Abstract
Background
Advances in genome sequencing technology, particularly restriction-site associated DNA sequence (RAD-seq) and whole-genome resequencing, have greatly aided the construction of cotton interspecific genetic maps based on single nucleotide polymorphism (SNPs), Indels, and other types of markers. High-density genetic maps can improve accuracy of quantitative trait locus (QTL) mapping, narrow down location intervals, and facilitate identification of the candidate genes.
Result
In this study, 249 individuals from an interspecific F2 population (TM-1 and Hai7124) were re-sequenced, yielding 6303 high-confidence bin markers spanning 5057.13 cM across 26 cotton chromosomes. A total of 3380 recombination hot regions RHRs were identified which unevenly distributed on the 26 chromosomes. Based on this map, 112 QTLs relating to agronomic and physiological traits from seedling to boll opening stage were identified, including 15 loci associated with 14 traits that contained genes harboring nonsynonymous SNPs. We analyzed the sequence and expression of these ten candidate genes and discovered that GhRHD3 (GH_D10G0500) may affect fiber yield while GhGPAT6 (GH_D04G1426) may affect photosynthesis efficiency.
Conclusion
Our research illustrates the efficiency of constructing a genetic map using binmap and QTL mapping on the basis of a certain size of the early-generation population. High-density genetic map features high recombination exchanges in number and distribution. The QTLs and the candidate genes identified based on this high-density genetic map may provide important gene resources for the genetic improvement of cotton.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-022-08528-2.
Keywords: Cotton, Re-sequencing, Genetic map, QTL mapping, Agronomic and physiological traits
Background
Gossypium hirsutum and G. barbadense are the most two important cultivated species of allotetraploid cotton in the world. Among the domesticated Gossypium species, G. hirsutum is the most widely cultivated, dominating modern cotton production due to its high lint yield and broad adaptability [1, 2]; meanwhile, G. barbadense provides excellent fiber that is finer, longer and stronger than fiber of G. hirsutum [3, 4]. Efficient and extensive transmission of valuable genes between G. barbadense and G. hirsutum is of extremely important practical significance for improving fiber quality while maintaining fiber yield, which is mainly limited by linkage drag.
Quantitative traits exhibit continuous variation and are generally controlled by multiple genes, hence having a complex genetic basis; moreover, they are readily affected by the environment. Genetic research on quantitative traits is therefore difficult, and investigating the inheritance and QTL mapping of cotton quantitative traits is of great significance to the advancement of cotton genetics and breeding. Since Shappley et al. [5] constructed the first genetic map of cotton, many studies have conducted QTL mapping for important cotton traits.
A high-density molecular genetic map is the foundation of plant genome research. Interspecific maps have been constructed for cotton, mainly between G. barbadense and G. hirsutum, and used to explore species differences such as in yield and quality traits [6–27]. These studies have provided very useful information for cotton molecular design and breeding. There are many QTL-enriched regions in the cotton genome, and there may be large numbers of related genes that play important roles in the plant’s growth and development [28]. Notably, QTLs for important traits are unevenly distributed among 26 different chromosomes of cotton. In interspecific populations, fiber quality QTLs are more typically located in the A subgenome, while in intraspecific populations, fiber yield and quality QTLs are more frequent in the D subgenome [29, 30]. Although DD diploid species do not have spinnable fibers, many studies have shown that the D subgenome of allotetraploid cotton contains many QTLs that control fiber quality [31, 32]. However, while previous studies have revealed these and other useful findings, the different groups and markers employed combined with the impact of environmental factors on QTL effects mean that the comparability of extant data is relatively poor. Therefore, QTL research on cotton is still advancing.
Recent advances in genome sequencing technology allow the construction of ultra-high-density genetic maps based on SNP loci. Consequently, more comprehensive and accurate map information can be used to analyze QTLs associated with important traits. Since bin genetic linkage maps based on SNP loci were first constructed in rice [33], it has been widely applied in other plants such as cotton [20], maize [34], soybean [35], Cucumis melo [36], radish [37] and so on. These genetic linkage maps have yielded many fine-mapped QTLs for which corresponding target genes were identified and cloned.
Recent high-quality assemblies of G. barbadense and G. hirsutum [2, 38–42] has provided good references for linkage map-based QTL identification. In light of these resources, we constructed an interspecific F2 population between G. hirsutum and G. barbadense and performed whole-genome sequencing of all 249 F2 individuals, achieving resequencing data on average over 5 × genome coverage of each material and generating a high-density genetic map containing 6303 bin markers. Based on the map, we subsequently identified 112 QTLs associated with an array of traits including plant type traits and physiological traits at the seedling stage, leaf chlorophyll content, plant type traits at flower and boll stage, yield traits, and fiber quality traits. Combining the SNPs located within the predicted genes in the target region and their expression pattern of the predicted genes, possible causable genes that are responsible for the mapping traits were identified. These QTLs and the related candidate genes are valuable in cotton breeding to improve plant biomass, physiological characteristics, and yield quality.
Methods
Plant materials and DNA extraction
The plant materials used consisted of G. hirsutum acc. TM-1 supplied by Dr. Kohel of Southern Plain Agricultural Research Center, USDA [43] and G. barbadense cv. Hai7124 which was selected by Cotton Research Institute of Nanjing Agricultural University for genetic research [17]. TM-1 is a genetic standard line of G. hirsutum developed through single plant selection. Hai7124, grown extensively in China, was also the offspring of a single plant selection before being used as a parent in the construction of the linkage map. Two highly homozygous parents, as well as 249 F2 individuals derived from a cross between TM-1 as the recipient and Hai7124 as the donor were planted in Pailou greenhouse of Nanjing Agricultural University, Jiangsu, China. Genomic DNA was extracted from young leaf tissues following the method cetyltriethylammnonium bromide (CTAB) described by Paterson [44] with increased RNase A and proteinase K treatment to prevent RNA and protein contamination. The isolated DNA was then subjected to Illumina sequencing technology.
To obtain the phenotypic data of two parents, F1, and all 249 F2 individual plants at different environments. All of them were cut off the trunk, transferred in the large nutrient bowls, and moved into the greenhouse in autumn. In the next spring, these materials were planted in the field for investigation of yield and fiber traits. The same operation was repeated twice in 2011 and 2012.
Phenotype data collection and evaluation
Plant type traits at seedling stage
The following plant type traits of the parents, F1, and F2 individual plants respectively were investigated at the cotton seedling stage: plant height (PH1, cm); cotyledonary node height (CNH, cm); first true leaf height (FTLH, cm); second true leaf height (STLH, cm); distance between the cotyledonary node and first true leaf (D1, cm); and distance between first true leaf and second true leaf (D2, cm). Each measurement was repeated three times and the average value was used in the analysis.
Physiological traits at seedling stage
Physiological characteristics such as leaf area and photosynthetic rate were measured in the parents, F1, and F2 individual plants at the cotton seedling stage. A portable leaf area meter (CI-202, Portable Laser Leaf Area Meter, USA) was used to measure the second true leaf area (SLA, cm2). At the same time, from 8:00 to 11:00 in the morning on a sunny day, a Li-6400 portable photosynthesis instrument was used to determine the photosynthesis ratio (Pn, μmol CO2·m−2·s−1) of the second true leaf. Also measured were intercellular CO2 concentration (Ci, μmol·mol−1), stomatal conductance (Cond, mmol·m−2·s−1), and transpiration rate (Tr, g·m−2·h−1). The intensity of the built-in light source was set to 1200 μmol·m−2·s−1, each leaf was measured three times, and the average value was used in the analysis. For instrument principle, sampling technique, and detailed settings, refer to "Using the LI-6400 Portable Photosynthesis System."
Determination of chlorophyll content in leaves
The leaf chlorophyll content of the parents, F1, and F2 individual plants was determined by UV/visible spectrophotometer. The main stem functional leaves were collected from each individual plant, and ten pieces were cut out with a 9-mm punch and weighed. About 0.1–0.2 g leaves were then placed in a 10-ml test tube, the fresh weight recorded, 10 ml of 95% ethanol added, and the tube sealed and stored for 48 h in the dark. Tubes were shaken in the middle of the incubation and mixed until the leaves were completely white. After the incubation, the extracted chlorophyll of each sample was placed in a spectrophotometer and the optical density was measured at 665 nm, 649 nm, and 470 nm to respectively determine chlorophyll a (Chl a), chlorophyll b (Chl b), and carotenoid (Car) content. Subsequently, the chlorophyll a/b ratio (Chl a/b) and total chlorophyll (Total Chl) were calculated. Each sample was repeated three times, and the average was taken as the result.
Pigment concentrations were calculated according to the following formulas:
| 1 |
in which Ca, Cb, and Cx•c represent the concentration in mg/L of chlorophyll a, chlorophyll b and carotenoids, respectively.
The pigment content of the leaves was then calculated as follows:
where C represents the pigment concentration (mg/L), V represents the total amount of extract (ml), W represents the fresh weight of the sample (g), and the subscript x represents the pigment: chlorophyll a or b, or carotenoids.
Plant type traits at flowering and boll stage
Plant height (PH2) and fruit branch number (FBN) were investigated at the first flowering and boll stage in the parents, F1, and F2 individual plants.
Yield traits
Yield constituent factors were assayed during the boll opening stage. The traits investigated consisted of boll number per plant (bolls/plant, BN), seed cotton yield (SCY), lint yield (LY), boll weight (BW), lint percentage (LP), lint index (LI), and seed index (SI).
Fiber quality traits
Middle and upper fibers were collected from the parents, F1, and F2 individual plants and sent for testing at the Cotton Quality Supervision, Inspection and Testing Center of the Ministry of Agriculture (HVI SPECTRUM 4.05.01 version, HVICC calibration level). Tested fiber quality properties included: fiber length (FL), fiber strength (FS), micronaire value (MIC), fiber length uniformity (FU), fiber elongation (FE). Due to high temperatures and too much rain in the summer of 2011, which caused abortion of pollen and super-separation of the sea-land hybrid population, some families failed to receive enough mature fiber, resulting in a lack of yield and quality trait data in some lines.
Population DNA preparation, resequencing, and genotyping
Sequencing libraries were constructed with an insert size of 150 bp and sequenced on the Illumina HiSeq 2000 platform (Illumina, San Diego, CA, USA). To construct paired-end libraries, DNA was fragmented by sonication, and DNA ends were blunted before adding an A base to each 3′ end. DNA adaptors with a single T-base 3′ end overhang were ligated to the above products. Ligation products were purified on 2% agarose gels that each targeted a specific range of insert sizes. Quantification and quality assessment were carried out by running 1 μL of the library on an Agilent DNA 1000 LabChip analyzer (Agilent Technology 2100 Bioanalyzer). All raw reads were processed for quality control and filtered using fastp (https://github.com/OpenGene/fastp) with default parameters. The clean reads were mapped to the TM-1 reference genome [38] using Burrows–Wheeler Aligner (BWA) with the parameters of ‘mem -t 20 -M -R’. The mapping results were sorted and duplicates marked using functions implemented in SAMtools and Picard (http://broadinstitute.github.io/picard/). Only reads that mapped uniquely to the reference genome sequence were used to call SNPs. Identification of SNPs between the parental lines and F2 individuals was performed with Genome Analysis Toolkit 4 (GATK4). High-quality SNPs were filtered following the best practices workflow developed by the GATK team. SNPs with minor allele frequency (MAF) < 5% and represented in less than 30% of the F2 population were excluded using VCFtools. Polymorphic markers between the two parental lines were retained if they had the aa × bb segregation pattern in F2 individuals.
Bin map construction
Recombinant breakpoints were identified using a slightly modified sliding window approach based on the ratio of SNP alleles derived from TM-1 and Hai7124 [38]. Consecutive 100-Kb intervals having the same genotype in the whole F2 population were merged as a recombination bin. Bins with significantly distorted segregation (P-value < 0.001) were filtered using the Chi-square test, and those remaining served as genetic markers for the construction of a genetic linkage map using Icimapping [45]. Collinearity between the genetic map and physical positions was visualized using ALLMAPS (https://github.com/tanghaibao/jcvi/wiki/ALLMAPS). A region containing three or more closely linked bins that exhibited significant segregation distortion (P < 0.001) was defined as an SDR.
Statistics of phenotypic traits
For all traits, ANOVA was used to test for significant differences between parents, F1, and F2 individuals, and correlation coefficients and phenotypic variation were also calculated using SPSS v18.0 (SPSS, Chicago, IL, USA). The heterosis (H) of each trait is expressed by two values, mid-parent heterosis and over-parent heterosis: MH = (F1-MP)/MP × 100%, where MP is the average value of the parents.
QTL mapping
IciMapping 3.0 (http://www.isbreeding.net) was used to detect the effects of QTLs in the F2 population. An LOD threshold of 2.5 was used to define significant additive QTLs; that is, when LOD ≥ 2.5 for a marker interval, it was considered to contain a significant QTL. At the same time, the additive effect (A), dominant effect (D), and contribution rate (R2) of each QTL on corresponding traits were calculated. The QTL genetic action mode uses the absolute value of D/A to judge the action effect of each QTL; a value greater than 1.20 indicates an over dominant effect, 0.81–1.20 a dominant effect, and 0.21–0.80 a partially dominant effect. Less than 0.20 indicates an additive effect. The method of naming QTLs follows that used for rice: QTL + traits + chromosome + QTL number.
Candidate gene identification and expression
The putative candidate genes for the QTLs were predicted as follows. First, we analyzed the SNP types located in QTLs based on our assembled genome sequence for TM-1. We focused on significantly associated nonsynonymous SNPs located in exons or SNPs in the upstream of the candidate genes. Second, based on expression profiling data for sixteen vegetative and reproductive tissues from TM-1 (cotton.zju.edu.cn). We checked whether these selected candidate genes were dominantly and/or specific expressed in a development stage that is critical for the target trait. We further narrowed down the candidate genes according to their expression levels between TM-1 and Hai7124 (cotton.zju.edu.cn).
Results
High-density genetic map construction and characteristics of the bin marker loci
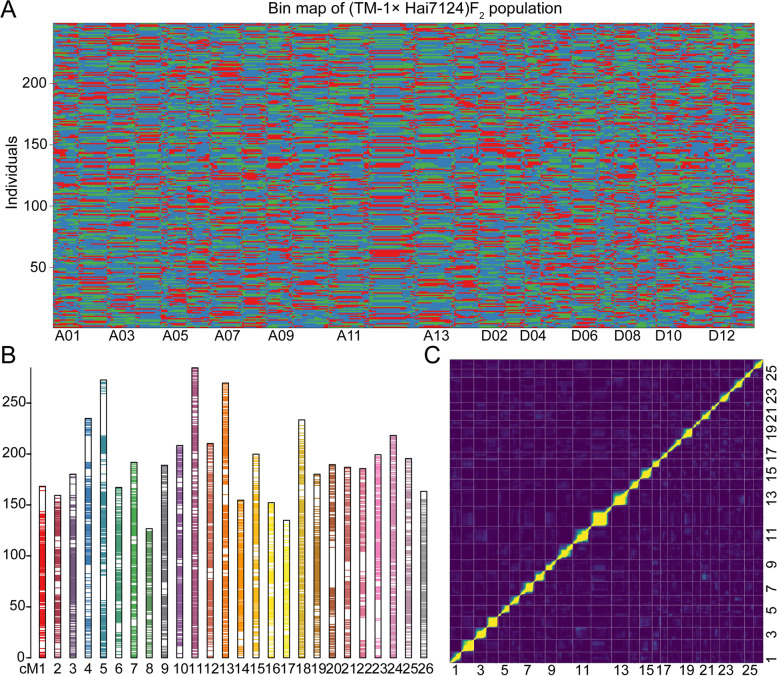
We developed an interspecific F2 population from a cross between G. hirsutum acc. TM-1 and G. barbadense cv. Hai7124, which contained 249 individuals in total. Whole-genome sequencing of all individuals was performed on an Illumina Hiseq2000. In total, 3.01 Tb clean reads were generated, with an average of 5.3 × depth genome coverage for each individual. For the parents ‘TM-1’ and ‘Hai7124’, we utilized clean data from our previous research totaling 185 Gb and 111.8 Gb respectively [20], with an average depth of over 50 × . All clean reads were mapped to the TM-1 as the reference genome. After filtering SNPs by established criteria, a total of 4,257,943 high-quality SNPs (Fig. 1) were retained and used to generate bin markers (a group of consecutive SNPs in the same block for genotyping) with a modified sliding window approach [33]. After filtering 1428 bins that exhibited significant segregation distortion (P < 0.001), a total of 6303 bin markers were generated, with an average length of 363.1 Kb (Table 1, Fig. 1). Finally, the high-density genetic map was constructed, covering 5057 cM with an average inter-bin genetic distance of 0.8 cM (Fig. 1, Table 1). The 26 linkage groups of the map was corresponding to 26 cotton chromosomes. Each of the linkage group contained 242.4 bins on average, ranging from 157 (D04) to 405 (A11), overall comprising 3,455 in the A subgenome and 2,848 in the D subgenome. The total length of the A subgenome was 2663.24 cM, and that for the D subgenome was 2393.89 cM. The longest linkage group was A11 of 284.58 cM, and the shortest one was A08 of 126.68 cM. The largest average distance between markers was 1.1 cM in the D07 linkage group, while the smallest average distance between markers in A10 was 0.64 cM (Table 1).
Fig. 1.
High-density genetic map construction of the (TM-1×Hai7124)F2 population. A Bin maps for the 241 scored F2 individual lines. Colored tracks represent the 241 individual lines of the THF2 population that were used for linkage map construction: red, alleles inherited from maternal parent (TM-1); green, alleles inherited from paternal parent (Hai7124); blue, alleles inherited from heterozygous genotype (TM-1 × Hai7124)F1. The horizontal scale indicates physical distance. B Distribution of markers across 26 chromosomes; ordinate is genetic distance, cM. C Genetic map quality as indicated by recombination fractions of all markers
Table 1.
Characteristics of the 26 linkage groups in allotetraploid cotton
| Chr | No. bins | Distance (cM) | Average distance | Average length of bin (Kb) | Gap number (> 10 cM) | Recombination rate (cM/Mb) | RHRs |
|---|---|---|---|---|---|---|---|
| A01 | 234 | 168.32 | 0.72 | 505.0 | 1 | 1.42 | 104 |
| A02 | 197 | 159.41 | 0.81 | 549.6 | 0 | 1.47 | 76 |
| A03 | 254 | 180.2 | 0.71 | 439.3 | 1 | 1.61 | 113 |
| A04 | 239 | 234.94 | 0.98 | 366.9 | 1 | 2.68 | 110 |
| A05 | 256 | 272.68 | 1.07 | 433.0 | 2 | 2.46 | 147 |
| A06 | 206 | 167.24 | 0.81 | 614.0 | 0 | 1.32 | 99 |
| A07 | 255 | 191.84 | 0.75 | 378.8 | 0 | 1.99 | 138 |
| A08 | 189 | 126.69 | 0.67 | 661.7 | 0 | 1.01 | 78 |
| A09 | 262 | 189.01 | 0.72 | 317.6 | 0 | 2.27 | 150 |
| A10 | 326 | 208.36 | 0.64 | 353.1 | 0 | 1.81 | 151 |
| A11 | 405 | 284.58 | 0.7 | 299.7 | 0 | 2.34 | 219 |
| A12 | 284 | 210.33 | 0.74 | 378.8 | 0 | 1.95 | 166 |
| A13 | 348 | 269.64 | 0.77 | 317.1 | 2 | 2.44 | 166 |
| At-Total | 3455 | 2663.24 | 0.78 | 431.89 | 7 | 1.78 | 1717 |
| D01 | 232 | 154.67 | 0.67 | 278.9 | 0 | 2.39 | 119 |
| D02 | 257 | 199.83 | 0.78 | 271.5 | 0 | 2.86 | 115 |
| D03 | 173 | 152.18 | 0.88 | 311.5 | 0 | 2.82 | 91 |
| D04 | 157 | 134.93 | 0.86 | 362.6 | 0 | 2.37 | 95 |
| D05 | 282 | 233.47 | 0.83 | 226.7 | 0 | 3.65 | 207 |
| D06 | 221 | 180.13 | 0.82 | 296.2 | 1 | 2.75 | 103 |
| D07 | 173 | 189.57 | 1.10 | 337.7 | 1 | 3.25 | 107 |
| D08 | 217 | 187.01 | 0.86 | 318.3 | 1 | 2.71 | 127 |
| D09 | 185 | 185.67 | 1.00 | 281.1 | 2 | 3.57 | 121 |
| D10 | 228 | 199.29 | 0.87 | 293.3 | 2 | 2.98 | 126 |
| D11 | 317 | 218.19 | 0.69 | 225.1 | 0 | 3.06 | 200 |
| D12 | 195 | 195.58 | 1.00 | 316.4 | 0 | 3.17 | 143 |
| D13 | 211 | 163.37 | 0.77 | 305.4 | 0 | 2.53 | 109 |
| Dt-Total | 2848 | 2393.89 | 0.86 | 294.21 | 7 | 2.85 | 1663 |
| Total | 6303 | 5057.13 | 0.80 | 363.1 | 14 | 2.21 | 3380 |
A total of fourteen gaps that larger than 10 cM were distributed across the all 26 chromosomes, seven at the A subgenome and seven at the D subgenome. The average ratio of bin marker interval (< 5 cM) for all linkage groups was more than 99%. A region containing three or more closely linked bins that exhibited significant segregation distortion (P < 0.001) was considered a segregation distortion region (SDR). There were 88 and 32 SDRs in the A and D subgenome, respectively (Table 1). The quality of the genetic map was further examined by comparing genetic and physical distances, which showed good collinearity (Supplementary Fig. 1).
Chi-square tests of the 6303 co-dominance bins identified 724 that do not conform to the 1:2:1 genetic law ratio of Mendelian theory. Among these 724 partial segregation bins, 86 were biased toward the parent TM-1, 638 toward the parent Hai7124, and none toward the heterozygote. In addition, significantly more of the partial segregation bins were located on the A subgenome (450) than on the D subgenome (274), and these bins comprised a higher proportion of the A subgenome (13.02%) than of the D subgenome (9.62%). Moreover, the partial segregation bins were unevenly distributed across the 26 chromosomes; the ratio of partial segregation bins to total bins in a given chromosome was more than 30% on chromosomes A05, A11, and D07 and more than 20% on A08, D09, and D10, but less than 1% on A01, A03, and D01. At the same time, some partial segregation bins exhibited an aggregation phenomenon; namely, bins distributed on four chromosomes (A05, A11, D07, and D08) account for 45% of all partial segregation bins (Supplementary Table 1).
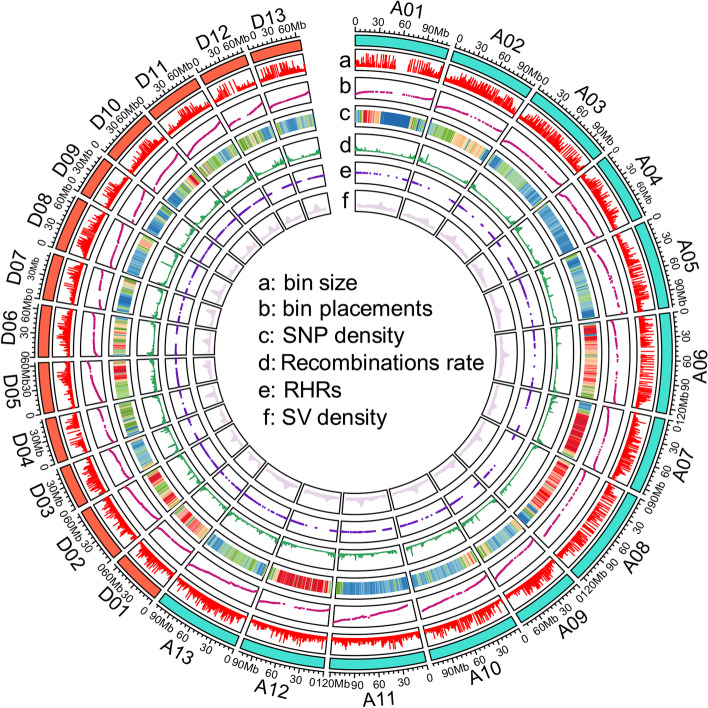
To provide a comprehensive overview of recombination in cotton, the recombination rate along each chromosome was estimated by comparing genetic and physical distances. Across the entire genome, the average recombination rate was 2.2 cM/Mb. High rates of recombination were observed in the telomere regions of all nine chromosomes, whereas recombination was suppressed in centromere regions (Fig. 2). Chromosomal regions with recombination rates greater than 1.0 cM/Mb [37] were defined as recombination hot regions (RHRs). A total of 3380 RHRs were identified, and were unevenly distributed on the 26 chromosomes (Table 1, Fig. 2).
Fig. 2.
Chromosomal features of (TM-1 × Hai7124)F2 population with genetic data. A The length of 6303 bins along each chromosome; B The bin marker placements in the genetic maps on the chromosome; C SNP (aa × bb) density of each chromosome; D Recombination rates of each chromosome; E Genetic positions of the RHRs in each chromosome; F Structural variations density of each chromosome
Analysis of 35 traits in parents and F1 and F2 generations
We surveyed 35 traits in the parents and F1 and F2 generations, including six plant type traits, ten leaf morphology and physiological traits at the seedling stage, five leaf chlorophyll content traits, two plant type traits at flower and boll stage, seven yield traits, and five fiber quality traits (Supplementary Table 2).
TM-1, Hai7124, and their F1 progeny differed to varying degrees in plant type, leaf morphology, physiology, yield, and fiber quality. Concerning plant type traits, TM-1 and F1 had extremely significant differences; TM-1 and Hai7124 likewise had extremely significant differences, except in CNH; but Hai7124 and F1 had no extremely significant differences in traits except for D1. Regarding leaf morphology and physiological traits, TM-1 exhibited extremely significant difference from Hai7124 and from F1 only in SLA and SPn; other traits were not significantly different among the three. In terms of chlorophyll content, TM-1 and F1 exhibited extremely significant differences; TM-1 and Hai7124 likewise had extremely significant differences in traits other than Chla; but Hai7124 and F1 did not differ significantly except in Chla/b. With regard to the 12 yield and fiber traits, G. hirsutum and G. barbadense are characterized by extremely significant differences; most of these characteristic differences were observed in comparisons of TM-1 and Hai7124 and of TM-1 and F1 individuals. When comparing Hai7124 and F1, only the five traits D1, SCY, LY, SI, and LI differed significantly, indicating that the F1 progeny of G. barbadense and G. hirsutum are more biased towards the G. barbadense phenotype. Taken together, these genetic differences provide a good basis for the screening of important trait QTLs (Supplementary Table 3).
In the F2 population, the average value and variance of each trait exhibited large changes relative to their parents, and the coefficients of variation differed between traits. Overall, physiology and yield traits featured the largest coefficients; the values for each yield component ranked as follows: BN > SCY > LY > LI > BW > SI > LP. This ranking indicates that in the offspring, different degrees of genetic variation are present for different traits, indicating that these traits are controlled by multiple genes (Table 2, Supplementary Fig. 2).
Table 2.
Phenotypic variation of 35 traits
| Traits | Number of individuals | Mean | Mean squared error | Min | Max | Standard deviation | variance | Skewness | kurtosis | CV (%) | mid-parent heterosis |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PH1 | 249 | 20.4 | 0.23 | 9.30 | 32.30 | 3.69 | 13.61 | -0.41 | 0.14 | 18.08 | -4.23 |
| CNH | 249 | 7.59 | 0.08 | 3.50 | 11.40 | 1.32 | 1.75 | 0.11 | 0.28 | 17.41 | 9.21 |
| FTLH | 249 | 15.42 | 0.17 | 8.20 | 23.00 | 2.73 | 7.47 | 0.11 | -0.2 | 17.73 | 6.53 |
| STLH | 247 | 18.62 | 0.2 | 8.90 | 30.50 | 3.14 | 9.84 | -0.2 | 0.48 | 16.85 | -2.10 |
| D1 | 249 | 7.83 | 0.14 | 1.90 | 14.10 | 2.15 | 4.63 | 0.36 | 0.09 | 27.49 | 4.05 |
| D2 | 247 | 3.19 | 0.11 | 0.20 | 9.80 | 1.79 | 3.21 | 0.64 | 0.25 | 56.12 | -29.89 |
| SLA | 249 | 30.6 | 0.54 | 7.93 | 61.66 | 8.54 | 72.87 | 0.43 | 0.68 | 27.90 | 4.42 |
| TLA | 233 | 25.07 | 0.64 | 4.99 | 54.89 | 9.80 | 96.08 | 0.34 | -0.19 | 39.10 | -3.00 |
| SPn | 249 | 9.24 | 0.25 | 0.74 | 18.64 | 3.95 | 15.64 | 0.05 | -0.71 | 42.79 | 6.39 |
| TPn | 249 | 11.62 | 0.25 | 3.24 | 20.89 | 3.98 | 15.85 | -0.18 | -0.79 | 34.27 | 1.18 |
| SCi | 249 | 235.77 | 3.14 | 84.24 | 325.24 | 49.53 | 2452.76 | -0.64 | 0.10 | 21.01 | 1.34 |
| TCi | 249 | 237.19 | 3.13 | 30.12 | 369.80 | 49.37 | 2437.74 | -0.59 | 0.78 | 20.82 | 3.63 |
| SCond | 249 | 0.13 | 0.01 | 0.01 | 0.37 | 0.08 | 0.01 | 0.56 | -0.51 | 63.78 | 23.81 |
| TCond | 249 | 0.16 | 0.01 | 0.02 | 0.44 | 0.09 | 0.01 | 0.42 | -0.5 | 54.37 | 14.29 |
| STr | 249 | 4.64 | 0.19 | 0.49 | 12.90 | 2.92 | 8.54 | 0.63 | -0.56 | 63.02 | 16.88 |
| TTr | 249 | 5.53 | 0.18 | 0.71 | 12.83 | 2.89 | 8.38 | 0.59 | -0.44 | 52.34 | 10.82 |
| Chl a | 240 | 0.78 | 0.01 | 0.37 | 1.27 | 0.13 | 0.02 | 0.30 | 1.49 | 16.42 | 4.00 |
| Chl b | 240 | 0.28 | 0.00 | 0.15 | 0.44 | 0.04 | 0.00 | 0.34 | 0.96 | 15.69 | 12.00 |
| Car | 240 | 0.16 | 0.00 | 0.07 | 0.27 | 0.03 | 0.00 | 0.25 | 1.19 | 17.30 | -5.88 |
| Chl a/b | 240 | 2.82 | 0.01 | 2.04 | 3.21 | 0.17 | 0.03 | -0.42 | 1.37 | 6.06 | -6.16 |
| Total Chl | 240 | 1.06 | 0.01 | 0.53 | 1.71 | 0.17 | 0.03 | 0.32 | 1.39 | 16.02 | 6.00 |
| PH2 | 238 | 144.01 | 2.26 | 48.00 | 220.00 | 34.94 | 1220.57 | -0.66 | 0.41 | 24.26 | 48.21 |
| FBN | 238 | 13.08 | 0.26 | 3.00 | 24.00 | 4.06 | 16.50 | -0.37 | -0.04 | 31.05 | -12.80 |
| BN | 224 | 13.62 | 0.52 | 0.00 | 42.00 | 7.84 | 61.40 | 1.14 | 1.13 | 57.55 | -34.09 |
| SCY | 215 | 48.81 | 1.87 | 4.08 | 142.83 | 27.45 | 753.45 | 1.00 | 0.67 | 56.24 | -36.10 |
| LY | 215 | 14.72 | 0.55 | 1.11 | 40.32 | 8.08 | 65.36 | 0.89 | 0.34 | 54.93 | -41.27 |
| BW | 220 | 3.56 | 0.06 | 1.57 | 7.47 | 0.92 | 0.85 | 0.75 | 1.18 | 25.93 | -13.59 |
| SI | 220 | 10.45 | 0.13 | 5.85 | 20.00 | 1.99 | 3.96 | 0.67 | 2.02 | 19.04 | -3.86 |
| LP | 220 | 30.65 | 0.36 | 20.79 | 52.38 | 5.38 | 28.98 | 0.97 | 1.63 | 17.56 | -7.49 |
| LI | 220 | 4.64 | 0.08 | 2.27 | 11.14 | 1.26 | 1.58 | 1.39 | 4.87 | 27.09 | -13.91 |
| FL | 77 | 29.69 | 0.24 | 25.41 | 35.43 | 2.06 | 4.26 | 0.21 | -0.09 | 6.95 | -3.49 |
| FS | 77 | 30.26 | 0.34 | 25.00 | 40.90 | 3.02 | 9.13 | 0.89 | 1.35 | 9.98 | -5.19 |
| MIC | 77 | 3.14 | 0.07 | 2.00 | 4.79 | 0.63 | 0.40 | 0.23 | -0.31 | 20.03 | -28.15 |
| FU | 77 | 83.9 | 0.21 | 78.8 | 88.20 | 1.81 | 3.29 | -0.32 | 0.19 | 2.16 | -1.53 |
| FE | 77 | 6.26 | 0.05 | 5.20 | 7.98 | 0.47 | 0.22 | 1.31 | 2.84 | 7.43 | -5.22 |
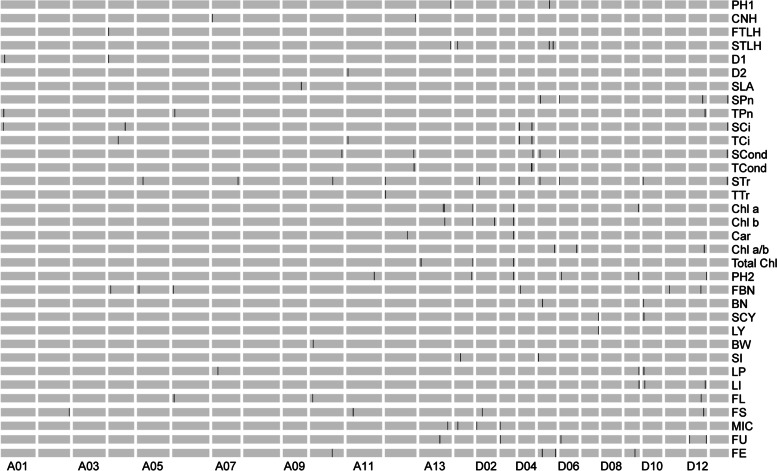
QTL mapping of important agronomic traits in cotton
A total of 112 QTLs, 41 in the A subgenome and 71 in the D subgenome, distributing across almost all 26 chromosomes except A03, A08, and D08, were assessed for association with 35 traits using ICIM analysis. The position, LOD score, additive effects, dominance effect, and percentage of phenotypic variance explained (PVE) of the QTLs are given in Table 3. Among them, 16 QTLs were located overlapped with the QTL regions in the previous studies (Table 4). PVE values ranged from 2.95 to 24.89%. The regions occupied by identified QTLs ranged in size from 0.20 to 8.45 Mb, with an average length of 0.78 Mb. With respect to traits, the number of QTLs per trait ranged from 0 to 10 with the most QTLs (up to 10) being detected for STr.
Table 3.
Analysis of QTLs for 35 traits
| Trait | QTL | Chromsome | Left Marker | Right Marker | LeftCI (cM) | RightCI (cM) | Start | End | LOD | PVE(%) | Add | Dom |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PH1 | qPH1-A13 | A13 | bin395 | bin396 | 248.5 | 252.5 | 107,859,745 | 108,097,137 | 3.16 | 6.89 | -1.09 | -0.8 |
| PH1 | qPH1-D05 | D05 | bin223 | bin229 | 155.5 | 167.5 | 39,478,283 | 41,627,412 | 3 | 6.47 | -1.21 | 0.7 |
| CNH | qCNH-A07 | A07 | bin3 | bin4 | 0 | 6.5 | 330,907 | 915,621 | 3.37 | 6.65 | -0.32 | 0.51 |
| CNH | qCNH-A12 | A12 | bin454 | bin455 | 181.5 | 186.5 | 104,326,658 | 104,861,614 | 3.95 | 8.35 | 0.5 | -0.1 |
| FTLH | qFTLH-A04 | A04 | bin2 | bin3 | 2.5 | 6.5 | 295,380 | 912,894 | 2.98 | 6.3 | -0.49 | -1.1 |
| STLH | qSTLH-A13 | A13 | bin393 | bin394 | 245.5 | 248.5 | 107,403,460 | 107,859,745 | 3.02 | 4.86 | -0.98 | -0.25 |
| STLH | qSTLH-D01 | D01 | bin57 | bin58 | 51.5 | 54.5 | 10,754,698 | 11,063,172 | 3.04 | 4.93 | 1.09 | -0.06 |
| STLH | qSTLH-D05-1 | D05 | bin221 | bin227 | 151.5 | 154.5 | 52,507,478 | 52,707,478 | 2.98 | 4.55 | -0.55 | 1.2 |
| STLH | qSTLH-D05-2 | D05 | bin250 | bin251 | 173.5 | 174.5 | 38,370,762 | 40,442,855 | 3.22 | 5.01 | -0.63 | 1.21 |
| D1 | qD1-A01 | A01 | bin58 | bin59 | 53.5 | 55.5 | 12,558,163 | 13,053,104 | 2.75 | 5.62 | 0.68 | 0.13 |
| D1 | qD1-A04 | A04 | bin2 | bin3 | 1.5 | 5.5 | 295,380 | 912,894 | 3.44 | 7.18 | -0.25 | -1.02 |
| D2 | qD2-A11 | A11 | bin7 | bin55 | 29.5 | 48.5 | 807,428 | 9,258,460 | 3.68 | 4.46 | -1.3 | -1.28 |
| SLA | qSLA-A09 | A09 | bin176 | bin177 | 103.5 | 105.5 | 63,066,211 | 63,735,059 | 2.62 | 7.16 | 0.38 | 3.73 |
| SPn | qSPn-D05 | D05 | bin59 | bin60 | 43.5 | 44.5 | 8,567,785 | 8,789,652 | 4.35 | 7.56 | 1.6 | 0.01 |
| SPn | qSPn-D06 | D06 | bin6 | bin7 | 0 | 1.5 | 617,114 | 818,140 | 2.87 | 4.89 | 0.28 | -1.78 |
| SPn | qSPn-D12 | D12 | bin138 | bin139 | 107.5 | 110.5 | 47,167,180 | 47,756,249 | 3.31 | 5.69 | -0.01 | 1.87 |
| SPn | qSPn-D13 | D13 | bin209 | bin211 | 135.5 | 139.5 | 61,357,809 | 61,953,285 | 3.24 | 5.37 | -1.2 | 0.71 |
| TPn | qTPn-A01 | A01 | bin48 | bin49 | 43.5 | 51.5 | 9,757,023 | 10,168,632 | 2.56 | 5 | -0.62 | 1.4 |
| TPn | qTPn-A06 | A06 | bin40 | bin41 | 43.5 | 44.5 | 8,721,887 | 9,167,458 | 3.33 | 6.91 | -0.12 | -1.88 |
| TPn | qTPn-D12 | D12 | bin187 | bin188 | 148.5 | 149.5 | 55,359,867 | 55,595,846 | 3.33 | 7.03 | 0.07 | 1.92 |
| SCi | qSCi-A01 | A01 | bin43 | bin44 | 42.5 | 45.5 | 8,036,084 | 8,969,646 | 2.63 | 4.43 | 6.78 | 18.33 |
| SCi | qSCi-A04 | A04 | bin182 | bin183 | 145.5 | 146.5 | 57,244,810 | 57,548,850 | 2.78 | 5.03 | 1.72 | -22.14 |
| SCi | qSCi-D04-1 | D04 | bin32 | bin33 | 28.5 | 32.5 | 4,450,078 | 4,794,145 | 3.19 | 5.67 | -7.03 | 21.69 |
| SCi | qSCi-D04-2 | D04 | bin131 | bin132 | 86.5 | 88.5 | 46,227,657 | 47,544,347 | 2.94 | 4.94 | -4.56 | -20.82 |
| SCi | qSCi-D13 | D13 | bin208 | bin209 | 135.5 | 139.5 | 61,187,337 | 61,554,710 | 2.5 | 4.3 | -14.5 | 1.89 |
| TCi | qTCi-A04 | A04 | bin103 | bin104 | 101.5 | 103.5 | 33,378,346 | 33,609,608 | 3.23 | 3.33 | -3.67 | -23.45 |
| TCi | qTCi-A11 | A11 | bin7 | bin55 | 14.5 | 53.5 | 807,428 | 9,258,460 | 2.75 | 12.52 | -21.49 | 36.36 |
| TCi | qTCi-D04-1 | D04 | bin32 | bin33 | 28.5 | 32.5 | 4,450,078 | 4,794,145 | 2.86 | 2.95 | -2.41 | 22.13 |
| TCi | qTCi-D04-2 | D04 | bin131 | bin132 | 86.5 | 88.5 | 46,227,657 | 47,544,347 | 3.32 | 3.21 | -5.82 | -21.48 |
| SCond | qSCond-A10 | A10 | bin352 | bin353 | 196.5 | 202.5 | 109,106,699 | 109,336,022 | 2.5 | 3.91 | 0.02 | 0.02 |
| SCond | qSCond-A12 | A12 | bin423 | bin425 | 158.5 | 159.5 | 98,555,400 | 99,115,052 | 3.91 | 6.42 | 0.03 | -0.01 |
| SCond | qSCond-D04 | D04 | bin142 | bin143 | 88.5 | 94.5 | 50,189,434 | 50,421,587 | 2.59 | 4.03 | -0.01 | -0.03 |
| SCond | qSCond-D05 | D05 | bin51 | bin52 | 39.5 | 40.5 | 7,536,477 | 7,742,595 | 5.19 | 8.67 | 0.03 | -0.01 |
| SCond | qSCond-D06 | D06 | bin6 | bin7 | 0 | 1.5 | 617,114 | 818,140 | 2.9 | 4.74 | 0.01 | -0.04 |
| SCond | qSCond-D13 | D13 | bin196 | bin197 | 121.5 | 123.5 | 59,385,048 | 59,708,159 | 3.57 | 5.9 | -0.03 | 0.01 |
| TCond | qTCond-A12 | A12 | bin427 | bin430 | 157.5 | 164.5 | 99,337,542 | 100,288,635 | 2.53 | 4.62 | 0.03 | -0.02 |
| TCond | qTCond-D04-1 | D04 | bin128 | bin129 | 81.5 | 85.5 | 45,564,063 | 45,808,186 | 3.42 | 6.18 | -0.01 | -0.04 |
| TCond | qTCond-D04-2 | D04 | bin131 | bin132 | 86.5 | 87.5 | 46,227,657 | 47,544,347 | 3.62 | 6.52 | -0.02 | -0.04 |
| STr | qSTr-A05 | A05 | bin119 | bin120 | 95.5 | 103.5 | 20,625,360 | 21,148,783 | 2.82 | 3.3 | 0.58 | 1.03 |
| STr | qSTr-A07 | A07 | bin285 | bin286 | 141.5 | 145.5 | 88,546,004 | 88,983,661 | 2.84 | 3.3 | 0.76 | -0.67 |
| STr | qSTr-A10 | A10 | bin228 | bin229 | 120.5 | 124.5 | 77,749,540 | 78,563,787 | 2.8 | 3.29 | 0.95 | 0.07 |
| STr | qSTr-A12 | A12 | bin7 | bin8 | 8.5 | 19.5 | 1,418,415 | 1,932,428 | 2.53 | 2.96 | 0.51 | -0.89 |
| STr | qSTr-D02 | D02 | bin66 | bin63 | 55.5 | 59.5 | 11,571,331 | 11,240,351 | 3.8 | 4.38 | -0.55 | -1.17 |
| STr | qSTr-D04 | D04 | bin23 | bin24 | 16.5 | 18.5 | 2,781,571 | 3,052,412 | 2.77 | 3.34 | -0.49 | 1.02 |
| STr | qSTr-D05 | D05 | bin51 | bin52 | 39.5 | 40.5 | 7,536,477 | 7,742,595 | 4.89 | 6.12 | 1.23 | -0.11 |
| STr | qSTr-D06 | D06 | bin6 | bin7 | 0 | 1.5 | 617,114 | 818,140 | 3.74 | 4.63 | 0.15 | -1.49 |
| STr | qSTr-D10 | D10 | bin7 | bin8 | 9.5 | 15.5 | 1,651,964 | 1,902,901 | 2.86 | 3.32 | -0.91 | -0.05 |
| STr | qSTr-D13 | D13 | bin196 | bin197 | 121.5 | 123.5 | 59,385,048 | 59,708,159 | 2.79 | 3.45 | -0.87 | 0.11 |
| TTr | qTTr-A12 | A12 | bin8 | bin9 | 12.5 | 21.5 | 1,656,702 | 2,140,812 | 3.56 | 6.6 | 0.69 | -1.07 |
| Chl a | qChl a-A13-1 | A13 | bin270 | bin278 | 145.5 | 159.5 | 85,560,449 | 86,656,939 | 2.73 | 4.64 | -0.04 | -0.03 |
| Chl a | qChl a-A13-2 | A13 | bin286 | bin287 | 173.5 | 184.5 | 80,629,132 | 83,806,916 | 2.69 | 5.21 | -0.04 | -0.01 |
| Chl a | qChl a-D01 | D01 | bin239 | bin240 | 136.5 | 146.5 | 62,926,804 | 63,199,901 | 3.05 | 5.09 | -0.02 | 0.05 |
| Chl a | qChl a-D03 | D03 | bin153 | bin154 | 114.5 | 119.5 | 48,191,318 | 48,417,933 | 4.2 | 7.97 | 0 | 0.07 |
| Chl a | qChl a-D09 | D09 | bin213 | bin214 | 158.5 | 160.5 | 47,673,829 | 48,167,059 | 2.53 | 4.43 | -0.04 | 0 |
| Chl b | qChl b-A13 | A13 | bin289 | bin290 | 173.5 | 184.5 | 87,115,053 | 87,473,165 | 2.7 | 5.01 | -0.01 | -0.01 |
| Chl b | qChl b-D01 | D01 | bin239 | bin240 | 136.5 | 146.5 | 62,926,804 | 63,199,901 | 2.79 | 5.22 | -0.01 | 0.02 |
| Chl b | qChl b-D02 | D02 | bin252 | bin253 | 145.5 | 153.5 | 62,524,672 | 62,831,343 | 2.53 | 4.78 | -0.01 | 0.02 |
| Chl b | qChl b-D03 | D03 | bin153 | bin154 | 114.5 | 119.5 | 48,191,318 | 48,417,933 | 3.97 | 8.1 | -0.01 | 0.02 |
| Car | qCar-A12 | A12 | bin314 | bin315 | 89.5 | 93.5 | 76,853,357 | 77,247,271 | 2.84 | 5.57 | 0 | 0.01 |
| Car | qCar-D03 | D03 | bin153 | bin154 | 114.5 | 119.5 | 48,191,318 | 48,417,933 | 2.92 | 6.46 | 0 | 0.01 |
| Chl a/b | qChl a/b-D05 | D05 | bin276 | bin277 | 184.5 | 194.5 | 57,297,699 | 57,671,573 | 2.9 | 6.81 | -0.05 | -0.05 |
| Chl a/b | qChl a/b-D06 | D06 | bin228 | bin229 | 123.5 | 125.5 | 59,098,820 | 59,707,362 | 2.82 | 6.74 | -0.05 | 0.04 |
| Chl a/b | qChl a/b-D12 | D12 | bin172 | bin173 | 125.5 | 138.5 | 52,342,567 | 53,232,613 | 2.56 | 6.27 | -0.05 | 0.05 |
| Total Chl | qTotal Chl-A13 | A13 | bin48 | bin49 | 47.5 | 49.5 | 5,904,987 | 6,263,087 | 2.51 | 4.46 | -0.05 | -0.01 |
| Total Chl | qTotal Chl-D01 | D01 | bin238 | bin239 | 136.5 | 147.5 | 62,826,804 | 63,033,940 | 2.67 | 4.49 | -0.03 | 0.06 |
| Total Chl | qTotal Chl-D03 | D03 | bin153 | bin154 | 114.5 | 118.5 | 48,191,318 | 48,417,933 | 4.99 | 9.49 | -0.02 | 0.1 |
| PH2 | qPH2-A11 | A11 | bin414 | bin439 | 258.5 | 264.5 | 92,237,505 | 97,642,814 | 2.66 | 4.32 | 0.32 | -15.33 |
| PH2 | qPH2-D01 | D01 | bin209 | bin210 | 111.5 | 117.5 | 59,207,021 | 59,470,354 | 2.73 | 4.04 | 7.05 | 10.07 |
| PH2 | qPH2-D03 | D03 | bin153 | bin154 | 114.5 | 119.5 | 48,191,318 | 48,417,933 | 4.06 | 6.47 | -13.14 | -0.61 |
| PH2 | qPH2-D06 | D06 | bin59 | bin60 | 60.5 | 66.5 | 7,355,757 | 7,642,365 | 2.66 | 3.94 | 9.93 | 4.05 |
| PH2 | qPH2-D09 | D09 | bin215 | bin216 | 158.5 | 165.5 | 48,167,059 | 48,412,567 | 2.54 | 3.77 | 4.15 | 11.91 |
| PH2 | qPH2-D12 | D12 | bin221 | bin222 | 189.5 | 192.5 | 60,587,231 | 60,897,033 | 4.86 | 7.87 | 14.21 | -1.36 |
| FBN | qFBN-A04 | A04 | bin39 | bin40 | 58.5 | 71.5 | 7,802,365 | 8,134,366 | 2.64 | 3.82 | 1.13 | -0.96 |
| FBN | qFBN-A05 | A05 | bin44 | bin45 | 34.5 | 35.5 | 7,019,176 | 7,291,976 | 3.58 | 5.29 | 0.47 | -1.89 |
| FBN | qFBN-A06 | A06 | bin22 | bin23 | 26.5 | 35.5 | 3,658,624 | 4,677,844 | 3.97 | 6.64 | 0.23 | -2.18 |
| FBN | qFBN-D04 | D04 | bin51 | bin52 | 45.5 | 48.5 | 7,626,894 | 7,826,894 | 2.8 | 3.88 | 0.72 | -1.34 |
| FBN | qFBN-D11 | D11 | bin89 | bin90 | 73.5 | 75.5 | 14,642,790 | 15,006,002 | 3.44 | 5.2 | -0.37 | -1.83 |
| FBN | qFBN-D12 | D12 | bin106 | bin107 | 77.5 | 78.5 | 41,087,553 | 41,476,816 | 3.4 | 5.05 | -1.33 | 0.37 |
| BN | qBN-D05 | D05 | bin104 | bin105 | 70.5 | 76.5 | 15,695,738 | 16,137,583 | 2.55 | 4.66 | -2.13 | -2.04 |
| BN | qBN-D10 | D10 | bin11 | bin22 | 19.5 | 43.5 | 2,290,240 | 4,417,087 | 2.85 | 5.34 | 2.58 | 1.96 |
| SCY | qSCY-D07 | D07 | bin231 | bin232 | 188.5 | 189 | 57,783,341 | 58,417,178 | 2.53 | 5.54 | 1.92 | 12.2 |
| SCY | qSCY-D10 | D10 | bin22 | bin21 | 21.5 | 45.5 | 4,200,666 | 4,417,087 | 2.67 | 5.85 | 8.64 | 7.69 |
| LY | qLY-D07 | D07 | bin231 | bin232 | 188.5 | 189 | 57,783,341 | 58,417,178 | 2.64 | 6.04 | 0.74 | 3.6 |
| BW | qBW-A10 | A10 | bin57 | bin58 | 48.5 | 52.5 | 10,608,267 | 10,844,347 | 3.55 | 7.6 | -0.32 | -0.22 |
| SI | qSI-D01 | D01 | bin101 | bin103 | 73.5 | 74.5 | 19,943,730 | 21,931,208 | 2.68 | 6.14 | 0.05 | 0.91 |
| SI | qSI-D05 | D05 | bin10 | bin11 | 8.5 | 10.5 | 2,123,451 | 2,323,451 | 3.7 | 8.56 | 0.8 | 0.13 |
| LP | qLP-A07 | A07 | bin107 | bin108 | 83.5 | 87.5 | 19,417,639 | 19,813,491 | 2.66 | 4.84 | -0.56 | -2.13 |
| LP | qLP-D09 | D09 | bin221 | bin222 | 164.5 | 165.5 | 49,209,126 | 49,409,127 | 3.48 | 6.77 | -1.94 | -0.37 |
| LP | qLP-D10 | D10 | bin22 | bin21 | 36.5 | 45.5 | 4,200,666 | 4,417,087 | 4.43 | 8.69 | -2.48 | 0.53 |
| LI | qLI-D09 | D09 | bin225 | bin226 | 168.5 | 169.5 | 49,640,725 | 49,892,967 | 3.41 | 6.69 | -0.44 | -0.05 |
| LI | qLI-D10 | D10 | bin21 | bin54 | 36.5 | 52.5 | 4,200,666 | 10,690,859 | 3.76 | 8.08 | -0.54 | 0.14 |
| LI | qLI-D12 | D12 | bin191 | bin192 | 149.5 | 155.5 | 55,816,594 | 56,150,659 | 2.83 | 5.17 | 0.32 | -0.3 |
| FL | qFL-A06 | A06 | bin26 | bin27 | 35.5 | 39.5 | 5,979,701 | 6,426,275 | 2.92 | 13.58 | -0.94 | 0.85 |
| FL | qFL-A10 | A10 | bin53 | bin50 | 49.5 | 51.5 | 9,611,144 | 9,322,125 | 2.82 | 14.17 | 0.99 | -0.94 |
| FL | qFL-D12 | D12 | bin109 | bin110 | 75.5 | 84.5 | 41,741,946 | 42,083,775 | 2.53 | 11.9 | 1.17 | -0.25 |
| FS | qFS-A02 | A02 | bin279 | bin280 | 152.5 | 156.5 | 104,826,173 | 105,601,237 | 5.37 | 17.11 | 0.21 | 2.46 |
| FS | qFS-A11 | A11 | bin129 | bin130 | 108.5 | 110.5 | 21,948,343 | 22,148,343 | 2.96 | 8.55 | 0.28 | -1.65 |
| FS | qFS-D02 | D02 | bin94 | bin95 | 68.5 | 75.5 | 19,742,795 | 20,003,289 | 3.74 | 11.13 | 0.3 | -1.95 |
| FS | qFS-D12 | D12 | bin154 | bin155 | 124.5 | 127.5 | 49,828,942 | 50,092,227 | 7.51 | 24.89 | 2.34 | -0.74 |
| MIC | qMIC-A13 | A13 | bin343 | bin344 | 206.5 | 208.5 | 96,488,822 | 97,053,406 | 2.93 | 14.4 | 0.31 | 0.05 |
| MIC | qMIC-D01 | D01 | bin58 | bin59 | 51.5 | 54.5 | 10,890,905 | 11,226,434 | 4.44 | 21.98 | -0.37 | 0.2 |
| MIC | qMIC-D02 | D02 | bin3 | bin4 | 5.5 | 7.5 | 620,059 | 1,091,799 | 2.63 | 11.75 | 0.03 | -0.39 |
| MIC | qMIC-D03 | D03 | bin10 | bin11 | 15.5 | 18.5 | 1,178,422 | 1,526,855 | 3.05 | 12.35 | 0.22 | -0.3 |
| FU | qFU-A13 | A13 | bin233 | bin234 | 134.5 | 136.5 | 69,500,824 | 70,270,368 | 6.86 | 20.51 | 0.26 | -1.6 |
| FU | qFU-D03 | D03 | bin13 | bin14 | 22.5 | 29.5 | 1,728,364 | 2,715,388 | 3.17 | 8.54 | 0.68 | 0.32 |
| FU | qFU-D06 | D06 | bin37 | bin38 | 40.5 | 47.5 | 4,733,957 | 4,933,957 | 3.82 | 10.62 | 0.08 | -1.24 |
| FU | qFU-D12-1 | D12 | bin10 | bin11 | 16.5 | 22.5 | 59,195,554 | 59,480,927 | 4.54 | 12.67 | 0.22 | -1.3 |
| FU | qFU-D12-2 | D12 | bin212 | bin213 | 170.5 | 175.5 | 1,425,534 | 1,957,383 | 5.15 | 15.29 | 0.12 | 1.38 |
| FE | qFE-A10 | A10 | bin217 | bin218 | 115.5 | 117.5 | 75,292,930 | 75,555,428 | 2.55 | 7.69 | 0.19 | -0.19 |
| FE | qFE-D05-1 | D05 | bin94 | bin95 | 69.5 | 74.5 | 60,252,767 | 60,452,767 | 4.08 | 11.71 | -0.26 | -0.37 |
| FE | qFE-D05-2 | D05 | bin295 | bin296 | 205.5 | 207.5 | 14,282,823 | 14,559,501 | 2.6 | 7.86 | -0.2 | -0.13 |
| FE | qFE-D09 | D09 | bin131 | bin151 | 86.5 | 99.5 | 33,095,130 | 37,366,030 | 3.76 | 20.3 | 0.39 | -0.44 |
Table 4.
Sixteen QTLs detected in this study overlapped with that identified in previous studies
| This Study | Previous Report | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Chr | Start | End | Reported | Start | End | QTL | LOD | R2 | Parents | Ref |
| FBN | D04 | 7,626,894 | 7,826,894 | FBN | 4,447,115 | 8,199,618 | qFBN-22–2 | 3.84 | 17.3 | TM-1*CSB22sh | [46] |
| BN | D05 | 15,695,738 | 16,137,583 | BN | 15,468,703 | 17,887,617 | qBN-D5-1 | 1.66 | 3.5 | (STV2B*Foster6)*(DPL15*CRI7) | [47] |
| LP | D10 | 4,200,666 | 4,417,087 | LP | 4,294,769 | 5,493,007 | Cs9_PF_20_(5.27 +)、Cs8_PF_20_(6.57 +) | 5.27 | 19.10 | Guazuncho2(Gh)*VH8-4602(Gb) | [48] |
| FL | A06 | 5,979,701 | 6,426,275 | FL | 4,440,391 | 6,580,618 | qFLchr6 | 4.54 | 7.80 | (Emian22*3–79)*Emian22 | [49] |
| FL | A06 | 5,979,701 | 6,426,275 | FL | 4,439,492 | 6,580,618 | qChr06FL | NA | NA | (Emian22*3–79)*Emian22 | [50] |
| FL | A10 | 9,611,144 | 9,322,125 | FL | 7,964,148 | 10,069,511 | qFL-A10-1* | 3.33 | 6.33 | Zhongmiansuo12*8891 | [51] |
| FL | A10 | 9,611,144 | 9,322,125 | FL | 7,964,148 | 10,069,511 | qFL-A10-1_ | 4.32 | 10.46 | Zhongmiansuo12*8891 | [52] |
| FS | A02 | 104,826,173 | 105,601,237 | FS | 104,227,599 | 105,227,815 | qFS-C2-1 | 3.95 | 5.69 | Lumianyan22*Luyuan343 | [53] |
| FS | D02 | 19,742,795 | 20,003,289 | FS | 19,710,781 | 20,711,127 | qFS-D2-1 | 8.12 | 14.4 | (Simian3*Sumian12)*(Zhong4133*8891) | [54] |
| FM | D02 | 620,059 | 1,091,799 | FM | 456,594 | 1,457,031 | qFM-D2-1 | 3.59 | 6.80 | (Simian3*Sumian12)*(Zhong4133*8891) | [54] |
| FM | D02 | 620,059 | 1,091,799 | FM | 991,691 | 3,224,078 | qFM-D2-2 | 12.82 | 21.92 | CRI12*J8891 | [55] |
| FM | D02 | 620,059 | 1,091,799 | FM | 456,594 | 3,223,772 | qFMIC-D2-1_ | 3.32 | 9.1 | Zhongmiansuo12*8891 | [52] |
| FM | D02 | 620,059 | 1,091,799 | FM | 456,594 | 3,224,164 | qFMIC-D2-1* | 8.01 | 12.49 | Zhongmiansuo12*8891 | [51] |
| FU | D03 | 1,728,364 | 2,715,388 | FU | 2,695,254 | 3,695,267 | TC-qFU-c17-2 | 3.11 | 5.7 | CRI36*Hai7124 | [56] |
| FE | A10 | 75,292,930 | 75,555,428 | FE | 72,032,342 | 79,020,626 | qFE-10-1b | 30.93 | 11.8 | HS427-10*TM-1 | [57] |
| FE | D05 | 14,282,823 | 14,559,501 | FE | 13,336,541 | 14,337,210 | qFE-D5-1 | 2.91 | 7.49 | Zhongmiansuo12*8891 | [52] |
Twelve QTLs were associated with plant type traits at seedling stage, most of which (75%) had positive effects and originated from TM-1, suggesting that G. hirsutum has a growth advantage in the seedling stage. Among these QTLs, the PVE varied from 4.46 to 8.35%; the QTL qCNH-A12 with the highest PVE (8.35%) had positive effects and came from Hai7124. Thirty-seven QTLs were detected for leaf morphology and physiological traits at seedling stage, featuring positive effects and coming from both TM-1 and Hai7127 (19 and 18 QTLs, respectively). We found that all nine QTLs associated with intracellular CO2 concentration had positive effects and originated with TM-1, and 7/9 demonstrated positive effects, which is the main component of heterosis. A total of 17 QTLs were identified for leaf chlorophyll, with PVE values ranging from 4.43 to 8.1%; both the additive and dominant effects of these QTLs were close to 0.
Twenty-six QTLs were identified for yield or yield-related traits. Most QTLs associated with qPH2 and qFBN, and all those with qSCY, qLY, and qSI, exhibited positive effects and came from Hai7124. Meanwhile, QTLs having positive effects associated with qBW, qLP, and qLI came from TM-1, suggesting that G. barbadense has a larger biomass but G. hirsutum has higher fiber yield. Of QTLs associated with fiber quality traits, 80% of those having positive effects came from Hai7124; only four QTLs (qFL-A06, qMIC-D01, qFE-D05-1, and qFE-D05-2) with positive effects originated from TM-1. This result indicated that the genetics governing excellent fiber quality come from G. barbadense. All QTLs and the corresponding location information, LOD, PVE, additive effect, and dominant effect values were presented in Table 3 and Fig. 3.
Fig. 3.
Chromosomal distribution of QTLs associated with 35 traits. Black lines indicate QTL positions on the chromosomes
Candidate gene identification and expression analysis
We identified ten genes that has nonsynonymous SNPs in exons or SNP in their upstream regions was located within the 15 loci of interest for 14 traits (D1, CHN, FLTH, PH1, Sci, TCi, Tcond, BW, SCY, LP, Li, SPn, TPn, TTr). We analyzed their expression in sixteen vegetative and reproductive tissues of TM-1 and compared values with those in Hai7124 (Supplementary Fig. 2, Supplementary Table 4, 5). Some SNP variants corresponding to QTLs associated with different traits were mapped to the same position or related to the same gene, such as GH_A04G0054/GB_A04G0055, GH_D04G1426/GB_D04G1512, and GH_D10G0500/GB_D06G1730 (Supplementary Table 4, 5).
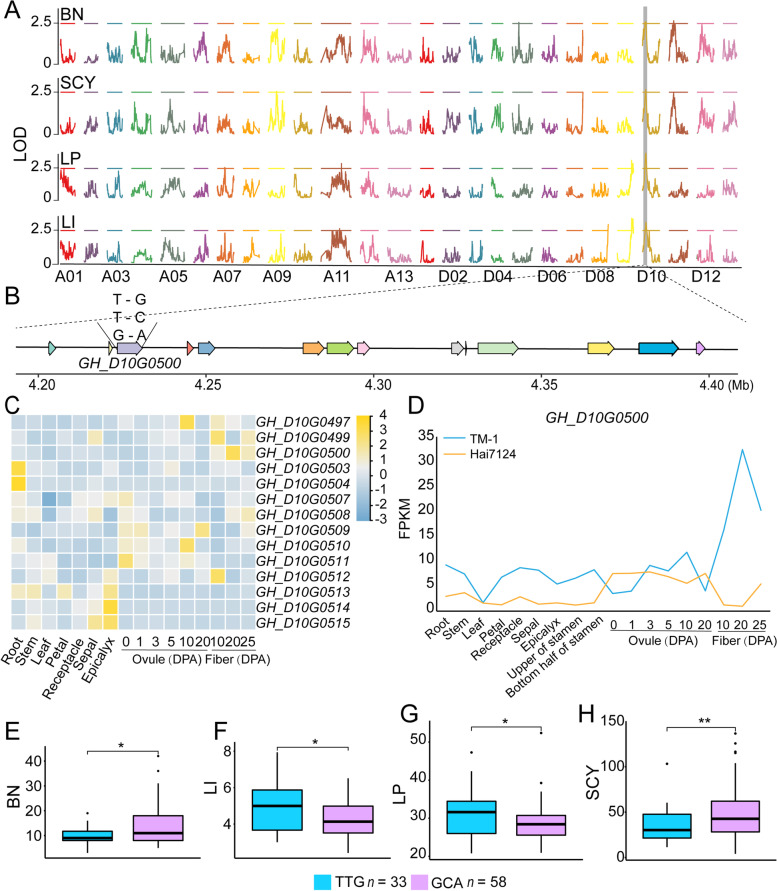
A representative QTL that related to multiple traits BN, SCY, LP, and LI was located on chromosome D10 (Fig. 4A). This locus encompassed fourteen genes harboring nonsynonymous SNPs. Considering the expression of these genes during fiber development, one was identified as a putative causal gene: root hair defective 3 GTP-binding protein (GhRHD3, GH_D10G0500), which was dominantly expressed during secondary cell-wall bio-synthesis (20 DPA) (Fig. 4B-C). Interestingly, its Hai7124 homolog showed high expression during fiber initiation (0, 1, and 3 DPA) (Fig. 4D). Three nonsynonymous SNPs in GH_D10G0500, D10Gh: 4,228,677/4228733/4229273 (TTG versus GCA 33:58), demonstrated significant associations with BN, SCY, LP, and LI (Fig. 4E-H). The orthologous gene in Arabidopsis thaliana was identified as involved in the regulation of cell expansion [58–60]
Fig. 4.
Functional haplotypes in associated loci from the TM-1 × Hai7124 F2 population on D10. A Genetic mapping of a QTL on the D10 chromosome identified as related to BN, SCY, LP, and LI. B Genes with nonsynonymous SNPs in the QTL region. C Transcriptomic expression of QTL-region genes with nonsynonymous SNPs in TM-1 tissues, based on FPKM values. D Transcriptomic expression of GH_D10G0500 in TM-1 and Hai7124 tissues, based on FPKM values. E–H Boxplot of GH_D10G0500 haplotypes. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5 × the interquartile range; dots, outliers (* P < 0.01, ** P < 0.001, two-tailed t-test)
Supplementary Fig. 3 illustrates a QTL, located on chromosome D04, which related to SCi, TCi, and Tcond. There were thirteen genes harboring nonsynonymous SNPs in this region. Combining their expression level during fiber development, one was identified as a putative causal gene: glycerol-3-phosphate acyltransferase 6 (GhGPAT6, GH_D04G1426), which was highly expressed in leaves. Within this gene, the nonsynonymous SNP D04Gh:47,064,565 (CC versus AA 65:50) was significant associated with SCi, TCi, and Tcond. As reported, its orthologous gene in tomato was involved in regulating the outer wall diameter of leaf epidermal cells [61].
Discussion
Bin markers are effective for constructing high-density genetic maps and QTL fine mapping in G. hirsutum and G. barbadense
In recent years, scientists have used specific-locus amplified fragment sequencing (SLAF-seq), genotyping by sequencing (GBS), and other sequencing methods to genotype the complex genome of cotton, and the resulting genetic map is based on SNP phasing. This method can identify markers with high throughput; in addition, the chromosome coverage is more uniform and the marker density greatly improved compared with traditional PCR-based markers. With the help of newly developed bioinformatics software, it is possible to complete genotyping and construct genetic maps in a very short time. Compared to the GBS-based enzyme digestion method, a binmap based on resequencing offers the following improvements: scanning of and mutation identification at all sites in the whole genome, without any prior marker information, yielding complete allelic variant information with higher accuracy than previous experimental methods. In this study, we obtained a total of 6303 high-confidence bin markers that not only extends the length of the cotton genetic map but also improve its resolution. In our previous studies, we constructed an SSR-based genetic map using a (TM-1 × Hai7124)F2 population [27] that spanned 3414 loci in 26 linkage groups, covering 3667.62 cM with an average inter-locus distance of 1.08 cM. The present 6303 bin markers expand that map to cover 5057.13 cM while also narrowing the average distance between adjacent markers on to an interval of 0.8 cM. The bin marker length varies from 0.64 to 1.10 Mb, which indicates that the final location of a QTL can be reduced to dozens of candidate genes. Thanks to the binmap algorithm, SNPs within a haplotype can be corrected, decreasing the false positive possibility of a single SNP. The binmap also allows obtaining the fragments of the tested samples on the whole chromosome to exchange recombination information.
Most traits show heterosis in F1 and F2
In most F2 populations, the traits exhibited by individual plants fall between those of their parents, while a few exceed their parents; thus, most traits demonstrate different degrees of over-parental segregation. Such phenotypic trait data exhibits an approximately normal distribution. Each pair of the 35 traits was evaluated for significant negative or positive correlations (Supplementary Fig. 2), and we also evaluated the heterosis of each trait in the F1 and F2 populations. In the F1 population, BN exhibited the highest mid-parent heterosis of all yield traits, at 59.7%; other yield traits ranked LY > SCY > LI > SI > LP, while the mid-parent heterosis of BW was negative. Accordingly, the mid-parent heterosis of BN contributes most to the heterosis of yield. With respect to plant type, morphology, and physiological traits, the mid-parent heterosis of SLA was the highest at 40.49%, followed by TLA at 36.16%, and then other traits in the range of 2.1%-23.81 except for D1, which had a negative value. Regarding fiber quality traits, these ranked in terms of mid-parent heterosis as FS > FL > FE, with the values for MIC and FU being negative (Supplementary Table 3). In the F2 population, both yield and quality traits exhibited negative mid-parent heterosis values. Among plant type, morphology, and physiological traits, most mid-parent heterosis values were positive, ranging from 1.18% to 23.81% except for the values associated with PH1, STLH, D2, TLA, Car, and Chl a/b. (Supplementary Table 3; Table 2).
Non-uniform distribution of QTLs in the A and D subgenomes
In this study, a total of 112 QTLs were detected, of which 71 were in the D subgenome, much more than the 41 in the A subgenome (Table S6). Of QTLs associated with the six plant type traits at seedling stage, more were sited in the A subgenome than in the D subgenome; in contrast, QTLs associated with the other ten leaf morphology and physiological traits at seedling stage, five traits reflecting leaf chlorophyll content, two plant type traits at flower and boll stage, seven yield traits, and five fiber quality traits were all less commonly located in the A subgenome than in the D subgenome. In particular, the D subgenome showed a strong advantage with regard to leaf chlorophyll content, yield traits, and fiber quality traits. This is consistent with previous reports that the D subgenome contributes more to the genetic control of fiber [62–64], and suggests that molecular marker selection in the D subgenome may be more efficient for breeding to improve yield and fiber quality.
QTLs and candidate genes may contribute to the improvement of cotton through breeding
Studies involving cotton QTL mapping and candidate gene identification generally focus on traits related to yield and fiber quality; considerably less research has been conducted concerning seedling traits, leaf physiology, and chlorophyll content. Nonetheless, these traits are also important for cotton growth: plant height and leaf area at the seedling stage determine growth vigor, which in turn affects adversity resistance; meanwhile, leaf physiological and chlorophyll content can enhance photosynthesis efficiency and solar energy utilization, eventually helping adaptation to dense planting and increasing production. Here, the candidate gene GH_D04G1426 demonstrated significant associations with SCi, TCi, and Tcond. Its orthologous gene in tomato has been reported to affect the outer wall diameter of leaf epidermal cells; such functionality may indirectly affect photosynthesis in cotton [61]. In looking beyond direct effects on yield and fiber quality, other QTL and candidate genes in our data may provide additional solutions for cotton molecular breeding.
Conclusions
In conclusion, we constructed a high-density genetic map based on the resequencing data of 249 individuals from an interspecific F2 population (TM-1 and Hai7124). This genetic map consists of 6303 high-confidence bin markers spanning 5057.13 cM across 26 chromosomes. Based on this map, 112 QTLs relating to agronomic and physiological traits from seedling to boll opening stage were identified. Through the analysis of sequence and expression of the candidate genes within the QTLs mapping regions, ten causal putative genes might responsible for the target traits. Of them, GhRHD3 (GH_D10G0500) was associated with fiber yield and GhGPAT6 (GH_D04G1426) might play important role in photosynthesis efficiency.
Supplementary Information
Additional file 1: Supplementary Figure 1. Comparisons of the TM-1 genome with (TM-1 × Hai7124) F2 genetic map. Supplementary Figure 2. Frequency distribution of phenotypic variation of 30 traits and correlation coefficients among the traits in the F2 population. Supplementary Figure 3. Functional haplotypes in associated loci from the TM-1×Hai7124 F2 population on D10. Supplementary Figure 4. Functional haplotypes in associated loci from the TM-1×Hai7124 F2 population on D04. Supplementary Table 1. The distribution characteristics of partial segregation markers. Supplementary Table 2. Shortened form and Unit of measurement of the 35 traits. Supplementary Table 3. Statistical analysis of 35 traits phenotypic differences in TM-1 Hai7124 and F1. Supplementary Table 4. Haplotype and gene ID of the 10 candidate genes in TM-1 and Hai7124. Supplementary Table 5. Expression of the 10 candidate genes in different tissues of TM-1 and Hai7124. Supplementary Table 6. The distribution of QTLs in the At and Dt subgenomes.
Additional file 2: Table S7. All markers in geneticmap.
Acknowledgements
We would like to thank Mr. Dai Fan of college of agriculture and biotechnology at Zhejiang University give the advice on data analysis and discussion; thank Dr. Wu Huaitong, Dr. Tian Shuhua and Dr. Chang Lijing of college of agriculture at Nanjing agricultural university for the material planting, transplanting and trait data collection.
Abbreviations
- RAD-seq
Restriction-site Associated DNA sequence
- SNPs
Single Nucleotide Polymorphism
- QTL
Quantitative Trait Locus
- CTAB
Cetyltriethylammnonium Bromide
- SSR-PCR
Simple Sequence Repeats PCR
- PH1
Plant Height
- CNH
Cotyledonary Node Height
- FTLH
First True Leaf Height
- STLH
Second True Leaf Height
- D1
Distance between the cotyledonary node and first true leaf
- D2
Distance between first true leaf and second true leaf
- Chl a
Chlorophyll a
- Chl b
Chlorophyll b
- Car
Carotenoid
- Chl a/b
Chlorophyll a/b ratio
- Total Chl
Total Chlorophyll
- PH2
Plant Height
- FBN
Fruit Branch Number
- BN
Boll Number per plant
- SCY
Seed Cotton Yield
- LY
Lint Yield
- BW
Boll Weight
- LP
Lint Percentage
- LI
Lint Index
- SI
Seed Index
- FL
Fiber Length
- FS
Fiber Strength
- MIC
Micronaire Value
- FU
Fiber length Uniformity
- FE
Fiber Elongation
- GATK4
Genome Analysis Toolkit 4
- MAF
Minor Allele Frequency
- H
Heterosis
- A
The additive effect
- D
Dominant effect
- R2
Contribution rate
Authors’ contributions
Z.F.S., Y.H. and T.Z.Z. conceived and designed the experiments. Z.F.S. and S.K.J. performed the experiments and wrote the manuscript. S.K.J. and J.D.C. analyzed the data. S.W., Y.H., L.F. and X.F.Z. participated in the experiments and analysis. Z.F.S., S.K.J. and T.Z.Z. edited the manuscript. All authors read and approved the last version.
Funding
This work was supported by grants from the Science Technology and Achievement Transformation Project of the Xinjiang Production and Construction Corps (2021AB008, 2020CB003), the Fundamental Research Funds for the Central Universities (2021QN81011), Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002).
Availability of data and materials
The raw sequencing data used in this study are available from the China National GenBank (CNGB) Nucleotide Sequence Archive (CNSA) under accession number sub026937.
Declarations
Ethics approval and consent to participate
Not applicable. No specific permits were required for the collection of specimens for this study. This research was carried out in compliance with the relevant laws of China.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhanfeng Si and Shangkun Jin share the first authorship.
References
- 1.ur-Rahman M, Shaheen T, Tabbasam N, Iqbal MA, Ashraf M, Zafar Y, Paterson AH. Cotton genetic resources A review. Agron Sustain Dev. 2012;32(2):419–432. doi: 10.1007/s13593-011-0051-z. [DOI] [Google Scholar]
- 2.Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 3.Fang L, Zhao T, Hu Y, Si ZF, Zhu XF, Han ZG, Liu GZ, Wang S, Ju LZ, Guo ML, et al. Divergent improvement of two cultivated allotetraploid cotton species. Plant Biotechnol J. 2021;19(7):1325–1336. doi: 10.1111/pbi.13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nie XH, Wen TW, Shao PX, Tang BH, Nuriman-guli A, Yu Y, Du XM, You CY, Lin ZX. High-density genetic variation maps reveal the correlation between asymmetric interspecific introgressions and improvement of agronomic traits in Upland and Pima cotton varieties developed in Xinjiang China. Plant J. 2020;103(2):677–689. doi: 10.1111/tpj.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shappley ZW, Jenkins JN, Watson CE, Kahler AL, Meredith WR. Establishment of molecular markers and linkage groups in two F-2 populations of Upland cotton. Theor Appl Genet. 1996;92(8):915–919. doi: 10.1007/BF00224030. [DOI] [PubMed] [Google Scholar]
- 6.Chandnani R, Kim C, Guo H, Shehzad T, Wallace JG, He D, Zhang Z, Patel JD, Adhikari J, Khanal S, et al. Genetic Analysis of Gossypium Fiber Quality Traits in Reciprocal Advanced Backcross Populations. The plantgenome. 2018;11(1):170057. [DOI] [PubMed]
- 7.Draye X, Chee P, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH. Molecular dissection of interspecific variation between Gossypium hirsutum and G-barbadense (cotton) by a backcross-self approach: II Fiber fineness. Theor Appl Genet. 2005;111(4):764–771. doi: 10.1007/s00122-005-2061-1. [DOI] [PubMed] [Google Scholar]
- 8.Gu QS, Ke HF, Liu ZW, Lv X, Sun ZW, Zhang M, Chen LT, Yang J, Zhang Y, Wu LQ, et al. A high-density genetic map and multiple environmental tests reveal novel quantitative trait loci and candidate genes for fibre quality and yield in cotton. Theor Appl Genet. 2020;133(12):3395–3408. doi: 10.1007/s00122-020-03676-z. [DOI] [PubMed] [Google Scholar]
- 9.He DH, Lin ZX, Zhang XL, Nie YC, Guo XP, Zhang YX, Li W. QTL mapping for economic traits based on a dense genetic map of cotton with PCR-based markers using the interspecific cross of Gossypium hirsutum x Gossypium barbadense. Euphytica. 2007;153(1–2):181–197. doi: 10.1007/s10681-006-9254-9. [DOI] [Google Scholar]
- 10.Lacape JM, Gawrysiak G, Cao TV, Viot C, Llewellyn D, Liu SM, Jacobs J, Becker D, Barroso PAV, de Assuncao JH, et al. Mapping QTLs for traits related to phenology, morphology and yield components in an inter-specific Gossypium hirsutum x G. barbadense cotton RIL population. Field Crop Res. 2013;144:256–267. doi: 10.1016/j.fcr.2013.01.001. [DOI] [Google Scholar]
- 11.Li SQ, Liu AY, Kong LL, Gong JW, Li JW, Gong WK, Lu QW, Li PT, Ge Q, Shang HH, et al. QTL mapping and genetic effect of chromosome segment substitution lines with excellent fiber quality from Gossypium hirsutum x Gossypium barbadense. Mol Genet Genomics. 2019;294(5):1123–1136. doi: 10.1007/s00438-019-01566-8. [DOI] [PubMed] [Google Scholar]
- 12.Ma JJ, Pei WF, Ma QF, Geng YH, Liu GY, Liu J, Cui YP, Zhang X, Wu M, Li XL, et al. QTL analysis and candidate gene identification for plant height in cotton based on an interspecific backcross inbred line population of Gossypium hirsutum x Gossypium barbadense. Theor Appl Genet. 2019;132(9):2663–2676. doi: 10.1007/s00122-019-03380-7. [DOI] [PubMed] [Google Scholar]
- 13.Reinisch AJ, Dong J, Brubaker CL, Stelly DM, Wendel JF, Paterson AH. A Detailed Rflp Map Of Cotton, Gossypium-Hirsutum X Gossypium Barbadense - Chromosome Organization And Evolution In a Disomic Polyploid Genome. Genetics. 1994;138(3):829–847. doi: 10.1093/genetics/138.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Said JI, Song MZ, Wang HT, Lin ZX, Zhang XL, Fang DD, Zhang JF. A comparative meta-analysis of QTL between intraspecific Gossypium hirsutum and interspecific G hirsutum x G barbadense populations. Mol Genet Genomics. 2015;290(3):1003–1025. doi: 10.1007/s00438-014-0963-9. [DOI] [PubMed] [Google Scholar]
- 15.Shi YZ, Zhang BC, Liu AY, Li WT, Li JW, Lu QW, Zhang Z, Li SQ, Gong WK, Shang HH, et al. Quantitative trait loci analysis of Verticillium wilt resistance in interspecific backcross populations of Gossypium hirsutum x Gossypium barbadense. BMC Genomics. 2016;17(1):877. doi: 10.1186/s12864-016-3128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Si ZF, Chen H, Zhu XF, Cao ZB, Zhang TZ. Genetic dissection of lint yield and fiber quality traits of G. hirsutum in G. barbadense background. Mol Breeding. 2017;37(1):9.
- 17.Song XL, Wang K, Guo WZ, Zhang J, Zhang TZ. A comparison of genetic maps constructed from haploid and BC1 mapping populations from the same crossing between Gossypium hirsutum L. and Gossypium barbadense L. Genome. 2005;48(3):378–390. doi: 10.1139/g04-126. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Zhang J, Chen Y, Zhang C, Gong J, Song Z, Zhou J, Wang J, Zhao C, Jiao M, et al. Identification of candidate genes for key fibre-related QTLs and derivation of favourable alleles in Gossypium hirsutum recombinant inbred lines with G. barbadense introgressions. Plant Biotechnol J. 2020;18(3):707–720. doi: 10.1111/pbi.13237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang P, Zhu Y, Song X, Cao Z, Ding Y, Liu B, Zhu X, Wang S, Guo W, Zhang T. Inheritance of long staple fiber quality traits of Gossypium barbadense in G. hirsutum background using CSILs. Theor Appl Genet. 2012;124(8):1415–1428. doi: 10.1007/s00122-012-1797-7. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Chen JD, Zhang WP, Hu Y, Chang LJ, Fang L, Wang Q, Lv FN, Wu HT, Si ZF, et al. Sequence-based ultra-dense genetic and physical maps reveal structural variations of allopolyploid cotton genomes. Genome Biol. 2015;16(1):108. doi: 10.1186/s13059-015-0678-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu JW, Zhang K, Li SY, Yu SX, Zhai HH, Wu M, Li XL, Fan SL, Song MZ, Yang DG, et al. Mapping quantitative trait loci for lint yield and fiber quality across environments in a Gossypium hirsutum x Gossypium barbadense backcross inbred line population. Theor Appl Genet. 2013;126(1):275–287. doi: 10.1007/s00122-012-1980-x. [DOI] [PubMed] [Google Scholar]
- 22.Yu JZ, Ulloa M, Hoffman SM, Kohel RJ, Pepper AE, Fang DD, Percy RG, Burke JJ. Mapping genomic loci for cotton plant architecture, yield components, and fiber properties in an interspecific (Gossypium hirsutum L. x G-barbadense L.) RIL population. Mol Genet Genomics. 2014;289(6):1347–1367. doi: 10.1007/s00438-014-0930-5. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Yuan D, Liang S, Li X, Wang X, Lin Z, Zhang X. Genome structure of cotton revealed by a genome-wide SSR genetic map constructed from a BC1 population between gossypium hirsutum and G. barbadense. BMC Genomics. 2011;12:15. doi: 10.1186/1471-2164-12-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, Guo W, Zhang T. Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. x Gossypium barbadense L.) with a haploid population. Theor Appl Genet. 2002;105(8):1166–1174. doi: 10.1007/s00122-002-1100-4. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Lin Z, Xia Q, Zhang M, Zhang X. Characteristics and analysis of simple sequence repeats in the cotton genome based on a linkage map constructed from a BC1 population between Gossypium hirsutum and G. barbadense. Genome. 2008;51(7):534–546. doi: 10.1139/G08-033. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Li JW, Jamshed M, Shi YZ, Liu AY, Gong JW, Wang SF, Zhang JH, Sun FD, Jia F, et al. Genome-wide quantitative trait loci reveal the genetic basis of cotton fibre quality and yield-related traits in a Gossypium hirsutum recombinant inbred line population. Plant Biotechnol J. 2020;18(1):239–253. doi: 10.1111/pbi.13191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao L, Yuanda L, Caiping C, Xiangchao T, Xiangdong C, Wei Z, Hao D, Xiuhua G, Wangzhen G. Toward allotetraploid cotton genome assembly: integration of a high-density molecular genetic linkage map with DNA sequence information. BMC Genomics. 2012;13:539. doi: 10.1186/1471-2164-13-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulloa M, Saha S, Jenkins JN, Meredith WR, Jr, McCarty JC, Jr, Stelly DM. Chromosomal assignment of RFLP linkage groups harboring important QTLs on an intraspecific cotton (Gossypium hirsutum L.) Joinmap. The Journal of heredity. 2005;96(2):132–144. doi: 10.1093/jhered/esi020. [DOI] [PubMed] [Google Scholar]
- 29.Qin HD, Guo WZ, Zhang YM, Zhang TZ. QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet. 2008;117(6):883–894. doi: 10.1007/s00122-008-0828-x. [DOI] [PubMed] [Google Scholar]
- 30.Shen XL, Guo WZ, Lu QX, Zhu XF, Yuan YL, Zhang TZ. Genetic mapping of quantitative trait loci for fiber quality and yield trait by RIL approach in Upland cotton. Euphytica. 2007;155(3):371–380. doi: 10.1007/s10681-006-9338-6. [DOI] [Google Scholar]
- 31.Jiang C, Wright RJ, Woo SS, DelMonte TA, Paterson AH. QTL analysis of leaf morphology in tetraploid Gossypium (cotton) Theor Appl Genet. 2000;100(3–4):409–418. doi: 10.1007/s001220050054. [DOI] [Google Scholar]
- 32.Rong J, Feltus EA, Waghmare VN, Pierce GJ, Chee PW, Draye X, Saranga Y, Wright RJ, Wilkins TA, May OL, et al. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics. 2007;176(4):2577–2588. doi: 10.1534/genetics.107.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang XH, Feng Q, Qian Q, Zhao Q, Wang L, Wang AH, Guan JP, Fan DL, Weng QJ, Huang T, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou ZQ, Zhang CS, Zhou Y, Hao ZF, Wang ZH, Zeng X, Di H, Li MS, Zhang DG, Yong HJ, et al. Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genomics. 2016;17:178. doi: 10.1186/s12864-016-2555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu X, Xiong Q, Cheng T, Li QT, Liu XL, Bi YD, Li W, Zhang WK, Ma B, Lai YC, et al. A PP2C-1 Allele Underlying a Quantitative Trait Locus Enhances Soybean 100-Seed Weight. Mol Plant. 2017;10(5):670–684. doi: 10.1016/j.molp.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 36.Hu ZY, Deng GC, Mou HP, Xu YH, Chen L, Yang JH, Zhang MF. A re-sequencing-based ultra-dense genetic map reveals a gummy stem blight resistance-associated gene in Cucumis melo. DNA Res. 2018;25(1):1–10. doi: 10.1093/dnares/dsx033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo XB, Xu L, Wang Y, Dong JH, Chen YL, Tang MJ, Fan LX, Zhu YL, Liu LW. An ultra-high-density genetic map provides insights into genome synteny, recombination landscape and taproot skin colour in radish (Raphanus sativus L.) Plant Biotechnol J. 2020;18(1):274–286. doi: 10.1111/pbi.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu Y, Chen JD, Fang L, Zhang ZY, Ma W, Niu YC, Ju LZ, Deng JQ, Zhao T, Lian JM, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–+. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 39.Li FG, Fan GY, Lu CR, Xiao GH, Zou CS, Kohel RJ, Ma ZY, Shang HH, Ma XF, Wu JY, et al. Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat Biotechnol. 2015;33(5):524–U242. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, Zhao B, Zheng HJ, Hu Y, Lu G, Yang CQ, Chen JD, Chen JJ, Chen DY, Zhang L, et al. Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci Rep. 2015;5:14139. doi: 10.1038/srep14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang MJ, Tu LL, Yuan DJ, Zhu D, Shen C, Li JY, Liu FY, Pei LL, Wang PC, Zhao GN, et al. Reference genome sequences of two cultivated allotetraploid cottons, Gossypium hirsutum and Gossypium barbadense. Nat Genet. 2019;51(2):224–+. doi: 10.1038/s41588-018-0282-x. [DOI] [PubMed] [Google Scholar]
- 42.Yuan DJ, Tang ZH, Wang MJ, Gao WH, Tu LL, Jin X, Chen LL, He YH, Zhang L, Zhu LF, et al. The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci Rep. 2015;5:1766. doi: 10.1038/srep17662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohel RJRT, Lewis CF. Texas Marker-1, a description of a genetic standard for Gossypium hirsutum L. Crop Sci. 1970;10(1):670–671. doi: 10.2135/cropsci1970.0011183X001000060019x. [DOI] [Google Scholar]
- 44.Paterson AH, Brubaker CL, Wendel JF. A rapid method for extraction of cotton ( Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Molecular Biol Rep. 1993;11(2):122–127. doi: 10.1007/BF02670470. [DOI] [Google Scholar]
- 45.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3(3):269–283. doi: 10.1016/j.cj.2015.01.001. [DOI] [Google Scholar]
- 46.Yao J, et al. Tagging QTLs of Yield-related Traits in Chromosome 22sh of Allotetraploid Cotton Using Substitution Line. Cotton Sci. 2010;22:521–526. [Google Scholar]
- 47.Zhang T, et al. Variations and Transmission of QTL Alleles for Yield and Fiber Qualities in Upland Cotton Cultivars Developed in China. PloS one. 2013;8:e57220. doi: 10.1371/journal.pone.0057220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lacape J, et al. Mapping QTLs for traits related to phenology, morphology and yield components in an inter-specific Gossypium hirsutum× G. barbadense cotton RIL population. Field Crops Res. 2013;144:256–267. doi: 10.1016/j.fcr.2013.01.001. [DOI] [Google Scholar]
- 49.Li X, et al. Increasing cotton genome coverage with polymorphic SSRs as revealed by SSCP. Genome. 2012;55:459–470. doi: 10.1139/g2012-032. [DOI] [PubMed] [Google Scholar]
- 50.Li X, Yuan D, Zhang J, Lin Z, Zhang X. Genetic mapping and characteristics of genes specifically or preferentially expressed during fiber development in cotton. PLoS One. 2013;8:e54444. doi: 10.1371/journal.pone.0054444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang B, et al. QTL mapping of fiber quality in an elite hybrid derived-RILpopulation of upland cotton. Euphytica. 2006;152:367–378. doi: 10.1007/s10681-006-9224-2. [DOI] [Google Scholar]
- 52.Wang B, et al. QTL analysis and epistasis effects dissection of fiber qualities in an elite cotton hybrid grown in second generation. Crop Sci. 2007;47:1384–92. doi: 10.2135/cropsci2006.10.0647. [DOI] [Google Scholar]
- 53.Wang F, et al. Genetic dissection of the introgressive genomic components from Gossypium barbadense L. that contribute to improved fiber quality in Gossypium hirsutum L. Molecular Breeding. 2013;32:547–62.
- 54.Qin H, Guo W, Zhang Y, Zhang T. QTL mapping of yield and fiber traits based on a four-way cross population in Gossypium hirsutum L. Theor Appl Genet. 2008;117:883–94. doi: 10.1007/s00122-008-0828-x. [DOI] [PubMed] [Google Scholar]
- 55.YongSheng Q, WenXue Y, RenZhong L, TianZhen Z, WangZhen G. QTL mapping for fiber quality properties in upland cotton (Gossypium hirsutum L.) Scientia Agricultura Sinica. 2009;42:4145–54. [Google Scholar]
- 56.Yu J, et al. Identification of quantitative trait loci across interspecific F2, F2: 3 and testcross populations for agronomic and fiber traits in tetraploid cotton. Euphytica. 2013;191:375–89. doi: 10.1007/s10681-013-0875-5. [DOI] [Google Scholar]
- 57.Shen X, et al. Molecularmapping of QTLs for _ber qualities in three diverse lines in Upland cotton using SSR markers. Mol Breeding. 2005;15:169–81. doi: 10.1007/s11032-004-4731-0. [DOI] [Google Scholar]
- 58.Hu Y, Zhong RQ, Morrison WH, Ye ZH. The Arabidopsis RHD3 gene is required for cell wall biosynthesis and actin organization. Planta. 2003;217(6):912–921. doi: 10.1007/s00425-003-1067-7. [DOI] [PubMed] [Google Scholar]
- 59.Sun JQ, Movahed N, Zheng HQ. LUNAPARK Is an E3 Ligase That Mediates Degradation of ROOT HAIR DEFECTIVE3 to Maintain a Tubular ER Network in Arabidopsis. Plant Cell. 2020;32(9):2964–2978. doi: 10.1105/tpc.18.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang HY, Lee MM, Schiefelbein JW. Regulation of the cell expansion gene RHD3 during Arabidopsis development. Plant Physiol. 2002;129(2):638–649. doi: 10.1104/pp.002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fawke S, Torode TA, Gogleva A, Fich EA, Sorensen I, Yunusov T, Rose JKC, Schornack S. Glycerol-3-phosphate acyltransferase 6 controls filamentous pathogen interactions and cell wall properties of the tomato and Nicotiana benthamiana leaf epidermis. New Phytol. 2019;223(3):1547–1559. doi: 10.1111/nph.15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guo LX, Shi YZ, Gong JW, Liu AY, Tan YN, Gong WK, Li JW, Chen TT, Shang HH, Ge Q, et al. Genetic analysis of the fiber quality and yield traits in G-hirsutum background using chromosome segments substitution lines (CSSLs) from Gossypium barbadense. Euphytica. 2018;214(5):82.
- 63.Ma Z, He S, Wang X, Sun J, Zhang Y, Zhang G, Wu L, Li Z, Liu Z, Sun G, et al. Resequencing a core collection of upland cotton identifies genomic variation and loci influencing fiber quality and yield. Nat Genet. 2018;50(6):803–813. doi: 10.1038/s41588-018-0119-7. [DOI] [PubMed] [Google Scholar]
- 64.Said JI, Lin ZX, Zhang XL, Song MZ, Zhang JF. A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genomics. 2013;14:776. doi: 10.1186/1471-2164-14-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplementary Figure 1. Comparisons of the TM-1 genome with (TM-1 × Hai7124) F2 genetic map. Supplementary Figure 2. Frequency distribution of phenotypic variation of 30 traits and correlation coefficients among the traits in the F2 population. Supplementary Figure 3. Functional haplotypes in associated loci from the TM-1×Hai7124 F2 population on D10. Supplementary Figure 4. Functional haplotypes in associated loci from the TM-1×Hai7124 F2 population on D04. Supplementary Table 1. The distribution characteristics of partial segregation markers. Supplementary Table 2. Shortened form and Unit of measurement of the 35 traits. Supplementary Table 3. Statistical analysis of 35 traits phenotypic differences in TM-1 Hai7124 and F1. Supplementary Table 4. Haplotype and gene ID of the 10 candidate genes in TM-1 and Hai7124. Supplementary Table 5. Expression of the 10 candidate genes in different tissues of TM-1 and Hai7124. Supplementary Table 6. The distribution of QTLs in the At and Dt subgenomes.
Additional file 2: Table S7. All markers in geneticmap.
Data Availability Statement
The raw sequencing data used in this study are available from the China National GenBank (CNGB) Nucleotide Sequence Archive (CNSA) under accession number sub026937.