It is now a commonplace to propose high-dimensional molecular characterization within the translational research objectives of contemporary clinical trials. Similarly, most of these methods are being applied to archived samples from completed clinical trials. The rationale is well understood—comprehensive molecular profiling should accelerate our goal of precision cancer medicine, especially when applied to the randomized clinical trials that incorporate current and emerging effective treatments. However, present barriers impede researchers from unlocking the full potential of these data sets and trials, and it is critical to solve these challenges. Currently, omics data generated from trials are largely decentralized: data are housed at a variety of sites, analyses take place locally, and other researchers do not have access until public deposition of data on repositories such as the database of Genotypes and Phenotypes (dbGaP) and National Cancer Institute's (NCI) Genomic Data Commons (GDC) at publication—often years after generation—and then potentially without adequate clinical annotation to correlate omics features with clinical outcomes. Furthermore, analyses vary widely in bioinformatics methods, including choice of tools, dependencies, file formats, parameterizations, data quality filtering thresholds, and other workflow elements, which makes integration across groups challenging.
Despite great strides toward improving clinical data acquisition (eg, iCARE initiative1) and storage (eg, NCI National Clinical Trials Network [NCTN] Data Archive2), the value of accumulated clinical data is yet to be fully unlocked. Historically, each clinical trial team defines their own trial vocabulary and designates their own data elements using an array of templates. Recent efforts such as the Clinical Data Interchange Standards Consortium (CDISC)3 standards for clinical trials elements are evidence that this paradigm is changing. Nevertheless, the lack of more standardized nomenclature hinders research and clinically important studies that require data aggregation and integration across trials in the domains of clinical data, biospecimen data, and statistical/omics meta-analyses. Currently, there is not a simple strategy for viewing the data landscape of all completed trials at a granular patient level in a single location. To enable complete data transparency, a technological solution must be explored for investigators not only to readily view information across trials but also to readily share data using the same vocabulary to facilitate analyses and interpretations. There is a critical need for both a unified vocabulary and a data clearinghouse that facilitates the harmonization of both prospective and retrospective clinical trials data. Such a system would synchronize data activities across clinical trials, facilitate the rapid advancement of correlative analyses, and allow future analysis of studies, within and across tumor type.
To address this within the NCI's NCTN framework, we have established the Alliance Standardized Translational Omics Resource (A-STOR) through planning by the Alliance Translational Research Program (TRP) and the Alliance Statistical and Data Management Center (SDMC) to develop a solution for this rate-limiting step. A-STOR is a single shared living repository for multiomics data and associated clinical data designed to facilitate rapid omics analyses and meta-analyses within and across studies.
The Challenge of Decentralization of Clinical Trial Omics Data
Two of the major gaps in clinicogenomic research that A-STOR aims to fill are (1) the decentralized nature of clinical trial omics data and (2) competing priorities of study teams, biostatisticians, correlative researchers, and data repositories. Data are owned by an individual study until publications and thus are siloed for years. Study teams aim to protect their own rights to the data, including first publication, without hindrance from other investigators. In an era of rapid advancement, this makes the data potentially less relevant in contemporary clinical contexts at the time they become available. Data repositories aim to accelerate data accessibility, but maintain extensive formatting guidelines for submission, and each repository has distinct content and requirements. For example,
dbGaP: Database of Genotypes and Phenotypes, maintained by the National Center for Biotechnology Information (NCBI). Primarily raw sequencing data with study-specific metadata, governed by complex data deposition requirements. Typically, deposit and public accessibility only occurs with publication of translational manuscripts.
GDC: Genomic Data Commons by NCI includes raw and processed sequencing data, curated clinical metadata, and even pathology image data. Only a subset of clinical trials are hosted (no pharmaceutical-supported trials), and typically, deposit and public accessibility only occurs with publication of translational manuscripts.
NCTN Data Archive: National Clinical Trials Network Data Archive includes detailed clinical trial data but not genomic data. Data deposit and public accessibility only occurs with publication of primary clinical trial end points.
NCI Cloud Resources: Components of the NCI Cancer Research Data Commons that allow users to deposit their own data, which can include raw or processed genomic data and other data types. Public data access defined by an individual or a research team that deposited data.
Effectively, these competing interests can create bottlenecks that further delay public data dissemination, whereas skepticism regarding the utility of data sharing persists.4 A-STOR will directly target these challenges by ingesting genomic and other high-dimensional (data with extensive, detailed features) data after generation to facilitate rapid, parallel analyses of translational data. To protect the rights of the study investigators, A-STOR–approved users will withhold presentation or publication of data until the initial presentation of the prespecified clinical and correlative end point(s) by the study Principal Investigator (PI). In these ways, A-STOR will accelerate the availability of clinical trial omics data while protecting the study investigators' rights.
A-STOR: Principles
A-STOR will accelerate, diversify, and coordinate a portfolio of analyses within and across clinical trials, including and not competing against the clinical trials' correlative science investigators. Specifically, A-STOR will provide a genomic data repository and standardized computational processing of sequencing data after primary clinical end point reporting, in conjunction with each clinical trial's correlative science investigators. This initiative will provide stable, secure, scalable storage for multiomic data; accessibility to approved investigators including controlled access in the prepublication window; rapid advancement of correlative analyses to enhance clinical trial design, grants, and publications; ability to perform rapid meta-analyses; seamless integration with existing data structures such as NCI GDC and dbGaP; and potential future growth, including increased computational analysis potential, integration with external validation data sets, and informative data visualization to speed the development of promising biomarkers. The overall structure of A-STOR focuses on developing a flexible, accessible system that can work with existing resources and within standing frameworks (Fig 1A).
FIG 1.
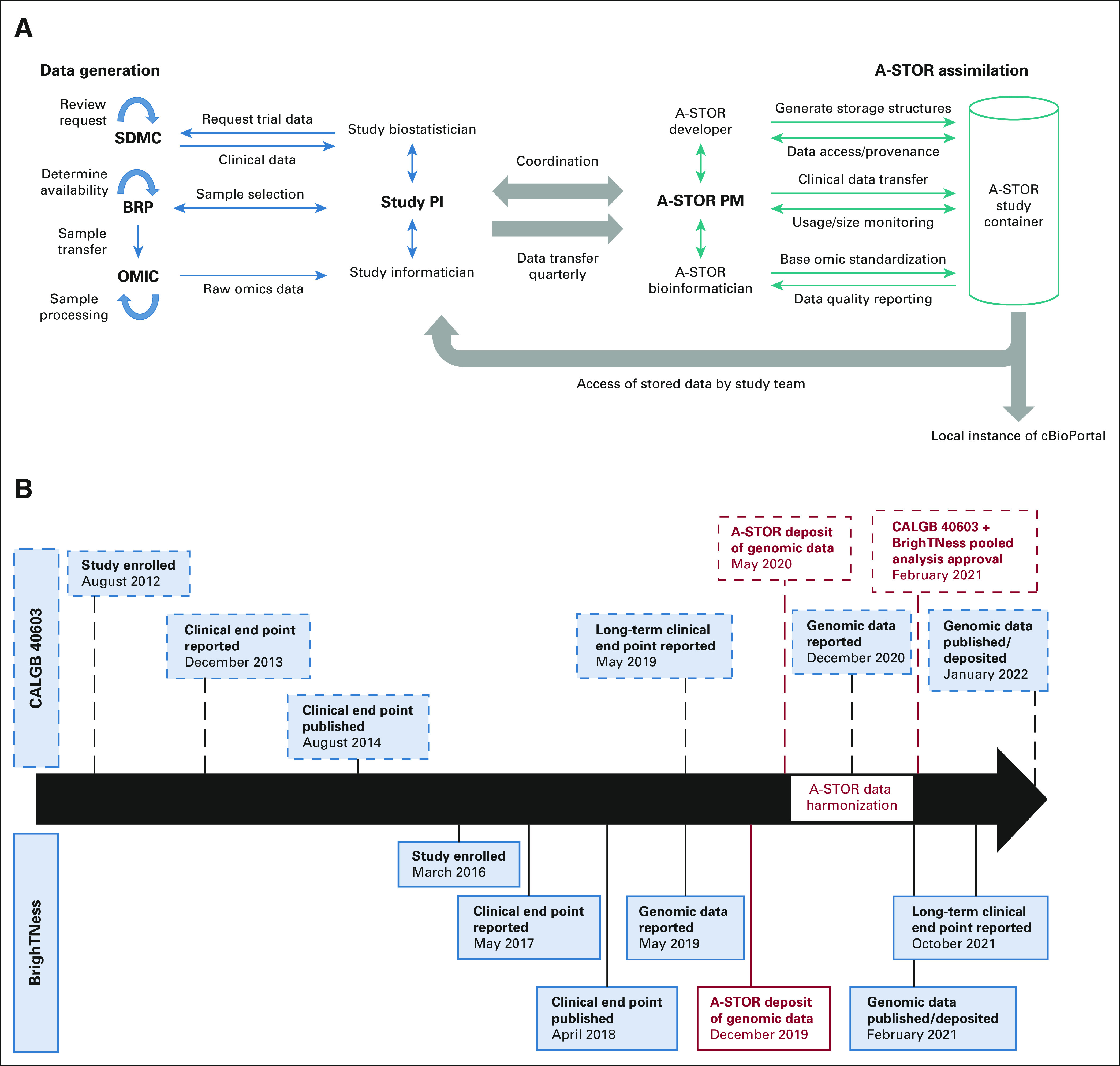
Alliance Standardized Translational Omics Resource (A-STOR) workflow and data timelines. (A) A-STOR workflow and roles. (B) Study and genomic data timelines for CALGB 40603 and BrighTNess, phase III clinical trials of neoadjuvant chemotherapy for triple-negative breast cancer. Dates of key study milestones (enrollment, end point reporting, and results publication) and genomic data milestones (genomic data reporting, publication, and public deposition) are noted. Dates of A-STOR milestones (indicated in red) demonstrate deposition, processing/harmonization, and pooled analysis approval at the accelerated timetable relative to public data deposition. CALGB 40603 milestones are indicated by dashed lines; BrighTNess milestones are indicated by solid lines. A-STOR, Alliance Standardized Translational Omics Resource; BRP, Biorepository; OMIC, Designated Sequencing Core; PI, Principal Investigator; PM, Project Manager; SDMC, Statistical and Data Management Center.
A-STOR Logistics
Study initiation and data deposition
First, the Alliance Study PI initiates a plan to sequence samples from an Alliance or Alliance Foundation Clinical Trial. The A-STOR Project Manager works with the study PI to create a trial-specific data bucket, confirms appropriate consent language, and coordinates upload of basic clinical metadata. Upon sequencing, raw or aligned sequence data files are deposited in A-STOR with initial access to the study PI only. No data processing or harmonization is needed at this point. The lead A-STOR bioinformatician coordinates data capture and ensures consistency and data integrity.
Data analyses
The Alliance Study PI provides A-STOR sequence data access to the study statistician(s) and computational biologist(s) for primary, study-specified analyses of the sequencing data. Concurrent with the primary analysis, the study PI may approve access to additional research team(s), who will be embargoed for presentation or publication until the primary study/end point presentation. This structure ensures that multiple analyses can be completed in parallel, rather than sequentially. The A-STOR project manager and bioinformatician will maintain access control throughout the data lifecycle.
Preparation for public data deposition
Upon completion and anticipated publication of the primary study end point(s), the study PI prepares necessary metadata for a NCI GDC/dbGaP submission. The A-STOR bioinformatician transfers sequencing data, facilitates provenance, and facilitates timely omics data release (NCTN archive/GDC) per mandates.
Presentation/publication of secondary analyses
After the first publication and approval by the study PI, parallel secondary or meta-analyses of the sequencing data via A-STOR may be presented and/or published. Processed data from the primary analysis and/or secondary analysis or meta-analyses, such as transcript abundance for RNA sequencing with clearly documented processing information, may also be uploaded and will be linked to the clinical metadata for that trial by the A-STOR bioinformatician.
A-STOR Omics Data Harmonization and Visualization
One major aim of A-STOR is harmonization of Alliance genomic data through standardized pipelines. A-STOR will implement standardized analytical pipelines, including alignment for DNA sequencing, transcript abundance for RNA sequencing, and circulating tumor DNA. All pipelines are maintained to ensure best practices with current scientific standards, versioned pipelines, systematized testing, and data provenance automatically logged throughout workflows. All elements of the workflows will be transparent with all tool, parameterization, and filtering choices documented on the project wiki. Containerized versions of the complete workflows will be made available for other researchers to explore, test, and analyze additional data using the A-STOR workflows. Stable, versioned pipelines will provide A-STOR users with uniform results, extending the reliability and accuracy of data.
Another major aim of A-STOR is to develop a data visualization tool for interaction by noninformaticians. As a proof of concept, A-STOR has leveraged cBioPortal as an interactive dashboard for users to interact with the clinical and genomic data from initial pilot studies.5 Currently, users can link phenotypic traits to the frequency of variants in a given gene within the study cohort. Furthermore, users can designate subpopulations of the cohort to analyze. RNA expression and DNA variation data are able to be explored in this manner. Future plans include incorporating all available Alliance studies into this platform, which will give investigators an in-depth and quick way to view genomic patterns among aggregated data with more samples and minimal analysis. The user-friendly cBioPortal interactive dashboard offers an avenue for noninformatician oncology practitioners and researchers to interact with clinical trial omic data, currently restricted to approved Alliance investigators.
Case Study: Neoadjuvant Chemotherapy for Triple-Negative Breast Cancers
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer that demonstrates greater chemosensitivity relative to hormone receptor–positive breast cancers but, paradoxically, has worse outcomes.6 Neoadjuvant chemotherapy (NAC) provides a window through which to evaluate the intrinsic chemosensitivity of breast cancer, as patients receive no treatments before NAC and there is a discrete end point—pathologic complete response (pCR) reflecting chemosensitivity or residual disease reflecting some level of chemoresistance. Patients with TNBC who do not achieve pCR to NAC have a very poor prognosis, with a 5-year overall survival of approximately 50% in a large meta-analysis.7 With this high risk of recurrence for patients with residual disease after NAC, there is interest in evaluating whether the addition of other agents, such as platinum chemotherapy,8,9 poly (ADP-ribose) polymerase inhibitors,10 or immune checkpoint inhibitors,11 can improve pCR rates and long-term outcomes.12 Among these were two phase III studies completed within Alliance: CALGB 40603, which evaluated the addition of carboplatin with or without bevacizumab, and BrighTNess, which evaluated the addition of carboplatin with or without poly (ADP-ribose) polymerase inhibitor veliparib.8,9 Each of these studies completed RNA sequencing of hundreds of primary tumors before NAC, 295 samples from CALGB 4060313 and 482 samples from BrighTNess,14 and there is great interest in determining underlying mechanisms of response and resistance to NAC in TNBC, including expression subtypes,15-21 expression-based markers of proliferation,16,22,23 immune expression signatures,22,24 and BRCAness biomarkers.25-27
Although these two studies reported their primary clinical end points many years before (CALGB 40603 in 2013 and BrighTNess in 2017), the deposition of these genomic data into publicly available repositories was delayed for both until 2021 (Fig 1B). This lag offered an opportunity to initiate translational omics analyses pending final public data deposition. To that end, starting in 2019, RNA sequencing data from these two studies were deposited into the Health Insurance Portability and Accountability Act (HIPAA)–compliant, encrypted A-STOR repository. In parallel and concurrent with the primary analyses by the study teams, the RNA sequencing data underwent harmonized processing to transcript abundance to facilitate meta-analyses. The analyses to determine outcome-agnostic features, such as the calculation of published signatures, were initiated in advance of final clinical data deposition. Upon public release of clinical end points, data analyses depending upon outcomes were initiated. In summary, despite lag of public data deposition of RNA sequencing data for these two phase III studies, the A-STOR approach accelerated harmonized transcriptome meta-analyses by at least a year—a time frame that is relevant to patients and the field.
Future Growth Opportunities
The immediate term goal of A-STOR is to provide a stable, efficient, and maintainable platform for data sharing for investigators performing discovery science and biomarker development via clinical trial samples. This in itself would be valuable, as it would give investigators earlier access to relevant data and speed discovery, provide opportunities for generating robust preliminary data for funding applications, and improve communication and collaboration among investigators and research teams. The intermediate term plan is to build out streamlined analytics capacity on the platform, where data can be interactively explored and basic biostatistical analyses are performed by investigators without computational experience using a web-based graphical user interface (under control of a strong and HIPAA-compliant authentication and authorization system). To support broad data access and usability, A-STOR does not have a cost to researchers. The development of A-STOR was funded through a grant from the Alliance Foundation, and in the future, our goal is to demonstrate value sufficient to warrant support through data storage contracts/subcontracts and independent and collaborative research grants.
The long-term vision is to integrate advanced analytics including machine learning, cross-platform feature integration, and automated workflows to optimize biomarker discovery and validation. This data repository will operate in conjunction with established Alliance resources including the Biospecimen Repository and Statistical Data Center with the goal of establishing a real-time dashboard to track specimens for ongoing clinical trials, emerging Alliance resources including digital pathology, observational genomic cohorts (such as The Cancer Genome Atlas and METABRIC), and broader resources such as NCI GDC and dbGaP. In addition to these opportunities, there is an opportunity to expand data hosting to other high-dimensional data types, such as single-cell sequencing data, digital pathology, and spatial pathology including high-dimensional proteomics such as cytometry by time of flight (CyTOF), cyclic/highly multiplexed immunofluorescence, or spatial genomics. Finally, A-STOR provides a framework and, in fact, infrastructure for other cooperative groups in oncology, collectively, independently, or through partnership with the NCI or other overarching organization(s).
In conclusion, A-STOR will accelerate discovery through data harmonization and accessibility while maintaining rigor, transparency, and provenance. Investigators will have prepublication accessibility through controlled access, thus protecting rights to the data. Finally, A-STOR ingestion of postprimary clinical analyses via standardized and validated bioinformatics pipelines will lead to improved reproducibility of results with quicker data access.
W. Fraser Symmans
Stock and Other Ownership Interests: ISIS Pharmaceuticals, Delphi Diagnostics, Eiger BioPharmaceuticals
Consulting or Advisory Role: Merck
Research Funding: Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Intellectual property, Intellectual Property (expired)
Uncompensated Relationships: Delphi Diagnostics
Open Payments Link: https://openpaymentsdata.cms.gov/physician/256534
Charles M. Perou
Leadership: GeneCentric
Stock and Other Ownership Interests: BioClassifier, GeneCentric, Reveal Genomics
Consulting or Advisory Role: BioClassifier, GeneCentric, NanoString Technologies, Veracyte, Reveal Genomics
Patents, Royalties, Other Intellectual Property: Royalties from PAM50 breast cancer gene patent application and from the lung gene signature patent
James S. Blachly
Consulting or Advisory Role: AstraZeneca, AbbVie, Kite, a Gilead company, AstraZeneca, Astellas Pharma, Innate Pharma
Patents, Royalties, Other Intellectual Property: Sequencing Technology patent pending (Inst)
Travel, Accommodations, Expenses: Oxford Nanopore Technologies
James Chen
Employment: Tempus
Consulting or Advisory Role: Syapse, Tempus
Speakers' Bureau: Foundation Medicine
Research Funding: Eisai
Patents, Royalties, Other Intellectual Property: MatchTX
Benjamin G. Vincent
Stock and Other Ownership Interests: GeneCentric
Consulting or Advisory Role: GeneCentric
Research Funding: Merck (Inst)
Daniel G. Stover
This author is a member of the JCO Clinical Cancer Informatics Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Novartis
No other potential conflicts of interest were reported.
SUPPORT
Supported by Alliance for Clinical Trials in Oncology Foundation (J.S.B., J.C., and D.G.S.) and NIH 1R21CA259985 (D.G.S.)
J.S.B., J.C., B.G.V., and D.G.S. codirected this work.
AUTHOR CONTRIBUTIONS
Conception and design: Charles M. Perou, James S. Blachly, James Chen, Benjamin G. Vincent, Daniel G. Stover
Administrative support: Yujia Wen
Collection and assembly of data: Sarah Asad, Kurt R. Mueller, Yujia Wen, James S. Blachly
Data analysis and interpretation: Kathryn Kananen, W. Fraser Symmans, James S. Blachly, James Chen
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
W. Fraser Symmans
Stock and Other Ownership Interests: ISIS Pharmaceuticals, Delphi Diagnostics, Eiger BioPharmaceuticals
Consulting or Advisory Role: Merck
Research Funding: Pfizer (Inst)
Patents, Royalties, Other Intellectual Property: Intellectual property, Intellectual Property (expired)
Uncompensated Relationships: Delphi Diagnostics
Open Payments Link: https://openpaymentsdata.cms.gov/physician/256534
Charles M. Perou
Leadership: GeneCentric
Stock and Other Ownership Interests: BioClassifier, GeneCentric, Reveal Genomics
Consulting or Advisory Role: BioClassifier, GeneCentric, NanoString Technologies, Veracyte, Reveal Genomics
Patents, Royalties, Other Intellectual Property: Royalties from PAM50 breast cancer gene patent application and from the lung gene signature patent
James S. Blachly
Consulting or Advisory Role: AstraZeneca, AbbVie, Kite, a Gilead company, AstraZeneca, Astellas Pharma, Innate Pharma
Patents, Royalties, Other Intellectual Property: Sequencing Technology patent pending (Inst)
Travel, Accommodations, Expenses: Oxford Nanopore Technologies
James Chen
Employment: Tempus
Consulting or Advisory Role: Syapse, Tempus
Speakers' Bureau: Foundation Medicine
Research Funding: Eisai
Patents, Royalties, Other Intellectual Property: MatchTX
Benjamin G. Vincent
Stock and Other Ownership Interests: GeneCentric
Consulting or Advisory Role: GeneCentric
Research Funding: Merck (Inst)
Daniel G. Stover
This author is a member of the JCO Clinical Cancer Informatics Editorial Board. Journal policy recused the author from having any role in the peer review of this manuscript.
Consulting or Advisory Role: Novartis
No other potential conflicts of interest were reported.
REFERENCES
- 1.Collaborative TSHR . ICAREdata® Project: Integrating Clinical Trials and Real-World Endpoints. http://icaredata.org/ [Google Scholar]
- 2.National Cancer Institute . National Clinical Trials Network and NCI Community Oncology Research Program Data Archive. https://nctn-data-archive.nci.nih.gov/ [Google Scholar]
- 3.Consortium CDIS . Clinical Data Interchange Standards Consortium. https://www.cdisc.org/ [Google Scholar]
- 4.Longo DL, Drazen JM.Data sharing N Engl J Med 374276–2772016 [DOI] [PubMed] [Google Scholar]
- 5. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry DA, Cirrincione C, Henderson IC, et al. Estrogen-receptor status and outcomes of modern chemotherapy for patients with node-positive breast cancer JAMA 2951658–16672006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis Lancet 384164–1722014 [DOI] [PubMed] [Google Scholar]
- 8.Loibl S, O'Shaughnessy J, Untch M, et al. Addition of the PARP inhibitor veliparib plus carboplatin or carboplatin alone to standard neoadjuvant chemotherapy in triple-negative breast cancer (BrighTNess): A randomised, phase 3 trial Lancet Oncol 19497–5092018 [DOI] [PubMed] [Google Scholar]
- 9.Sikov WM, Berry DA, Perou CM, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance) J Clin Oncol 3313–212015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rugo HS, Olopade OI, DeMichele A, et al. Adaptive randomization of veliparib-carboplatin treatment in breast cancer N Engl J Med 37523–342016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid P, Cortés J, Dent R, et al. KEYNOTE-522: Phase 3 study of pembrolizumab (pembro) + chemotherapy (chemo) vs placebo (pbo) + chemo as neoadjuvant treatment, followed by pembro vs pbo as adjuvant treatment for early triple-negative breast cancer (TNBC) Ann Oncol 30v851–v9342019suppl 5 [Google Scholar]
- 12.Stover DG, Winer EP.Tailoring adjuvant chemotherapy regimens for patients with triple negative breast cancer Breast 24S132–S1352015suppl 2 [DOI] [PubMed] [Google Scholar]
- 13. Shepherd JH, Hyslop T, Fan C, et al. Genomic analysis of the CALGB 40603 (Alliance) neoadjuvant trial in TNBC identifies immune features associated with pathological complete response and event-free survival. Cancer Res. 2021;81 suppl 4; abstr PD9-03. [Google Scholar]
- 14.Filho OM, Stover DG, Asad S, et al. Association of immunophenotype with pathologic complete response to neoadjuvant chemotherapy for triple-negative breast cancer: A secondary analysis of the BrighTNess phase 3 randomized clinical trial JAMA Oncol 7603–6082021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours Nature 406747–7522000 [DOI] [PubMed] [Google Scholar]
- 16.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes J Clin Oncol 271160–11672009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies J Clin Invest 1212750–27672011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lehmann BD, Jovanovic B, Chen X, et al. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer Clin Cancer Res 211688–16982015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers Nature 486395–3992012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups Nature 486346–3522012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stover DG, Coloff JL, Barry WT, et al. The role of proliferation in determining response to neoadjuvant chemotherapy in breast cancer: A gene expression-based meta-analysis Clin Cancer Res 226039–60502016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prat A, Lluch A, Albanell J, et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer Br J Cancer 1111532–15412014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S-R, Gavin PG, Pogue-Geile KL, et al. A surrogate gene expression signature of tumor infiltrating lymphocytes (TILs) predicts degree of benefit from trastuzumab added to standard adjuvant chemotherapy in NSABP (NRG) trial B-31 for HER2+ breast cancer. Cancer Res. 2015;75 suppl 15; abstr 2837. [Google Scholar]
- 25.Telli ML, Jensen KC, Vinayak S, et al. A phase II study of gemcitabine, carboplatin and iniparib as neoadjuvant therapy for triple-negative and BRCA1/2 mutation-associated breast cancer with assessment of a tumor-based measure of genomic instability (PrECOG 0105) J Clin Oncol 331895–19012015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. von Minckwitz G, Timms K, Untch M, et al. Prediction of pathological complete response (pCR) by Homologous Recombination Deficiency (HRD) after carboplatin-containing neoadjuvant chemotherapy in patients with TNBC: Results from GeparSixto. J Clin Oncol. 2015;33 suppl; abstr 1004. [Google Scholar]
- 27.Telli ML, Stover DG, Loi S, et al. Homologous recombination deficiency and host anti-tumor immunity in triple-negative breast cancer Breast Cancer Res Treat 17121–312018 [DOI] [PubMed] [Google Scholar]


