Abstract
Background
Chicken intramuscular fat (IMF) content is closely related to meat quality and performance, such as tenderness and flavor. Abdominal fat (AF) in chickens is one of the main waste products at slaughter. Excessive AF reduces feed efficiency and carcass quality.
Results
To analyze the differential deposition of IMF and AF in chickens, gene expression profiles in the breast muscle (BM) and AF tissues of 18 animals were analyzed by differential expression analysis and weighted co-expression network analysis. The results showed that IMF deposition in BM was associated with pyruvate and citric acid metabolism through GAPDH, LDHA, GPX1, GBE1, and other genes. In contrast, AF deposition was related to acetyl CoA and glycerol metabolism through FABP1, ELOVL6, SCD, ADIPOQ, and other genes. Carbohydrate metabolism plays an essential role in IMF deposition, and fatty acid and glycerol metabolism regulate AF deposition.
Conclusion
This study elucidated the molecular mechanism governing IMF and AF deposition through crucial genes and signaling pathways and provided a theoretical basis for producing high-quality broilers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08538-0.
Keywords: Abdominal fat, Chicken, Intramuscular fat, Transcriptomics, WGCNA
Introduction
Fat deposition in broilers is affected by genetic factors (breed, sex, and genotype) and nongenetic factors (nutrition, age, and environmental factors). The rate of fat deposition varies across tissues in broilers. In addition, glucose and lipid metabolism and hormone sensitivity vary in adipocytes from different tissues [1–4]. The deposition of abdominal fat (AF) and intramuscular fat (IMF) is regulated by different mechanisms in chickens [5], and this characteristic allows the genetic selection of broilers with high IMF and low AF [6].
Previous studies showed a significant and positive correlation between IMF and AF [7, 8]. In contrast, a study found that the chicken with lower AF had higher IMF. The discrepancy may be caused by breeding characteristics, environmental factors, genetic selection, and sampling methods [6]. A balanced selection population (increasing IMF, decreasing AF percentage (AFP)) was more effective than selecting for IMF alone in Jing-xing yellow chicken [9]. Various genetic mechanisms regulate lipid deposition in tissues. Genes encoding perilipin and long-chain acyl CoA dehydrogenase are highly expressed in subcutaneous adipocytes, whereas genes encoding bone morphogenetic proteins 4 and 7 are abundantly expressed in intramuscular adipocytes [10]. With the development of cost-effective sequencing technologies, a large amount of transcriptome data has been generated under different biological back scenes. Currently, based on these high-throughput sequencing data, Weighted correlation network analysis (WGCNA) has been successfully applied to identify gene expression networks and biomarkers of interest in various biological domains [11–16]. Studies have shown that the transcription factor CREB3L1 was associated with low AF, whereas the transcription factor L3MBTL1 and the cofactor TNIP1 were related to high IMF and low AF in the Jing-xing Huang chicken line [17]. These results underscore the need to study the genetic mechanisms regulating fat deposition in broiler chickens. Wenchang breed is mainly distributed in Hainan, the lowest altitude in south China. It is an excellent local breed with more than 400 years of breeding history. Wenchang chicken is usually marketed in 100–110 days and weighs about 1.4–1.8 kg. Wenchang chicken has the characteristics of thin and tender skin, fragrant meat, and high IMF. However, the AF content of Wenchang chicken is also high, which will cause waste in production and reduce the uniformity of population. Therefore, it is necessary to understand the characteristics of fat deposition in Wenchang chickens, which is beneficial to improve Wenchang chickens’ quality and production efficiency. Few studies jointly evaluated IMF and AF in broilers. This study evaluates differential fat deposition in breast muscle (BM) and AF tissues of Wenchang chicken, gene expression profiles these tissues based on transcriptome data, and genes and signaling pathways involved in IMF and AF deposition in chicken to reveal the difference of molecular regulation mechanisms.
Results
Evaluation of sequencing data
Transcriptomic data were obtained from the BM and AF of 18 Wenchang chickens. Thirty-six RNA-sequencing (RNA-seq) libraries were constructed and sequenced (Additional file: Table S1). A total of 23,910 genes were detected, of which 17,179 genes were expressed in BM, and 18,912 genes were expressed in AF. In addition, 72.24% (16,753/23,910) of the genes were expressed in both BM and AF (Fig. 1). Raw sequence reads from each library are shown in Additional files: Tables S2 and S3. The most genes abundantly expressed in BM and AF were glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and fatty acid-binding protein (FABP4), respectively.
Fig. 1.
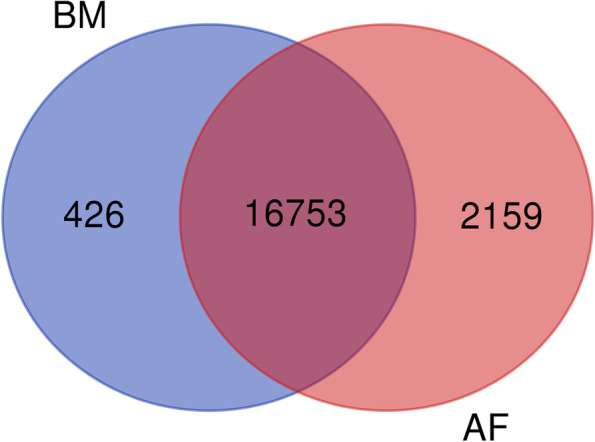
Venn diagrams of genes differentially expressed in the breast muscle and abdominal fat of broiler chickens
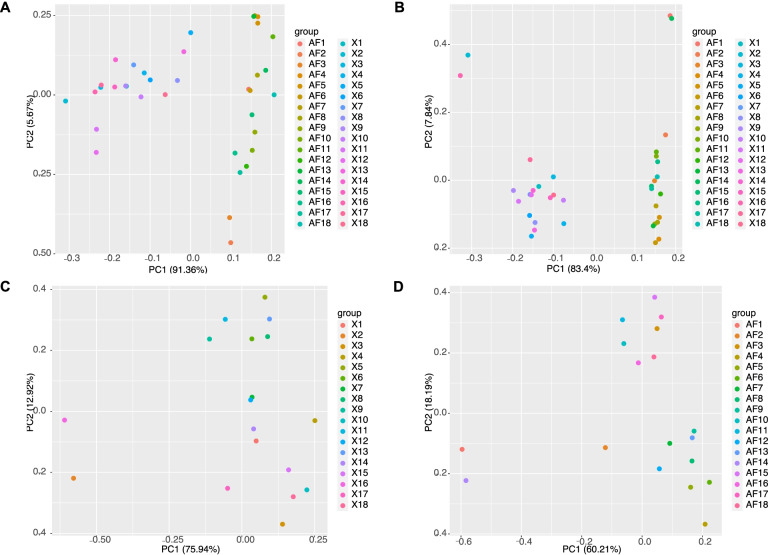
To explore differences in IMF and AF deposition (Table 1), Principal component analysis (PCA) of fatty acid composition was performed to assess differences in fat deposition in BM and AF tissue (Fig. 2A) [18].
Table 1.
Phenotypes of Wenchang chicken in different tissues
| Phenotypes | Breast Muscle | Abdominal Fat |
|---|---|---|
| IMF/AFP | 1.2944 ± 0.1089a | 4.7266 ± 0.3988b |
| TG | 4.9691 ± 0.5710a | 23.5572 ± 1.8791b |
| C10:0 | ND | 0.0093 ± 0.0005 |
| C12:0 | 0.0364 ± 0.0023 | 0.0280 ± 0.0009 |
| C14:0 | 0.4470 ± 0.0145 | 0.6762 ± 0.0117 |
| C14:1 | 0.0788 ± 0.0052 | 0.1531 ± 0.0075 |
| C15:0 | 0.0706 ± 0.0016a | 0.08967 ± 0.0033 |
| C16:0 | 26.1824 ± 0.1856a | 27.2354 ± 0.3255b |
| C16:1 | 2.8540 ± 0.1824 | 5.1318 ± 0.2777 |
| C17:0 | 0.1328 ± 0.0062 | 0.1417 ± 0.0060 |
| C18:0 | 11.5981 ± 0.2874 | 6.9239 ± 0.2392 |
| C18:1n9c | 28.9134 ± 0.7301a | 39.0340 ± 0.3500b |
| C18:2n6c | 17.6387 ± 0.1924a | 18.7408 ± 0.4955b |
| C18:3n3 | 0.4663 ± 0.0158 | 0.8446 ± 0.0158 |
| C20:0 | 0.1898 ± 0.0083 | 0.1227 ± 0.0056 |
| C20:1 | 0.2493 ± 0.0079 | 0.3452 ± 0.0129 |
| C20:2 | ND | 0.0132 ± 0.0007 |
| C20:3n3 | 0.1106 ± 0.0080a | 0.0082 ± 0.0007b |
| C20:3n6 | 0.9901 ± 0.0583a | 0.0846 ± 0.0042b |
| C20:4n6 | 7.8473 ± 0.4910a | 0.1625 ± 0.0135b |
| C20:5n3 | 0.1186 ± 0.0073a | 0.0082 ± 0.0009b |
| C21:0 | 0.4080 ± 0.0460a | 0.1433 ± 0.0074b |
| C22:0 | 0.1949 ± 0.0152a | 0.0254 ± 0.0015b |
| C22:1n9 | 0.1233 ± 0.0098a | 0.0214 ± 0.0008b |
| C22:2 | ND | 0.0039 ± 0.0009 |
| C22:6n3 | 0.8751 ± 0.0648a | 0.0139 ± 0.0016b |
| C23:0 | 0.0872 ± 0.0053a | 0.0233 ± 0.0009b |
| C24:0 | 0.1540 ± 0.0084a | 0.0131 ± 0.0013b |
| C24:1 | 0.2333 ± 0.0130a | 0.0027 ± 0.0008b |
Note: Means differences of fatty acids among treatments were determined with Duncan’s multiple range test. a, b Means in the same rows with different superscripts differ (P < 0.05) (n = 18). ND means not detected
Fig. 2.
Principal component analysis (PCA) results of BM and AF tissue. A. PCA of phenotypes in BM and AF. B. PCA of genes expressed in both BM and AF. C. PCA of genes expressed in BM. D. PCA of genes expressed in AF
PCA analysis was performed on 16,753 genes co-expressed in BM and AF (Fig. 2B). PCA analysis was performed on all expressed genes in BM and AF, respectively (Fig. 2C and 2D).
WGCNA identified module genes significantly related to phenotypes
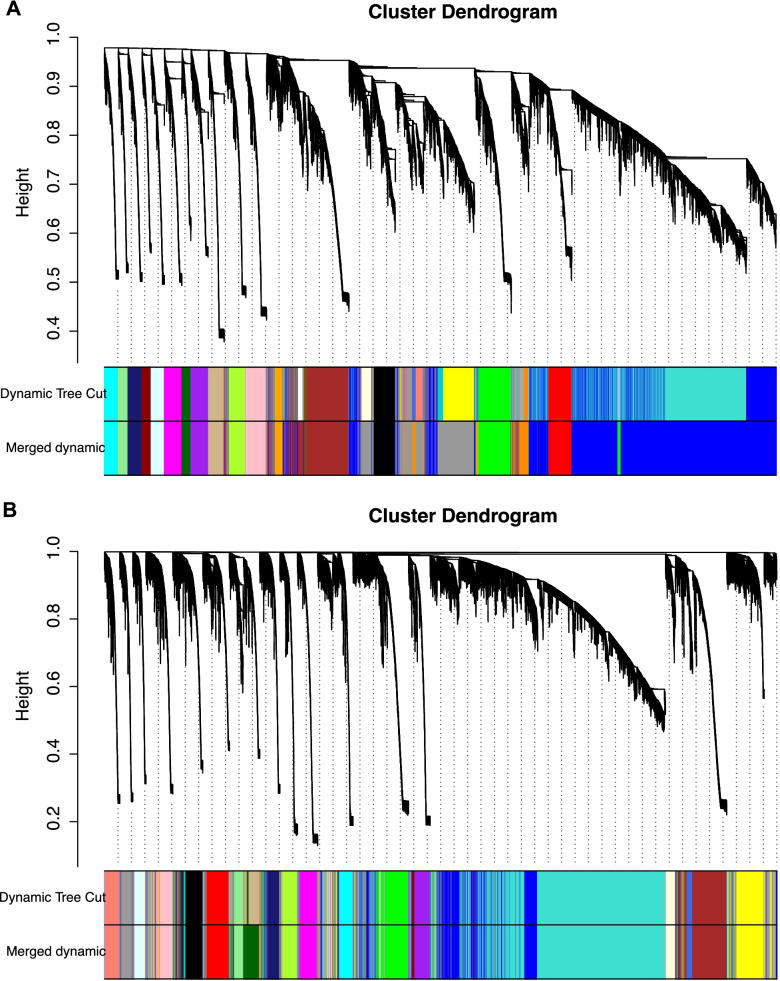
Genes with fragments per kilobase of transcript per million fragments mapped values of less than one were excluded. Gene co-expression networks were constructed using 18,295 expressed genes from BM and 19,778 expressed genes from AF, and the coordination matrix was converted to a topology matrix. The similarity in gene co-expression between modules was quantified using a dynamic tree-cutting algorithm, and modules with height values of less than 0.25 were merged (the minimum number of genes in the module was set to 30) (Fig. 3).
Fig. 3.
Clustering of gene modules. A. Breast muscle tissue microarray. B. Abdominal fat tissue microarray. Upper panel: genes were clustered into different groups. Lower panel: genes were assigned to modules after dynamic tree-cutting and merging
Twenty and twenty-seven expression modules were detected in BM (Additional file: Table S4) and AF (Additional file: Table S5), respectively (Fig. 4), and the number of genes in each network module was counted. The largest module in BM (blue) and AF (turquoise) contained 6972 and 5755 genes, respectively, and the smallest module in BM (dark brown) and AF (white) contained 113 and 47 genes, respectively.
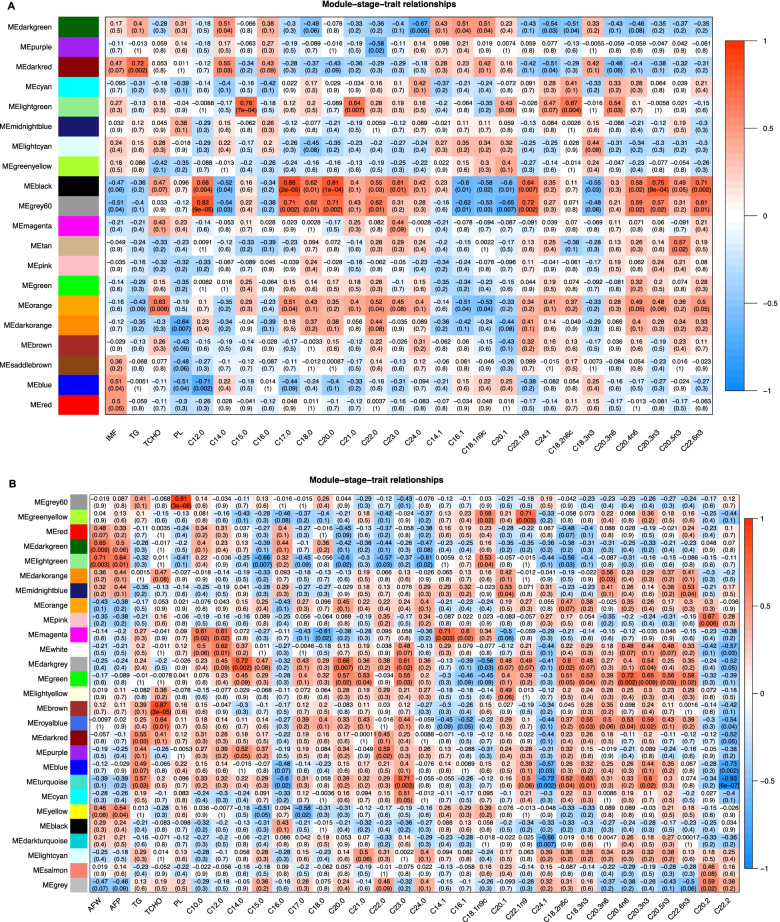
Fig. 4.
Relationship between gene modules and traits in breast muscle tissue (A) and abdominal fat tissue (B). Note: The upper value in each module is the correlation coefficient between the module and the lower character, and the lower value is the p-value of the coefficient
Gene modules significantly associated with genetic traits were identified. Absolute values of 0 and 1 indicated weak and strong correlation between the module and the trait, respectively. Among the evaluated traits, IMF was significantly and positively correlated with the blue module (r = 0.51, P = 0.04), triglycerides (TG) was significantly and positively associated with the darkred module (r = 0.72, P = 0.002), total cholesterol (TCHO) was significantly and positively related to the darkorange module (r = 0.63, P = 0.008), phospholipids (PLIP) was significantly and positively associated with the orange and blue modules (r = -0.64, P = 0.007; r = -0.51, P = 0.04) and traits such as fatty acids composition showed significant correlations with related modules. AF weight showed a significant and positive correlation with the darkgreen and lightgreen modules (r = 0.65, P = 0.009; r = 0.71, P = 0.003), and AFP showed a significant and positive association with the lightgreen and yellow modules (r = 0.64, P = 0.01; r = 0.54, P = 0.04). In AF tissue, TG was significantly and positively correlated with the darkred and turquoise modules (r = 0.55, P = 0.03; r = 0.57, P = 0.03). TCHO showed a significant and positive association with the brown and blue modules (r = 0.87, P = 3e-05; r = 0.64, P = 0.01). PLIP was significantly and positively related to the grey60 module (r = 0.91, P = 3e-06), and traits such as fatty acids composition showed a significant and positive correlations with related modules.
Identification of DEGs in BM and AF
We selected three individuals with high IMF content and three individuals with low IMF content for BM tissue's difference analysis. And we selected three individuals with high AFP and three individuals with low AFP for AF tissue's difference analysis (Additional file: Table S6). Analysis using Deseq2 detected 461 DEGs (163 upregulated and 298 downregulated) in the high and low IMF groups (Additional file: Table S7). Pathway enrichment analysis revealed that these genes were enriched in 29 pathways, mainly including metabolic pathways, MAPK signaling pathways, arachidonic acid metabolism, glycolysis/gluconeogenesis, starch, and sucrose metabolism, fructose and mannose metabolism, and other signaling pathways (Additional file: Table S8). GO analysis showed that DEGs in this group were enriched in 27 biological processes, 16 cellular components, and 12 molecular functions (Additional file: Table S9). A total of 2010 genes (1301 upregulated and 700-downregulated) were detected in the high and low AF groups (Additional file: Table S10). These genes were enriched in 59 KEGG pathways, mainly including metabolic pathways, PPAR signaling pathway, MAPK signaling pathway, tyrosine metabolism, glycerolipid metabolism, phenylalanine metabolism, glycerophospholipid metabolism, pyruvate metabolism, unsaturated fatty acid biosynthesis, fatty acid metabolism, and other signaling pathways (Additional file: Table S11). GO analysis showed that DEGs in this group were enriched in 66 biological processes, 21 cellular components, and 18 molecular functions (Additional file: Table S12).
Identification of pathways in BM and AF
The genes expressed in the modules significantly associated with all phenotypes were selected from BM and AF tissues, respectively. The significantly expressed genes in the modules significantly associated with the traits were pooled, and a total of 3818 expressed genes were identified in BM tissue, while a total of 5826 expressed genes were identified in AF tissue.
These genes were analyzed separately in association with differentially expressed genes, in which 114 co-expressed genes were obtained in BM tissue (Fig. 5A), while a total of 1229 co-expressed genes were obtained in AF tissue (Fig. 5B).
Fig. 5.
Venn of module characteristic genes and differential expression genes. A. Breast muscle tissue. B. Abdominal fat tissue
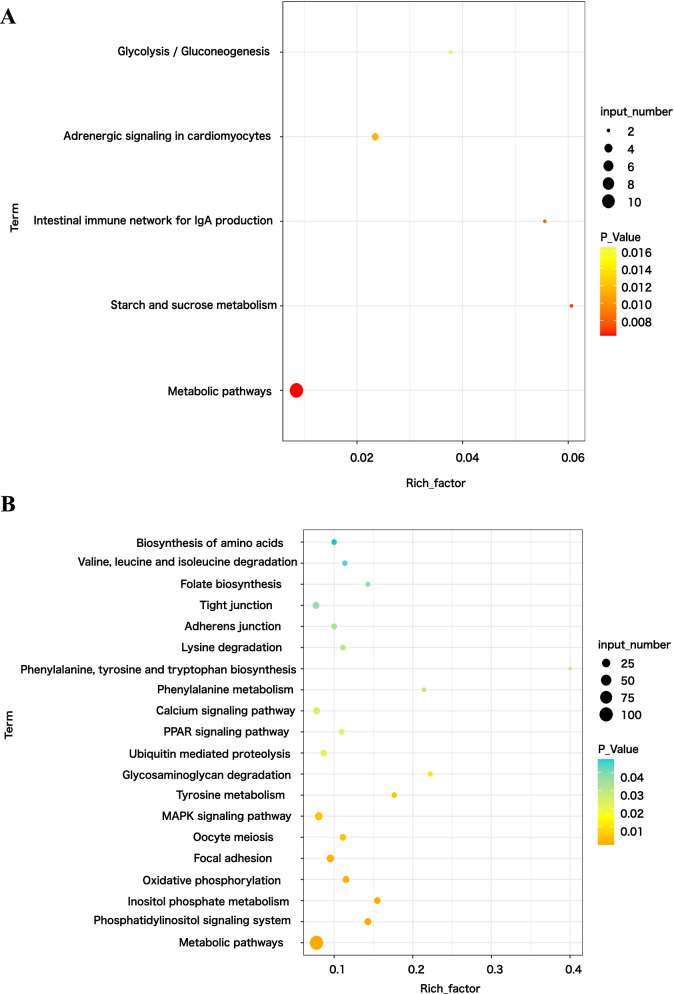
The differential module genes obtained from the two tissues were analyzed separately for KEGG pathway enrichment. In BM tissue, 114 genes were assigned to 5 KEGG pathways (P < 0.05) (Fig. 6A), and genes were enriched in MAPK signaling pathway, ECM-receptor interaction, Gap junction, Tight junction, Vascular smooth muscle contraction, GnRH signaling pathway, and Focal adhesion. In AF tissue, 1229 genes were assigned to 41 KEGG pathways (P < 0.05) (Fig. 6B), and genes were mainly enriched in biological processes, including Oxidative phosphorylation, ABC transporters, C-type lectin receptor signaling pathway, Phosphatidylinositol signaling system.
Fig. 6.
Pathway enrichment analysis of co-expression genes in breast muscle (A) and abdominal fat (B)
Identification of co-expressed genes
The molecular expression networks between pathway and co-expressed genes in BM (Fig. 7A) and AF (Fig. 7B) were performed, respectively. The genes more abundantly expressed in BM with high IMF were related to muscle development (TPM2, MTMR7) and glycolipid metabolism (LDHA, AMY1A, ST3GAL6, GBE1, SGPL1, GPX1, COX5A, ALDH5A1). The genes more significantly expressed in the high AF group were related to fatty acid metabolism (ELOVL6, SCD, FABP1, ME3, ADIPOQ, HMGCS2, PDGFA, ACAT2).
Fig. 7.
Network of pathways and genes in breast muscle tissue (A) and abdominal fat tissue (B)
Discussion
Yellow feather broiler chickens are widely consumed in China, and chicken meat production has increased in recent years. IMF and AF contents strongly affect the quality of chicken meat. Fasting before slaughter is essential in the experimental sampling process [19]. The fasting method can reduce the volume of stomach contents and the risk of visceral microbial contamination during slaughter [20]. In general, moderate fasting (no more than 24 h) is suitable for meat quality because it accelerates glycogen consumption [21, 22]. Especially in poultry, fasting can decrease breast muscle glycogen and increase initial pH [23–25]. Studies have shown that fasting for a certain period before slaughter can control the muscle glycogen content of beef cattle without affecting the marbling score [26–28]. Studies have shown that fasting for 12 h has less influence on the structural changes and plasma corticosterone level and less on the energy metabolism level of chest muscle tissue [29]. Therefore, fasting for 12 h before slaughter would not significantly impact subsequent meat quality and energy metabolism levels, and the effect of glucose metabolism in food on the chicken could be ruled out.
Although the mechanisms underlying fat deposition in broiler chickens are well known, differences in the rate of fat deposition in BM and AF tissues have not been determined.
PCA results based on phenotypes showed a strong separation of breast muscle tissue from abdominal fat tissue, as expected. This suggests that BM tissue and AF tissue have different regulatory roles in phenotype. Many DEGs can be identified by RNA-seq analysis, however, the characterization of gene expression patterns and correlation with phenotypes are challenging, underscoring the need to perform weighted co-expression network analyses. In the study, DEGs were identified in BM and AF by RNA-seq analysis and joint analysis with WGCNA to identify specifically expressed genes and signaling pathways for IMF and AF deposition. The molecular regulatory mechanisms of fat deposition in different tissues at the same age were elucidated in chicken.
Based on the differentially expressed genes, the results of KEGG signaling pathway analysis showed that DEGs from BM and AF tissues were jointly enriched in the AGE-RAGE signaling pathway, amino acid biosynthesis, cytokine-cytokine receptor interactions, local adhesion, GnRH signaling pathway, MAPK signaling pathway, metabolic pathway, in diabetic complications. Tight junctions and other signaling pathways. Studies have shown that MAPK signaling pathway is involved in lipid deposition [30, 31], which can regulate PPAR pathway [32]. Moreover, PPAR signaling pathway also has the function of regulating lipid metabolism [33, 34].
In contrast, DEGs in BM tissue were specifically enriched in pathways related to gluconeogenesis (glycolysis/gluconeogenesis, starch, sucrose metabolism, fructose, mannose metabolism, pentose, and glucuronide). LDHA and LDHB in the glycolysis/gluconeogenesis signaling pathway are involved in pyruvate metabolism and tricarboxylic acid cycle, facilitating the glycolytic process by converting pyruvate to lactate [35]. In addition, GBE1 is essential in starch and glycogen formation metabolism, and there is a strong link between the hexokinase family genes HK1, HK2, HK3 in this pathway and the metabolic process of glucose. These data suggest that IMF deposition in BM tissue mainly depends on glucose metabolism, with some energy metabolism involved in the process.
DEGs in BM tissue were specifically enriched in pathways related to fatty acid syntheses (PPAR signaling, glycolipid metabolism, glycerophospholipid metabolism, and unsaturated fatty acid biosynthesis). Sixteen genes were enriched in the PPAR signaling pathway, including HMGCS2 (catalyze ketogenesis) [36, 37], ACOX1 (lipid degradation) [38], ADIPOQ (adipocyte differentiation) [39], APOA1 and ME3 (cholesterol metabolism) [40], SCD (fatty acid transporter proteins) [41], PLIN1 and PLIN2 (lipid droplet protection) [42–44], ACSL1 [45], FABP7, FABP1, FADS2. The formation of intracellular lipid droplets is a highly conserved process, including fatty acid transport and activation, neutral lipid synthesis, and lipid droplet formation, regulated by many factors and pathways [46]. These results suggest that AF deposition is dependent on fatty acid synthesis and transport and lipid droplet formation.
WGCNA was performed using RNA-seq data to identify expressed genes in different modules and predict the role of genes in lipid deposition. Several modules were significantly associated with IMF/AFW/AFP, lipid composition, and fatty acids metabolism in BM and AF, and the expressed genes significantly associated with the phenotypes in the modules were aggregated together and analyzed jointly with DEGs, which could further identify the functional genes that play essential roles in the IMF and AF deposition. Among all expressed genes, GAPDH has the highest expression level in breast muscle tissue. GAPDH is a key enzyme in glycolysis. It plays an important role in glycolysis by catalyzing the first step of this pathway by converting D-glyceraldehyde 3-phosphate (G3P) to 3-phospho-d-glyceroylphosphate [47, 48]. FABP4 gene in the lipid-binding protein family regulates fatty acid uptake and transport [41].
We identified 114 expressed genes associated with Metabolic pathways, Starch and sucrose metabolism, Intestinal immune network for IgA production, Adrenergic signaling in cardiomyocytes, and Glycolysis/Gluconeogenesis in BM. These pathways are regulated by genes such as ALDH5A1, LDHA, GAPDH, GBE1, and GPX1 and may play roles in IMF deposition. A total of 1229 expressed genes associated with 41 signaling pathways were identified in AF, of which 20 pathways that may play roles in AF deposition via PPAR signaling pathway, amino acid biosynthesis, oxidative phosphorylation, may be controlled by several genes, including ADIPOQ, ELOVL6, HMGCS2, ME3, DGKE, AOX1, UBB, SCD, APOC3, and FABP1. The results revealed that IMF deposition in BM tissue was regulated by gluconeogenesis-related pathways (glycolysis/gluconeogenesis, starch and sucrose metabolism signaling pathways), and by several genes, including GAPDH, LDHA, GPX1, ALDH5A1, and GBE1, among which, the key differential gene ALDH5A1 can catalyze a step in the degradation of the inhibitory neurotransmitter γ-aminobutyric acid [49]. The protein encoded by LDHA is involved in pyruvate metabolism by catalyzing the conversion of lactate to pyruvate in anaerobic glycolysis. GAPDH plays an important role in glycerol metabolism. These data further suggest that IMF deposition in BM is dependent on gluconeogenesis and energy metabolism.
While expressed genes in AF tissue are enriched to PPAR signaling pathway, oxidative phosphorylation, amino acid biosynthesis, and other signaling pathways, acting through genes such as FABP1, ELOVL6, SCD, and ADIPOQ. A PPAR signaling pathway is involved in lipid droplets formation and mitochondrial metabolism, and fatty acid oxidation and lipid synthesis are necessary for cellular signaling [50]. FABP1 plays a crucial role in the PPAR pathway, PPARG expression, and fatty acid uptake, transport, and metabolism in vivo [51]. In addition, PPARϒ can be regulated by modulating SCD1 expression to control fatty acid synthesis in adipocytes [52]. ELOVLs, which encode long-chain and extra-long-chain fatty acid elongases, play an important role in synthesizing fatty acids and can limit elongation. ELOVL6 is involved in synthesizing fatty acid enzymes in vivo, promoting fatty acid elongation [53]. ACAT2 encodes acetyl-coenzyme A acetyltransferase 2, which is involved in acetyl-coenzyme A metabolism [54]. These results suggest that AF deposition may result from changes in fatty acid synthesis through mitochondrial activity.
The results suggest that different signaling pathways regulate fat deposition in BM and AF tissues. IMF deposition in BM was associated with pyruvate and citrate metabolism through GAPDH, LDHA, GPX1, GBE1, and other genes, whereas AF deposition was related to acetyl-coenzyme A and glycerophospholipid metabolism through FABP1, ELOVL6, SCD, and ADIPOQ. The transcriptional regulation of genes in network modules associated with traits and metabolic pathway analysis can provide new insights into the genetic mechanisms governing fat deposition in broiler chickens.
Conclusion
The analysis of transcriptome data from BM and AF tissues, combined with differential expression analysis and WGCNA, showed that the genes GAPDH, LDHA, GPX1, and GBE1 were involved in regulating IMF deposition, and the genes FABP1, ELOVL6, SCD, and ADIPOQ determined AF deposition. Glycolysis/gluconeogenesis and other signaling pathways play an important role in IMF deposition, whereas PPAR metabolism controls AF deposition. Therefore, we hypothesized that IMF deposition in BM tissue may affect energy metabolism during myocyte gluconeogenesis while fatty acid synthesis pathways may affect AF deposition.
Materials and methods
Animals and Sample Collection
Wenchang chickens (18 females) of 98 days were obtained from the Institute of Poultry Research of the Chinese Academy of Agricultural Sciences (Yangzhou, China). The diet (Additional file: Table S13) was formulated according to the feeding standard (NY/T33-2004). Under the same standard conditions of light (20 lx), temperature (35℃ ~ 37℃), humidity (not less than 50%), and immunization schedule, after a 12-h fast, the chickens were stunned by electric shock and killed by cervical dislocation at 98 days. After slaughter, breast muscle (BM) and AF were collected and immediately stored at − 80 °C until use.
Measurement of Biochemical Indices [18]
The determination of fatty acids in BM and AF tissues refers to the national standard《GB/T 5413.27–2010 Food Safety National Standard for infant food and dairy products determination of fatty acids》in the first method: acetyl chloride—methanol methyl esterification method. The determination of intramuscular fat in BM tissue refers to the national standard《GB/T 5009.6–2016 National Standard for Food Safety determination of fat in food》in the first method: Soxhlet extraction. The TG, TCHO, and PLIP contents in BM and AF tissues were measured using assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China, and Beijing Leichuang Biotechnology Co., LTD, China). We weighed 2.0 ± 0.01 g breast muscle and abdominal fat samples and mixed them with 18 mL anhydrous ethanol in 50 ml centrifuge tubes. Each sample was homogenized by a hand-held high-speed homogenizer (35,000 r/min, 15 s/ time). The samples were crushed, centrifuged at 2500 r/min for 10 min. After centrifugation, the supernatant was used for measurement. Refer to the instruction for specific sample loading quantity. After incubation, the absorbance value (510 nm) was measured using a microplate reader (Tecan Infinite 200 Pro, Switzerland).
Transcriptome Profiling
Transcriptome profiling of 18 breast muscle and 18 abdominal fat tissues was performed on the Illumina PE150 platform (Berry Genomics; Beijing, China) (https://www.berrygenomics.com). RNA was extracted as described previously [55]. Adapter sequences and other low-quality data were removed using Cutadapt. Reads were aligned to the reference genome using HISAT, while clean reads obtained by filtering were compared to the reference genome according to the gene position information specified in the genome annotation file gtf, and the total comparison rate was approximately 90%.
Differential Expression Analysis
Based on phenotypic data, DEGs were identified in BM tissue with high IMF and low IMF (from three animals each) and AF tissue with high AF and low AF (from three animals each) using DESeq2 in R [56]. Adjusted p-values (q-values) were calculated using Benjamin and Hochberg’s approach for controlling the false discovery rate. Genes with | log2(fold change) |≥ 0.58 and P < 0.05 were considered differentially expressed.
Weighted Gene Co-expression Network Analysis (WGCNA)
WGCNA was performed using the WGCNA package [57] with default settings and minor modifications. For the analysis of genes expressed in BM tissue (n = 16), the minModuleSize was set to 50, and mergeCutHeight was set to 0.25 for tissue- or stage-specific module detection (soft threshold = 4). For the analysis of genes expressed in AF tissue (n = 15), the minModuleSize was set to 30, and mergeCutHeight was set to 0.25 for tissue- or stage-specific module detection (soft threshold = 10). Dendrograms were created using topology overlap matrix, module detection, and similar module merging functions. Module-trait relationships were calculated, and correlations were adjusted using Benjamin-Hochberg correction.
Pathway Enrichment Analyses
Gene ontology (GO) enrichment analysis was performed with the DAVID database (https://david.ncifcrf.gov/) to identify gene classes and categories. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using KOBAS version 3.0 (http://kobas.cbi.pku.edu.cn). The significance level was set at P < 0.05. The gene-pathway interaction networks for the candidate genes were visualized with Cytoscape 3.8.0 (http://www.cytoscape.org/) [58].
Statistical Analyses
The significance of differences between means was assessed using Student’s t-test in SPSS 22.0 (IBM Corp, Armonk, NY, USA). P-values less than 0.05 were considered statistically significant. Data are presented as the mean ± standard error of the mean.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- ACAT2
Acetyl-CoA Acetyltransferase 2
- ACOX1
Acyl-CoA Oxidase 1
- ACSL1
Acyl-CoA Synthetase Long Chain Family Member 1
- ADIPOQ
Adiponectin, C1Q And Collagen Domain Containing; AF: Abdominal fat; AFW: Abdominal fat weight
- AFP
Abdominal fat percentage
- ALDH5A1
Aldehyde Dehydrogenase 5 Family Member A1
- AMY1A
Amylase Alpha 1A
- AOX1
Aldehyde Oxidase 1
- APOA1
Apolipoprotein A1
- APOC3
Apolipoprotein C3
- BM
Breast Muscle
- COX5A
Cytochrome C Oxidase Subunit 5A
- CREB3L1
CAMP Responsive Element Binding Protein 3 Like 1
- DEGs
Differential Expressed Genes
- DGKE
Diacylglycerol Kinase Epsilon
- ECM
Extracellular Matrix
- ELOVL6
ELOVL Fatty Acid Elongase 6
- FABP1
Fatty Acid Binding Protein 1
- FABP4
Fatty Acid Binding Protein 4
- FABP7
Fatty Acid Binding Protein 7
- FADS2
Fatty Acid Desaturase 2
- GAPDH
Glyceraldehyde-3-Phosphate Dehydrogenase
- GBE1
1,4-Alpha-Glucan Branching Enzyme 1
- GO
Gene Ontology
- GPX1
Glutathione Peroxidase 1
- HK1
Hexokinase 1
- HK2
Hexokinase 2
- HK3
Hexokinase 3
- HMGCS2
3-Hydroxy-3-Methylglutaryl-CoA Synthase 2
- IMF
Intramuscular Fat
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- L3MBTL1
Histone Methyl-Lysine Binding Protein 1
- LDHA
Lactate Dehydrogenase A
- ME3
Malic Enzyme 3
- PCA
Principal Component Analysis
- PDGFA
Platelet Derived Growth Factor Subunit A
- PLIN1
Perilipin 1
- PLIN2
Perilipin 2
- PLIP
Phospholipid
- RNA-Seq
RNA-sequencing
- SCD
Stearoyl-CoA Desaturase
- SGPL1
Sphingosine-1-Phosphate Lyase 1
- ST3GAL6
ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 6
- TCHO
Total Cholesterol
- TG
Triglycerides
- TNP1
Transition Protein 1
- UBB
Ubiquitin B
- WGCNA
Weighted Gene Co-expression Network Analysis
Authors’ contributions
LN and SJT performed the study, analyzed the data, and drafted the manuscript. YXY and JYX collected samples and analyzed the data. HXC contributed to the design of the study, acquiring the funding, and modifying the manuscript. GPZ contributed to providing resources and contributed to acquiring the funding. JW designed the study and followed up on the overall project. All authors submitted comments on drafts and read and approved the final manuscript.
Funding
The research was funded by grants from the National Natural Science Foundation of China (32172723), the State Key Laboratory of Animal Nutrition (2004DA125184G2109), the Agricultural Science and Technology Innovation Program (ASTIP-IAS04), and the Earmarked Fund for Modern Agro-Industry Technology Research System (CARS-41).
Availability of data and materials
The RNA sequencing raw data reported in this paper have been deposited in the Genome Sequence Archive [59] in BIG Data Center [60] under accession number CRA006031, which can be publicly accessed at http://bigd.big.ac.cn/gsa. All data analyzed during this study are included in the article and additional files.
Declarations
Ethics approval and consent to participate
This study was conducted in accordance with the Guide for the Care and Use of Laboratory Animals published by the Ministry of Science and Technology (Beijing, China). The study was approved by the Research Ethics Committee of the Institute of Animal Sciences of the Chinese Academy of Agricultural Sciences (Beijing, China) (Protocol No. IAS2019-21). All methods were carried out in accordance with relevant guidelines and regulations. This study was carried out in compliance with the ARRIVE guidelines (Animal Research: Reporting of In Vivo Experiments).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Na Luo and Jingting Shu contributed equally to this work.
References
- 1.Chu W, Wei W, Han H, Gao Y, Liu K, Tian Y, Jiang Z, Zhang L, Chen J. Muscle-specific downregulation of GR levels inhibits adipogenesis in porcine intramuscular adipocyte tissue. Sci Rep. 2017;7(1):510. doi: 10.1038/s41598-017-00615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han H, Wei W, Chu W, Liu K, Tian Y, Jiang Z, Chen J. Muscle Conditional Medium Reduces Intramuscular Adipocyte Differentiation and Lipid Accumulation through Regulating Insulin Signaling. Int J Mol Sci. 2017;18(8):1799. doi: 10.3390/ijms18081799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith SB, Crouse JD. Relative contributions of acetate, lactate and glucose to lipogenesis in bovine intramuscular and subcutaneous adipose tissue. J Nutr. 1984;114(4):792–800. doi: 10.1093/jn/114.4.792. [DOI] [PubMed] [Google Scholar]
- 4.Wang S, Zhou G, Shu G, Wang L, Zhu X, Gao P, Xi Q, Zhang Y, Yuan L, Jiang Q. Glucose utilization, lipid metabolism and BMP-Smad signaling pathway of porcine intramuscular preadipocytes compared with subcutaneous preadipocytes. Cell Physiol Biochem. 2013;31(6):981–996. doi: 10.1159/000350116. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Li F, Ma XF, Li WT, Jiang RR, Han RL, Li GX, Wang YB, Li ZY, Tian YD, et al. Identification of differentially expressed genes and pathways between intramuscular and abdominal fat-derived preadipocyte differentiation of chickens in vitro. BMC Genomics. 2019;20(1):743. doi: 10.1186/s12864-019-6116-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leng L, Zhang H, Dong JQ, Wang ZP, Zhang XY, Wang SZ, Cao ZP, Li YM, Li H. Selection against abdominal fat percentage may increase intramuscular fat content in broilers. J Anim Breed Genet. 2016;133(5):422–428. doi: 10.1111/jbg.12204. [DOI] [PubMed] [Google Scholar]
- 7.Chen JL, Zhao GP, Zheng MQ, Wen J, Yang N. Estimation of genetic parameters for contents of intramuscular fat and inosine-5’-monophosphate and carcass traits in Chinese Beijing-You chickens. Poult Sci. 2008;87(6):1098–104. [DOI] [PubMed]
- 8.Ge K, Ye P, Yang L, Kuang J, Chen X, Geng Z. Comparison of slaughter performance, meat traits, serum lipid parameters and fat tissue between Chaohu ducks with high- and low-intramuscular fat content. Anim Biotechnol. 2020;31(3):245–255. doi: 10.1080/10495398.2019.1664565. [DOI] [PubMed] [Google Scholar]
- 9.Jiang M, Fan WL, Xing SY, Wang J, Li P, Liu RR, Li QH, Zheng MQ, Cui HX, Wen J, et al. Effects of balanced selection for intramuscular fat and abdominal fat percentage and estimates of genetic parameters. Poult Sci. 2017;96(2):282–287. doi: 10.3382/ps/pew334. [DOI] [PubMed] [Google Scholar]
- 10.Zhou G, Wang S, Wang Z, Zhu X, Shu G, Liao W, Yu K, Gao P, Xi Q, Wang X, et al. Global comparison of gene expression profiles between intramuscular and subcutaneous adipocytes of neonatal landrace pig using microarray. Meat Sci. 2010;86(2):440–450. doi: 10.1016/j.meatsci.2010.05.031. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Kikuchi M, Li X, Gao Q, Xiong Z, Ren Y, Zhao R, Mao B, Kondo M, Irie N, et al. Weighted gene co-expression network analysis reveals potential genes involved in early metamorphosis process in sea cucumber Apostichopus japonicus. Biochem Biophys Res Commun. 2018;495(1):1395–1402. doi: 10.1016/j.bbrc.2017.11.154. [DOI] [PubMed] [Google Scholar]
- 12.Wang W, Jiang W, Hou L, Duan H, Wu Y, Xu C, Tan Q, Li S, Zhang D. Weighted gene co-expression network analysis of expression data of monozygotic twins identifies specific modules and hub genes related to BMI. BMC Genomics. 2017;18(1):872. doi: 10.1186/s12864-017-4257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin L, Cai Z, Zhu B, Xu C. Identification of Key Pathways and Genes in the Dynamic Progression of HCC Based on WGCNA. Genes. 2018;9(2):92. doi: 10.3390/genes9020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Z, Ding R, Zhai X, Wang Y, Chen Y, Yang CX, Du ZQ. Common Gene Modules Identified for Chicken Adiposity by Network Construction and Comparison. Front Genet. 2020;11:537. doi: 10.3389/fgene.2020.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tian W, Zhang B, Zhong H, Nie R, Ling Y, Zhang H, Wu C. Dynamic Expression and Regulatory Network of Circular RNA for Abdominal Preadipocytes Differentiation in Chicken (Gallus gallus) Frontiers in cell and developmental biology. 2021;9:761638. doi: 10.3389/fcell.2021.761638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan H, Lu J. Consensus module analysis of abdominal fat deposition across multiple broiler lines. BMC Genomics. 2021;22(1):115. doi: 10.1186/s12864-021-07423-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xing S, Liu R, Zhao G, Liu L, Groenen MAM, Madsen O, Zheng M, Yang X, Crooijmans R, Wen J. RNA-Seq Analysis Reveals Hub Genes Involved in Chicken Intramuscular Fat and Abdominal Fat Deposition During Development. Front Genet. 2020;11:1009. doi: 10.3389/fgene.2020.01009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Na L, Huanxian C, Xiaoya Y, Yuxi J, Jingting S, Guiping Z, Jie W. Study on the Differences in the Composition of Intramuscular Fat and Abdominal Fat in Wenchang Chicken. China Poultry. 2021;43(09):6–11. [Google Scholar]
- 19.Beattie VE, Burrows MS, Moss BW, Weatherup RN. The effect of food deprivation prior to slaughter on performance, behaviour and meat quality. Meat Sci. 2002;62(4):413–418. doi: 10.1016/S0309-1740(02)00031-1. [DOI] [PubMed] [Google Scholar]
- 20.Eikelenboom G, Bolink AH, Sybesma W. Effects of feed withdrawal before delivery on pork quality and carcass yield. Meat Sci. 1991;29(1):25–30. doi: 10.1016/0309-1740(91)90020-Q. [DOI] [PubMed] [Google Scholar]
- 21.Crouse JD, Smith SB, Prior RL. Bovine muscle glycogen as affected by fasting and refeeding. J Anim Sci. 1984;59(2):384–387. doi: 10.2527/jas1984.592384x. [DOI] [PubMed] [Google Scholar]
- 22.McVeigh JM, Tarrant PV. Glycogen content and repletion rates in beef muscle, effect of feeding and fasting. J Nutr. 1982;112(7):1306–1314. doi: 10.1093/jn/112.7.1306. [DOI] [PubMed] [Google Scholar]
- 23.Kotula KL, Wang Y. Characterization of Broiler Meat Quality Factors as Influence by Feed Withdrawal Time. J Appl Poultry Res. 1994;3(2):103–110. doi: 10.1093/japr/3.2.103. [DOI] [Google Scholar]
- 24.Savenije B, Lambooij E, Gerritzen MA, Venema K, Korf J. Effects of feed deprivation and transport on preslaughter blood metabolites, early postmortem muscle metabolites, and meat quality. Poult Sci. 2002;81(5):699–708. doi: 10.1093/ps/81.5.699. [DOI] [PubMed] [Google Scholar]
- 25.van Schalkwyk SJ, Hoffman LC, Cloete SW, Mellett FD. The effect of feed withdrawal during lairage on meat quality characteristics in ostriches. Meat Sci. 2005;69(4):647–651. doi: 10.1016/j.meatsci.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Jones SDM, Schaefer AL, Robertson WM, Vincent BC. The effects of withholding feed and water on carcass shrinkage and meat quality in beef cattle. Meat Sci. 1990;28(2):131–139. doi: 10.1016/0309-1740(90)90037-7. [DOI] [PubMed] [Google Scholar]
- 27.del Campo M, Brito G. Soares de Lima J, Hernández P, Montossi F: Finishing diet, temperament and lairage time effects on carcass and meat quality traits in steers. Meat Sci. 2010;86(4):908–914. doi: 10.1016/j.meatsci.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 28.Clariget J, Banchero G, Luzardo S, Fernández E, Pérez E, La Manna A, Saravia A, Del Campo M, Ferrés A, Canozzi MEA. Effect of pre-slaughter fasting duration on physiology, carcass and meat quality in beef cattle finished on pastures or feedlot. Res Vet Sci. 2021;136:158–165. doi: 10.1016/j.rvsc.2021.02.018. [DOI] [PubMed] [Google Scholar]
- 29.Wang S, Li C, Xu X, Zhou G. Effect of fasting on energy metabolism and tenderizing enzymes in chicken breast muscle early postmortem. Meat Sci. 2013;93(4):865–872. doi: 10.1016/j.meatsci.2012.11.053. [DOI] [PubMed] [Google Scholar]
- 30.Shen Y, Zhao Y, Zheng D, Chang X, Ju S, Guo L. Effects of orexin A on GLUT4 expression and lipid content via MAPK signaling in 3T3-L1 adipocytes. J Steroid Biochem Mol Biol. 2013;138:376–383. doi: 10.1016/j.jsbmb.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Gubern A, Barceló-Torns M, Barneda D, López JM, Masgrau R, Picatoste F, Chalfant CE, Balsinde J, Balboa MA, Claro E. JNK and ceramide kinase govern the biogenesis of lipid droplets through activation of group IVA phospholipase A2. J Biol Chem. 2009;284(47):32359–32369. doi: 10.1074/jbc.M109.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capobianco E, Martínez N, Fornes D, Higa R, Di Marco I, Basualdo MN, Faingold MC, Jawerbaum A. PPAR activation as a regulator of lipid metabolism, nitric oxide production and lipid peroxidation in the placenta from type 2 diabetic patients. Mol Cell Endocrinol. 2013;377(1–2):7–15. doi: 10.1016/j.mce.2013.06.027. [DOI] [PubMed] [Google Scholar]
- 33.Moreno M, Lombardi A, Silvestri E, Senese R, Cioffi F, Goglia F, Lanni A, de Lange P: PPARs: Nuclear Receptors Controlled by, and Controlling, Nutrient Handling through Nuclear and Cytosolic Signaling. PPAR Res. 2010; 2010. [DOI] [PMC free article] [PubMed]
- 34.Cui HX, Liu RR, Zhao GP, Zheng MQ, Chen JL, Wen J. Identification of differentially expressed genes and pathways for intramuscular fat deposition in pectoralis major tissues of fast-and slow-growing chickens. BMC Genomics. 2012;13:213. doi: 10.1186/1471-2164-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y, Xiong Y, Qiao T, Li X, Jia L, Han Y. Lactate dehydrogenase A: A key player in carcinogenesis and potential target in cancer therapy. Cancer Med. 2018;7(12):6124–6136. doi: 10.1002/cam4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shukla SK, Liu W, Sikder K, Addya S, Sarkar A, Wei Y, Rafiq K. HMGCS2 is a key ketogenic enzyme potentially involved in type 1 diabetes with high cardiovascular risk. Sci Rep. 2017;7(1):4590. doi: 10.1038/s41598-017-04469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee YE, He HL, Shiue YL, Lee SW, Lin LC, Wu TF, Chang IW, Lee HH, Li CF. The prognostic impact of lipid biosynthesis-associated markers, HSD17B2 and HMGCS2, in rectal cancer treated with neoadjuvant concurrent chemoradiotherapy. Tumour Biol. 2015;36(10):7675–7683. doi: 10.1007/s13277-015-3503-2. [DOI] [PubMed] [Google Scholar]
- 38.He A, Dean JM, Lu D, Chen Y, Lodhi IJ. Hepatic peroxisomal β-oxidation suppresses lipophagy via RPTOR acetylation and MTOR activation. Autophagy. 2020;16(9):1727–1728. doi: 10.1080/15548627.2020.1797288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Li F, Zhang Y, Dai L, Jiang H, Liu H, Zhang S, Chen C, Zhang J. Silencing of ADIPOQ efficiently suppresses preadipocyte differentiation in porcine. Cell Physiol Biochem. 2013;31(2–3):452–461. doi: 10.1159/000343381. [DOI] [PubMed] [Google Scholar]
- 40.Ray A, Ghosh A, Chakraborty R, Upadhyay SK, Maiti S, Sengupta S, Thukral L. Specific Cholesterol Binding Drives Drastic Structural Alterations in Apolipoprotein A1. J Phys Chem Lett. 2018;9(20):6060–6065. doi: 10.1021/acs.jpclett.8b02042. [DOI] [PubMed] [Google Scholar]
- 41.Jin S, Lee SH, Lee DH, Manjula P, Lee SH, Lee JH. Genetic association of DEGS1, ELOVL6, FABP3, FABP4, FASN and SCD genes with fatty acid composition in breast and thigh muscles of Korean native chicken. Anim Genet. 2020;51(2):344–345. doi: 10.1111/age.12908. [DOI] [PubMed] [Google Scholar]
- 42.MacPherson RE, Ramos SV, Vandenboom R, Roy BD, Peters SJ. Skeletal muscle PLIN proteins, ATGL and CGI-58, interactions at rest and following stimulated contraction. Am J Physiol Regul Integr Comp Physiol. 2013;304(8):R644–650. doi: 10.1152/ajpregu.00418.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng YZ, Lund J, Li Y, Knabenes IK, Bakke SS, Kase ET, Lee YK, Kimmel AR, Thoresen GH, Rustan AC, et al. Loss of perilipin 2 in cultured myotubes enhances lipolysis and redirects the metabolic energy balance from glucose oxidation towards fatty acid oxidation. J Lipid Res. 2017;58(11):2147–2161. doi: 10.1194/jlr.M079764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bosma M, Hesselink MK, Sparks LM, Timmers S, Ferraz MJ, Mattijssen F, van Beurden D, Schaart G, de Baets MH, Verheyen FK, et al. Perilipin 2 improves insulin sensitivity in skeletal muscle despite elevated intramuscular lipid levels. Diabetes. 2012;61(11):2679–2690. doi: 10.2337/db11-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian W, Wang D, Wang Z, Jiang K, Li Z, Tian Y, Kang X, Liu X, Li H. Evolution, expression profile, and regulatory characteristics of ACSL gene family in chicken (Gallus gallus) Gene. 2021;764:145094. doi: 10.1016/j.gene.2020.145094. [DOI] [PubMed] [Google Scholar]
- 46.Walther TC, Chung J, Farese RV., Jr Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol. 2017;33:491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ercolani L, Florence B, Denaro M, Alexander M. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J Biol Chem. 1988;263(30):15335–15341. doi: 10.1016/S0021-9258(19)37593-3. [DOI] [PubMed] [Google Scholar]
- 48.Tisdale EJ. Glyceraldehyde-3-phosphate dehydrogenase is phosphorylated by protein kinase Ciota /lambda and plays a role in microtubule dynamics in the early secretory pathway. J Biol Chem. 2002;277(5):3334–3341. doi: 10.1074/jbc.M109744200. [DOI] [PubMed] [Google Scholar]
- 49.Escalera GI, Ferrer I, Marina LC, Sala PR, Salomons GS, Jakobs C, Pérez-Cerdá C. Succinic semialdehyde dehydrogenase deficiency: decrease in 4-OH-butyric acid levels with low doses of vigabatrin. An Pediatr (Barc) 2010;72(2):128–132. doi: 10.1016/j.anpedi.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19(3):380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discovery. 2008;7(6):489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Risérus U, Tan GD, Fielding BA, Neville MJ, Currie J, Savage DB, Chatterjee VK, Frayn KN, O’Rahilly S, Karpe F. Rosiglitazone increases indexes of stearoyl-CoA desaturase activity in humans: link to insulin sensitization and the role of dominant-negative mutation in peroxisome proliferator-activated receptor-gamma. Diabetes. 2005;54(5):1379–84. [DOI] [PubMed]
- 53.Junjvlieke Z, Khan R, Mei C, Cheng G, Wang S, Raza SHA, Hong J, Wang X, Yang W, Zan L. Effect of ELOVL6 on the lipid metabolism of bovine adipocytes. Genomics. 2020;112(3):2282–2290. doi: 10.1016/j.ygeno.2019.12.024. [DOI] [PubMed] [Google Scholar]
- 54.Wang YJ, Bian Y, Luo J, Lu M, Xiong Y, Guo SY, Yin HY, Lin X, Li Q, Chang CCY, et al. Cholesterol and fatty acids regulate cysteine ubiquitylation of ACAT2 through competitive oxidation. Nat Cell Biol. 2017;19(7):808–819. doi: 10.1038/ncb3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L, Liu X, Cui H, Liu R, Zhao G, Wen J. Transcriptional insights into key genes and pathways controlling muscle lipid metabolism in broiler chickens. BMC Genomics. 2019;20(1):863. doi: 10.1186/s12864-019-6221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, Tang B, Dong L, Ding N, Zhang Q, et al. GSA: Genome Sequence Archive<sup/>. Genomics Proteomics Bioinformatics. 2017;15(1):14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Database Resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019; 47(D1):D8-d14. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing raw data reported in this paper have been deposited in the Genome Sequence Archive [59] in BIG Data Center [60] under accession number CRA006031, which can be publicly accessed at http://bigd.big.ac.cn/gsa. All data analyzed during this study are included in the article and additional files.