Abstract
Background
Pneumocystis jirovecii is an opportunistic fungus that causes Pneumocystis pneumonia (PCP) in immunocompromised hosts. Over an 11-month period, we observed a rise in cases of PCP among kidney-transplant recipients (KTR), prompting an outbreak investigation.
Methods
Clinical and epidemiologic data were collected for KTR diagnosed with PCP between July 2019 and May 2020. Pneumocystis strain typing was performed using restriction fragment length polymorphism analyses and multilocus sequence typing in combination with next-generation sequencing. A transmission map was drawn, and a case-control analysis was performed to determine risk factors associated with PCP.
Results
Nineteen cases of PCP in KTR were diagnosed at a median of 79 months post-transplantation; 8 received monthly belatacept infusions. Baseline characteristics were similar for KTR on belatacept versus other regimens; the number of clinic visits was numerically higher for the belatacept group during the study period (median 7.5 vs 3). Molecular typing of respiratory specimens from 9 patients revealed coinfection with up to 7 P. jirovecii strains per patient. A transmission map suggested multiple clusters of interhuman transmission. In a case-control univariate analysis, belatacept, lower absolute lymphocyte count, non-White race, and more transplant clinic visits were associated with an increased risk of PCP. In multivariate and prediction power estimate analyses, frequent clinic visits was the strongest risk factor for PCP.
Conclusions
Increased clinic exposure appeared to facilitate multiple clusters of nosocomial PCP transmission among KTR. Belatacept was a risk factor for PCP, possibly by increasing clinic exposure through the need for frequent visits for monthly infusions.
Keywords: Pneumocystis jirovecii, kidney transplant, outbreak, next-generation sequencing, belatacept
In a Pneumocystis jiroveci pneumonia (PCP) outbreak among 19 kidney-transplant recipients in the United States, strain genotyping suggested coinfection and interhuman transmission of multiple P. jiroveci strains. Belatacept was a risk factor for PCP, potentially via increased clinic exposure.
Pneumocystis jirovecii is an opportunistic fungus that causes pneumonia primarily in hosts with compromised cell-mediated immunity and is associated with high mortality in solid organ transplant recipients [1]. Whereas disease may result from reactivation after prolonged quiescence, de novo acquisition of Pneumocystis is now thought to be a more common occurrence in susceptible hosts [2, 3]. Interhuman transmission has been documented and is thought to result from inhalation of aerosols generated by an infected individual [4]. Studies in animal models suggest that close contact for 24 hours may be sufficient for transmission to others [5], and studies of air-samples from rooms of patients with Pneumocystis pneumonia (PCP) have identified aerosolized organisms genotypically-identical to those in clinical specimens [6]. PCP outbreaks with evidence of interhuman transmission have been reported among heart [7], liver, and kidney [8] transplant recipients (KTR) in Europe, North America (Canada), and Asia but not convincingly in the United States [9–13]. In accordance with consensus guidelines [14], our transplant center institutes PCP prophylaxis for a 6-month period post-kidney transplantation and following lymphocyte-depleting therapy and had previously observed a low incidence of PCP (<1 case per year) with this strategy. However, in mid-2019, we noted a rise in the incidence of PCP among KTR, including in several patients on belatacept, prompting an outbreak investigation.
METHODS
Transplant Center Activity and Protocol
The Yale New Haven Hospital (YNHH) transplant center is located in New Haven, Connecticut, United States, and performs approximately 150 kidney transplantations annually. Protocol specifics are found in Supplementary Materials. PCP prophylaxis is initiated after transplantation with trimethoprim-sulfamethoxazole (TMP-SMX) and in those who develop acute T-cell-mediated rejection (TCMR) treated with lymphocyte depleting therapy for a period of 6 months. Routine long-term outpatient transplant clinic follow-up of KTR is performed weekly for the first month post-transplant, biweekly for months 2–3, monthly for months 4–6, every 3 months until 1 year, then biannually at the outpatient transplant center clinic. The outpatient clinic includes a common waiting room where KTR wait for appointments in proximity to other KTR.
Outbreak Investigation and Case-Control Analysis
Between 2012 and 2018, 45 cases of proven and probable PCP occurred at YNHH including 21 human immunodeficiency virus (HIV)-positive patients and 24 HIV-negative patients [15]. During this 7-year period, there were 4 cases of PCP in KTR. Starting July 2019, an increase in the number of cases of PCP among KTR was anecdotally noted by treating providers, prompting an outbreak investigation. Between July 2019 and May 2020 respiratory specimens (sputum, bronchoalveolar lavage [BAL] and/or formalin-fixed paraffin-embedded [FFPE] transbronchial lung biopsy) were prospectively saved in the pathology laboratory for future molecular analysis. Furthermore, a case-control analysis was undertaken to determine factors associated with Pneumocystis infection. For each incident case, 2 PCP-negative KTR controls with functioning grafts were randomly selected after matching by decade of age and transplant month/year. For controls, the absolute lymphocyte count (ALC) at time of PCP diagnosis was the value at the time their matched case was diagnosed with PCP. A chart review was performed to collect relevant data for cases and controls. Consensus definitions for PCP were employed [16]. Statistical analyses are detailed in the Supplementary Materials. The Yale Institutional Review board approved this study (IRB 2000023859).
P. jirovecii Genotyping
Respiratory specimens from cases with PCP were initially stored at the YNHH microbiology laboratory, then sent to the National Institutes of Health (NIH), Bethesda, MD, USA, for genotypic analysis after de-identification. P. jirovecii strain typing was performed using restriction fragment length polymorphism (RFLP) analyses, multilocus sequence typing (MLST) and next-generation sequencing (NGS) using Illumina sequencing technology to determine strain relatedness (Supplementary Table 1). Detailed typing methods are reported in the Supplementary Materials. A strain of P. jirovecii was defined as a population of P. jirovecii organisms within a clinical specimen with a unique genetic profile distinguished from other populations within the same or other specimens; the genetic profile is represented by single-nucleotide polymorphisms (SNPs) or other genetic polymorphisms determined by MLST and RFLP in this study. A cluster was defined as a set of clinical specimens containing the same strain.
Transmission Map
A record of outpatient transplant clinic visits at the YNHH transplant center was compiled in order to determine visit overlaps (on the same day) that may have facilitated transmission events. These data were used to construct a transmission map to delineate potential pathways of transmission between cases taking into account genotyping results. Outpatient transplant clinic visits from 6 months prior to the first case of PCP (15 January 2019) until the most recent PCP case (18 May 2020) were included in the analysis.
Outbreak Mitigation Measures
Once an outbreak was suspected, PCP prophylaxis with TMP-SMX was recommended for all KTR with either ALC ≤500/μL or with ALC ≤1000/μL and absolute CD4 count ≤200 cell/mm3 using TMP-SMX. Alternative agents were offered only if TMP-SMX was contraindicated. Prophylaxis was instituted over time by transplant nephrologists during follow-up outpatient clinic visits.
RESULTS
Patient Characteristics and Outcomes
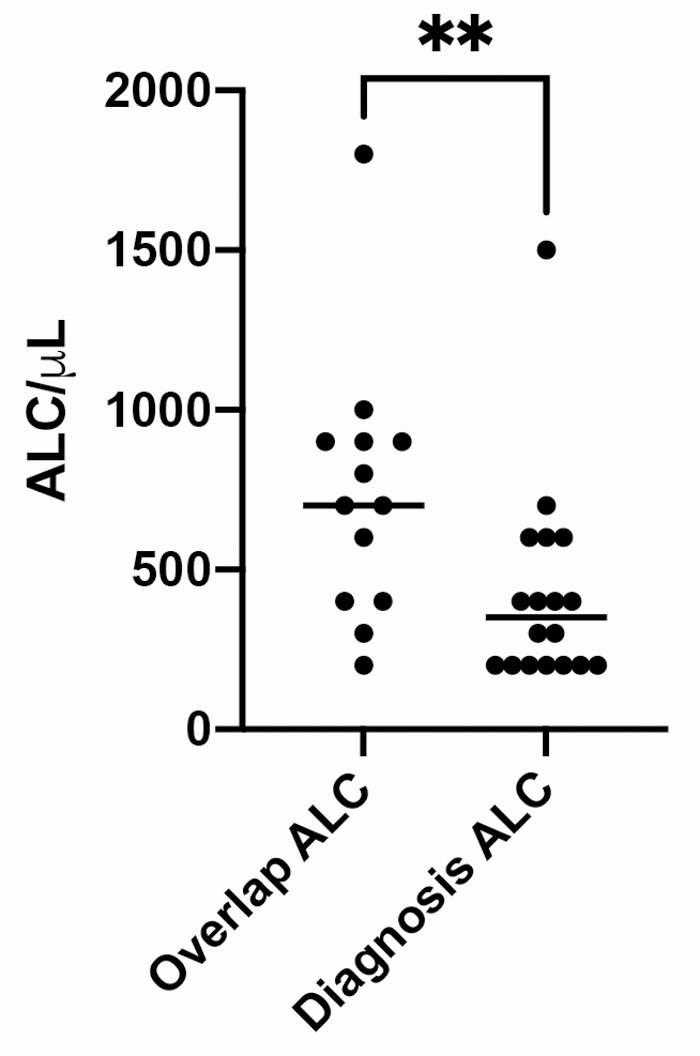
Between 1 July 2019 and 18 May 2020, 19 cases of PCP in KTR were reported, including 11 proven and 8 probable cases (Table 1). The median age was 57 years (range: 25–82), 8 (42%) were female, and 7 (37%) were Black. A calcineurin inhibitor was used for maintenance immunosuppression in 11 versus belatacept in 8. Other patient characteristics are found in Supplementary Table 2. The median time to PCP onset post-kidney transplantation was 79 months (range: 9–199). Two patients were diagnosed within 1-year post-transplant. None were receiving PCP prophylaxis at the time of PCP diagnosis. Two patients experienced TCMR treated with high-dose corticosteroids within 90 days of PCP diagnosis (since no lymphocyte depleting therapy was used, PCP prophylaxis was not instituted) while another 2 patients received ≥20mg prednisone for ≥2 weeks prior to PCP diagnosis (1 for TCMR and 1 for neurosarcoidosis). The median ALC at PCP diagnosis was 400 cells/μL (range: 200–5400). The median ALC at diagnosis was lower than in the initial observation period when several clinic overlaps occurred (500 cells/μL vs 800 cells/μL; P = .005; Figure 1). One KTR with an ALC of 5400 cells/μL (case 11) had underlying lymphoma. The median CD4 count at time of PCP diagnosis was 192 cells/mm3 (range: 27–975).
Table 1.
Baseline Characteristics of Kidney Transplant Recipients With Pneumocystis jirovecii Pneumonia (Cases) and Matched Controls
| Characteristics | Cases (N = 19) | Controls (N = 38) | Total (N = 57) | P-value |
|---|---|---|---|---|
| Sex | .45 | |||
| Female | 8 (42%) | 20 (53%) | 28 (49%) | |
| Male | 11 (59%) | 18 (47%) | 29 (51%) | |
| Race | .004 | |||
| White | 7 (37%) | 24 (63%) | 31 (55%) | |
| Black | 7 (37%) | 6 (16%) | 13 (23%) | |
| Asian | 1 (5%) | 2 (5%) | 3 (5%) | |
| Hispanic | 4 (21%) | 0 (0%) | 4 (7%) | |
| Other | 0 (0%) | 5 (13%) | 5 (9%) | |
| CMV Donor/Recipient serotype | .053 | |||
| D+/R- | 1 (5%) | 8 (21%) | 9 (16%) | |
| R+ | 14 (74%) | 14 (37%) | 28 (49%) | |
| D-/R- | 3 (16%) | 8 (21%) | 11 (19%) | |
| Unknown | 1 (5%) | 8 (21%) | 9 (16%) | |
| Donor type | .64 | |||
| Deceased donor | 12 (63%) | 21 (55%) | 33 (59%) | |
| Living donor | 7 (37%) | 16 (42%) | 23 (41%) | |
| Induction immunosuppression | .43 | |||
| Alemtuzumab | 6 (32%) | 8 (21%) | 14 (27%) | |
| Basiliximab | 2 (11%) | 10 (26%) | 12 (24%) | |
| Daclizumab | 1 (5%) | 0 (0%) | 1 (2%) | |
| Steroids | 3 (16%) | 6 (16%) | 9 (18%) | |
| Thymoglobulin | 5 (26%) | 10 (26%) | 15 (29%) | |
| Belatacept | .021 | |||
| Yes | 8 (42%) | 5 (13%) | 13 (23%) | |
| No | 11 (58%) | 33 (87%) | 44 (77%) | |
| mTOR inhibitor | .20 | |||
| Sirolimus | 0 (0%) | 3 (8%) | 3 (5%) | |
| Everolimus | 1 (5%) | 0 (0%) | 1 (2%) | |
| None | 18 (95%) | 35 (92%) | 53 (93%) | |
| Antiproliferative agent | .26 | |||
| Mycophenolate mofetil | 17 (89%) | 26 (68%) | 43 (75%) | |
| Azathioprine | 0 (0%) | 4 (11%) | 4 (7%) | |
| None | 2 (11%) | 8 (21%) | 10 (18%) | |
| Mycophenolate daily dose (mg) | .42 | |||
| 0 | 3 (16%) | 12 (32%) | 15 (26%) | |
| 500 | 0 (0%) | 2 (5%) | 2 (4%) | |
| 1000 | 7 (37%) | 9 (24%) | 16 (28%) | |
| 1500 | 3 (16%) | 4 (11%) | 7 (12%) | |
| 2000 | 5 (26%) | 11 (29%) | 16 (28%) | |
| 2500 | 1 (5%) | 0 (0%) | 1 (2%) | |
| Prednisone daily dose (mg) | .46 | |||
| 0 | 0 (0%) | 5 (13%) | 5 (9%) | |
| 2.5 | 0 (0%) | 1 (3%) | 1 (2%) | |
| 5 | 18 (95%) | 28 (74%) | 46 (81%) | |
| 7.5 | 0 (0%) | 1 (3%) | 1 (2%) | |
| 10 | 1 (5%) | 3 (8%) | 4 (7%) | |
| Duration between transplant and PCP diagnosis in months (median, range) | 79 (9–199) | N/A | N/A | N/A |
| TCMR within 90 days of PCP diagnosis | .59 | |||
| Yes | 2 (11%) | 2 (5%) | 4 (7%) | |
| No | 17 (89%) | 36 (95%) | 53 (93%) | |
| Receipt of prednisone ≥ 20mg/day for ≥ 2 weeks within 90 days of PCP diagnosis | .59 | |||
| Yes | 2 (11%) | 2 (5%) | 4 (7%) | |
| No | 17 (89%) | 36 (95%) | 53 (93%) | |
| Number of transplant clinic visits within 180 days of PCP diagnosis (median, range) | 6(0–16) | 2 (0– 15) | 2 (0–16) | .003 |
| CMV viremia within 90 days of PCP diagnosis | .011 | |||
| Yes | 4 (21%) | 0 (0%) | 4 (7%) | |
| No | 15 (79%) | 37 (97%) | 52 (93%) | |
| Absolute lymphocyte count (cells/μL) at PCP diagnosis (median, range) | 400 (200–5400) | 1200 (100–3800) | 800 (100–5400) | .044 |
| CD4 T-cell count at PCP diagnosis (cells/mm3) | 192 (27–975)a | N/A | N/A | N/A |
Abbreviations: CMV, cytomegalovirus; N/A, not applicable; PCP, Pneumocystis jirovecii pneumonia; TCMR, T-cell-mediated rejection.
aAvailable for 8 patients.
Figure 1.
Median absolute lymphocyte count was significantly higher at time of initial patient clinic overlap than at time of diagnosis with PCP (800 cells/μL vs 500 cells/μL; P = .005). One outlier (ALC 4500 cells/μL) is not represented in the future in order to maintain a readable scale. Abbreviations: ALC, absolute lymphocyte count; PCP, Pneumocystis pneumonia.
Baseline characteristics were similar for 8 KTR on belatacept and 11 KTR on non-belatacept regimens (Table 2), but the number of clinic visits was numerically higher for the belatacept group (median 7.5 vs 3). The median time from transplant to belatacept initiation was 38 months (range: 5–104). The time from belatacept initiation to PCP diagnosis ranged from 15 days to 4 years (Supplementary Table 2). The median ALC at belatacept initiation was 700 cells/μL (range: 200–3600) and the median ALC at PCP diagnosis in the belatacept group was 300 cells/μL (range: 200–5400) compared to 400 cells/μL (range: 200–1500) for the non-belatacept group.
Table 2.
Baseline Characteristics of Kidney Transplant Recipients on Belatacept Versus Non-Belatecept Maintenance Immunosuppression Regimens
| Non-Belatecept (n = 11) | Belatecept (n = 8) | P value | |
|---|---|---|---|
| Age, average ± SD | 58.3 ± 15.6 | 56.1 ± 16.3 | .7743 |
| Race | .0635 | ||
| White, n (%) | 2 (18.2%) | 5 (62.5%) | |
| Black, n (%) | 5 (45.5%) | 2 (25.0%) | |
| Hispanic, n (%) | 4 (36.4%) | 0 (0%) | |
| Asian, n (%) | 0 (0%) | 1 (12.5%) | |
| Rejection within 90 days of PCP diagnosis, n (%) | 2 (18.2%) | 0 (0%) | .4854 |
| CMV viremia within 90 days, n (%) | 2 (18.2%) | 2 (25.0%) | 1.000 |
| Number of clinic visits within 180 days of PCP diagnosis, median (IQR) | 3 (1–11) | 7.5 (4.5–9.75) | .4461 |
| ALC/µL at PCP diagnosis, median (IQR) | 400 (200–600) | 300 (200–600) | .8878 |
Abbreviations: ALC, absolute lymphocyte count; CMV, cytomegalovirus; IQR, interquartile range; PCP, Pneumocystis jirovecii pneumonia; SD, standard deviation.
TMP-SMX was the most common treatment regimens for PCP (n = 8); other agents included atovaquone and clindamycin/primaquine. Of 19 KTR, 3 died within 4 weeks of PCP diagnosis. All 16 survivors were placed on lifelong secondary prophylaxis with TMP-SMX or atovaquone.
Genotyping Results and Transmission Map
We performed comprehensive genotypic analysis of P. jirovecii DNA extracted from 3 BAL specimens (cases 8, 11, and 15) and 6 FFPE specimens (cases 1, 3, 6, 13, 14, and 16) for individual patients (Supplemental Text 4). Initial Sanger sequencing identified homogenous sequences in most (7/9) specimens, resulting in 6 distinct P. jirovecii strains based on MLST profiles (Supplemental Table 3). Subsequent NGS identified extensive variations within and between all specimens and revealed co-infection with up to 7 P. jirovecii strains in all patients (Supplementary Tables 4–7). Coinfection complicated strain type determination, especially for specimens showing coinfection with ≥3 strains.
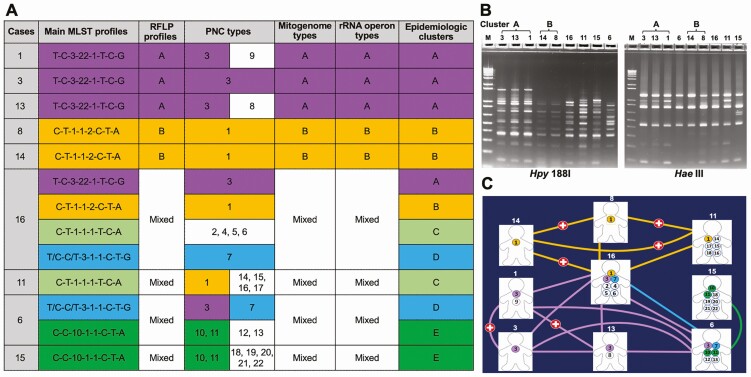
The combination of MLST and RFLP profiles allowed identification of 5 clusters (Figure 2 and Supplementary Figure 1). The dominant P. jirovecii strains from cases 1, 3, and 13 showed identical MLST and RFLP profiles (designated as cluster A), as did the dominant strains from cases 8 and 14 (cluster B). The identity of the dominant strains within both clusters was further supported by sequence analysis of the complete or large portions of the P. jirovecii mitogenome and nuclear rDNA sequences (Figure 2 and Supplementary Materials). All remaining patients appeared to be coinfected with multiple strains, some of which were shared among multiple patients in the cohort, forming 3 additional clusters (C, D, and E). Remarkably, 1 patient (case 16) was likely coinfected with 7 P. jirovecii strains, 4 of which were shared with 7 other patients including the 5 in clusters A and B (Figure 2 and Supplementary Tables 5 and 7).
Figure 2.
Molecular epidemiology of P. jirovecii in 9 kidney transplant recipients. A, Summary of molecular genotyping and epidemiologic clusters. Each MLST profile represents a combination of SNPs (or genotypes) at mtLSU, PNC, ITS1-ITS2, 26S rRNA, soda, and dhps as detailed in Supplementary Figure 2. In cases 1, 3, 13, 8, and 14, the profiles shown represent the dominant strains. In the remaining patients (each coinfected with 4–7 strains), the profiles shown represent putative matching profiles between patients; profiles without matches are not shown to enhance visualization. Epidemiologic clusters are defined according to the consensus of the main MLST and RFLP profiles, and PNC, mitogenome and rRNA operon types. Some PNC types shared between different patients but lacking support by MLST profiles are excluded in the cluster determination. For example, PNC type 1 was shared among patients 8, 14, 16, and 11 but patient 11 did not contain a putative MLST profile shared with patients 8 and 14; thus patient 11 was not considered to be related to cluster B. Multiple strains unable to be differentiated reliably are indicated as mixed profiles or types. B, RFLP analysis of P. jirovecii samples. PCR products from the first-run were digested with restriction enzyme Hpy 188I while the semi-nested PCR products were digested with Hae III. Both gels were stained with SYBR green. Numbers at the top represent the individual patient IDs. M at the top represents DNA size markers. Samples showing the same pattern for both the first-run and semi-nested PCR products are considered the same cluster. Samples 3, 13, and 1 exhibit the same RFLP pattern (designated as cluster A) and so do samples 14 and 8 (cluster B). All the remaining samples have a unique RFLP pattern. C, Distribution of the 22 PNC types in transplant patients. Case numbers are indicated above the image of each patient; 22 PNC types (Supplementary Tables 5 and 8) are represented by numbers 1 to 22 in the circles. PNC types shared between 2 or more patients are color-coded. Patients infected with the same PNC types are connected by lines with the same colors for the PNC types. Of note, every patient shows a direct connection with at least 1 other patient, whereas patient 16 shows a direct connection with all other patients except for patient 15. Red cross indicates overlaps in clinic visit between patients. Human images are from https://www.hyglossproducts.com/people-shape-cut-outs-white-19131.html. Abbreviations: MLST, multilocus sequence typing; PCR, polymerase chain reaction; PNC, polymorphic non-coding; RFLP, restriction fragment length polymorphism; SNP, single-nucleotide polymorphism.
To complement complete mitogenome and rRNA open sequence analyses, we examined NGS data for a ~100-bp polymorphic non-coding (PNC) region of the P. jirovecii mitogenome and identified extensive SNPs and indels, yielding a total of 22 unique genotypes from all 9 patients (Supplementary Tables 5 and 8). Four of these patients showed coinfection with 6–7 different PNC genotypes. Based on the distribution patterns of these 22 PNC types (Figure 2), every patient was infected with ≥1 strain that were detected in at least 1 other patient, although 1 or more strains from case 16 were detected in all other patients except for case 15.
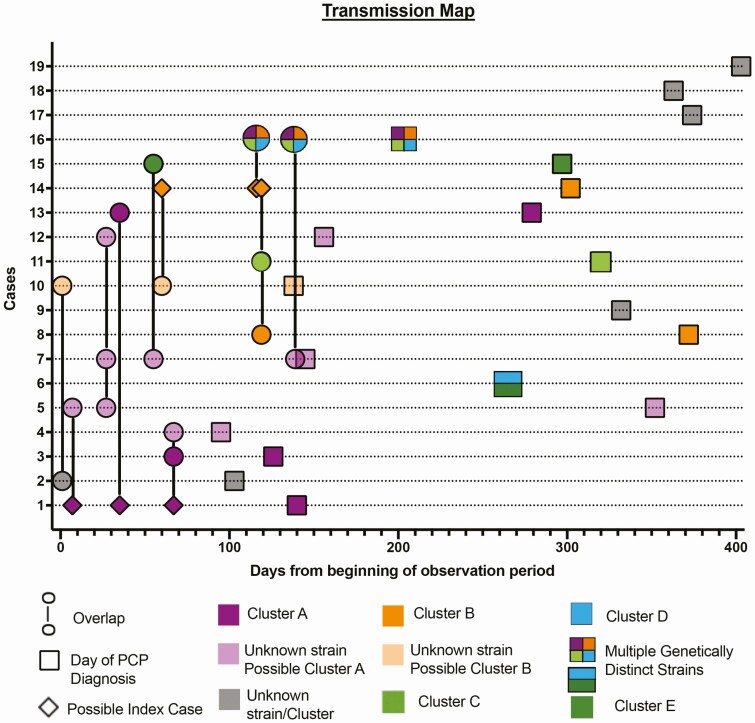
Dates of PCP diagnoses, clinic visits and overlaps, and results of genetic analyses were compiled to form a proposed transmission map (Figure 3 and Supplementary Materials).
Figure 3.
Transmission map depicting clinic overlaps (illustrated by a connecting vertical line) and times of PCP diagnosis for 19 cases of PCP during the study time period. These include genotypically characterized clusters A (cases 1, 3, 13, and 16) and B (cases 8, 14, and 16), C (cases 11 and 16), D (cases 6 and 16), and E (cases 6 and 15). Cases 4, 5, 7, and 12, for which no strain genotyping was done, were considered as possibly belonging to cluster A because of multiple overlaps with cluster A cases and each other, whereas case 10 (also not genotyped) was considered as possibly belonging to cluster B because of overlaps with cluster B cases. Case 16 had 4 cluster strains, each belonging to a different cluster (A through D), whereas case 6 had 2 cluster strains (D and E). Cases 1 and 14 were considered to be possible index patients for clusters A and B, respectively, due to multiple clinic-overlaps with other cases in the corresponding clusters and their severe immunodeficiency. Abbreviation: PCP, Pneumocystis pneumonia.
Case-Control Analysis
Among 38 non-PCP matched controls, 18 (47%) were males, 24 (63%) were White, the median age was 60 years (range: 26–84), and 33 were on calcineurin inhibitors, whereas 5 were on belatacept (Table 1). Two controls were on high-dose corticosteroids, and 2 had TCMR within 90 days of PCP diagnosis of their matched case. CMV viremia was detected in 4 cases versus no controls (P = .011). The median ALC during this time period was 400 cells/μL for cases versus 1200 cells/μL for controls (P = .044). The median number of transplant clinic visits within 180 days prior to diagnosis was 6 for cases versus 2 for controls (P = .003). Other variables are found in Table 1.
In a univariate conditional analysis, use of belatacept (odds ratio [OR] 4.67 [95% confidence interval {CI}, 1.21, 18.05], P = .026), lower ALC at PCP diagnosis (OR 1.11 [95% CI 1.01, 1.22], P = .028), and more transplant clinic visits (OR 1.31 [95% CI 1.06, 1.63], P = .012) were associated with an increased odds of PCP while White race (OR 0.18 [95% CI .04, .85], P = .033) was associated with a lower rate of PCP as compared to other races (Table 3). For multivariate analyses, the stepwise model selection procedure indicated the number of clinic visits was the strongest predictor of PCP infection. Given the strong correlation between number of clinics visits, belatacept and TMCR within 90 days of PCP diagnosis, we repeated the model selection process after removing the number of clinic visits from the candidate variables list and noted that race, belatacept and TMCR within 90 days of PCP diagnosis were significantly associated with PCP (Table 4). When a prediction power estimate analysis was performed, the log likelihood and Briar score change indicated that belatacept, TCMR and ALC contributed to predicting the outcome of developing PCP. However, the negligible amount of change using Akaike’s information criterion suggested that the number of clinic visit was the strongest predictor for this outcome (Supplementary Table 9). Furthermore, the number of clinic visits contributed the biggest variance (R2 = 0.35, Supplementary Table 10) to the outcome of developing PCP.
Table 3.
Simple Logistic Regression Analysis Evaluating Risk of PCP
| Variable | Comparison | Odds Ratios (95% CI) | P value |
|---|---|---|---|
| Age | Increase by 1 year | 0.93 (.81, 1·07) | .30 |
| Sex | Female vs male | 0.67 (.22, 1.99) | .47 |
| Race | White vs other | 0.18 (.04, .87) | .033 |
| Duration between transplant and PCP diagnosis | Increase by 1 month | 1.20 (.91, 1.58) | .20 |
| Donor Type | Deceased donor vs living donor | 1.32 (.36, 4.80) | .67 |
| Induction immunosuppression | Non-steroids vs steroids | 1.00 (.05, 18·91) | 1.00 |
| TCMR within 90 days of PCP diagnosis | Yes vs no | 2.00 (.28, 14.20) | .49 |
| Belatacept | Yes vs no | 4.67 (1.21, 18·05) | .026 |
| mTOR inhibitor | Yes vs no | 0.67 (.07, 6.41) | .73 |
| Antiproliferative agent | Yes vs no | 2.14 (.43, 10.71) | .36 |
| MMF total daily dose | Increase by 500 mg | 1.24 (.87, 1.77) | .24 |
| Number of transplant clinic visits within 180 days of PCP diagnosis | Increase by 1 day | 1.31 (1.06, 1.63) | .012 |
| Receipt of prednisone ≥ 20mg/day for ≥ 2 weeks within 90 days of PCP diagnosis | Yes vs no | 2.00 (.28, 14.20) | .49 |
| Absolute Lymphocyte Count (cells/μL) at PCP diagnosis | Decrease by 100 cells/μL | 1.11 (1.01, 1.22) | .028 |
Abbreviations: CI, confidence interval; MMF, mycofenolate mofetil; mTOR, mechanistic target of rapamycin; PCP, Pneumocystis jirovecii pneumonia; TCMR: T-cell-mediated rejection.
Table 4.
Multiple Logistic Regression Analysis Evaluating Risk of PCP
| Variable | Comparison | Odds Ratios (95% CI) | P value |
|---|---|---|---|
| Race | White vs other | 0.05 (.00, .48) | .010 |
| Belatacept | Yes vs no | 15.21 (1.70, 135.91) | .015 |
| TCMR within 90 days of PCP diagnosis | Yes vs no | 26.68 (1.07, 665.05) | .045 |
Race, Belatacept use, and TCMR within 90 days of PCP diagnosis were included in the multivariate analysis. Number of transplant clinic visits within 180 days was excluded from this multivariate analysis to evaluate the effect of other variables on the risk of PCP since these are rendered nonsignificant when the number of transplant clinic visits is included in the model.
Abbreviations: CI, confidence interval; PCP, Pneumocystis jirovecii pneumonia; TCMR, T-cell-mediated rejection.
DISCUSSION
In this study, we report an outbreak of PCP among KTR in our transplant center that occurred over the span of a year. An epidemiologic investigation using in-depth genomic analysis, transmission mapping and a case-control analysis was suggestive of multiple clusters of interhuman transmission of genotypically related P. jirovecii strains between transplant recipients, the first such report in the United States. Although an outbreak of genotypically distinct strains of P. jirovecii that included 3 clusters was described in kidney and liver transplant recipients in Denmark [8], our findings are in contrast to several other PCP outbreak analyses that suggested transmission of a single common P. jirovecii strain [7, 8, 11]. Notably, genotyping in some studies was based on polymorphism analysis in a small number of genes, a method that may have overlooked minor strains and coinfection.
In this study, the high coverage of NGS data for the selected genetic loci allowed detection of coinfection in all patients, the majority of which were undetected by initial Sanger sequencing. Although the short NGS reads did not permit construction of haplotypes for all minor strains, sequence analysis of nuclear rDNA and mitochondrial genomes as well as ~100 bp mitochondrial PNC types offered deeper insights into coinfection. Our analyses identified co-infection with up to 7 P. jirovecii strains in all patients. These observations raise the possibility of transmission of multiple strains, a finding that may have implications for other outbreak analyses. It is possible that after co-infection of newly infected patients, some strains preferentially survived due to pre-existing immune responses, strain fitness or other factors. Our results also argue that NGS has a critical role in epidemiologic studies of PCP given the high prevalence of coinfection [17] and that PNC is potentially a novel, highly discriminatory genetic marker for P. jirovecii strain typing.
Interhuman transmission of P. jirovecii between transplant recipients has been well documented in solid organ transplant recipients, but underlying host and environmental conditions that facilitate a chain of nosocomial transmission in this population are not well defined [7–9]. In a case-control analysis to identify factors underlying the outbreak, a greater number of clinic visits was most strongly associated with PCP risk. In the context of this outbreak, frequent exposures to sub-clinically infected KTR who constituted a reservoir for the fungus may have placed heretofore uninfected patients at risk of PCP acquisition, facilitating nosocomial transmission. Patient-to-patient transmission potentially occurred through inhalation of airborne droplets produced by colonized or infected KTR, a phenomenon not assessed here, but demonstrated in other studies through air sampling [4]. A transmission map supported the hypothesis of multiple transmission chains originating from at least 2 immunosuppressed index patients who likely harbored a high organism burden (as evidenced by positive BAL silver stains in both cases) that could be transmitted to others.
In addition to clinic exposure, lower ALC, belatacept use and race as well as recent TCMR and CMV viremia (suggestive of a higher net state of immunosuppression) may have influenced the risk of PCP development. In a case-control analysis, ALC at diagnosis was lower in cases than in controls. In addition to a greater likelihood of developing PCP, lymphopenia has been associated with a higher burden of Pneumocystis organisms, which may pose a greater transmission risk to others [18]. Additionally, the ALC of KTRs appeared to decrease from >500 cells/μL around the time of potential exposure (early on in the outbreak) to <500 cells/μL at time of PCP diagnosis. Therefore, lymphopenia may have contributed both to transmission and development of PCP.
Notably, maintenance immunosuppression with belatacept was associated with a greater likelihood of PCP. Belatacept, a selective co-stimulation blocker, has been linked to an increased risk for PCP in a handful of studies, but a causative relationship has not been demonstrated [19, 20]. In a case-control study of 23 KTR with PCP at a single transplant center, patients on belatacept had an increased risk for PCP. However, the risk was attributed to other co-occurring risk factors including older age, higher rates of lymphopenia, and lower estimated glomerular filtration rate in the belatacept group rather than a direct effect on host immune function [20]. Although ALC and other factors were similar across belatacept and non-belatacept groups, the median number of clinic visits was numerically higher in the belatacept group (median 7.5 vs 3). This is likely because belatacept administration requires monthly clinic visits for infusions whereas other maintenance immunosuppression regimens are self-administered orally in the home setting. Although belatacept use was significantly associated with PCP in univariate analyses and in multivariate analyses that excluded the number of clinic visits, it was no longer significant when the number of clinic visits was included in the model, suggesting that the risk conferred by belatacept may be largely encompassed by the number of clinic visits. Taken together, these data suggest that belatacept was possibly a proxy for increased clinic visits and conferred a higher risk of PCP more so by increasing the likelihood of exposure to the clinic setting than by its impact on cell-mediated immunity.
A notable observation in this study was the extended period (median of 79 months) between transplantation and eventual diagnosis of PCP. Although underlying reasons are unclear, the longer interval could be related to development of graft dysfunction and switch to belatacept in the later post-transplant period (median of 38 months post-transplantation). Although most treatment guidelines recommend limited post-kidney transplant Pneumocystis prophylaxis, both sporadic and epidemic PCP cases have been documented in KTR many years after transplantation, prompting calls for universal prophylaxis after kidney transplantation [21]. A better delineation of the safety and tolerability of lifelong prophylactic strategies is needed but indefinite prophylaxis strategies have already been implemented in KTR with good success [21].
This study has several limitations. Due to the single-center retrospective design, robust causal inference cannot be established with certainty and results may not be generalizable to other centers. Based on the institutional protocol, Pneumocystis prophylaxis was not instituted when steroids were used for TCMR treatment, which may have impacted the incidence of PCP. Probable cases of PCP were included. Genotyping was performed for only 9 of 19 cases, precluding a more comprehensive genotypic transmission network analysis. In addition, we did not collect or perform genotyping of control strains. Other factors that were unaccounted for may have led to increased clinic visits.
In conclusion, this outbreak investigation suggested multiple clusters of interhuman transmission of PCP among KTR as well as coinfection with multiple strains. Belatacept was a risk factor for PCP, potentially via increased clinic exposure. Infection prevention measures and long-term chemoprophylaxis for PCP should be considered in transplant clinics given the possibility of person-to-person transmission, particularly when higher than usual incidences are observed.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This study was support in part by intramural research funds from the National Institute of Health Clinical Center.
Potential conflicts of interest. J. K. was a principal investigator on a Cooperative Research and Development Agreements (CRADA), now expired, between the National Institutes of Health and Matinas BioPharma Inc. to support clinical trials of encochleated drugs. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Iriart X, Bouar ML, Kamar N, Berry A.. Pneumocystis pneumonia in solid-organ transplant recipients. J Fungi (Basel) 2015; 1:293–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wakefield AE, Lindley AR, Ambrose HE, Denis CM, Miller RF. Limited asymptomatic carriage of Pneumocystis jiroveci in human immunodeficiency virus-infected patients. J Infect Dis 2003; 187:901–8. [DOI] [PubMed] [Google Scholar]
- 3. Morris A, Norris KA. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev 2012; 25:297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Le Gal S, Pougnet L, Damiani C, et al. Pneumocystis jirovecii in the air surrounding patients with Pneumocystis pulmonary colonization. Diagn Microbiol Infect Dis 2015; 82:137–42. [DOI] [PubMed] [Google Scholar]
- 5. Dumoulin A, Mazars E, Seguy N, et al. Transmission of Pneumocystis carinii disease from immunocompetent contacts of infected hosts to susceptible hosts. Eur J Clin Microbiol Infect Dis 2000; 19:671–8. [DOI] [PubMed] [Google Scholar]
- 6. Choukri F, Menotti J, Sarfati C, et al. Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis 2010; 51:259–65. [DOI] [PubMed] [Google Scholar]
- 7. Vindrios W, Argy N, Le Gal S, et al. Outbreak of Pneumocystis jirovecii infection among heart transplant recipients: molecular investigation and management of an interhuman transmission. Clin Infect Dis 2017; 65:1120–6. [DOI] [PubMed] [Google Scholar]
- 8. Rostved AA, Sassi M, Kurtzhals JA, et al. Outbreak of Pneumocystis pneumonia in renal and liver transplant patients caused by genotypically distinct strains of Pneumocystis jirovecii. Transplantation 2013; 96:834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Desoubeaux G, Dominique M, Morio F, et al. Epidemiological outbreaks of Pneumocystis jirovecii pneumonia are not limited to kidney transplant recipients: genotyping confirms common source of transmission in a liver transplantation unit. J Clin Microbiol 2016; 54:1314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Boer MG, de Fijter JW, Kroon FP. Outbreaks and clustering of Pneumocystis pneumonia in kidney transplant recipients: a systematic review. Med Mycol 2011; 49:673–80. [DOI] [PubMed] [Google Scholar]
- 11. Mulpuru S, Knoll G, Weir C, et al. Pneumocystis pneumonia outbreak among renal transplant recipients at a North American transplant center: risk factors and implications for infection control. Am J Infect Control 2016; 44:425–31. [DOI] [PubMed] [Google Scholar]
- 12. Hardy AM, Wajszczuk CP, Suffredini AF, Hakala TR, Ho M. Pneumocystis carinii pneumonia in renal-transplant recipients treated with cyclosporine and steroids. J Infect Dis 1984; 149:143–7. [DOI] [PubMed] [Google Scholar]
- 13. Mejia CD, Malat GE, Boyle SM, Ranganna K, Lee DH. Experience with a six-month regimen of Pneumocystis pneumonia prophylaxis in 122 HIV-positive kidney transplant recipients. Transpl Infect Dis 2020: e13511. [DOI] [PubMed] [Google Scholar]
- 14. Fishman JA, Gans H; AST Infectious Diseases Community of Practice . Pneumocystis jiroveci in solid organ transplantation: guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 2019; 33:e13587. [DOI] [PubMed] [Google Scholar]
- 15. Azar MM, Slotkin R, Abi-Raad R, Liu Y, Grant MH, Malinis MF. Gomori methenamine silver stain on bronchoalveolar lavage fluid is poorly sensitive for diagnosis of Pneumocystis jiroveci pneumonia in HIV-negative immunocompromised patients and may lead to missed or delayed diagnoses. Arch Pathol Lab Med 2020. [DOI] [PubMed] [Google Scholar]
- 16. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis 2019; 71:1367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ma L, Cisse OH, Kovacs JA. A molecular window into the biology and epidemiology of Pneumocystis spp. Clin Microbiol Rev 2018; 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu CJ, Lee TF, Ruan SY, Yu CJ, Chien JY, Hsueh PR. Clinical characteristics, treatment outcomes, and prognostic factors of Pneumocystis pneumonia in non-HIV-infected patients. Infect Drug Resist 2019; 12:1457–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haidinger M, Hecking M, Memarsadeghi M, et al. Late onset Pneumocystis pneumonia in renal transplantation after long-term immunosuppression with belatacept. Transpl Infect Dis 2009; 11:171–4. [DOI] [PubMed] [Google Scholar]
- 20. Brakemeier S, Dürr M, Bachmann F, Schmidt D, Gaedeke J, Budde K. Risk evaluation and outcome of Pneumocystis jirovecii pneumonia in kidney transplant patients. Transplant Proc 2016; 48:2924–30. [DOI] [PubMed] [Google Scholar]
- 21. Goto N, Takahashi-Nakazato A, Futamura K, et al. Lifelong prophylaxis with trimethoprim-sulfamethoxazole for prevention of outbreak of Pneumocystis jirovecii pneumonia in kidney transplant recipients. Transplant Direct 2017; 3:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.