 (n = 24; Table 4) or the rdxA mutant, SS1-11 (n = 25), which had been harvested directly from 48-h plate cultures into peptone-trypsin broth (Organotéchnique, La Courneuve, France). SS1-11 is a mouse-derived H. pylori SS1 isolate that is resistant to metronidazole (MIC of 32 μg/ml) and whose rdxA gene contains frameshift mutations at positions 90 and 159, resulting in the creation of two translational stop codons within the gene (26, 27). Each animal was administered a single 100-μl aliquot of an inoculating suspension of 105 CFU/ml (equivalent to 100 times the 100% infectious dose [14]) on two consecutive days. This was administered with polyethylene catheters (Biotrol, Paris, France) attached to 1-ml disposable syringes. A control group of mice (n = 10) was given peptone-trypsin broth alone.
TABLE 4.
Comparison of regimens with metronidazole and nitrofurantoin for the eradication of metronidazole-sensitive and -resistant H. pylori from mice
| Group | No. of mice | Inoculating suspension | Treatmenta | No. of mice infected with:
|
||
|---|---|---|---|---|---|---|
| H. pylorib | MTZ-resistant H. pylori | NF-resistant H. pylorid | ||||
| Control | 10 | PTB | PTB | 0 | 0 | 0 |
| 1 | 10 | H. pylori SS1 | PTB | 10 | 0 | 0 |
| 2 | 10 | H. pylori SS1-11 | PTB | 10 | 10 | 0 |
| 3 | 8 | H. pylori SS1 | OCM | 0e | 0 | 0 |
| 4 | 8 | H. pylori SS1-11 | OCM | 7e | 7 | 0 |
| 5 | 6 | H. pylori SS1 | OCNf | 6e | 0 | 0 |
| 6 | 7 | H. pylori SS1-11 | OCNf | 6 | 6 | 0 |
PTB, peptone-trypsin broth; OCM, omeprazole, clarithromycin, and metronidazole; OCNf, omeprazole, clarithromycin, and nitrofurantoin.
Number of mice infected with H. pylori 1 month after the completion of treatment.
Number of mice from which metronidazole (MTZ)-resistant strains were isolated.
Number of mice from which nitrofurantoin (NF)-resistant strains were isolated.
Statistically significant difference (P < 0.001).
Antimicrobial chemotherapy.
Mice were administered antimicrobial chemotherapy 7 weeks after infection (Table 4). All solutions were administered intragastrically in a final volume of 100 μl via polyethylene catheters as previously described. The H. pylori SS1-colonized mice in group 1 (n = 10) and the H. pylori SS1-11-colonized mice in group 2 (n = 10) were treated for 7 days with peptone-trypsin broth. The H. pylori SS1-colonized mice in group 3 (n = 8) and the H. pylori SS1-11-colonized mice in group 4 (n = 8) were treated for 7 days with the mouse body weight equivalent of a recommended H. pylori eradication regimen of 20 mg of omeprazole (0.0086 mg; Astra Hassle AB, Mölndal, Sweden), 250 mg of clarithromycin (0.107 mg; Abbott Laboratories, Saint-Rémy-sur-Avre, France), and 400 mg of metronidazole (0.171 mg; Rhône-Poulenc Rorer, Vitry sur Seine, France) twice daily for 1 week (12). The H. pylori SS1-colonized mice in group 5 (n = 6) and the H. pylori SS1-11-colonized mice in group 6 (n = 7) were treated for 7 days with the mouse body weight equivalent of 20 mg of omeprazole (0.0086 mg), 250 mg of clarithromycin (0.107 mg), and 200 mg of nitrofurantoin (0.086 mg; Proctor & Gamble Pharmaceuticals, Staines, United Kingdom) twice daily for 1 week.
Assessment of H. pylori infection in mice.
Colonization with H. pylori was assessed 1 month after the completion of each treatment regimen as recommended by recent guidelines (56). The animals were sacrificed, the stomach of each mouse was removed, and serum was recovered in microtubes (Sarstedt France, Orsay, France). The presence of H. pylori infection was determined by biopsy urease, quantitative culture, and serology. Stomachs were washed in physiological buffered saline and divided longitudinally into tissue fragments so that each fragment contained the cardia, body, and antrum. For each stomach, one fragment was immediately placed in urea-indole medium and another was immediately placed in peptone-trypsin broth. The presence of urease activity in tissue fragments was detected in urea-indole medium incubated for 24 h at room temperature (14). For the performance of quantitative bacterial cultures on stomach samples, tissue fragments were homogenized in peptone-trypsin broth using disposable plastic grinders and tubes (PolyLabo, Strasbourg, France). The homogenates were serially diluted in sterile saline and plated directly onto blood and serum plates for enumeration and onto selective plates containing 8 μg of either metronidazole or nitrofurantoin/ml. To increase the sensitivity of detection of metronidazole- and nitrofurantoin-resistant strains, all colonies that grew on the two enumeration plates were pooled and subcultured onto plates containing 8 μg of either metronidazole or nitrofurantoin/ml, respectively. H. pylori colonies were identified using standard criteria and were enumerated as described above.
Serum samples were tested for the H. pylori antigen-specific immunoglobulin antibody by a previously described enzyme-linked immunosorbent assay technique (14). Briefly, 96-well Maxisorb plates (Nunc, Kamstrup, Denmark) were coated with 25 μg of a sonicated whole-cell extract of H. pylori SS1. Serum samples were diluted 1:100 and were added in 100-μl aliquots to coated microtiter wells. To allow for nonspecific antibody binding, samples were also added to uncoated wells. Bound H. pylori-specific antibodies were detected by using biotinylated goat anti-mouse immunoglobulin and streptavidin-peroxidase conjugate (Amersham, Les Ulis, France). The readings for uncoated wells were subtracted from those for the respective test samples. A cutoff value was determined from the mean optical density value ± 2 standard deviations for the corresponding samples from naive uninfected mice. Samples with optical density readings greater than this cutoff value were considered positive for H. pylori-specific antibodies.
Statistical analysis.
Differences in the eradication rates between the groups of mice were determined by Fisher's exact probability test. Differences in bacterial loads were determined by the Mann-Whitney U test (two-sided). A P value of ≤0.05 was considered significant.
RESULTS
Construction of the H. pylori rdxA mutant.
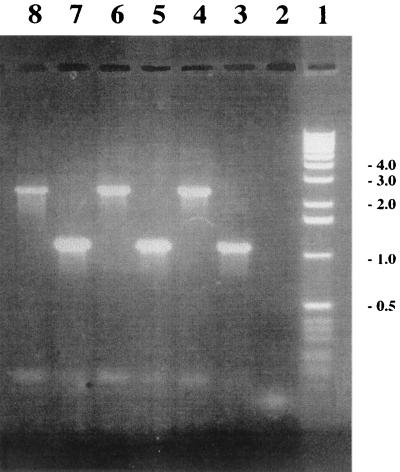
In order to determine whether the oxygen-insensitive NADPH nitroreductase of H. pylori, RdxA, was responsible for susceptibility to nitrofurantoin as well as metronidazole, an isogenic mutant in the rdxA gene was constructed in three strains. To do this, a plasmid with the aphA3 kanamycin resistance gene (50) inserted in rdxA was constructed in E. coli. H. pylori rdxA mutants derived from strains SS1, G27, and HAS-141 were then produced by allelic exchange following natural transformation with a concentrated preparation of the recombinant plasmid. The genotypes of the constructed mutants were verified by performing PCR with primers that flanked the point of insertion of the antibiotic resistance cassette (HP0954-1 and HP0954-4). The PCR products obtained with genomic DNA of these strains were of the correct size and consistent with insertion of the kanamycin cassette (1,400 bp) and an engineered deletion of 114 bp within rdxA: for parental strains SS1, G27, and HAS-141, 1,114 bp; for mutant strains SS1-rdxA, G27-rdxA, and HAS-141-rdxA, 2,400 bp (Fig. 1).
FIG. 1.
Genotypic confirmation of the H. pylori rdxA mutant by PCR. Lane 1, 1-kb molecular weight marker (Gibco BRL); lane 2, negative control; lanes 3 to 8, SS1, SS1-rdxA, G27, G27-rdxA, HAS-141, and HAS-141-rdxA amplification products, respectively, with primers HP0954-1 and HP0954-4.
In vitro activity of nitrofurantoin against metronidazole-sensitive and -resistant H. pylori.
To determine if there was cross-resistance between nitrofurantoin and metronidazole in H. pylori, the MICs of nitrofurantoin and metronidazole for the rdxA mutants and a series of matched metronidazole-sensitive and -resistant strains were determined using an agar dilution method (Table 3). The metronidazole MICs for rdxA mutant strains SS1-rdxA, G27-rdxA, and HAS-141-rdxA were significantly higher than those for the respective parental strains (Table 3). Although the MIC for SS1 rose from 0.0625 μg/ml for the parent strain to 2 μg/ml for the mutant, the MIC for the mutant was not sufficiently high for this strain to be considered resistant by standard criteria (56). The susceptibilities to nitrofurantoin of the rdxA mutants and respective parental strains were identical (Table 3).
TABLE 3.
In vitro activity of metronidazole and nitrofurantoin against H. pylori
| Strain | MICa (μg/ml) of:
|
|
|---|---|---|
| Metronidazole | Nitrofurantoin | |
| SS1 | 0.0625 | 0.5 |
| SS1-rdxA | 2 | 0.5 |
| G27 | 1 | 1 |
| G27-rdxA | 64 | 1 |
| HAS-141 | 1 | 1 |
| HAS-141-rdxA | 64 | 1 |
Mean obtained from three independent experiments.
The metronidazole and nitrofurantoin MICs for 10 mouse-derived metronidazole-sensitive SS1 isolates (SS1-1 to SS1-10) (27) were between 0.0625 and 0.125 μg/ml and between 0.5 and 1 μg/ml, respectively. The MICs of metronidazole and nitrofurantoin for 20 mouse-derived metronidazole-resistant SS1 isolates (SS1-11 to SS1-30) (27) were between 8 and 64 μg/ml and between 0.5 and 2 μg/ml, respectively. These strains included 10 in which the rdxA gene had previously been sequenced; in 9 the rdxA gene contained one or more mutations, and in 1 the gene sequence was identical to that of the parental SS1 strain (26). The susceptibilities to nitrofurantoin and metronidazole of 10 paired metronidazole-sensitive and -resistant clinical strains were also determined (48). The metronidazole and nitrofurantoin MICs for the 10 metronidazole-sensitive strains (T1S to T10S) were between 0.5 and 2 μg/ml and between 0.5 and 4 μg/ml, respectively. The MICs of metronidazole and nitrofurantoin for the 10 corresponding metronidazole-resistant strains (T1R to T10R) were between 8 and 64 μg/ml and between 0.5 and 4 μg/ml, respectively.
In vivo activity of nitrofurantoin against metronidazole-sensitive and -resistant H. pylori.
In the control group, none of the 10 mice inoculated with peptone-trypsin broth were infected with H. pylori 1 month after the completion of treatment (Table 4). In contrast, all 10 SS1-inoculated mice in group 1 that were treated with peptone-trypsin broth (Table 4) were infected, with bacterial counts of between 2.2 × 104 and 7.0 × 106 CFU/g of tissue. In group 2, quantitative cultures of gastric tissue samples taken from all 10 rdxA mutant-inoculated mice 1 month after completion of treatment were positive for H. pylori (Table 4). The bacterial counts obtained varied from 2.9 × 103 to 9.1 × 105 CFU/g of tissue. Although the bacterial loads recovered from mice infected with the rdxA mutant (group 2) were approximately 10-fold lower than those recovered from mice infected with SS1, this difference was not statistically significant.
A recommended triple-therapy regimen (omeprazole, clarithromycin, and metronidazole) eradicated infection from 100% of mice inoculated with H. pylori SS1 (group 3) and from one of eight (12.5%) mice inoculated with the rdxA mutant (group 4; P < 0.001; Table 4). The bacterial counts in the six mice still infected with the rdxA mutant after treatment were similar to those observed in nontreated, rdxA mutant-inoculated mice (between 3.5 × 103 and 6.7 × 105 CFU/g of tissue).
In contrast, when metronidazole was replaced by nitrofurantoin, the regimen failed to eradicate infection from any of the SS1-inoculated mice in group 5 (P < 0.001; Table 4). In group 6, the regimen with nitrofurantoin eradicated infection in one of seven (14%) mice inoculated with the rdxA mutant (Table 4). The bacterial counts in the mice in the groups still infected with either H. pylori SS1 or the rdxA mutant after treatment were similar to those observed in nontreated mice (between 5.0 × 103 and 5.9 × 106 CFU/g of tissue and between 4.0 × 104 and 6.3 × 105 CFU/g of tissue, respectively).
Metronidazole-resistant H. pylori cells were isolated from all rdxA mutant-inoculated mice still infected 1 month after the completion of treatment (groups 2, 4, and 6; Table 4). In contrast, none of the SS1-inoculated mice in groups 1, 3, and 5 were infected with metronidazole-resistant isolates (Table 4). None of the mice still infected 1 month after the completion of treatment harbored nitrofurantoin-resistant isolates (Table 4). At the time of sacrifice (1 month), serological testing was not predictive of successful eradication of H. pylori (results not shown).
DISCUSSION
Currently the most effective regimens for the eradication of H. pylori combine a proton pump inhibitor with two of the following antibiotics: metronidazole, clarithromycin, and amoxicillin (11, 12). It is, however, increasingly recognized that the rising prevalence of resistant H. pylori strains, particularly those resistant to metronidazole, threatens to compromise the efficacy of these regimens. Although there has been controversy regarding the clinical relevance of metronidazole resistance, it is now generally accepted that there is a global decrease in the efficacies of treatment regimens involving metronidazole when strains are resistant to this agent (37). This problem has led to the evaluation of a number of compounds with properties similar to those of metronidazole but without the problems of resistance (7, 33, 38). Like that of metronidazole, the bactericidal mechanism of action of the nitrofurans involves enzymatic reduction of the parent compound to generate electrophilic radicals (2, 36). These compounds have good in vitro activity against H. pylori (16, 40, 47), and antimicrobial combinations that included nitrofurantoin have been shown to have a greater in vitro bactericidal effect against a metronidazole-sensitive and a metronidazole-resistant strain of H. pylori than those with metronidazole (7). In addition, H. pylori does not appear to readily acquire resistance to this group of antimicrobial agents (22).
In E. coli, resistance to the nitrofuran derivatives occurs in a stepwise manner and results from mutations in genes encoding oxygen-insensitive nitroreductases (nfsA and nfsB) (35). First-step resistance results from an nfsA mutation, while the increased resistance associated with second-step mutants is due to mutation of nfsB (55). While there is no homolog of NfsA in the genome sequences of H. pylori, NfsB has 22.9% amino acid sequence identity with RdxA, the nitroreductase responsible for reductive activation of metronidazole in H. pylori (1, 18, 49, 55). The identity between NfsB and RdxA is particularly high (71%) in a conserved 14-residue region corresponding to Ser-37 to Val-50 (1, 49, 55). As well as being responsible for susceptibility to metronidazole, the activity of the oxygen-insensitive NADPH nitroreductase encoded by the rdxA gene might also be associated with reduction of, and hence susceptibility to, nitrofurantoin in H. pylori. In order to test this hypothesis, we constructed an isogenic rdxA deletion mutant from three different strains of H. pylori. Although the metronidazole MICs for strains SS1, G27, and HAS-141 carrying the mutant rdxA were significantly higher than those for the respective parental strains, the MIC for mutant SS1-rdxA was not sufficiently raised for this strain to be considered resistant by standard criteria (56). This observation is unlikely to result from residual RdxA activity and provides indirect evidence for additional mechanisms of metronidazole resistance in H. pylori which may increase the degree of resistance in an additive, stepwise fashion. The nitrofurantoin MICs for the rdxA mutants and respective parental strains were identical, suggesting that this enzyme is not responsible for the reductive activation of nitrofurantoin in H. pylori and that inactivation of rdxA does not result in resistance to this antimicrobial agent. Whether nitrofurantoin is reduced by one of the other putative nitroreductases identified in H. pylori remains to be determined. Alternatively, the mechanism of action in this organism may not require production of reactive nitrofurantoin metabolites by a bacterial reductase.
To confirm these observations and to determine whether other mechanisms of cross-resistance to metronidazole and nitrofurantoin might exist in H. pylori, we examined the MICs of metronidazole and nitrofurantoin for a series of well-characterized strains of H. pylori. These included a group of 10 metronidazole-sensitive and 20 metronidazole-resistant isolates generated in vivo by treating mice infected with the metronidazole-sensitive SS1 H. pylori strain with various regimens involving metronidazole (27). Of the 20 resistant isolates, 9 were known to contain mutations within the rdxA gene. In one, the rdxA gene was intact, suggesting that other mechanisms were responsible for the resistant phenotype of this isolate (26). In addition a series of 10 paired metronidazole-sensitive and -resistant clinical strains (48) were also tested for susceptibility to nitrofurantoin. The nitrofurantoin MICs for all strains tested were within a range (0.5 to 4 μg/ml) that would be considered susceptible for a comparative antimicrobial, such as metronidazole. There were no significant differences between the nitrofurantoin MICs for the metronidazole-sensitive and -resistant SS1 isolates, regardless of whether the rdxA gene was intact or not, or between MICs for the sensitive and resistant isolates of each individual pair of clinical strains. These data suggest that nitrofurantoin has comparable in vitro activities against metronidazole-sensitive and -resistant strains of H. pylori and that there is no cross-resistance between metronidazole and nitrofurantoin in this organism.
Clinical trials have demonstrated that triple therapies involving furazolidone (including omeprazole, clarithromycin, and furazolidone regimens) are able to achieve a high cure rate of H. pylori, and such regimens may prove particularly useful in areas where the prevalence of metronidazole-resistant strains is high (10, 33, 46, 57). The clinical evaluation of nitrofurantoin has been limited to studies in which the agent was given either as monotherapy or in combination with bismuth subsalicylate and not as part of a triple-agent regimen (4, 21, 24, 39, 44). Similarly, an assessment of the ability of various antimicrobial agents to eradicate H. pylori-infected gnotobiotic piglets only examined nitrofurantoin monotherapy (29). We therefore wanted to compare the abilities of two regimens to eradicate metronidazole-sensitive and -resistant strains from mice: the first regimen was a standard triple therapy involving metronidazole (12), and the second was the same regimen with nitrofurantoin substituted for metronidazole. To establish the infections, mice were inoculated with H. pylori SS1 and an SS1-derived rdxA mutant (27). Although the bacterial loads recovered from mice infected with the rdxA mutant were approximately 10-fold lower than those from mice infected with SS1, this difference was not statistically significant. The apparent ability of the rdxA mutant to colonize mice at levels similar to those for the parental strain was observed despite a reported decreased fitness of the mutant in the stationary phase of in vitro growth (J. Y. Jeong, W. W. Su, P. S. Hoffmann, and D. E. Berg, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. D/B-182, 1999).
In previous work, we have demonstrated that prior exposure of H. pylori to metronidazole had a considerable negative influence on eradication of the organism by a regimen involving metronidazole (27). In this study, the efficacy of the regimen with metronidazole was significantly reduced in mice infected with the metronidazole-resistant rdxA mutant (eradicated in one of eight mice) compared to its efficacy in mice infected with the susceptible SS1 strain (eradicated in all mice). The magnitude of this effect is likely to reflect the fact that the stomachs of the rdxA mutant-infected mice contained a population of bacteria entirely made up of resistant isolates rather than the mixed population of sensitive and resistant strains that is frequently observed in clinical practice (20, 28). This observation provides compelling evidence for the role of metronidazole resistance in determining the successful outcome of regimens with metronidazole. When metronidazole was replaced by nitrofurantoin, the regimen failed to eradicate infection from any of the SS1-inoculated mice and eradicated infection in only one of seven mice inoculated with the rdxA mutant. Nitrofurantoin therefore appears to be similar to a number of antimicrobial agents that have been found to be ineffective in eradicating H. pylori in clinical practice despite good in vitro activity (19). The failure of the regimen with nitrofurantoin to eradicate H. pylori may be due to poor delivery of the drug, resulting in an insufficient concentration of the antimicrobial to exert an effective antibacterial activity in vivo (17). It is also possible that therapy was unsuccessful because of intrinsic differences between the activity of nitrofurantoin in mice and in humans. However, the limited data available suggest that nitrofurans are capable of therapeutic activity in murine models of infection (9, 15). Alternatively, it is possible that H. pylori has a low level of metabolic activity within the stomach and is thus relatively resistant to certain bactericidal agents.
These data demonstrate that, despite good in vitro activity and lack of induction of resistance (and particularly cross-resistance to metronidazole), nitrofurantoin is unable to eradicate H. pylori from mice when included as a component of a triple-therapy regimen. The H. pylori SS1 mouse model appears to be a suitable system for assessing novel anti-H. pylori agents and for determining the ability of new regimens to eradicate resistant strains.
ACKNOWLEDGMENTS
P. J. Jenks was supported by a Research Training Fellowship in Medical Microbiology from the Wellcome Trust, United Kingdom (Ref. 044330). Financial support was provided in part by Pasteur-Mérieux-Connaught (Lyon, France) and OraVax Inc. (Boston, Mass.).
We are grateful to Proctor & Gamble Pharmaceuticals, Rhône-Poulenc Rorer, Astra Hassle AB, and Abbott Laboratories for the gift of the pharmaceutical agents used in the treatment protocols.
REFERENCES
- 1.Alm R A, Ling L-S L, Moir D T, King B L, Brown E D, Doig P C, Smith D R, Noonan B, Guild B C, deJonge B L, Carmel G, Tummino P J, Caruso A, Uria-Nickelsen M, Mills D M, Ives C, Gibson R, Merberg D, Mills S D, Jiang Q, Taylor D E, Vovis G F, Trust T J. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature. 1999;397:176–180. doi: 10.1038/16495. [DOI] [PubMed] [Google Scholar]
- 2.Asnis R E. The reduction of furacin by cell-free extracts of furacin-resistant and parent-susceptible strains of Escherichia coli. Arch Biochem Biophys. 1957;66:208–216. doi: 10.1016/0003-9861(57)90551-9. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M J. Helicobacter pylori: microbiology of a ‘slow’ bacterial infection. Trends Microbiol. 1993;1:255–260. doi: 10.1016/0966-842x(93)90047-u. [DOI] [PubMed] [Google Scholar]
- 4.Börsch G, Mai U, Müller K M. Monotherapy or polychemotherapy in the treatment of Campylobacter pylori-related gastroduodenal disease. Scand J Gastroenterol. 1988;23:101–106. doi: 10.3109/00365528809091722. [DOI] [PubMed] [Google Scholar]
- 5.Buckley M J M, Xia H X, Hyde D M, Keane C T, O'Morain C A. Metronidazole resistance reduces efficacy of triple therapy and leads to secondary clarithromycin resistance. Dig Dis Sci. 1997;42:2111–2115. doi: 10.1023/a:1018882804607. [DOI] [PubMed] [Google Scholar]
- 6.Casadaban M, Cohen S N. Analysis of gene control signals by DNA fusions and cloning in E. coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 7.Coudron P E, Stratton C W. In-vitro evaluation of nitrofurantoin as an alternative agent for metronidazole in combination antimicrobial therapy against Helicobacter pylori. J Antimicrob Chemother. 1998;42:657–660. doi: 10.1093/jac/42.5.657. [DOI] [PubMed] [Google Scholar]
- 8.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N, Rappuoli R. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc Natl Acad Sci USA. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz C C, Ferrari L, Sogayar R. A therapeutic trial in Giardia muris infection in the mouse with metronidazole, tinidazole, secnidazole and furazolidone. Rev Soc Bras Med Trop. 1997;30:223–228. doi: 10.1590/s0037-86821997000300009. [DOI] [PubMed] [Google Scholar]
- 10.Dani R, Queiroz D M, Dias M G, Franco J M, Magalhaes L C, Mendes G S, Moreira L S, Castro L D, Toppa N H, Rocha G A, Cabral M M, Salles P G. Omeprazole, clarithromycin and furazolidone for the eradication of Helicobacter pylori in patients with duodenal ulcer. Aliment Pharmacol Ther. 1999;13:1647–1652. doi: 10.1046/j.1365-2036.1999.00653.x. [DOI] [PubMed] [Google Scholar]
- 11.Dunn B E, Cohen H, Blaser M J. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–741. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Helicobacter pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Gut. 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Study Group on Antibiotic Susceptibility of Helicobacter pylori. Results of a multicentre European survey in 1991 of metronidazole resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1992;11:777–781. [PubMed] [Google Scholar]
- 14.Ferrero R L, Thiberge J M, Huerre M, Labigne A. Immune responses of specific-pathogen-free mice to chronic Helicobacter pylori (strain SS1) infection. Infect Immun. 1998;66:1349–1355. doi: 10.1128/iai.66.4.1349-1355.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster R, Pringle G, King D F, Paris J. The therapeutic activity of some nitrofurans in experimental filariasis and trypanosomiasis. Ann Trop Med Parasitol. 1969;63:95–107. doi: 10.1080/00034983.1969.11686604. [DOI] [PubMed] [Google Scholar]
- 16.Glupczynski Y, Delmee M, Bruck C, Labbe M, Avesani V, Burette A. Susceptibility of clinical isolates of Campylobacter pylori to 24 antimicrobial and anti-ulcer agents. Eur J Epidemiol. 1988;4:154–157. doi: 10.1007/BF00144743. [DOI] [PubMed] [Google Scholar]
- 17.Goddard A F. Getting to the route of Helicobacter pylori treatment. J Antimicrob Chemother. 1998;42:1–3. [PubMed] [Google Scholar]
- 18.Goodwin A, Kersulyte D, Sisson G, van Zanten S J O V, Berg D E, Hoffman P S. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin C S. Antimicrobial treatment of Helicobacter pylori infection. Clin Infect Dis. 1997;25:1023–1026. doi: 10.1086/516078. [DOI] [PubMed] [Google Scholar]
- 20.Graham D Y, de Boer W A, Tytgat G N. Choosing the best anti-Helicobacter pylori therapy: effect of antimicrobial resistance. Am J Gastroenterol. 1996;91:1072–1076. [PubMed] [Google Scholar]
- 21.Graham D Y, Klein P D, Evans D G, Evans D J, Alpert L C, Opekun A, Jerdack G R, Morgan D R. Simple noninvasive method to test efficacy of drugs in the eradication of Helicobacter pylori infection: the example of combined bismuth subsalicylate and nitrofurantoin. Am J Gastroenterol. 1991;86:1158–1162. [PubMed] [Google Scholar]
- 22.Haas C E, Nix D E, Schentag J J. In vitro selection of resistant Helicobacter pylori. Antimicrob Agents Chemother. 1990;34:1637–1641. doi: 10.1128/aac.34.9.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris A W, Pryce D I, Gabe S M, Karim Q N, Walker M M, Langworthy H, Baron J H, Misiewicz J J. Lansoprazole, clarithromycin and metronidazole for seven days in Helicobacter pylori infection. Aliment Pharmacol Ther. 1996;10:1005–1008. [PubMed] [Google Scholar]
- 24.Hunter F M, Correa P, Fontham E, Ruiz B, Sobhan M, Samloff I M. Serum pepsinogens as markers of response to therapy for Helicobacter pylori gastritis. Dig Dis Sci. 1993;38:2081–2086. doi: 10.1007/BF01297088. [DOI] [PubMed] [Google Scholar]
- 25.Janvier B, Grignon B, Audibert C, Pezennec L, Fauchère J L. Phenotypic changes of Helicobacter pylori components during experimental infection in mice. FEMS Immunol Med Microbiol. 1999;24:27–33. doi: 10.1111/j.1574-695X.1999.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 26.Jenks P J, Ferrero R L, Labigne A. The role of the rdxA gene in the evolution of metronidazole resistance in Helicobacter pylori. J Antimicrob Chemother. 1999;43:753–758. doi: 10.1093/jac/43.6.753. [DOI] [PubMed] [Google Scholar]
- 27.Jenks P J, Labigne A, Ferrero R L. Exposure to metronidazole in vivo readily induces resistance in Helicobacter pylori and reduces the efficacy of eradication therapy in mice. Antimicrob Agents Chemother. 1999;43:777–781. doi: 10.1128/aac.43.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jorgensen M, Daskalopoulos G, Warburton V, Mitchell H M, Hazell S L. Multiple strain colonization and metronidazole resistance in Helicobacter pylori-infected patients: identification from sequential and multiple biopsy specimens. J Infect Dis. 1996;174:631–635. doi: 10.1093/infdis/174.3.631. [DOI] [PubMed] [Google Scholar]
- 29.Krakowka S, Eaton K A, Leunk R D. Antimicrobial therapies for Helicobacter pylori infection in gnotobiotic piglets. Antimicrob Agents Chemother. 1998;42:1549–1554. doi: 10.1128/aac.42.7.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labigne A, Cussac V, Courcoux P. Shuttle cloning and nucleotide sequence of Helicobacter pylori genes responsible for urease activity. J Bacteriol. 1991;173:1920–1931. doi: 10.1128/jb.173.6.1920-1931.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Labigne-Roussel A, Courcoux P, Tompkins L. Gene disruption and replacement as a feasible approach for mutagenesis of Campylobacter jejuni. J Bacteriol. 1988;170:1704–1708. doi: 10.1128/jb.170.4.1704-1708.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 33.Liu W Z, Xiao S D, Shi Y, Wu S M, Zhang D Z, Xu W W, Tytgat G N. Furazolidone-containing short-term therapies are effective in the treatment of Helicobacter pylori infection. Aliment Pharmacol Ther. 1999;13:317–322. doi: 10.1046/j.1365-2036.1999.00492.x. [DOI] [PubMed] [Google Scholar]
- 34.Lopez-Brea M, Domingo D, Sanchez I, Alarcon T. Evolution of resistance to metronidazole and clarithromycin in Helicobacter pylori clinical isolates from Spain. J Antimicrob Chemother. 1997;40:279–281. doi: 10.1093/jac/40.2.279. [DOI] [PubMed] [Google Scholar]
- 35.McCalla D R, Kaiser C, Green M H L. Genetics of nitrofurazone resistance in Escherichia coli. J Bacteriol. 1978;133:10–16. doi: 10.1128/jb.133.1.10-16.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McOsker C C, Fitzpatrick P M. Nitrofurantoin: mechanism of action and implications for resistance development in common uropathogens. J Antimicrob Chemother. 1994;33(Suppl. A):23–30. doi: 10.1093/jac/33.suppl_a.23. [DOI] [PubMed] [Google Scholar]
- 37.Mégraud F, Doermann H P. Clinical relevance of resistant strains of Helicobacter pylori: a review of current data. Gut. 1998;43(Suppl. 1):S61–S65. doi: 10.1136/gut.43.2008.s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mégraud F, Occhialini A, Rossignol J F. Nitazoxanide, a potential drug for eradication of Helicobacter pylori with no cross-resistance to metronidazole. Antimicrob Agents Chemother. 1998;42:2836–2840. doi: 10.1128/aac.42.11.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morgan D, Kraft W, Bender M, Pearson A. Nitrofurans in the treatment of gastritis associated with Campylobacter pylori. Gastroenterology. 1988;95:1178–1184. doi: 10.1016/0016-5085(88)90348-4. [DOI] [PubMed] [Google Scholar]
- 40.Morgan D R, Fitzpatrick P M, David K L, Kraft W G. Susceptibility patterns of Campylobacter pyloridis. FEMS Microbiol Lett. 1987;42:245–248. [Google Scholar]
- 41.Parsonnet J, Friedman G D, Vandersteed D P, Chang Y, Vogelman J H, Orentreich N, Sibley R K. Helicobacter pylori infection and the risk of gastric cancer. N Engl J Med. 1991;325:1127–1129. doi: 10.1056/NEJM199110173251603. [DOI] [PubMed] [Google Scholar]
- 42.Parsonnet J, Hansen S, Rodriguez L, Gelb A B, Warnke R A, Jellum E, Orentreich N, Vogelman J H, Friedman G D. Helicobacter pylori infection and gastric lymphoma. N Engl J Med. 1994;330:1267–1271. doi: 10.1056/NEJM199405053301803. [DOI] [PubMed] [Google Scholar]
- 43.Rautelin H, Seppala K, Renkonen O V, Vainio U, Kosunen T U. Role of metronidazole in therapy of Helicobacter pylori infections. Antimicrob Agents Chemother. 1992;36:163–166. doi: 10.1128/aac.36.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ruiz B, Rood J C, Fontham E T H, Malcom G T, Hunter F M, Sobhan M, Johnson W D, Correa P. Vitamin C concentration in gastric juice before and after anti-Helicobacter pylori treatment. Am J Gastroenterol. 1994;89:533–539. [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 46.Segura A M, Gutierrez O, Otero W, Angel A, Genta R M, Graham D Y. Furazolidone, amoxycillin, bismuth triple therapy for Helicobacter pylori infection. Aliment Pharmacol Ther. 1997;11:529–532. doi: 10.1046/j.1365-2036.1997.00172.x. [DOI] [PubMed] [Google Scholar]
- 47.Simor A E, Ferro S, Low D E. Comparative in vitro activities of six new fluoroquinolones and other oral antimicrobial agents against Campylobacter pylori. Antimicrob Agents Chemother. 1989;33:108–109. doi: 10.1128/aac.33.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tankovic J, Lamarque D, Delchier J C, Soussy C J, Labigne A, Jenks P J. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44:608–613. doi: 10.1128/aac.44.3.608-613.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomb J F, White O, Kervalage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, Nelson K, Quackenbush J, Zhou L, Kirkness E F, Peterson S, Loftus B, Richardson D, Dodson R, Khalak H G, Glodek A, McKenney K, Fitzegerald L M, Lee N, Adams M D, Hickey E K, Berg D E, Gocayne J D, Utterback T R, Peterson J D, Kelley J M, Cotton M D, Weidman J M, Fujii C, Bowman C, Watthey L, Wallin E, Hayes W S, Borodovsky M, Karp P D, Smith H O, Fraser C M, Ventor J C. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 50.Trieu-Cout P, Gerbaud G, Lambert T, Courvalin P. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 1985;4:3583–3587. doi: 10.1002/j.1460-2075.1985.tb04120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van de Hulst R W M, van der Ende A, Homan A, Roorda P, Dankert J, Tytgat G N J. Influence of metronidazole resistance on efficacy of quadruple therapy for Helicobacter pylori eradication. Gut. 1998;42:166–169. doi: 10.1136/gut.42.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Wouden E J, van Zwet M, Vosmaer G D, Oom J A, de Jong A, Kleibeuker J H. Rapid increase in the prevalence of metronidazole-resistant Helicobacter pylori in the Netherlands. Emerg Infect Dis. 1997;3:385–389. doi: 10.3201/eid0303.970320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, Roos K P, Taylor D E. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J Gen Microbiol. 1993;39:2485–2493. doi: 10.1099/00221287-139-10-2485. [DOI] [PubMed] [Google Scholar]
- 54.Warren J R. Unidentified curved bacilli on gastric epithelium in chronic active gastritis. Lancet. 1983;i:1273. [PubMed] [Google Scholar]
- 55.Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert I B. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Working Party of the European Helicobacter pylori Study Group. Guidelines for clinical trials in Helicobacter pylori infection. Gut. 1997;41(Suppl. 2):S1–S23. [PubMed] [Google Scholar]
- 57.Xiao S D, Liu W Z, Hu P J, Xia D H, Tytgat G N. High cure rate of Helicobacter pylori infection using tripotassium bismuthate, furazolidone and clarithromycin triple therapy for 1 week. Aliment Pharmacol Ther. 1999;13:311–315. doi: 10.1046/j.1365-2036.1999.00487.x. [DOI] [PubMed] [Google Scholar]