Abstract
To benchmark the activity of moxifloxacin (a newer fluoroquinolone), a U.S. study comprising 16,141 contemporary isolates of Streptococcus pneumoniae (5,640), Haemophilus influenzae (6,583), and Moraxella catarrhalis (3,648) referred from 377 institutions during 1998 is described. For S. pneumoniae the modal MIC and MIC at which 90% of the isolates were inhibited (MIC90) for moxifloxacin were 0.12 and 0.25 μg/ml, respectively, independent of susceptibility to other drug classes, geography, or site of infection. Eleven isolates were intermediate or resistant to levofloxacin and grepafloxacin; of these isolates, 1 remained susceptible to sparfloxacin, 2 remained susceptible to moxifloxacin, and 4 remained susceptible to trovafloxacin. All 11 isolates possessed classic mutations in gyrA and/or parC known to confer reduced susceptibility to fluoroquinolones. Four isolates (originating from four separate states) belonging to a multidrug-resistant, fluoroquinolone-resistant clone were identified by pulsed-field gel electrophoresis. For moxifloxacin and trovafloxacin, at least 87% of isolates demonstrated MICs ≥3 twofold concentrations below the susceptibility breakpoints, in contrast to no more than 15% for levofloxacin, grepafloxacin, and sparfloxacin. Of the isolates that were multidrug resistant (7.4%), >98% remained susceptible to moxifloxacin. The modal MIC and MIC90 for M. catarrhalis (both 0.06 μg/ml) and for H. influenzae (both 0.03 μg/ml) were independent of β-lactamase production. These data demonstrate the in vitro activity of moxifloxacin and establish a baseline for future studies.
In recent years, increasing antibiotic resistance among bacteria causing infections within both the hospital and community environments has severely compromised our ability to successfully treat patients empirically. Perhaps nowhere is this more apparent than with patients presenting with community-acquired respiratory tract infections (CA-RTI), in which Streptococcus pneumoniae, Moraxella catarrhalis, and Haemophilus influenzae are common pathogens (6). The emergence and dissemination of penicillin-resistant pneumococci are now global phenomena (1, 4, 14, 22, 36) and continue to increase. Compounding the problem is the tendency for organisms to also be refractory to some cephalosporins and macrolides (11, 42). Similarly, resistance caused by the widespread acquisition of plasmid-encoded BRO1 and/or BRO2 β-lactamase in M. catarrhalis and of TEM-1 enzyme in H. influenzae (13, 35) has effectively removed ampicillin and amoxicillin used alone as therapeutic choices for β-lactamase-producing isolates, neither being stable in the presence of those enzymes. Together, these factors have created a need for alternative oral candidate drugs for use as empiric therapies for patients with CA-RTI.
Recently, the further evolution of the fluoroquinolone class of drugs has resulted in a number of new compounds with an expanded spectrum of activity compared with earlier compounds such as ciprofloxacin and ofloxacin, most significantly against S. pneumoniae and other gram-positive pathogens. Several previous studies have demonstrated that some new fluoroquinolone compounds, such as sparfloxacin and levofloxacin, have better in vitro activity against S. pneumoniae, M. catarrhalis, and H. influenzae than previous fluoroquinolones (3, 34, 35, 44) with a very low prevalence of resistance. In addition, previous reports have suggested that the newer compounds are mostly unaffected by decreased susceptibility to β-lactam compounds, even those with elevated penicillin MICs of ≥1 μg/ml (35, 42, 44).
Moxifloxacin is a new 8-methoxyquinolone shown to be active against a considerable spectrum of pathogens (23, 39, 46). Although a previous U.S. study has demonstrated the activity of moxifloxacin against pathogens associated with CA-RTI (7), the surveillance study described here is the first extensive U.S. multisite study designed to benchmark the activity of moxifloxacin against clinical isolates of S. pneumoniae, H. influenzae, and M. catarrhalis prior to or concomitant with the release of the drug into clinical use. Since the medical community has great concern about antimicrobial resistance, the comprehensive data set derived through this study will permit concise tracking of any future changes in susceptibility to moxifloxacin.
MATERIALS AND METHODS
Organism collection.
During the winter period 1997 to 1998, isolates of S. pneumoniae, H. influenzae, and M. catarrhalis were collected from 377 participant hospital laboratories distributed throughout the nine Centers for Disease Control and Prevention-designated regions of the United States. From each laboratory, one isolate per patient, accompanied by strain-specific patient data, was referred for study. Isolates were collected from clinical samples derived from various upper and lower respiratory tract sites, blood, ears, and eyes. Following shipment to the MRL central testing laboratory, each isolate was subcultured onto blood agar (or chocolate agar for H. influenzae) and reidentified using standard methods (5). Only pure culture isolates were included in the final study and subcultured for immediate susceptibility testing prior to banking at −70°C.
Antibiotic susceptibility testing.
All isolates were tested for susceptibility to amoxicillin-clavulanate, ceftriaxone, cefuroxime, clarithromycin, azithromycin, erythromycin, trimethoprim-sulfamethoxazole (SXT), trovafloxacin, grepafloxacin, levofloxacin, sparfloxacin, and moxifloxacin using drug concentrations extending at least 1 twofold concentration above and below the breakpoints used in this study. In addition, all S. pneumoniae isolates were tested for susceptibility to penicillin and all H. influenzae and M. catarrhalis isolates were tested for susceptibility to ampicillin. For S. pneumoniae and H. influenzae, antibiotic susceptibility testing using broth microdilution and breakpoint interpretation was conducted according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS) (26), with the exception of moxifloxacin, for which no NCCLS breakpoints exist. For moxifloxacin, U.S. Food and Drug Administration (FDA) breakpoints were used to interpret data for S. pneumoniae (susceptible, ≤1 μg/ml; intermediate, 2 μg/ml; resistant, ≥4 μg/ml). For M. catarrhalis NCCLS standards for broth microdilution susceptibility testing are not defined by the NCCLS. S. pneumoniae ATCC 49619 and H. influenzae ATCC 49247 were used as controls throughout. Tests for β-lactamase production in H. influenzae and M. catarrhalis were performed with DrySlide nitrocefin (Difco Laboratories, Detroit, Mich.).
Nucleotide sequence determination of gyrA and parC and molecular typing using PFGE.
High-quality chromosomal DNA was extracted directly from single bacterial colonies using a standard procedure (2). Prepared chromosomal DNA was used as templates for PCR amplification of target quinolone resistance-determining regions (QRDRs) within gyrA and parC using previously defined primers (17, 27) and methodologies (37). Sequenced products were resolved and automatically analyzed using an ABI PRISM 377 DNA sequencer. Amplified fragments were sequenced in both directions to control for accuracy of sequence data obtained. Wild-type sequences with no mutations were identified on the basis of being identical to the published sequences of the gyrA and parC genes (17, 27). Mutations within these genes were identified by comparison. Strains of S. pneumoniae were distinguished using pulsed-field gel electrophoresis (PFGE) according to established methodologies (24), and molecular types were assigned according to the criteria established by Tenover et al. (41).
RESULTS
Organism referral.
A total of 5,640 S. pneumoniae, 6,583 H. influenzae, and 3,648 M. catarrhalis isolates were included in the final study analysis. Isolates were distributed roughly equally among the participating hospital laboratories, with an average frequency of 15 S. pneumoniae isolates, 18 H. influenzae isolates, and 9 M. catarrhalis isolates per laboratory. All regions yielded a representative sampling of organisms, with the lowest number of organisms derived from New England (278 S. pneumoniae isolates, 314 H. influenzae isolates, and 278 M. catarrhalis isolates) and the highest number of organisms derived from the east north central region (1,249 S. pneumoniae isolates, 1,490 H. influenzae isolates, and 718 M. catarrhalis isolates). For S. pneumoniae, H. influenzae, and M. catarrhalis, respectively, 1,641, 148, and 36 isolates were derived from blood, 3,482, 5,630, and 2,983 isolates were derived from respiratory sites, and 517, 810, and 211 isolates were derived from other sources.
S. pneumoniae susceptibility.
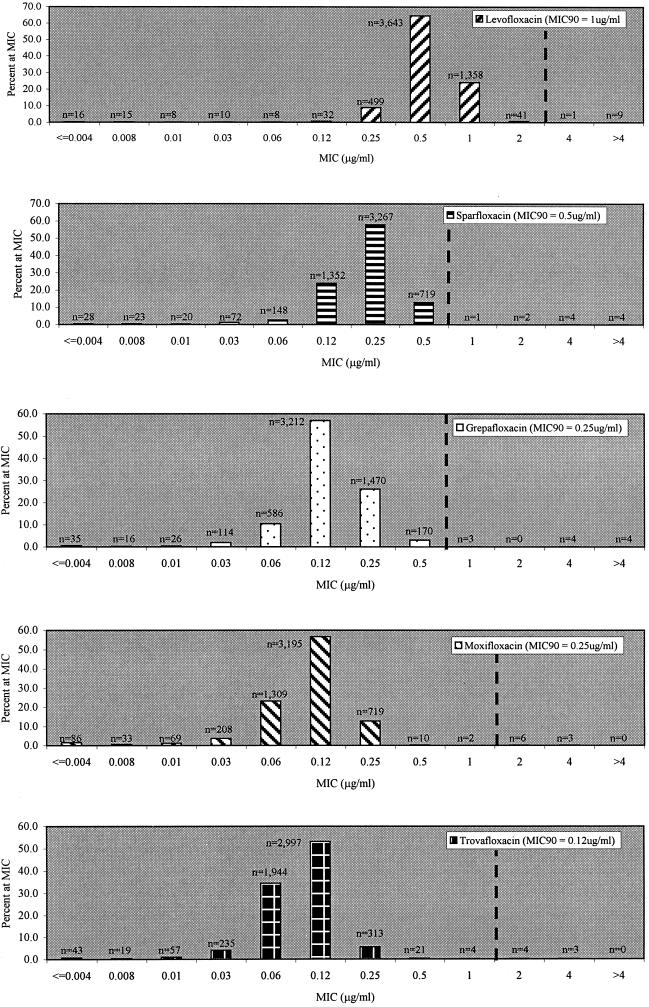
For S. pneumoniae, the susceptibility profiles of isolates for all drugs tested, categorized according to penicillin susceptibility, are shown in Table 1. For pooled data, overall nonsusceptibility (intermediate and resistant categories combined) was recorded as 36.2% for penicillin, 18.5% for amoxicillin-clavulanate, 28.2% for cefuroxime, 15.3% for ceftriaxone, 24.1% for erythromycin, 24.0% for both azithromycin and clarithromycin, and 32.9% for SXT. MIC distributions for each of the fluoroquinolones tested are shown in Fig. 1. For pooled data derived from each study region, the MIC at which 90% of the isolates were inhibited (MIC90) and modal MIC, respectively, for each fluoroquinolone studied were as follows: moxifloxacin, 0.25 and 0.12 μg/ml; trovafloxacin, 0.12 and 0.12 μg/ml; grepafloxacin, 0.25 and 0.12 μg/ml; sparfloxacin, 0.50 and 0.25 μg/ml; levofloxacin, 1.0 and 0.5 μg/ml. The MIC90 and modal MIC of moxifloxacin remained the same (0.25 and 0.12 μg/ml, respectively) regardless of penicillin or macrolide susceptibility, geographic region, specimen source, or patient age.
TABLE 1.
Susceptibility of S. pneumoniae to all antimicrobials tested
| Antimicrobial agent and phenotypea (no. of isolates) | MIC (μg/ml)
|
% of isolates that were:
|
||||
|---|---|---|---|---|---|---|
| Range | Mode | 90% | S | I | R | |
| Penicillin | ||||||
| All | ≤0.03–>8 | ≤0.03 | 2 | 63.9 | 22.5 | 13.7 |
| Pen S | ≤0.03–0.06 | ≤0.03 | 0.06 | 100.0 | 0.0 | 0.0 |
| Pen I | 0.12–1 | 1 | 1 | 0.0 | 100.0 | 0.0 |
| Pen R | 2–>8 | 2 | 4 | 0.0 | 0.0 | 100.0 |
| Amoxicillin-clavulanate | ||||||
| All | ≤0.01–>16 | ≤0.01 | 1 | 81.5 | 9.3 | 9.2 |
| Pen S | ≤0.01–0.5 | ≤0.01 | 0.03 | 100.0 | 0.0 | 0.0 |
| Pen I | ≤0.01–4 | 0.5 | 1 | 76.1 | 19.7 | 4.2 |
| Pen R | 0.25–>16 | 2 | 4 | 4.2 | 35.6 | 60.3 |
| Cefuroxime | ||||||
| All | ≤0.12–64 | ≤0.12 | 4 | 71.9 | 3.4 | 24.8 |
| Pen S | ≤0.12–1 | ≤0.12 | ≤0.12 | 100.0 | 0.0 | 0.0 |
| Pen I | ≤0.12–16 | 2 | 4 | 35.5 | 14.8 | 49.7 |
| Pen R | ≤0.12–64 | 4 | 8 | 0.3 | 0.1 | 99.6 |
| Ceftriaxone | ||||||
| All | ≤0.01–8 | ≤0.01 | 1 | 84.6 | 11.2 | 4.1 |
| Pen S | ≤0.01–0.5 | ≤0.01 | ≤0.06 | 100.0 | 0.0 | 0.0 |
| Pen I | ≤0.01–8 | 0.5 | 1 | 35.5 | 14.8 | 49.7 |
| Pen R | ≤0.01–8 | 1 | 2 | 13.9 | 60.3 | 25.8 |
| Erythromycin | ||||||
| All | ≤0.03–>4 | ≤0.03 | 4 | 75.9 | 0.1 | 24.0 |
| Pen S | ≤0.03–>4 | ≤0.03 | 0.03 | 93.9 | 0.0 | 6.1 |
| Pen I | ≤0.03–>4 | ≤0.03 | >4 | 54.1 | 0.5 | 45.5 |
| Pen R | ≤0.03–>4 | 4 | >4 | 27.5 | 0.1 | 72.3 |
| Azithromycin | ||||||
| All | ≤0.03–>4 | ≤0.03 | 4 | 75.9 | 3.4 | 20.6 |
| Pen S | ≤0.03–>4 | ≤0.03 | 0.06 | 93.9 | 0.8 | 5.3 |
| Pen I | ≤0.03–>4 | ≤0.03 | >4 | 54.3 | 7.8 | 37.9 |
| Pen R | ≤0.03–>4 | >4 | >4 | 27.5 | 8.4 | 64.0 |
| Clarithromycin | ||||||
| All | ≤0.01–>32 | 0.03 | 4 | 75.9 | 3.4 | 20.6 |
| Pen S | ≤0.01–>32 | 0.03 | 0.03 | 93.9 | 0.4 | 5.7 |
| Pen I | ≤0.01–>32 | 0.03 | 32 | 54.2 | 2.1 | 43.7 |
| Pen R | ≤0.01–>32 | 2 | >32 | 27.5 | 1.4 | 71.0 |
| Levofloxacin | ||||||
| All | ≤0.004–>8 | 0.5 | 1 | 99.8 | 0.0 | 0.2 |
| Pen S | ≤0.004–>8 | 0.5 | 1 | 93.9 | 0.0 | 0.0 |
| Pen I | ≤0.004–>8 | 0.5 | 1 | 99.8 | 0.0 | 0.2 |
| Pen R | 0.12–>8 | 0.5 | 1 | 99.4 | 0.0 | 0.6 |
| Grepafloxacin | ||||||
| All | ≤0.002–>4 | 0.12 | 0.25 | 99.8 | 0.1 | 0.1 |
| Pen S | ≤0.002–1 | 0.12 | 0.25 | 99.9 | 0.1 | 0.0 |
| Pen I | ≤0.004–>4 | 0.12 | 0.25 | 99.8 | 0.0 | 0.2 |
| Pen R | ≤0.01–>4 | 0.12 | 0.25 | 99.2 | 0.1 | 0.6 |
| Sparfloxacin | ||||||
| All | ≤0.002–>4 | 0.25 | 0.5 | 99.8 | 0.0 | 0.2 |
| Pen S | ≤0.002–2 | 0.25 | 0.5 | 99.9 | 0.0 | 0.1 |
| Pen I | 0.004–>4 | 0.25 | 0.5 | 99.8 | 0.0 | 0.2 |
| Pen R | 0.03–>4 | 0.25 | 0.5 | 99.2 | 0.1 | 0.6 |
| Moxifloxacinb | ||||||
| All | ≤0.002–4 | 0.12 | 0.25 | 99.8 | 0.1 | 0.1 |
| Pen S | ≤0.002–2 | 0.12 | 0.25 | 100.0 | 0.0 | 0.0 |
| Pen I | ≤0.002–4 | 0.12 | 0.25 | 99.8 | 0.1 | 0.2 |
| Pen R | 0.01–4 | 0.12 | 0.25 | 99.4 | 0.5 | 0.1 |
| Trovafloxacin | ||||||
| All | ≤0.002–4 | 0.12 | 0.12 | 99.9 | 0.1 | 0.1 |
| Pen S | ≤0.002–1 | 0.12 | 0.12 | 100.0 | 0.0 | 0.0 |
| Pen I | 0.004–4 | 0.12 | 0.12 | 99.8 | 0.1 | 0.2 |
| Pen R | 0.01–4 | 0.12 | 0.12 | 99.5 | 0.4 | 0.1 |
| SXT | ||||||
| All | ≤0.01–>4 | 0.12 | 4 | 67.1 | 16.5 | 16.4 |
| Pen S | ≤0.01–>4 | 0.12 | 1 | 89.5 | 4.9 | 5.6 |
| Pen I | ≤0.01–>4 | 2 | 4 | 40.2 | 35.3 | 24.5 |
| Pen R | 0.06–>4 | 4 | >4 | 6.6 | 39.9 | 53.5 |
Pen, penicillin; S, susceptible; I, intermediate; R, resistant. Of the 5,640 S. pneumoniae isolates, 3,603 were Pen S, 1,267 were Pen I, and 770 were Pen R.
Moxifloxacin interpretive breakpoints based on U.S. FDA breakpoints.
FIG. 1.
MIC distributions for study fluoroquinolones among S. pneumoniae isolates. Dashed lines, susceptibility breakpoints.
For moxifloxacin and trovafloxacin, 87 and 94% of isolates, respectively, had MICs that were ≥3 times lower than the susceptibility breakpoint used in this study. This is in contrast to grepafloxacin, levofloxacin, and sparfloxacin, whose MICs for 14, 10, and 5% of isolates, respectively, were ≥3 twofold concentrations below their respective susceptibility breakpoints. Taking into consideration the number of organisms with MICs at least 2 twofold concentrations below the susceptibility breakpoints used, these values increased to >99% for trovafloxacin and moxifloxacin, compared with 71, 75, and 29% for grepafloxacin, levofloxacin, and sparfloxacin, respectively.
Of the 5,640 S. pneumoniae isolates referred, only 11 (0.20%) were not susceptible to any one of the fluoroquinolones tested (Table 2). Four of these isolates remained susceptible to trovafloxacin, two remained susceptible to moxifloxacin, and one remained susceptible to sparfloxacin. All 11 isolates were intermediate or resistant to grepafloxacin and levofloxacin.
TABLE 2.
Origin and phenotypic and genotypic characteristics of 11 S. pneumoniae isolates requiring MICs of ≥4 μg/ml for any fluoroquinolone tested
| Isolate | Statea | MIC (μg/ml) (interpretive categoryb) of:
|
MDR phenotypec | Molecular PFGE type | Amino acid alterations in:
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sparfloxacin | Levofloxacin | Grepafloxacin | Trovafloxacin | Moxifloxacin | GyrA | ParC | ||||
| 1 | CA | 4 (R) | 8 (R) | 4 (R) | 1 (S) | 2 (I) | Yes | A | Ser-81-Phe | Lys-137-Asn |
| 2 | HI | >4 (R) | >8 (R) | >4 (R) | 4 (R) | 4 (R) | No | D | WTd | Ser-16-Gly; Ser-79-Phe; Asn-91-Asp; Glu-125-Asp |
| 3 | HI | >4 (R) | >8 (R) | >4 (R) | 4 (R) | 2 (I) | Yes | A | Ser-81-Phe | Ser-79-Phe; Lys-137-Asn |
| 4 | IL | 2 (R) | >8 (R) | 1 (I) | 1 (S) | 2 (I) | No | E | Ser-81-Tyr | WT |
| 5 | MA | 4 (R) | 8 (R) | 4 (R) | 4 (R) | 4 (R) | No | B | Ser-81-Phe | Ser-79-Phe |
| 6 | MN | >4 (R) | >8 (R) | >4 (R) | 2 (I) | 2 (I) | Yes | G | Ser-81-Phe | Ser-79-Phe |
| 7 | MN | 4 (R) | >8 (R) | 4 (R) | 2 (I) | 2 (I) | Yes | A | Ser-81-Phe | Ser-79-Phe; Lys-137-Asn |
| 8 | NJ | 2 (R) | 4 (I) | 1 (I) | 0.25 (S) | 1 (S) | No | F | Ser-81-Phe | WT |
| 9 | PA | 4 (R) | >8 (R) | 4 (R) | 2 (I) | 2 (I) | Yes | A | Ser-81-Phe | Ser-79-Phe; Lys-137-Asn |
| 10 | VA | >4 (R) | 8 (R) | >4 (R) | 2 (I) | 4 (R) | Yes | H | Ser-81-Tyr | Ser-79-Tyr |
| 11 | VA | 1 (S) | 8 (R) | 1 (I) | 0.25 (S) | 1 (S) | No | C | Ser-81-Phe | Ser-79-Phe; Lys-137-Asn |
Molecular characterization of fluoroquinolone resistance and clonality studies.
Alterations in GyrA previously shown to be associated with reduced susceptibility to fluoroquinolone compounds were identified in 10 of the 11 fluoroquinolone-resistant strains at position Ser-81, altered in each case to either a Phe or Tyr residue. No GyrA alterations were detected in strain 2, although this strain was idiosyncratic in possessing multiple mutations in parC conferring alterations Ser-16→Gly, Ser-79→Phe, Asn-91→Asp, and Glu-125→Asp. Of the 11 fluoroquinolone-resistant isolates, 7 possessed alteration Ser-79→Phe in ParC. Alteration Lys-137→Asn in ParC was identified in five isolates, each in combination with GyrA Ser-81→Phe and (except for isolate 1) with ParC Ser-79→Phe (Table 2). We did not look for mutations in parE or gyrB, since these are considered not to play a significant role in conferring reduced susceptibility to the fluoroquinolones studied (18).
The 11 strains demonstrating reduced susceptibility to fluoroquinolones were typed using PFGE to investigate clonality (Table 2). Eight distinct PFGE molecular types were identified on the basis of being different by three or more bands (41). The four PFGE type A isolates detected were derived from geographically diverse states (Virginia, Hawaii, Minnesota, and California), and, except for no detected ParC Ser79→Phe alteration in isolate 1, each carried identical mutant gyrA and parC gene loci and a multidrug-resistant (MDR) phenotype characterized by nonsusceptibility to all drugs tested while remaining intermediate or susceptible to ceftriaxone, designated MDR types J, H, and L (see next section).
Multidrug resistance among isolates of S. pneumoniae.
For the purposes of this study, an MDR phenotype was defined as resistance to three or more of the following drugs: penicillin, ceftriaxone, erythromycin, SXT, and any fluoroquinolone (Table 3). Of all MDR isolates, 98.3% remained susceptible to fluoroquinolones, with moxifloxacin MICs ranging from 0.01 to 0.5 μg/ml. Of these, the most common MDR phenotype (type A: penicillin resistant, ceftriaxone intermediate, erythromycin resistant, SXT resistant, and fluoroquinolone susceptible) comprised nearly 50% of all MDR isolates. No isolate was resistant to all compounds tested, although four isolates (types H and K) with all susceptibilities in either intermediate or resistant categories were recovered. Seven MDR isolates (types H through L) demonstrated resistance to at least one fluoroquinolone, although only one of these (from type H) was resistant to moxifloxacin (MIC, 4 μg/ml), the others being intermediate (types H, J, K, and L) or susceptible (type I) to moxifloxacin. In contrast, all seven fluoroquinolone-resistant MDR type H, I, J, K, and L strains were resistant to levofloxacin (MIC, ≥4 μg/ml).
TABLE 3.
Frequency of MDRa S. pneumoniae phenotypes
| MDR phenotype (no. of isolates) | % | Interpretive category for:
|
Moxifloxacin MIC range (μg/ml) | ||||
|---|---|---|---|---|---|---|---|
| Penicillin | Ceftriaxone | Erythromycin | SXT | Fluoroquinolones | |||
| A (205) | 48.3 | R | I | R | R | S | 0.03–0.5 |
| B (65) | 15.3 | R | R | R | R | S | 0.03–0.25 |
| C (60) | 14.2 | R | R | R | I | S | 0.06–0.25 |
| D (46) | 10.8 | R | S | R | R | S | 0.01–0.05 |
| E (31) | 7.3 | R | R | S | R | S | 0.06–0.25 |
| F (6) | 1.4 | R | R | R | S | S | 0.06–0.25 |
| G (4) | 0.9 | I | R | R | R | S | 0.06–0.25 |
| H (3) | 0.6 | R | I | R | R | R | 2–4 |
| I (1) | 0.2 | R | I | S | R | R | 1 |
| J (1) | 0.2 | I | S | R | R | R | 2 |
| K (1) | 0.2 | R | I | R | I | R | 2 |
| L (1) | 0.2 | R | S | R | R | R | 2 |
Defined as resistance to three or more drug classes. Total number of isolates, 424.
H. influenzae and M. catarrhalis susceptibilities.
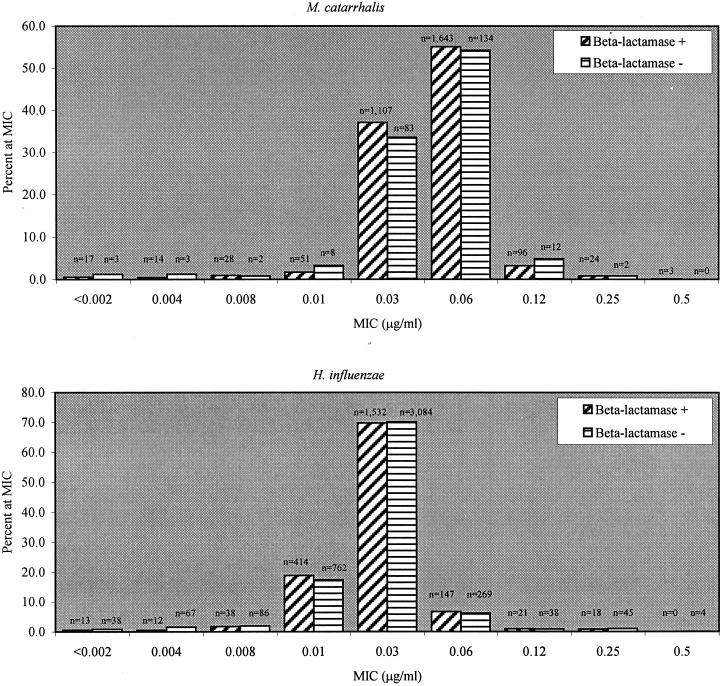
For moxifloxacin and all other fluoroquinolone compounds tested, all isolates of H. influenzae remained susceptible and no isolate of M. catarrhalis required an MIC of >0.5 μg/ml (modal MIC and MIC90 were both 0.06 μg/ml), irrespective of geographic region or specimen source (data not shown). A total of 33.3% (2,195 of 6,588) of the H. influenzae isolates and 92.4% (2,983 of 3,230) of the M. catarrhalis isolates produced β-lactamase; the moxifloxacin MIC distribution and modal MIC for both organisms remained independent of β-lactamase production (Fig. 2). For H. influenzae, no isolates that were β-lactamase negative and ampicillin resistant were detected; eight isolates (0.1%) were resistant to amoxicillin-clavulanate, and only one isolate was resistant to cefuroxime. Azithromycin and clarithromycin were active against 99.9 and 92.7%, respectively, of all isolates, in contrast with SXT, to which 20.9% of isolates (1,377) were not susceptible. Apart from resistance to ampicillin, 100% of M. catarrhalis isolates demonstrated low MICs to all drugs tested. With the exception of 4 isolates (0.1%; all β-lactamase positive) requiring MICs of cefuroxime of >4 μg/ml and 16 isolates (0.5%; 13 β-lactamase positive and 3 β-lactamase negative) requiring MICs of SXT of 8 to 16 μg/ml, all M. catarrhalis isolates demonstrated MICs below the susceptibility breakpoint defined by NCCLS for Staphylococcus aureus for all drugs tested (data not shown).
FIG. 2.
MIC distributions for moxifloxacin among M. catarrhalis and H. influenzae isolates.
DISCUSSION
This study was undertaken in order to benchmark the in vitro activity of moxifloxacin against clinically significant pathogens associated with respiratory tract infections derived from a large number of hospital laboratories across the United States, including the characterization of all resistant isolates and MDR organisms, enabling future comparative studies and trend analyses. In contrast with other drug classes tested, moxifloxacin and the other fluoroquinolones demonstrated overall high activity against the pathogens studied. Moxifloxacin inhibited the growth of 99.8% of S. pneumoniae isolates and 100% of H. influenzae isolates at concentrations well below the susceptibility breakpoint criteria. For M. catarrhalis no isolate required an MIC of moxifloxacin of >0.5 μg/ml. For moxifloxacin, as is evident from Table 1, in S. pneumoniae there is no significant relationship between the degree of resistance to penicillin and the modal MIC MIC90, both remaining at 0.12 and 0.25 μg/ml, respectively, for each penicillin susceptibility category. This is consistent with other reports from the United States and Europe demonstrating no relationship between penicillin susceptibility and the activity of moxifloxacin or other newer fluoroquinolones (8, 11, 32, 33, 35, 42–44). These observations are in contrast to recent reports from Canada and Ireland, which observed a relationship between penicillin nonsusceptibility and reduced susceptibility to ciprofloxacin (9, 16), perhaps explained by the lower activity of ciprofloxacin against S. pneumoniae. Additionally, the modal moxifloxacin MIC and MIC90 are unchanged when considered in relation to the susceptibility categories of the other drugs studied (such as erythromycin), geographic region, and site of infection, parameters not previously explored. In contrast with results from other reports concerning children (11, 43), we detected no relationship between moxifloxacin susceptibility and age. The clear-cut association between penicillin resistance and resistance to other classes of antibiotics, in particular other β-lactams, the macrolides, and SXT (Table 1), confirms the findings of several previous studies (11, 32, 35, 42–44). In addition, the prevalence of penicillin resistance (12.7% resistant and 22.5% intermediate) remains similar to that in data derived from surveillance studies reported from the 1996 to 1997 respiratory winter season (12, 42, 43) but is clearly increased from the 9.5% resistant and 14% intermediate reported from the 1994 to 1995 winter season (11).
The higher activities of moxifloxacin and trovafloxacin compared with those of other test fluoroquinolones, coupled with the fact that these hydrophobic compounds are resilient to efflux (15, 38), mean that in isolates with decreased susceptibilities, perhaps through the acquisition of single point mutations in gyrA or parC, MICs can still remain below the susceptible breakpoint. However, the widespread use of newer fluoroquinolones may be accompanied by an accumulation of additional mutations in QRDRs, resulting in upward “MIC creep.” Should this happen, the extent to which the drugs' modal MICs are below the currently used breakpoints becomes relevant. At least for trovafloxacin and moxifloxacin, the majority of strains (approximately 90%) remained at least 3 twofold concentrations lower than the FDA breakpoint and almost 100% were 2 twofold concentrations lower.
In S. pneumoniae (18), as in S. aureus (37, 38), mutations in the DNA gyrase subunit A (gyrA) and topoisomerase IV (parC or grlA in S. aureus) are mostly responsible for conferring reduced susceptibility to fluoroquinolone compounds. Most previous reports suggest that mutations in gyrA (Ser-81→Phe or Tyr) and parC (Ser-79→Tyr) are the most significant (17, 18, 25, 27–29, 31, 40, 45). In concordance with this, the 11 fluoroquinolone-resistant isolates detected in this study possessed classic single or combination mutations in gyrA and parC responsible for conferring fluoroquinolone-resistant phenotypes. It is worth noting, however, that the effects of mutations in QRDRs are not always clear-cut and that considerable biovariation clearly occurs in clinical isolates of S. pneumoniae (18). This is apparent from strain 11, which, despite possessing ParC alterations Lys-137→Asn and Ser-79→Phe and GyrA alteration Ser-81→Phe, maintained a moxifloxacin-susceptible phenotype, although the MIC was close to the breakpoint. MICs for other isolates with similar gyrA and parC mutational combinations of moxifloxacin were 4 μg/ml (resistant). In addition, strain 4 carries only the GyrA Ser-81→Tyr alteration, remaining wild type at ParC, but nevertheless requires raised MICs of the fluoroquinolones tested, including moxifloxacin. Since moxifloxacin is a hydrophobic fluoroquinolone and not readily removed by efflux from the cell, there are probably other physiological factors affecting fluoroquinolone susceptibility such as changes in outer membrane proteins. Of note is isolate 2, which, despite being wild type at gyrA, was highly resistant to all the fluoroquinolone compounds tested, including trovafloxacin. In addition to the classic Ser-79→Phe alteration, this isolate possessed a number of ParC mutations to our knowledge not previously reported (Ser-16→Gly, Asn-91→Asp, and Glu-125→Asp) whose impact on fluoroquinolone susceptibility is unknown. These may warrant further study. This isolate may have possessed mutations in gyrB and parE which have been implicated previously (21, 25) as playing a role in fluoroquinolone resistance, although their role is still debatable (18, 27, 30). Previous studies have shown moxifloxacin to manifest a low mutation rate of <1.4 × 10−9 at 4 times the MIC, compared with 2.2 × 10−7 for ciprofloxacin (10), providing at least circumstantial evidence that the emergence of fluoroquinolone resistance during appropriate moxifloxacin therapy is unlikely. Of course such benefits are negated should the widespread use of less-active compounds with a greater tendency to select for mutations occur, especially since we know that key mutations will have at least some effect on the activity of all quinolones (18) and appear to be stable over time (19).
The emergence of MDR (resistance to any three drug classes tested) in S. pneumoniae is of major concern since it severely limits treatment options. During the 1997 to 1998 season, 424 (7.5%) isolates studied demonstrated an MDR phenotype; of these, almost 80% were resistant or intermediate to all drug classes tested (Table 3) except for moxifloxacin and/or other fluoroquinolones, which all had MICs in the susceptible category. Overall, 98.2% of all MDR strains remained susceptible to at least one of the new fluoroquinolones tested in this study. This clearly positions the new fluoroquinolones as viable therapeutic options for treating infections with MDR pneumococci, particularly since in the vast majority of cases, the use of fluoroquinolones, unlike the use of other drug classes, is currently unlikely to select for the emergence or clonal expansion of organisms with MDR phenotypes, since the vast majority of MDR types remain fluoroquinolone susceptible. Nonetheless, it is important to note that 6 of the 11 fluoroquinolone-resistant isolates detected demonstrated MDR phenotypes, which confirms the need for careful monitoring of MDR phenotypes, including the complete molecular characterization of all fluoroquinolone-resistant organisms. Three of these MDR phenotypes (types J, H, and L) comprised PFGE clonal type A. Additionally, each PFGE type A strain, except for one with Ser-79→Phe alteration at ParC, possessed identical mutant gyrA and parC gene loci. It appears that this study provides the first evidence for the emergence of an MDR clone of S. pneumoniae that includes resistance to the fluoroquinolones, although such MDR resistance is very rare. A longitudinal comparative determination of MDR status using similar representations of drug classes, combined with characterization of clonality and QRDRs, will play a crucial role in tracking the further evolution of MDR and fluoroquinolone resistance.
For clinical isolates of H. influenzae and M. catarrhalis, the incidence of β-lactamase production (33.3 and 92.4%, respectively) remains almost identical to that found in 1996 to 1997 data and previously reported by our group (33.4 and 92.7%, respectively) (42). The incidence of β-lactamase production in both species remained mostly constant, irrespective of geographical location within the United States (MRL, unpublished data). This is in contrast to the situation in Europe and Asia, in which far greater regional differences have been reported (13, 20, 35). This is especially true for H. influenzae, which in some regional populations produces β-lactamase at an incidence of less than 5% (13, 35). Apart from a 20.9% incidence of nonsusceptibility to SXT among H. influenzae isolates, β-lactamase production remains the only significant cause of resistance in isolates for both this species and M. catarrhalis. A recent U.S. study reported “fluoroquinolone-resistant” isolates of both H. influenzae and M. catarrhalis (D. J. Biedenbach, R. N. Jones, J. Dipersio, K. C. Kugler, and W. W. Wilke, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 54, p. 141, 1999). However, in this U.S.-wide study, similar to a recent European study (20), no isolates of either species required MICs of any of the fluoroquinolones tested of >1.0 μg/ml, MIC90s for moxifloxacin were 0.03 μg/ml for H. influenzae and 0.06 μg/ml for M. catarrhalis, and there was no association with β-lactamase status for either species.
Resistance to antibacterial agents commonly prescribed as empiric therapies for the treatment of CA-RTI is a major health care concern. For H. influenzae and M. catarrhalis, the likely production of β-lactamase must be considered when choosing a therapy; for S. pneumoniae, the frequent occurrence of penicillin resistance and associated MDR is of even greater concern. In this respect, the positive features of the fluoroquinolones include their relatively high activity, their lack of association with the common MDR phenotypes, and the fact that emergence of resistance during therapy with proper dosing seems unlikely. It is important to note however that the apparent benefits of the newer quinolone antibiotics cannot be extended to the pediatric patient population in which these drugs are not currently an option.
In future years, geographically widespread surveillance studies incorporating a broad selection of antimicrobial agents and queriable parameters, including quantitative MIC data for extended-range antibiotic concentrations, in addition to the molecular characterization of resistant phenotypes, will enable the detection and monitoring of any changes in moxifloxacin susceptibilities among RTI pathogens, possibly enabling the timely implementation of preventive measures.
ACKNOWLEDGMENTS
We express our appreciation to the many microbiologists and other laboratory personnel in the participating institutions whose cooperation made this study possible. We are also grateful to the personnel at MRL for their work and their support of the study.
We thank Bayer Pharmaceutical, Inc., which provided financial support for this work.
REFERENCES
- 1.Appelbaum P C. World-wide development of antibiotic resistance in pneumococci. Eur J Clin Microbiol. 1987;6:367–377. doi: 10.1007/BF02013089. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 3.Ballow C H, Jones R N, Johnson D M, Deinhart J A, Schentag J J the SPAR Study Group. Comparative in vitro assessment of the activity and spectrum using results from over 14,000 pathogens isolated from 190 centers in the U.S. Diagn Microbiol Infect Dis. 1997;29:173–186. doi: 10.1016/s0732-8893(97)81807-x. [DOI] [PubMed] [Google Scholar]
- 4.Baquero F. Trends in antibiotic resistance of respiratory pathogens: an analysis and commentary on a collaborative surveillance study. J Antimicrob Chemother. 1996;38(Suppl. A):117–132. doi: 10.1093/jac/38.suppl_a.117. [DOI] [PubMed] [Google Scholar]
- 5.Baron E J, Murray P R. Bacteriology. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: ASM Press; 1995. pp. 246–662. [Google Scholar]
- 6.Bartlett J G, Mundy L M. Community-acquired pneumonia. N Engl J Med. 1995;333:1619–1624. doi: 10.1056/NEJM199512143332408. [DOI] [PubMed] [Google Scholar]
- 7.Brueggemann A B, Kugler K C, Doern G V. In vitro activity of BAY 12-8039, a novel 8-methoxyquinolone, compared to activities of six fluoroquinolones against Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis. Antimicrob Agents Chemother. 1997;41:1594–1597. doi: 10.1128/aac.41.7.1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buxbaum A, Straschil U, Moser C, Graninger W, Georgopoulos A. Comparative susceptibility to penicillin and quinolones of 1385 Streptococcus pneumoniae isolates. Austrian bacterial surveillance network. J Antimicrob Chemother. 1999;43(Suppl. B):13–18. doi: 10.1093/jac/43.suppl_2.13. [DOI] [PubMed] [Google Scholar]
- 9.Chen D K, McGeer A, de Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolones in Canada. Canadian bacterial surveillance network. N Engl J Med. 1999;341:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 10.Dalhoff A, Petersen U, Endermann E. In vitro activity of BAY 12-8039, a new 8-methoxyquinolone. Chemotherapy. 1996;42:410–425. doi: 10.1159/000239474. [DOI] [PubMed] [Google Scholar]
- 11.Doern G V, Brueggeman A, Holley H P, Rauch A M. Antimicrobial resistance of Streptococcus pneumoniae recovered from outpatients in the United States during the winter months of 1994–1995: results of a 30-center national surveillance study. Antimicrob Agents Chemother. 1996;40:1208–1213. doi: 10.1128/aac.40.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doern G V, Pfaller M A, Kugler K, Freeman J, Jones R N. Prevalence of antimicrobial resistance among respiratory tract isolates of Streptococcus pneumoniae in North America: 1997 results from the SENTRY antimicrobial surveillance program. Clin Infect Dis. 1998;27:764–770. doi: 10.1086/514953. [DOI] [PubMed] [Google Scholar]
- 13.Felmingham D, Washington J. Trends in the antimicrobial susceptibility of bacterial respiratory tract pathogens: findings of the Alexander project 1992–1996. J Chemother. 1999;11(Suppl. 1):5–21. doi: 10.1179/joc.1999.11.Supplement-2.5. [DOI] [PubMed] [Google Scholar]
- 14.Fluit A C, Schmitz F J, Jones M E, Acar J, Gupta R, Verhoef J the SENTRY Participants Group. Antimicrobial resistance among community-acquired pneumonia isolates in Europe: first results from the SENTRY Antimicrobial Surveillance Program 1997. Int J Infect Dis. 1999;3:153–156. doi: 10.1016/s1201-9712(99)90037-1. [DOI] [PubMed] [Google Scholar]
- 15.Gill M J, Brenwald N P, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:187–189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith C E, Moore J E, Murphy P G, Ambler J E. Increased incidence of ciprofloxacin resistance in penicillin-resistant pneumococci in Northern Ireland. J Antimicrob Chemother. 1998;41:420–421. doi: 10.1093/jac/41.3.420. [DOI] [PubMed] [Google Scholar]
- 17.Janoir C, Zeller V, Kitzis M D, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutations in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones M E, Sahm D F, Martin N, Scheuring S, Heisig P, Thornsberry C, Kohrer K, Schmitz F J. Prevalence of gyrA, gyrB, parC, and parE mutations in clinical isolates of Streptococcus pneumoniae with decreased susceptibilities to different fluoroquinolones and originating from Worldwide Surveillance Studies during the 1997–1998 respiratory season. Antimicrob Agents Chemother. 2000;44:462–466. doi: 10.1128/aac.44.2.462-466.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones M E, Boenink N M, Verhoef J, Köhrer K, Schmitz F-J. Multiple mutations conferring ciprofloxacin resistance in Staphylococcus aureus demonstrate long-term stability in an antibiotic-free environment. J Antimicrob Chemother. 2000;45:353–356. doi: 10.1093/jac/45.3.353. [DOI] [PubMed] [Google Scholar]
- 20.Jones M E, Staples A M, Critchley I, Thornsberry C, Heinze P, Engler H D, Sahm D F. Benchmarking the activity of moxifloxacin against recent clinical isolates of Streptococcus pneumoniae, Moraxella catarrhalis and Haemophilus influenzae. A European multi-center study. Diagn Microbiol Infect Dis. 2000;37:203–211. doi: 10.1016/s0732-8893(00)00128-0. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen J H, Weigel L M, Ferraro M J, Swenson J M, Tenover F C. Activities of newer fluoroquinolones against Streptococcus pneumoniae clinical isolates including those with mutations in the gyrA, parC, and parE loci. Antimicrob Agents Chemother. 1999;43:329–334. doi: 10.1128/aac.43.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klugman K. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGowan A P, Bowker K E, Holt H A, Wootton M, Reeves D S. BAY12-8039, a new 8-methoxy-quinolone: comparative in vitro activity with nine other antimicrobials against anaerobic bacteria. J Antimicrob Chemother. 1997;40:503–509. doi: 10.1093/jac/40.4.503. [DOI] [PubMed] [Google Scholar]
- 24.McEllistrem M C, Stout J E, Harrison L H. Simplified protocol for pulsed-field gel electrophoresis analysis of Streptococcus pneumoniae. J Clin Microbiol. 2000;38:351–353. doi: 10.1128/jcm.38.1.351-353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational supplement. Approved standard M100-S8. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 27.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X-S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pestova E, Beyer R, Cianciotto N P, Noskin G A, Peterson L R. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob Agents Chemother. 1999;43:2000–2004. doi: 10.1128/aac.43.8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaller M A, Jones R N, Doern G, Kugler K. Bacterial pathogens isolated from patients with bloodstream infection: frequencies of occurrence and antimicrobial susceptibility patterns from the SENTRY antimicrobial surveillance program. Antimicrob Agents Chemother. 1998;42:1763–1770. doi: 10.1128/aac.42.7.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinert R R, Schlaeger J J, Lütticken R. Moxifloxacin: a comparison with other antimicrobial agents of in vitro activity against Streptococcus pneumoniae. J Antimicrob Chemother. 1998;42:803–806. doi: 10.1093/jac/42.6.803. [DOI] [PubMed] [Google Scholar]
- 34.Richard M P, Aguado A, Mattina R, Marre R the SPAR Study Group. Sensitivity to sparfloxacin and other antibiotics of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis strains isolated from adults with community acquired lower respiratory tract infections: a European multi centre study. J Antimicrob Chemother. 1998;41:207–214. doi: 10.1093/jac/41.2.207. [DOI] [PubMed] [Google Scholar]
- 35.Sahm D F, Jones M E, Hickey M L, Diakun D R, Mani S, Thornsberry C. Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis isolated in Asia and Europe, 1997–1998. J Antimicrob Chemother. 2000;45:457–466. doi: 10.1093/jac/45.4.457. [DOI] [PubMed] [Google Scholar]
- 36.Schito G C, Manelli S, Pesce A the Alexander Project. Trends in the activity of macrolide and β-lactam antibiotics and resistance development. J Chemother. 1997;9(Suppl. 3):18–28. [PubMed] [Google Scholar]
- 37.Schmitz F J, Jones M E, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Fluit A, Verhoef J, Hadding U, Heinz H-P, Köhrer K. Characterization of grlA, grlB, gyrA, and gyrB in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmitz F J, Lückefahr M, Engler B, Hoffman B, Hansen B, et al. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro activity of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J Antimicrob Chemother. 1998;42:807–810. doi: 10.1093/jac/42.6.807. [DOI] [PubMed] [Google Scholar]
- 39.Souli M, Wennersten C B, Eliopoulos G M. In vitro activity of BAY 12-8039, a new fluoroquinolone, against species representative of respiratory tract pathogens. Int J Antimicrob Agents. 1998;10:23–30. doi: 10.1016/s0924-8579(98)00020-x. [DOI] [PubMed] [Google Scholar]
- 40.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thornsberry C, Ogilvie P, Kahn J, Mauriz Y. Surveillance of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the United States in 1996–1997 respiratory season. The Laboratory Investigator Group. Diagn Microbiol Infect Dis. 1997;29:249–257. doi: 10.1016/s0732-8893(97)00195-8. [DOI] [PubMed] [Google Scholar]
- 43.Thornsberry C, Ogilvie P T, Holley H P, Sahm D F. In vitro activity of grepafloxacin and 25 other antimicrobials against Streptococcus pneumoniae: correlation with penicillin resistance. Clin Ther. 1998;20:1179–1190. doi: 10.1016/s0149-2918(98)80113-6. [DOI] [PubMed] [Google Scholar]
- 44.Thornsberry C, Jones M E, Hickey M, Mauriz Y, Kahn J, Sahm D F. Resistance surveillance of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis in the United States, 1997–1998. J Antimicrob Chemother. 1999;44:749–759. doi: 10.1093/jac/44.6.749. [DOI] [PubMed] [Google Scholar]
- 45.Varon E, Janoir C, Kitzis M-D, Gutmann L. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:302–306. doi: 10.1128/aac.43.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woodcock J M, Andrews J M, Boswell F J, Brenwald N P, Wise R. In vitro activity of BAY 12-8039, a new fluoroquinolone. Antimicrob Agents Chemother. 1997;41:101–106. doi: 10.1128/aac.41.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]