Abstract
Four-dimensional (4D) bioprinting is promising to build cell-laden constructs (bioconstructs) with complex geometries and functions for tissue/organ regeneration applications. The development of hydrogel-based 4D bioinks, especially those allowing living cell printing, with easy preparation, defined composition, and controlled physical properties is critically important for 4D bioprinting. Here, a single-component jammed micro-flake hydrogel (MFH) system with heterogeneous size distribution, which differs from the conventional granular microgel, has been developed as a new cell-laden bioink for 4D bioprinting. This jammed cytocompatible MFH features scalable production and straightforward composition with shear-thinning, shear-yielding, and rapid self-healing properties. As such, it can be smoothly printed into stable 3D bioconstructs, which can be further crosslinked to form a gradient in crosslinking density when a photoinitiator and a UV absorber are incorporated. After being subject to shape morphing, a variety of complex bioconstructs with well-defined configurations and high cell viability were obtained. Based on this system, 4D cartilage-like tissue formation was demonstrated as a proof-of-concept. The establishment of this versatile new 4D bioink system may open up a number of applications in tissue engineering.
Keywords: bioink, crosslinking gradient, 4D printing, shape morphing, tissue engineering
Graphical Abstract
Single-component jammed micro-flake hydrogels (MFHs) were developed as cell-laden bioinks for 4D bioprinting. Cytocompatible MFH bioinks without the need of additional fillers are rheologically favorable for bioprinting via smooth direct ink writing (DIW). A controllable crosslinking gradient in the 3D printed bioconstructs was achieved, enabling predefined shape transformations. Ultimately, 4D tissue engineering was demonstrated in a proof-of-concept 4D cartilage-like tissue regeneration study.
1. Introduction
Four-dimensional (4D) printing is a rapidly emerging field originating from 3D printing,[1] where the printed structures undergo a transformation in shape, property or functionality[2] when exposed to a predetermined stimulus such as humidity,[3] temperature,[4] light,[5] current,[6] magnetic field,[7] and other energy sources.[8] 4D processes, such as shape memory, reconfiguration, shape shifting, self-adaptability, and self-assembly, have been demonstrated in a wide range of 4D printing systems,[9] including polymers,[10] alloys,[11] ceramics,[12] liquid crystal elastomers,[13] hydrogels,[14] and composite materials.[15] As such, this new technology has quickly found widespread investigation and application in the fields of actuation,[16] robotics,[17] functional devices,[18] and tissue engineering and regenerative medicine.[19] In particular, one of the aims of 4D bioprinting, a cutting-edge sub-field of 4D printing, is to enable printing of biocompatible materials into functional 3D geometrically complex cell-laden constructs (bioconstructs) to engineer and regenerate tissues or organs.[20, 21] In contrast to traditional 4D printing, bioinks for 4D bioprinting must be cytocompatible and the printed bioconstructs must be capable of executing the 4D process under physiological conditions.[22] However, most materials used for 4D printing are non-cytocompatible and non-biocompatible and/or require a harsh stimulus to induce deformation, such as high temperature, an organic solvent, extreme pH, or an abrupt change in ionic strength.[23] Responsive biomaterials (or bioinks) for bioprinting that permit living cell printing (cell-laden bioinks) are essential for 4D biomedical applications, but there remains a dearth of options.
The use of hydrogels as bioinks has multiple intrinsic advantages over other types of materials, including facile processability, live-cell embedding, and importantly, capacity for biomimicry of native tissue extracellular matrix structure.[24] Moreover, hydrogels also exhibit high compatibility with extrusion-based 4D printing techniques.[25] However, common hurdles encountered in extrusion-based free-standing printing, including inadequate printing resolution, unsatisfactory shape fidelity, and frequent nozzle obstruction, still impede the development of hydrogel-based bioinks.[2] Particularly, specific rheological properties of hydrogel-based bioinks need to be fulfilled to facilitate printability. For example, appropriate viscosity and shear thinning properties are critically important to afford smooth extrudability, while rapid self-healing and sufficient mechanical rigidity are critically important to afford rapid structural restoration and high shape fidelity after printing.[25] To obtain inks meeting above requirements, multi-component materials containing a filler to endow high viscosity and shear thinning properties are typically employed.[26] The incorporation of fillers presents complexity in ink design and could result in uncontrolled or undesired bioactive and mechanical properties of the final generated hydrogel constructs. In addition, it may also be necessary to generate structural anisotropy (heterogeneous structure) within the 3D printed constructs to enable geometric transformations.[27] Due to these demanding criteria, the development of single-component 4D bioinks with straightforward fabrication, defined composition, and the ability to successfully incorporate and maintain living cells remains a significant challenge.
Among hydrogel-based materials, microgels, such as granular hydrogels, have been widely employed as building blocks for hydrogel construction.[28, 29] Compared to bulk hydrogels, microgels possess numerous unique properties that make them attractive as printable materials. For example, since the size of microgels is small (1~1000 μm), they can be easily extruded through a needle. Moreover, physical interactions between the particles can result in shear-thinning during extrusion.[30]-[31] Nonetheless, to maintain the printed structure of microgels, a supporting bath[31] or additional engineering of inter-particle interactions is often needed.[30] Granular microgels are specially processed into a “jammed state” under sufficient conditions of stress and temperature to improve the printability.[32] Microgels in the jammed state are densely packed and move along with the adjacent microgels,[33] leading to a solid-like mass while solid-to-liquid phase transition can be achieved by external stress. Solid granular microgels with a spherical morphology can be packed to reach a theoretical maximum particle-volume fraction (ϕ) of 0.64, which is known as “random close packing”.[34] Even though these closely packed microgels can maintain simple bulk geometries after printing if left unperturbed, the particles can be readily reorganized and displaced by a minimal external force.[28] Therefore, a higher ϕ is desirable to allow microgels to pack more closely and enhance the stability of 3D printed structures. It is possible that jammed microgels with non-spherical shapes, heterogeneous sizes, and large deformability contribute to strong interparticular interfacial friction and can boost the packing density to reach a ϕ higher than 0.74.[30, 35] Hence, we conjecture that jammed microgels with non-spherical and non-uniform morphology could provide better bioprinted construct shape stability. Although microgels have been used as cell-laden bioinks for 3D bioprinting,[36] the use of microgels for 4D bioprinting has not been investigated.
In this study, a jammed heterogeneous single-component micro-flake hydrogel (MFH) system consisting of only ionically crosslinked oxidized and methacrylate alginate (OMA) hydrogels was developed as a cell-laden bioink for 4D living cell bioprinting. This MFH can be easily printed into a stable 3D (bio)construct and can be further crosslinked to form a more robust hydrogel construct with a crosslinking gradient within the hydrogel when a photoinitiator (PI) and a UV absorber are incorporated. The crosslinking gradient of the MFHs in the printed 3D (bio)constructs permits controlled morphing into defined geometries after being cultured in cell culture media (Scheme 1). With this system, bioconstructs with complex structures and high cell viability were obtained through a shape transformation of the 3D printed counterparts. Finally, 4D cartilage-like tissue, as a proof-of-concept, was engineered to demonstrate the potential of the MFH in the field of tissue engineering and regenerative medicine. The combination of gradient formation with printable jammed microgels to enable 4D bioprinting is new and unique. To the best of our knowledge, this is the first example of a jammed microgel system as the cell-laden bioink for 4D living cell bioprinting.
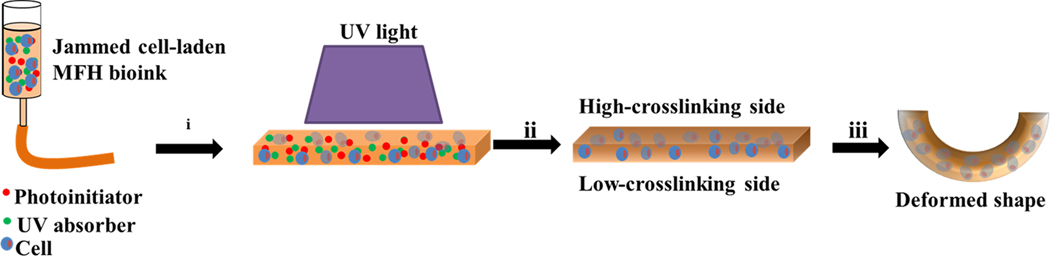
Scheme 1.
Scheme of the PI and UV absorber incorporated 4D MFH bioprinting: i) printing the jammed cell-laden MFH bioinks into a bioconstruct, ii) UV crosslinking to generate a crosslinking gradient within the 3D printed bioconstruct, iii) culturing in media to drive shape morphing.
2. Results and discussion
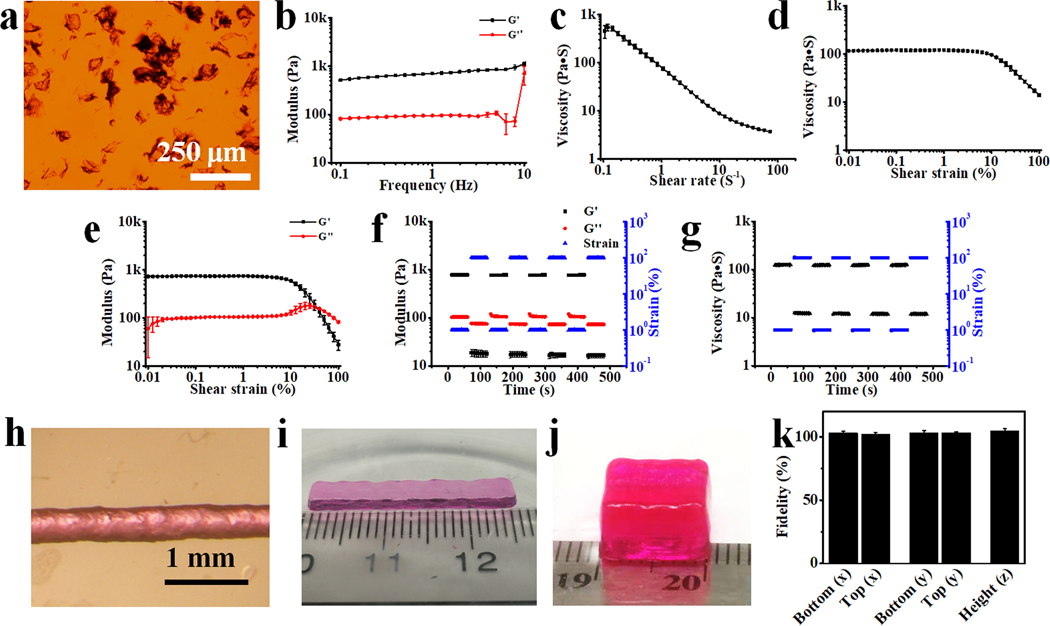
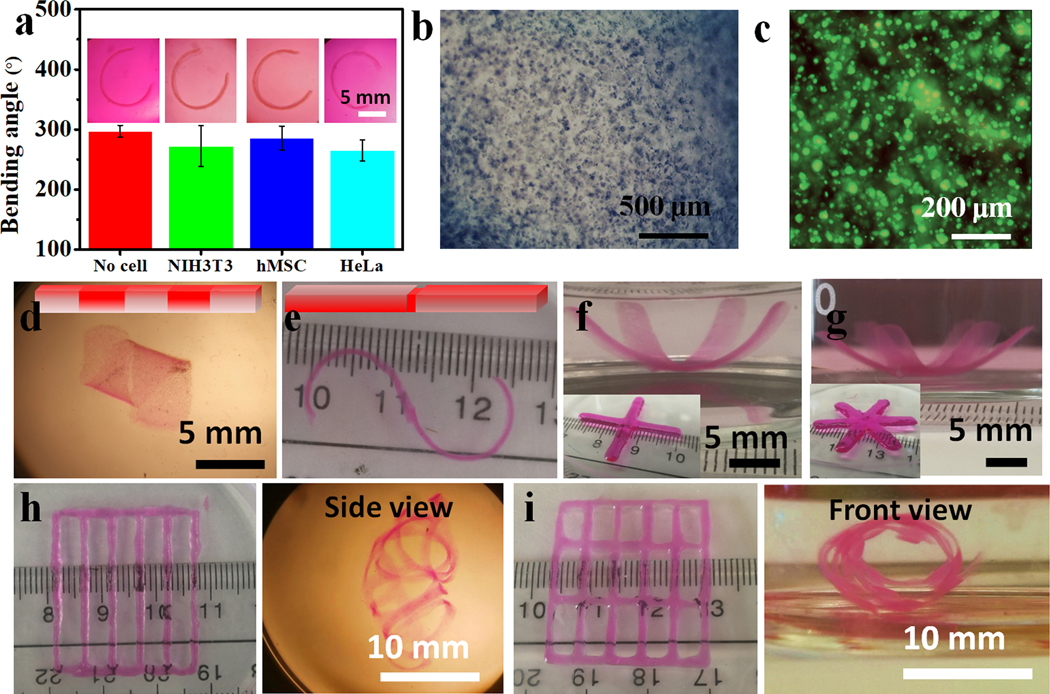
OMA with theoretical oxidation and methacrylation of 1% and 30%, respectively, were synthesized and fully crosslinked with calcium ion (Ca2+) to form ionically crosslinked hydrogels by dropping the OMA solution (2%) into a Ca2+ solution (0.2 M), and then MFH precursors were fabricated by simple blending using a household blender (Supporting Information).[37] The as-prepared MFH precursors could be stably stored in 70% ethanol at −20 °C for few months. The MFH precursors turned into the jammed state with a flake morphology (41.7 ± 19.8 μm, Figure 1a and S3) after reconstitution by washing with the PI- and UV absorber-containing media, most likely due to the volume expansion during reconstitution (Figure S2). These jammed microgels exhibited solid-like behavior at low shear strain (Figure 1b). Once the MFHs receive an increasing shear rate (Figure 1c) or a shear strain > 10% (Figure 1d and 1e), they displayed typical shear-thinning and shear-yielding behaviors. Importantly, the MFHs underwent rapid and repeatable phase transitions upon receiving alternating shear strains between 1% and 100%, demonstrating its capacity for rapid self-healing (Figure 1f and 1g). The rheological behaviors of this newly developed MFH agree with our previous findings with different OMA microgel systems.[31, 37] Possessing these properties, the MFHs could be readily extruded evenly through a needle (Figure S4), form a uniform filament with high printing accuracy (Figure 1h, 117% of the inner diameter of the needle), and be printed into stable freeform 3D constructs with various shapes (Figure 1i, 1j, and Movie S1) with high fidelity (Figure 1k) and stability (Movie S2).
Figure 1.
(a) Photomicrograph of safranin O stained MFHs. (b) Schematics showing lower packing density of granular microgels (upper) and more highly packed irregular MFHs (bottom). (c) Storage (G’) and loss (G”) moduli of MFHs as a function of frequency. Material viscosity decreases while continuously increasing (d) shear rate and (e) shear strain over 10% strain. (f) Crossover of G’ and G” with increasing shear strain indicative of shear yielding. Rapid recovery of MFHs’ (g) modulus and (h) viscosity by alternating the applied strain between 1% and 100%. (i) Photomicrograph of a filament printed through a 22-gauge (22G) needle (inner diameter 413 μm). Photographs of 3D printed (j) hydrogel bar (25 × 4 × 1 mm3) and (k) hydrogel cuboid (10 × 8 × 6 mm3). (l) Fidelity of the as-printed 3D construct in j before UV crosslinking. Data are presented as mean ±standard deviation (± SD), N = 3.
Due to the presence of the methacrylate groups, the microgels can be further stabilized by photocrosslinking under UV light in the presence of a PI (Figure S5). The incorporation of a UV absorber results in the generation of a light attenuation pathway within the hydrogel and subsequent formation of a gradient in the crosslinking density (structural anisotropy) (Scheme 1), which has been demonstrated in our previous work.[38] As a result, crosslinked bulk gradient MFH constructs showed significantly lower elastic modulus than crosslinked non-gradient MFH constructs (Figure S5). This novel post-printing anisotropization approach to generate structural heterogeneity within a 3D printed construct differs from the widely adopted synchronous-programming approach in the current 4D printing field, by which the structural heterogeneity is generated during printing,[39] thus making it a more facile and more flexible approach to design a 3D printable (bio)ink for formation of 4D constructs. The printed construct is then able to morph into a predefined shape after culturing in media.
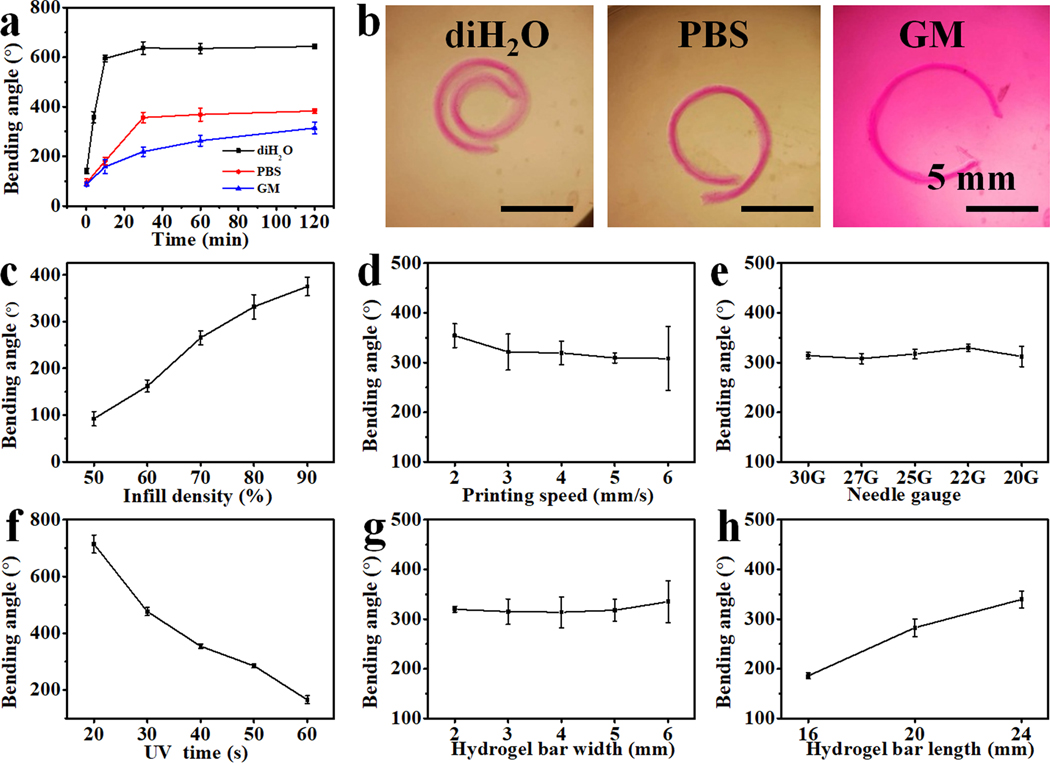
To study the shape-morphing behaviors of the 4D constructs, hydrogel bars with a gradient crosslinking density throughout their thickness were used as prototypes. Unless specified, hydrogel bars with dimensions of 24 × 4 × 0.6 mm3 were printed at 80% infill density and 4 mm/s printing speed using a 22G needle. Note that the infill density herein correlates to extruded material volume in a construct and not the spacing between filaments, as printed filaments would merge under this infill density. The resulting deformations, which were quantified by bending angles as described in the supporting information (Figure S6), depend on the structure dimensions, printing parameters, UV crosslinking time, as well as the incubation media. As expected, the hydrogel bars bent to the high-crosslinked side, forming a closed or open hydrogel ring, in the three types of media (i.e., deionized water (diH2O, Movie S3), PBS (pH 7.4), and cell growth media (GM)) (Figure 2a and 2b). The hydrogel bars in diH2O exhibited much faster bending kinetics and much larger bending angles than those in PBS and GM, and hydrogel bars in PBS showed slightly higher bending kinetics and angles compared to GM. The distinct variations in bending angles are caused by the swelling differences of the hydrogel bars in the respective medias; that is, a higher swelling ratio, S, led to a larger bending angle (SdiH2O > SPBS > SGM, Figure S7). For those printing parameters determined herein (Figure 2c, 2d, and 2e), only the infill density obviously influenced the bending angles, while the printing speed and needle gauge imposed negligible effects. Higher filling density brought about larger crosslinking-gradient range due to increased light attenuation, resulting in greater bending.[38] Varying UV irradiation time resulted in a dramatic impact on the bending of the hydrogel bars (Figure 2f). The bending angle decreased along with the increase of the irradiation time as a higher light dose reduces the crosslinking gradient range throughout the thickness of the hydrogel bar.[38] Additionally, the bending angle increased with the length of the hydrogel bar but did not change with the width (Figure 2g and 2h), which aligned with our previous investigations.[38] The bending behaviors of these crosslinked printed hydrogel bars under different conditions can be explained by Timoshenko’s theory, a thermal expansion bilayer beam model describing the bending of a bilayer based on mismatched strain in the two layers,[40] which is also widely employed as a empirical theory to interpret the bending behaviors of bilayer hydrogel systems.[41] According to equation (1) presented in experimental section, if we approximately view the gradient hydrogel as a bilayer with a high crosslinking layer and a low crosslinking layer, the resultant bending angle (θ) is proportional to the mismatch of expansion strain (Δε), which is attributable to differential hydrogel swelling resulting from the differences in microgel crosslinking density (Dc). Thus, it is understood that those parameters contributing to a larger Δε, such as enhanced swelling media, increased infill density, and lowered overall crosslinking, could effectively increase the bending angle. However, Timoshenko’s theory cannot be used to explain the impact of the length on the bending angle because it assumes bending curvature is independent to beam length.[42] Interestingly, it was observed that increasing in aspect ratio (length/width) of samples with fixed width could result in larger bending (Figure 2h), which is consistent with the results in reported literature,[43] while the change in aspect ratio in samples with varying width but fixed length did not give rise to observable changes in the bending angle (Figure 2g). The length of the hydrogel bar is much longer than the width, thereby those hydrogel bars tend to bend perpendicularly with the longitudinal axis to reach a thermodynamically stable state. Since the strain only varies in the radical direction (er) but keeps relatively constant in the tangential direction (eθ) (Figure S7), the bending curvature κ does not depend on the aspect ratio.[44] According to equation (2) in experimental section, the bending angle only correlates with hydrogel length (L) and curvature (κ). That is the reason why length change rather than width change influences the bending angle in these systems.[45] These results indicate that deformation extent can be readily adjusted by tuning the printing parameters and/or hydrogel dimensions. Thus, by varying these parameters, we may obtain a pre-programmable tailored deformation.
Figure 2.
4D shape-morphing behaviors of hydrogel bars in different incubation solutions or fabricated with different parameters. (a) Hydrogel bending angle kinetics in diH2O, PBS (pH 7.4), and GM at room temperature. (b) Photomicrographs of deformed hydrogel bars in diH2O, PBS (pH 7.4), and GM after swelling for 2 h. Effects of (c) infill density, (d) printing speed, (e) needle gauge, (f) UV irradiation time, (g) hydrogel bar width, and (h) hydrogel bar length on the bending angles of hydrogel bars cultured in PBS (pH 7.4) for 2 h at room temperature. UV absorber: 0.02% 4’-hydroxy-3’-methylacetophenone (HMAP), 0.005% methacryloxyethyl thiocarbamoyl rhodamine B (RhB) was incorporated to impart the hydrogel with red color for better clarity. Data are presented as mean ± SD, N = 3.
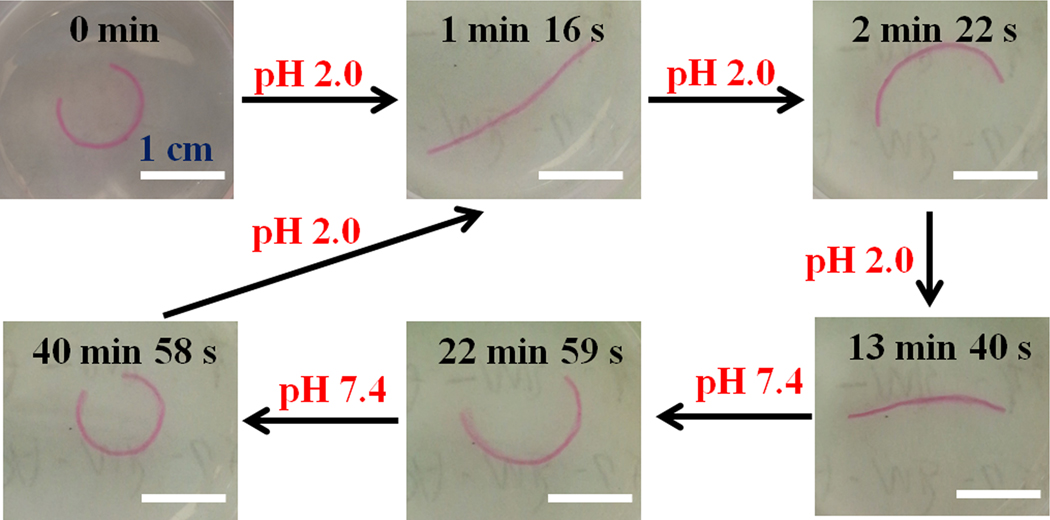
In addition to programmable deformation, external stimulation can be applied to further manipulate the shape of a printed construct on demand. To demonstrate the capacity to control shape changes of the MFH-based constructs by external stimuli, a gradient hydrogel bar was first incubated in PBS (pH 7.4) to form a curve (Figure 3, 0 min) and then subsequently transferred to another PBS solution with a different pH to record the shape changes over time (Figure 3, Movie S4 and S5). The hydrogel bar rapidly stretched after incubation at a low pH of 2.0 for 1 min 16 s and bent to form a backward-facing curve upon further treatment to 2 min 22 s. Interestingly, this curve re-stretched and contracted at a much slower rate until reaching an equilibrium state at 13 min 40 s. After switching the pH back to 7.4, the hydrogel bar reverted to the initial state in a much slower manner (from 13 min 40 s to 40 min 58 s). This may be explained by the differential swelling properties of alginate hydrogel in solutions of different pH. Alginate is a polyelectrolyte that contains both weak acidic and weak basic groups on the polymer chains and these groups can respond to the environmental pH via protonation or deprotonation, leading to a volume change in the way of swelling/shrinkage.[46] For instance, the MFH hydrogels exhibited much smaller swelling ratios at pH 2.0 than at pH 7.4 (Figure S7). In the initial state (0 min, Figure 3), the outer side (low-crosslinking side) of the “unclosed” ring is the high-swelling side and has larger pore sizes than the inner side.[47] Therefore, the protons in the solution surrounding the hydrogels diffuse into this side at a faster speed.[48] Thus, the outer side shrank faster than the inner side (high-crosslinking side) due to the better access to the carboxyl groups on the outer side. As a result, the hydrogel bar rapidly stretched and bent to the opposite direction in the first 2 min 22s. However, since the available carboxyl groups, the reactive moiety receiving the protons, are homogeneously distributed inside the hydrogel, the inverted hydrogel curve at 2 min 22s re-stretched over time, and by 13 min 40s was stabilized as a straightened hydrogel at pH 2.0, showing no further shape change. After altering the pH back to 7.4, the straight hydrogel bar in the shrunken phase releases the bound protons to the surrounding solution in a much slower manner due to the smaller pores compared to the hydrogel bar in the fully swelled state at pH 7.4, thereby exhibiting a much slower shape recovery process. This is the first example of a single-component and single-layer hydrogel showing multiple shape transitions with only pH stimulation, and this multiple-shape transition can be realized in multiple cycles without showing signs of fatigue (Figure S9). Although the low pH (2.0) is not applicable for live cell culture, the reversible shape conversion suggests the potential to use this 4D printed hydrogel as an environment-controlled actuator/robot in some specific conditions, such as the gastric environment (pH 1.5~3.5).
Figure 3.
Video snapshots showing the shape changes of a hydrogel bar in response to different pH treatments at room temperature.
Hydrogels fabricated with covalently crosslinked and/or ionically crosslinked OMA have been extensively used as cell scaffolding materials for tissue engineering.[49] The HMAP is a highly efficient and cytocompatible UV absorber (Figure S10) for crosslinking gradient generation.[38] To demonstrate the feasibility to use the jammed MFHs as cell-laden bioinks for 4D living cell bioprinting, three types of cells were examined: a fibroblast cell line (NIH3T3), a cancer cell line (HeLa), and primary stem cells (human bone marrow-derived mesenchymal stem cells, hMSCs). These cells were individually mixed with the PI- and UV absorber-containing MFHs (5 × 106 cells/mL MFH, Supporting Information) and printed into hydrogel bars as described earlier, which were then cultured in GM to investigate the resulting shape changes. The printed cell-embedded hydrogel bars showed comparable bending with the cell-free counterparts in all cases (Figure 4a), and the encapsulated cells (Figure 4b, S11i and S11ii) remained highly viable after 24 h culture (Figure 4c, S11iii~S11vi). The results indicate this 4D system is highly compatible for inclusion of live cells and may be useful for fabricating other bioconstructs with more sophisticated geometries.
Figure 4.
(a) Bending behaviors of MFH gradient hydrogel bars with and without embedded cells. Insets show representative photomicrographs of the bent hydrogel bars. (b) Photomicrograph of a NIH3T3-laden MFH-based construct. (c) Representative live/dead image of NIH3T3 fibroblasts in the gradient hydrogel bar. (d) Photomicrograph of a cell-laden “biohelix” structure. (e) Photograph of a cell-laden “bioS” structure. Photographs of (f) a cell-laden “pseudo-four-petal” and (g) a cell-laden “pseudo-six-petal” flower. Inset photographs show the corresponding as-printed structures. Kirigami-based structures and the deformed configurations: hydrogels in bar-grid patterns (h) without and (i) with inner horizontal bars. For cell-laden gradient hydrogel bars (insets in a), only 0.02% HMAP was used as the UV absorber, they were cultured in GM overnight at 37 °C, and then imaged and live/dead stained. For complex cell-laden hydrogel configuration formation, a mixture of 0.02% HMAP and 0.005% RhB UV absorbers was used. These bioconstructs were obtained after culturing in GM at 37 °C overnight, and images were immediately taken after replacing the GM with FBS for clarity. Data are presented as mean ± SD, N = 3.
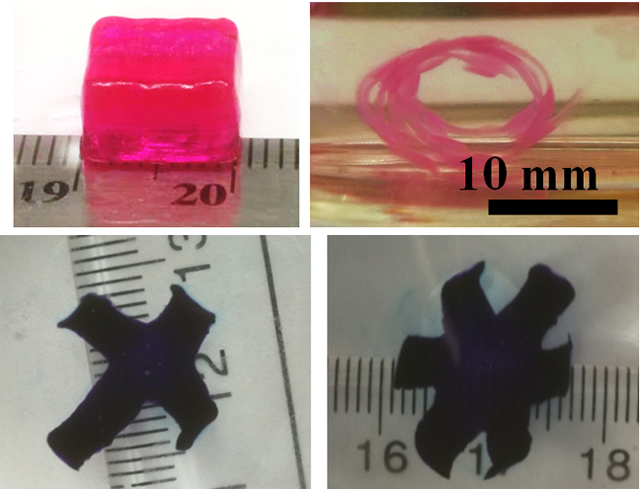
Inspired by this, we sought to fabricate various bioconstructs with more complex geometries by integrating gradient formation with a mask-based photolithography or intricate bioprinted geometric designs. With masked-based photolithography, we locally photocrosslinked the cell-laden hydrogel bar using photolithography and cultured the hydrogel bar in GM at 37 °C to elicit more complex shape transformations beyond unidirectional bending. For example, a pre-formed gradient hydrogel bar with a further discrete local photocrosslinking (schematic in Figure 4d, Figure S12) or an as-printed hydrogel bar subsequently photocrosslinked with two separate gradients in opposite directions (schematic in Figure 4e, Figure S13) turned into a “biohelix” (Figure 4d) or a “bioS” structure (Figure 4e), respectively, after deformation. Alternatively, by printing MFH bioinks into specific geometries, cell-laden hydrogels with more complex structures can be obtained. Printed multi-arm gradient hydrogels morphed into “pseudo-four petal” and “pseudo-six petal” flowers (Figures 4f, S13a, 4g, and S14b). When the cell-laden MFH bioinks were printed into specific “kirigami-based” structures[50] displaying bar-grid patterns, the bioconstruct with no inner horizontal bars self-curled into a curved cage that crudely resembles the human rib cage (Figure 4h and S14c), and the construct with inner horizontal bars self-curled into a “net tube” (Figure 4i and S14d). It is worth mentioning that these 4D-engineered bioconstructs are very robust and can maintain their geometry even when exposed to strong agitation, with manipulation of the “net tube” presented as an example (Movie S6).
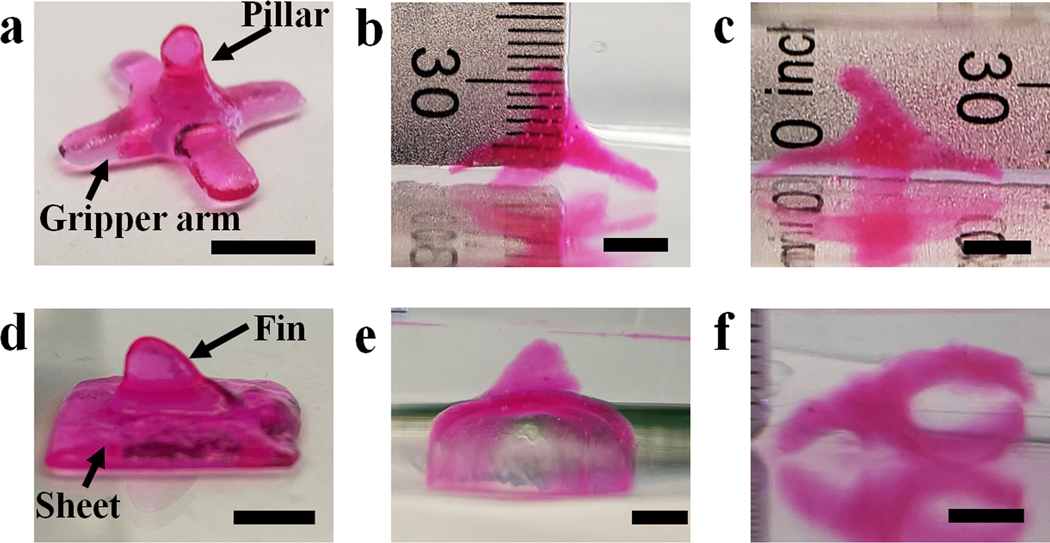
Currently, all previous 4D bioprinting work only presents shape transformation from 2D and/or 2.5D (2D structure with a certain addition in the z-direction) to 3D.[51] To the best of our knowledge, 3D-to-3D shape morphing of cytocompatible biomaterials with encapsulated cells, enabled by 4D bioprinting or any other means, has not been reported. 3D-to-3D morphing is particularly challenging for hydrogel materials due to the difficulty in obtaining a stable printed 3D structure with effective structural anisotropy incorporation. Since our system allows 3D printing and independent anisotropy generation, it is possible to achieve 3D-to-3D transformations of constructs fabricated in a single print in a controllable manner. 3D architectures such as a “pillar gripper” (Figure 5a) and a “shark-fin sheet” (figure 5d) were readily printed. Multiple location-specific crosslinking gradients in the two representative 3D constructs were then created by controlling the photoirradiation direction and the regions exposed to light. For example, gradients in the bases of the gripper and the sheet were created by applying UV irradiation from the bottom of the constructs, while the gradients in the pillar and shark-fin were created by applying UV irradiation from the side of the pillar and shark-fin (Figure S15). With this unique structural anisotropy, complex 3D-to-3D shape morphing with controlled location-specific deformations was then achieved (Figure 5b, 5c, 5e, and 5f). With advanced designs, 3D constructs with more sophisticated structures, such as a “double shark-fin” sheet (Figure S16a) and a “double pillar gripper” (Figure S16c) both presenting a crosslinking gradients from the inner sides (low-crosslinking sides) to the outer sides (high-crosslinking sides) within the “fins” or “pillars” and a separate different crosslinking gradient from bottom to top within the “sheet” or “gripper”, demonstrate the capacity for more complex shape morphing (Figure S16b, S16d, and S16e). These 3D-to-3D shape transformations suggest the feasibility and reliability of this system for developing more transformable 3D structures due to excellent printability and ease of anisotropy incorporation.
Figure 5.
Demonstration of 3D-to-3D shape morphing. A pillar gripper (a) before being subject to deformation, after being submerged in media (b) less than 10 s and (c) for 60 s. A shark-fin sheet: (d) as-printed shape, (e) front-view image of deformed shape, and (f) image of deformed shape with construct on its side. Scale bars indicate 5 mm.
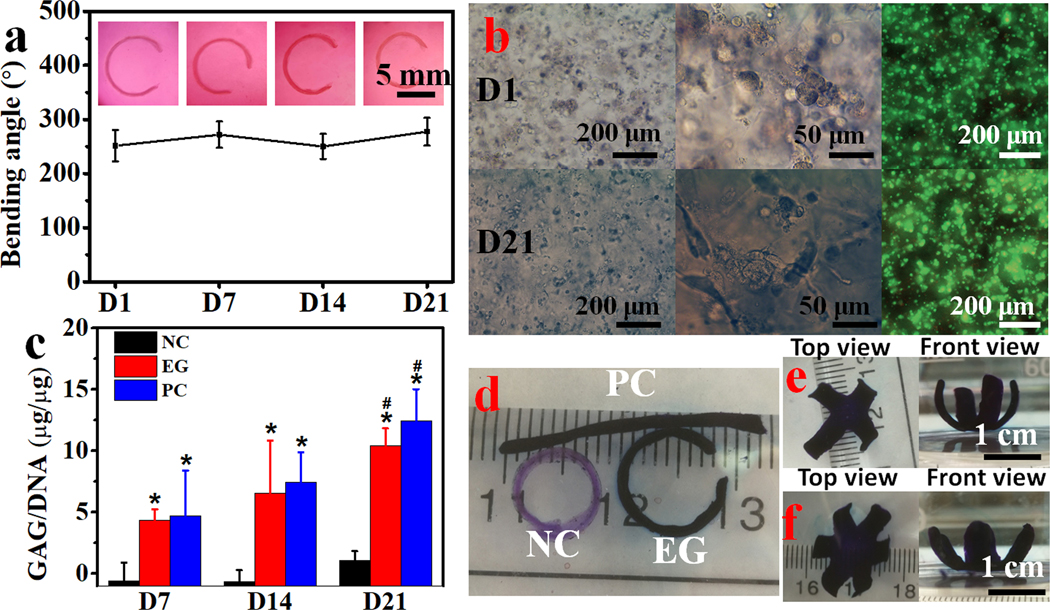
To utilize this type of shape morphing strategy for tissue engineering applications, it is important that the 4D cell-laden constructs enable and/or drive encapsulated cell differentiation and formation and maturation of new tissue.[52–54] The 4D bioprinting system reported here enables fabrication of architecturally complex bioconstructs while at the same time facilitating the engineering of functional tissues. Since the hMSC is a multipotent stem cell with the capacity to differentiate down multiple connective tissue lineages when provided with appropriate environmental cues, it is a promising cell source for engineering tissues such as cartilage, bone and fat.[55] Hence, we cultured 4D bioprinted hMSC-incorporated MFHs in chondrogenic media (CM) to induce the formation of cartilage-like tissue with relatively predefined final configurations. The chondrogenesis of the 4D hMSC-laden hydrogel bars along with their shape changes was continuously monitored over a course of 21 days. The initially straight gradient hydrogel bars bent into “C” shapes in CM within 2 h and the shapes of the bent hydrogel bars changed very little during the course of chondrogenesis (Figure 6a), suggesting good stability of the 4D bioconstructs. Meanwhile, the morphologically round cells on day 1 (D1) maintained a predominantly round morphology and high cell viability after 21 days of culture (Figure 5b, left and right panels). However, elongation of some of the cells that resided near the hydrogel surface was observed on D21 (Figure 6b, middle panel). To quantify the chondrogenesis, levels of DNA and the primary cartilage extracellular matrix component, glycosaminoglycan (GAG), were analyzed. The DNA content manifested a relatively constant level over time, with no significant difference found when comparing the experimental groups (EG, 4D bioprinted hydrogel bars cultured in CM) to the negative control (NC, 4D bioprinted hydrogel bars cultured in GM) and positive control (PC, 3D bioprinted hydrogel bar photocrosslinked in the absence of UV absorber cultured in CM) (Figure S17). In contrast, GAG production steadily increased during the 21 days of culture and was similar to the PC group but significantly higher than the NC group (Figure 6c and S18). This difference in GAG production was corroborated by the intense positive toluidine blue O (TBO) staining only in EG and PC samples (Figure 6d). The results imply that the structural self-remodeling is independent of cell differentiation and tissue maturation, and vice versa. Therefore, this decoupling of shape morphing with tissue maturation enables flexible 4D design for 4D tissue engineering. With respect to the formation of 4D cartilage-like tissues with more complex shapes, four- and six-petal flower-shaped cartilage-like tissues were also fabricated (Figure 6e and 6f). Results from this study demonstrate that this jammed MFH system satisfies two critically important criteria for its use in 4D bioprinting for tissue engineering: i) controlled shape morphing capacity and ii) support of new tissue formation by incorporated cells.
Figure 6.
MFH based 4D bioprinting for application in tissue engineering. (a) The bending angles of hydrogel bars in EG and the corresponding photomicrographs depicting the shape changes of 4D bioprinted cell-laden hydrogel bars in CM over time. (b) Photomicrographs depicting cell morphology and distribution and live/dead stained cells within the EG hydrogel bars in CM on D1 and D21. (c) Biochemical quantification of GAG production normalized to DNA, *p < 0.05 compared to NC at the same time point, #p < 0.05 compared to D7 within a group. TBO stained (d) hydrogel bars and (e) four- and (f) six-petal flower-shaped hydrogels after chondrogenesis for 21 days in CM. NC: negative control, 4D bioprinted cell-laden hydrogel bars cultured in GM; EG: experimental group, 4D bioprinted cell-laden hydrogel bars cultured in CM; PC: positive control, 3D bioprinted (without incorporation of UV absorber) cell-laden hydrogel bars cultured in CM. Data are presented as mean ± SD, N = 3.
4D bioprinting opens new avenues to fabricate cell-laden constructs with dynamic shape morphing capabilities and complex configurations, which are beyond the capacity of conventional 3D bioprinting.[20, 53, 54] The bioprinted constructs can be further induced to form specific tissues when culturing in an appropriate environment. Thus, this newly emerging technology also enables morphodynamical tissue engineering, bearing the potential to biomimic the conformational evolutions occurring during tissue development and healing.[38, 53, 54] So far, a few reports have prepared shape-morphing constructs through 3D printing to fabricate scaffolds with sophisticated structures and investigate and/or modulate cell behaviors, such as proliferation,[56] alignment,[57] and differentiation.[58] However, the cells involved in those studies were only seeded on the post-printed scaffolds rather than encapsulated within the constructs due to the lack of or limited cytocompatibility of materials, harsh printing process, and/or extreme stimulation conditions applied to induce shape change. Hence, those studies could not meet the critical requirement of loading and maintaining viable cells within 4D bioprinted constructs.[21] Although live-cell 4D bioprinting was realized using alginate-, silk-, and gelatin-derived cytocompatible materials,[59, 60] limitations were still present in terms of either shape morphing or capacity to use the materials as cell-laden bioinks for direct ink writing (DIW). For example, the cell-free parts rather than the cell-laden parts in the constructs were the active parts driving the morphing of the constructs,[59] or the inks were just simple macromer solutions lacking appropriate rheological properties for deposition to form stable freestanding 3D objects.[60]
Here, we report the first 4D DIW bioprinting system enabling live cell encapsulation using microgels and cells as bioinks. The single-component jammed MFHs by themselves, without the need of other filler components or a slurry support bath, are rheologically favorable for excellent printability (Figure 1). Specifically, anisotropy (gradient crosslinking) was generated using a post-printing anisotropization approach, which liberates the anisotropy generation from printing. With this approach, the anisotropy formation is tunable, enabling facile user-adjustable shape morphing (Figure 2). In addition, anisotropy can be incorporated in multiple ways within a single construct to produce complex 3D geometries (Figure 4d and 4e). With this system, 4D tissue maturation was also demonstrated using hMSCs and MFHs in a proof-of-concept 4D cartilage regeneration study (Figure 5). Cartilage-like tissues with complex geometries, such as “C” shape and four-/six-petal flowers were engineered. It is noteworthy that curved, bent, folded, and rolled structures often emerge in tissue morphogenesis during processes such as development of gut villi and mammary epithelial acini. Our results suggest important progress for 4D live-cell bioprinting, which would benefit morphodynamic tissue engineering.
3. Conclusion
In this study, a new single-component jammed MFH system with heterogeneous size distribution has been developed as a cell-laden bioink for 4D living cell bioprinting. This new bioink showed desirable shear-thinning, shear-yielding, and rapid self-healing properties, and was directly deposited into various 3D bioconstructs with high resolution and high fidelity in the absence of a support bath. 4D shape changes were achieved under physiological conditions and high cell viability was maintained after an effective generation of a crosslinking gradient within the hydrogels using a specific post-printing anisotropization method. In addition, it was also demonstrated that multiple-shape transformations (multiple bending and stretching cycles) could be elicited by a singular, although non-biocompatible, stimulation (low pH) in this single-component and single-layer system, which has not been reported in other existing systems. By utilizing this bioink, shape morphing cell-laden bioconstructs with well-defined configurations were fabricated by combining photomask-based photolithography and/or intricate geometric designs. Ultimately, proof-of-concept 4D cartilage-like tissue formation was demonstrated in curved hydrogel bars and folded four- and six-petal flowers. We anticipate this unique 4D bioprinting system will have promising applications in 4D tissue and organ engineering and potentially aid in the study of developmental processes.
4. Experimental Section
MFH preparation:
To make the stock MFHs, O1M30A (1.2 g) was dissolved in diH2O (60 mL) and then slowly dispensed (approximately 20–30 mL/min) into a gelling bath containing an aqueous solution of CaCl2 (600 mL, 0.2 M) under fast stirring with a magnetic stir bar. After being fully ionically crosslinked overnight, the resultant O1M30A beads were collected, washed with 40 mL of 70% ethanol (EtOH)/water (H2O) once, and then blended using a household blender (Osterizer MFG, at “pulse” speed) for 2 min with 120 mL of 70% EtOH/H2O. Then, the OMA microgels were loaded into 50 mL conical tubes and centrifuged at 2000×g (Sorvall ST40R centrifuge, ThermoScientific, Waltham, MA) for 5 min and stored in 70% EtOH at 4 °C for future use.
To make the jammed MFHs, the as-prepared microgels above (5 mL) were washed 3 times by replacing the previous media with 25 mL of diH2O containing PI (0.05% w/v) and UV absorber (0.02% HMAP or 0.02% HMAP/0.005% RhB w/v), while vortexing (Fisher Scientific, 10× speed) for 2 min between washes, and then washed 2 times with 25 mL of DMEM-LG containing PI and UV absorber while vortexing (10× speed) for 1 min each time between washes.
To evaluate the morphology and measure the size of the jammed MFHs, 100 μL of MFHs were stained with 1 mL of 0.1% safranin O solution for 2 h. After staining, the MFH mixture was vortexed for 10 s to disperse homogeneously in the media, and then 200 μL of the stained samples were added into 3 mL of PBS (pH 7.4) and imaged using a fluorescence microscope equipped with a 14MP APTNA Color CMOS Microscope Camera (AmScope, Irvine, CA) under a bright field. The average diameter of the MFHs was determined by measuring 86 microgels in a representative image (Figure 1a) using ImageJ.
Rheological properties of the MFHs:
Dynamic rheological examination of the MFHs was performed to evaluate shear-thinning, shear yielding, and self-healing properties with a Kinexus ultra+ rheometer (Malvern Panalytical). In oscillatory mode, a parallel plate (8 mm diameter) geometry measuring system was employed, and the gap was set to 1 mm. MFHs were placed between the plates. All the tests were carried out at 25 °C. Oscillatory frequency sweep (0.1~100 Hz at 1% strain) tests were performed to measure storage moduli (G’), loss moduli (G”), and viscosity. Oscillatory strain sweep (0.01~100% strain at 1 Hz) tests were performed to show the shear-thinning characteristics of the MFHs and to determine the shear-yielding points at which the jammed MFHs behave fluid-like. To demonstrate the self-healing properties, cyclic deformation tests were performed at 100% strain with recovery at 1% strain, each for 1 min at 1 Hz.
4D bioprinting:
The printing of the cell-free and cell-laden bioinks was performed using a 3D printer (PrintrBot Simple Metal 3D Printer, Printrbot) modified with a syringe-based extruder. More information about this printer can be found in the literature.[31, 61] The STL files for printing were generated from www.tinkercad.com under open license.
To load the cells into the bioink, MFHs and cells (5 × 106 cells/mL bioink) were separately loaded into two 3 mL syringes. After the two syringes were connected with a female-female luer lock coupler (Value Plastics), the MFHs and cells were thoroughly mixed, and this cell-laden bioink was ready to use.
The cell-free and cell-laden bioinks were separately loaded into 1 mL glass syringes (Hamilton, Reno, NV), which were connected to a stainless-steel needle (McMaster-Carr, Elmhurst, IL) and mounted into the syringe pump extruder on the 3D printer. A petri dish was placed on the building platform. The tip of the needle was positioned at the center and near the bottom of the dish, and the print instructions were sent to the printer using the host software (Cura Software, Ultimaker, Geldermalsen, the Netherlands), which is an open-source 3D printer host software. After 3D printing of the bioinks, the resulting constructs were immediately photocured under UV (EXFO OmnicureR S1000–1B, Lumen Dynamics Group, Ontario, Canada) at 12 mW/cm2. Then the cell-free or cell-laden constructs were carefully transferred into the wells of 6-well tissue culture plates with 8 mL of media and further cultured to record shape changes. The hydrogels were imaged, and the bending angles were quantified according to the previous literature.[54] Briefly, as shown in Figure S6, a circle was drawn to match well with the shape of the bent hydrogel curve. The bending angle (θ) is defined as the central angle generated by drawing two lines between the endpoints of the hydrogel curve and the circle center, respectively. Unless otherwise specified, MFH bioinks were printed using a 22G needle under 4 mm/s printing speed and 80% infill density and subsequently photocured for 40 s.
The resultant bending angle (θ) can be described using the equation below:
| (1) |
where κ denotes the resultant curvature of the bent hydrogel bar, h denotes the total thickness of the hydrogel, Δε denotes the mismatch of expansion strains between the two layers, and m and n denote the thickness ratio and the modulus ratio of the two layers, respectively.
To describe the relationship between the bending angle and the curvature (κ). κ is defined as 1/r, where r is the radius. Therefore, the length of a hydrogel (L) arc can be described as L = θπ/180 κ, and the bending angle can be expressed as:
| (2) |
Data presentation and statistical analysis:
All quantitative data was expressed as mean ± standard deviation. Statistical analysis was performed with one-way analysis of variance (ANOVA) with Tukey honestly significant difference post hoc tests using Origin software (OriginLab Corporation, Northampton, MA). A value of p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health’s National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01AR069564, and R01AR066193) and National Institute of Biomedical Imaging and Bioengineering (R01EB023907). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The authors also thank Susan R. Ross at the University of Illinois at Chicago for generously providing the NIH3T3 cells used in this study.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Aixiang Ding, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
Oju Jeon, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
David Cleveland, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
Kaelyn Gasvoda, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
Derrick Wells, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
Sang Jin Lee, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA..
Eben Alsberg, Richard and Loan Hill Department of Biomedical Engineering, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA.; Departments of Mechanical & Industrial Engineering, Orthopaedics, and Pharmacology, University of Illinois at Chicago, 909 S. Wolcott Ave., Chicago, IL 60612, USA.
References
- [1].Tibbits S, Archit. Des. 2014, 84, 116. [Google Scholar]
- [2].Momeni F, Mehdi Hassani SM.Liu N,X, Ni J, Mater. Des. 2017, 122, 42. [Google Scholar]
- [3].Gladman AS, Matsumoto EA, Nuzzo RG, Mahadevan L, Lewis JA, Nat. Mater. 2016, 15, 413. [DOI] [PubMed] [Google Scholar]
- [4].Bakarich SE, Gorkin Iii R, Panhuis MIH, Spinks GM, Macromol. Rapid Commun. 2015, 36, 1211. [DOI] [PubMed] [Google Scholar]
- [5].del Barrio J, Sánchez-Somolinos C, Adv. Opt. Mater. 2019, 7, 1900598. [Google Scholar]
- [6].Le Duigou A, Chabaud G, Scarpa F, Castro M, Adv. Func. Mater. 2019, 29, 1903280. [Google Scholar]
- [7].Zhang F, Wang L, Zheng Z, Liu Y, Leng J, Composites, Part A 2019, 125, 105571. [Google Scholar]
- [8].González-Henríquez CM, Sarabia-Vallejos MA, Rodriguez-Hernandez J, Prog. Polym. Sci. 2019, 94, 57. [Google Scholar]
- [9].Li Y, Zhang F, Liu Y, Leng J, Sci. Chin. Technol. Sci. 2020, 63, 545. [Google Scholar]
- [10].Zhang Z, Corrigan N, Bagheri A, Jin J, Boyer C, Angew. Chem. Int. Ed. 2019, 58, 17954. [DOI] [PubMed] [Google Scholar]
- [11].Lu HZ, Yang C, Luo X, Ma HW, Song B, Li YY, Zhang LC, Mater. Sci. Eng. A 2019, 763, 138166. [Google Scholar]
- [12].Liu G, Zhao Y, Wu G, Lu J, Sci. Adv. 2018, 4, eaat0641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ren L, Li B, He Y, Song Z, Zhou X, Liu Q, Ren L, ACS Appl. Mater. Interfaces 2020, 12, 15562. [DOI] [PubMed] [Google Scholar]
- [14].Guo J, Zhang R, Zhang L, Cao X, ACS Macro Lett. 2018, 7, 442. [DOI] [PubMed] [Google Scholar]
- [15].Wang Q, Tian X, Huang L, Li D, Malakhov AV, Polilov AN, Mater. Des. 2018, 155, 404. [Google Scholar]
- [16].Zolfagharian A, Denk M, Bodaghi M, Kouzani AZ, Kaynak A, Acta Mech. Solida Sin. 2020, 33, 418. [Google Scholar]
- [17].de Marco C, Pané S, Nelson BJ, Sci. Robot 2018, 3, 287. [DOI] [PubMed] [Google Scholar]
- [18].Lee J, Kim H-C, Choi J-W, Lee IH, Int. J. Precis. Eng. Manuf.-Green Technol. 2017, 4, 373. [Google Scholar]
- [19].Zhou W, Qiao Z, Nazarzadeh Zare E, Huang J, Zheng X, Sun X, Shao M, Wang H, Wang X, Chen D, J. Med. Chem. 2020, 63, 8003. [DOI] [PubMed] [Google Scholar]
- [20].Gao B, Yang Q, Zhao X, Jin G, Ma Y, Xu F, Trends Biotechnol. 2016, 34, 746. [DOI] [PubMed] [Google Scholar]
- [21].Yang Q, Gao B, Xu F, Biotechnol. J. 2020, 15, 1900086. [DOI] [PubMed] [Google Scholar]
- [22].Ashammakhi N, Ahadian S, Zengjie F, Suthiwanich K, Lorestani F, Orive G, Ostrovidov S, Khademhosseini A, Biotechnol. J. 2018, 13, 1800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Miao S, Castro N, Nowicki M, Xia L, Cui H, Zhou X, Zhu W, Lee S.-j., Sarkar K, Vozzi G, Mater. Today 2017, 20, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].a) Ahmed K, Shiblee MDNI, Khosla A, Nagahara L, Thundat T, Furukawa H, J. Electrochem. Soc. 2020, 167, 037563; [Google Scholar]; b) Elkhoury K, Morsink M, Sanchez-Gonzalez L, Kahn C, Tamayol A, Arab-Tehrany E, Bioact. Mater. 2021, 6, 3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Champeau M, Heinze DA, Viana TN, de Souza ER, Chinellato AC, Titotto S, Adv. Funct. Mater. 2020, 1910606. [Google Scholar]
- [26].Wan X, Luo L, Liu Y, Leng J, Adv. Sci. 2020, 7, 2001000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chimene D, Lennox KK, Kaunas RR, Gaharwar AK, Ann. Biomed. Eng. 2016, 44, 2090. [DOI] [PubMed] [Google Scholar]
- [28].Riley L, Schirmer L, Segura T, Curr. Opin. Biotechnol. 2019, 60, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].a) Tong Z, Jin L, Oliveira JM, Reis RL, Zhong Q, Mao Z, Gao C, Bioact. Mater. 2021, 6, 1375; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhang H, Cong Y, Osi AR, Zhou Y, Huang F, Zaccaria RP, Chen J, Wang R, Fu J, Adv. Funct. Mater. 2020, 30, 1910573. [Google Scholar]
- [30].Daly AC, Riley L, Segura T, Burdick JA, Nat. Rev. Mater. 2020, 5, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jeon O, Lee YB, Hinton TJ, Feinberg AW, Alsberg E, Mater. Today Chem. 2019, 12, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Highley CB, Song KH, Daly AC, Burdick JA, Adv. Sci. 2019, 6, 1801076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Torquato S, Stillinger FH, Rev. Mod. Phys. 2010, 82, 2633. [Google Scholar]
- [34].Weeks ER, Statistical physics of complex fluids, Eds Maruyama S, Tokuyama M, pag 2007, 2, 87. [Google Scholar]
- [35].van Hecke M, Phys J.: Condens. Matter 2009, 22, 033101. [DOI] [PubMed] [Google Scholar]
- [36].Newsom JP, Payne KA, Krebs MD, Acta Biomater. 2019, 88, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jeon O, Lee YB, Jeong H, Lee SJ, Wells D, Alsberg E, Mater. Horiz. 2019, 6, 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ding A, Lee SJ, Ayyagari S, Tang R, Huynh CT, Alsberg E, Bioact. Mater. 2022, 7, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yao T, Wang Y, Zhu B, Wei D, Yang Y, Han X, Smart Mater. Struct. 2020, 30, 015018. [Google Scholar]
- [40].Timoshenko S, Josa 1925, 11, 233. [Google Scholar]
- [41].Shiblee MDNI, Ahmed K, Kawakami M, Furukawa H, Adv. Mater. Technol. 2019, 4, 1900071. [Google Scholar]
- [42].Albenga G, Scientia 1954, 48, 140. [Google Scholar]
- [43].Kim J, Kim C, Song Y, Jeong S-G, Kim T-S, Lee C-S, Chem. Eng. J. 2017, 321, 384. [Google Scholar]
- [44].Shojaeifard M, Baghani M, Meccanica 2019, 54, 841. [Google Scholar]
- [45].Morimoto T, Ashida F, Int. J. Solids Struct. 2015, 56–57, 20. [Google Scholar]
- [46].Ghobashy MM, Bassioni G, Adv. Polym. Technol. 2018, 37, 2123. [Google Scholar]
- [47].Hoo SP, Loh QL, Yue Z, Fu J, Tan TTY, Choong C, Chan PPY, J. Mater. Chem. B 2013, 1, 3107. [DOI] [PubMed] [Google Scholar]
- [48].Wu Y, Joseph S, Aluru NR, J. Phys. Chem. B 2009, 113, 3512. [DOI] [PubMed] [Google Scholar]
- [49].a) Pawar SN, Edgar KJ, Biomaterials 2012, 33, 3279; [DOI] [PubMed] [Google Scholar]; b) Jeon O, Shin J-Y, Marks R, Hopkins M, Kim T-H, Park H-H, Alsberg E, Chem. Mater. 2017, 29, 8425; [Google Scholar]; c) Jeon O, Marks R, Wolfson D, Alsberg E, J. Mater. Chem. B 2016, 4, 3526; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Jeon O, Wolfson DW, Alsberg E, Adv. Mater. 2015, 27, 2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ning X, Wang X, Zhang Y, Yu X, Choi D, Zheng N, Kim DS, Huang Y, Zhang Y, Rogers JA, Adv. Mater. Interfaces 2018, 5, 1800284. [Google Scholar]
- [51].Lee AY, Zhou A, An J, Chua CK, Zhang Y, Virtual Phys. Prototyp. 2020, 15, 481. [Google Scholar]
- [52].Wan Z, Zhang P, Liu Y, Lv L, Zhou Y, Acta Biomater. 2020, 101, 26. [DOI] [PubMed] [Google Scholar]
- [53].Lee YB, Jeon O, Lee SJ, Ding A, Wells D, Alsberg E, Adv. Func. Mater. 2021, 31, 2010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ding A, Jeon O, Tang R, Lee YB, Lee SJ, Alsberg E, Adv. Sci. 2021, 8, 2004616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].a) Pountos I, Corscadden D, Emery P, Giannoudis PV, Injury 2007, 38, S23; [DOI] [PubMed] [Google Scholar]; b) Alsberg E, von Recum HA, Mahoney MJ, Expert Opin. Biol. Ther. 2006, 6, 847. [DOI] [PubMed] [Google Scholar]
- [56].Miao S, Zhu W, Castro NJ, Nowicki M, Zhou X, Cui H, Fisher JP, Zhang LG, Sci. Rep. 2016, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].a) Constante G, Apsite I, Alkhamis H, Dulle M, Schwarzer M, Caspari A, Synytska A, Salehi S, Ionov L, ACS Appl. Mater. Interfaces 2021, 13, 12767; [DOI] [PubMed] [Google Scholar]; b) Wang Y, Cui H, Wang Y, Xu C, Esworthy TJ, Hann SY, Boehm M, Shen Y-L, Mei D, Zhang LG, ACS Appl. Mater. Interfaces 2021, 13, 12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].a) Cui H, Miao S, Esworthy T, Lee S.-j., Zhou X, Hann SY, Webster TJ, Harris BT, Zhang LG, Nano Res. 2019, 12, 1381; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Miao S, Cui H, Esworthy T, Mahadik B, Lee S. j., Zhou X, Hann SY, Fisher JP, Zhang LG, Adv. Sci. 2020, 7, 1902403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].a) Yang GH, Kim W, Kim J, Kim G, Theranostics 2021, 11, 48; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Luo Y, Lin X, Chen B, Wei X, Biofabrication 2019, 11, 045019. [DOI] [PubMed] [Google Scholar]
- [60].a) Kirillova A, Maxson R, Stoychev G, Gomillion CT, Ionov L, Adv. Mater. 2017, 29, 1703443; [DOI] [PubMed] [Google Scholar]; b) Kim SH, Seo YB, Yeon YK, Lee YJ, Park HS, Sultan MT, Lee JM, Lee JS, Lee OJ, Hong H, Biomaterials 2020, 260, 120281. [DOI] [PubMed] [Google Scholar]
- [61].Hinton Thomas J, Jallerat Q, Palchesko Rachelle N, Park Joon H, Grodzicki Martin S, Shue H-J, Ramadan Mohamed H, Hudson Andrew R, Feinberg Adam W, Sci. Adv, 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.