Abstract
Purpose
The spread of New Delhi metallo-β-lactamase (NDM) encoded by the blaNDM gene has been a global health crisis for many years. Most of blaNDM-harboring bacteria commonly carry various antimicrobial resistance (AMR) genes on their chromosomes or plasmids, leading to limited treatment options. Thus, we aimed to evaluate the synergistic effects of fosfomycin in combination with other antimicrobial agents against blaNDM-harboring carbapenem-resistant Escherichia coli (CREC) and to characterize the whole-genome and plasmid sequences of these pathogens.
Methods
Thirty-eight CREC isolates were collected from patients in the Medicine Ward, Songklanagarind Hospital, Thailand. The activity of fosfomycin in combination with other antimicrobial agents against CREC isolates harboring blaNDM on the plasmid was evaluated using the checkerboard method. In this method, the serial dilutions of two antibiotics were mixed with the cultured CREC, the mixtures were incubated, and FICI was calculated to interpret the synergistic activity of the combination. The whole-genome and particular plasmids of these pathogens were sequenced using next-generation sequencing. Sequence analysis, especially on antimicrobial resistance (AMR) genes, mobile-genetic elements (MGEs), and virulence genes was performed using many bioinformatics tools.
Results
Of the E. coli 38 isolates, only 3 isolates contained the blaNDM-1 gene, which is located on the IncN2 plasmid. The combinations of fosfomycin with aminoglycosides, colistin, tigecycline, sitafloxacin, and ciprofloxacin were synergies against blaNDM-1-harboring CREC isolates. Genomic analysis revealed that these isolates harbored many β-lactam resistance genes and other AMR genes that may confer resistance to aminoglycoside, fluoroquinolone, rifampicin, trimethoprim, sulfonamide, tetracycline, and macrolide. Also, various MGEs, especially the blaNDM-1-bearing IncN2 plasmid, were present in these isolates.
Conclusion
Our study demonstrated some synergistic effects of antimicrobial combination against CREC isolates harboring blaNDM-1 on the IncN2 plasmid. Also, our data on the whole-genome and plasmid sequences might be beneficial in the control of the spread of blaNDM-1-harboring CREC isolates. The linkages between blaNDM-1-carrying plasmid, patient information, and time of collection will be elucidated to track the horizontal gene transfer in the future.
Keywords: antimicrobial resistance gene, checkerboard method, next-generation sequencing, bioinformatics tool, mobile genetic element
Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) has been a serious public health concern for many decades. Carbapenems were an important, life-saving mainstay treatment for many years, but due to widespread over- and inappropriate use, in recent years widespread resistance to this antibiotic has developed.1 Importantly, as CRE can hydrolyze almost all β-lactam antibiotics, the treatments are limited, resulting in a high mortality rate.
CREC resists carbapenem through various mechanisms, especially the production of carbapenemase enzymes.2 The carbapenemase-encoding genes are usually located on mobile genetic elements (MGEs) that can transfer these particular genes to other pathogenic bacteria. Among them, the blaNDM gene is one of the most prevalent and strong carbapenemase genes with high hydrolysis activity.3 The outbreak of blaNDM gene has been reported in many parts of the world such as Australia (1.6%), America (11%), Africa (11%), Europe (17%), and Asia (58%) and the evidence therefore confirm the global spread of the blaNDM gene.4 Moreover, the infection caused by this pathogen seriously threaten the patient with a high mortality rate.5 The blaNDM gene can be detected on many types of plasmids and transposons (Tn), especially Tn125.6 A previous study reported that Acinetobacter spp. acted as the intermediate host to disseminate the blaNDM-1 into Enterobacteriaceae.7 The blaNDM-1 gene is mostly found within the bracket of two copies of ISAba125 in the form of Tn125, which provides a −35 region promoter for the expression of blaNDM-1. However, the genetic content of Tn125 can be truncated due to the insertion of various insertion sequence (IS) elements.8 Outbreaks of blaNDM have been reported on almost all continents. Twenty replicon types of blaNDM-bearing plasmids, IncHI2, IncHI3, IncN, IncN2, IncC, IncB/O/K/Z, IncFIA, IncFIB, IncFIC, IncFIII, IncHI1, IncL/M, IncP, IncR, IncT, IncX1, IncX3, IncX4, IncY, and ColE10, have been identified.3 The co-existence of other antimicrobial resistance (AMR) genes located on these plasmids has led to the current crisis and challenging clinical situation.
In the treatment of CREC infections, the widespread use of carbapenems as monotherapy has resulted in increasing resistance to these antibiotics and other β-lactams. A combination of antibiotics therefore could be an optional therapy to reduce further resistance development and/or enhance the antimicrobial activity. A previous study reported that the combination of a carbapenem (e.g., ertapenem) with fosfomycin resulted in a synergistic effect of increased antimicrobial activity. This theory could lead to effective alternative treatments for infections caused by carbapenem-resistant Enterobacteriaceae (CRE).9
Although some antibiotics such as fosfomycin, tigecycline, and colistin are considered as a treatment option for the CRE infections, fosfomycin is mostly considered for severe patients due to its broad-spectrum bactericidal activity and less toxicity than other antibiotics of choice. However, many studies suggested that fosfomycin should be combined with other antibiotics to avoid the fosfomycin resistance that might be rapidly developed during the therapy.10,11 Additionally, there are few studies reporting the fosfomycin-based combination against blaNDM-1-harboring CREC. Thus, the objective of this study was to assess the synergistic activities of fosfomycin in combination with other antimicrobial agents against blaNDM-1-harboring CREC, and to genetically characterize the entire genome of these pathogens.
Materials and Methods
Bacterial Isolation and Identification
The bacterial isolates were obtained from colonized patients in the Medicine Ward, Songklanagarind Hospital, between March and April 2017. One µg/mL of meropenem containing MacConkey agar plates were used to screen and select carbapenem-resistant Gram-negative bacteria.12 The strain of Escherichia coli was identified by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF).
Antimicrobial Susceptibility Testing (AST) and Detection of the blaNDM Gene
Thirty-eight E. coli isolates were confirmed to be carbapenem-resistant isolates by evaluation of the minimum inhibitory concentrations (MICs) of imipenem and meropenem through the broth microdilution method. E. coli ATCC 25922 was used as the quality control.13 The blaNDM gene was detected among these CREC isolates by a PCR method using the NDM-F primer (GGTTTGGCGATCTGGTTTTC) and the NDM-R primer (CGGAATGGCTCATCACGATC).10,14 The PCR products were then checked using agarose gel electrophoresis and the expected size was 621 bp. In addition, the MIC values of other 9 antibiotics (fosfomycin, ciprofloxacin, doripenem, levofloxacin, tigecycline, sitafloxacin, colistin, gentamicin, and amikacin) and 1 β-lactamase inhibitor (sulbactam) (Sigma-Aldrich) were evaluated against blaNDM-harboring isolates by the broth microdilution method, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines, 2016 or US Food and Drug Administration (FDA) breakpoints for tigecycline or European Committee on Antimicrobial Susceptibility Testing (EUCAST) for colistin.13,15,16
Evaluation of Synergistic Effects
All blaNDM-harboring CREC isolates were assessed for the synergistic activity of fosfomycin in combination with imipenem, meropenem, doripenem, gentamicin, amikacin, ciprofloxacin, levofloxacin, sitafloxacin, colistin, tigecycline, and sulbactam (Sigma-Aldrich, USA) using the checkerboard method. As we used fosfomycin, the Clinical and Laboratory Standards Institute (CLSI) recommended adding 25 mg/mL of glucose-6-phosphate (G-6-P) in the 96-well plates containing Mueller-Hinton broth (MHB). The Fractional Inhibitory Concentration Index (FICI) was used to interpret the synergism (FICI ≤ 0.5), indifference (0.5 < FICI ≤ 4.0), or antagonism (FICI > 4.0).
DNA Extraction and Whole-Genome Sequencing (WGS)
The genomic DNA (gDNA) of all blaNDM-harboring CREC isolates was extracted using a TIANamp Bacterial DNA Kit (Tiangen, Beijing, China), following the manufacturer’s instructions. Then, the quality of the gDNA was checked in terms of concentration and purity with a Thermo Scientific™ NanoDrop 2000 and gel electrophoresis, respectively. The qualified DNA samples were then sent to Beijing Genomics Institute (BGI), Beijing, China, for conducting short-read sequencing (WGS) with the use of a BGISEQ-500 sequencer (Beijing Genomics Institute, Beijing, China).
Plasmid Extraction and Sequencing
Plasmid DNA was extracted by the alkaline lysis method, as previously described.17 Briefly, the bacteria were treated with a base solution (0.2 N NaOH-1% SDS) to denature the double-strand DNA. An acid solution (4M potassium acetate-2M acetic acid) was immediately added to neutralize the mixture, which was then centrifuged at high speed to separate the plasmid DNA from the chromosomal DNA. The purity of the plasmid DNA was visualized by agarose gel electrophoresis. The qualified plasmids were then sequenced using NovaSeq 6000 sequencer (Beijing Headquarters Novogene Co., Ltd. Beijing, China).
Data Analysis of WGS and Plasmids
All sequence reads were de novo assembled using Unicycler v0.4.7.18 The assembled sequences were then annotated using Prokka v1.12.19 For analysis of these sequence reads, the contigs of all CREC isolates were uploaded to the Center for Genomic Epidemiology (CGE) (http://www.genomicepidemiology.org/), a free online bioinformatics service allowing scientists to analyze sequencing data in relation to infectious diseases. Here, we used ResFinder 4.1 (https://cge.cbs.dtu.dk/services/ResFinder/)20 and PlasmidFinder 2.1 (https://cge.cbs.dtu.dk/services/PlasmidFinder/),21 with a set of 95% identity and 80% minimum length for the detection of acquired AMR genes and plasmids, respectively. Meanwhile, VirulenceFinder 2.0 (https://cge.cbs.dtu.dk/services/VirulenceFinder/)22 with a set of 90% identity and 60% minimum length and MLST 2.0 (Multi-Locus Sequence Typing) (https://cge.cbs.dtu.dk/services/MLST/)23 were used to identify virulence genes and sequence types (STs), respectively. In addition to the CGE website, we also used other web-based bioinformatics tools. For other mobile-genetic elements (MGEs), insertion sequences (ISs) and integrons were also investigated using blastn against the IS database and integron_finder, respectively. The bacteriophage genomes were explored using phaster (https://phaster.ca/),24 while CRISPR-Cas regions were detected using CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr/CrisprCasFinder/Index). The bacteriocin-encoding genes were also investigated using BAGEL4 (http://bagel4.molgenrug.nl/index.php).25
Pan-Genome and Phylogenetic Analysis
The pan-genome profiles of blaNDM-harboring CREC isolates were compared to published genomes from Africa (n = 11), Austria (n = 39), Germany (n = 120), and Thailand (n = 12), which are available from the National Center for Biotechnology Information (NCBI).26–29 The pan-genome analysis was performed using Roary v3.13.0,30 with minimum blastx identity and core definition threshold at 95% and 99%, respectively. A phylogenetic tree was generated from the pan-genome alignment, based on accessory genes, using a maximum likelihood method (1,000 bootstraps). The interactive visualization of the phylogenetic tree was performed using the Geneious R10.2631 and Phandango website (https://jameshadfield.github.io/phandango/).32 Additionally, a pan-genome frequency plot, a pie chart of the pan-genome, and a presence and absence matrix against a phylogenetic tree were also created using roary_plots script (https://github.com/sanger-pathogens/Roary/tree/master/contrib/roary_plots).
Data Availability
The assembled genomes of the blaNDM-1-harboring CREC isolates from this study have been submitted to the NCBI GenBank under BioProject number PRJNA780210 with BioSample numbers SAMN23132882, SAMN23132883, and SAMN23132884.
Results
Bacterial Isolates and Clinical Data
A total of 38 E. coli isolates were collected from the patients and were classified the carbapenem resistance. Due to a screening method can be used to collect the isolates that provide non-susceptible to the carbapenem (intermediate resistance and resistance), only 6 (15.8%) out of 38 isolates were then identified as CREC by phenotypic method and a half of the CREC isolates (n = 3) were positive for the blaNDM gene. These 3 blaNDM-harboring isolates were obtained from the rectums of 2 female and 1 male patients, who had cardiac arrest or pulmonary edema as common underlying disease. Importantly, all 3 patients have previously received carbapenem and other antibiotics, which might be the reason for carbapenem-resistance isolates.
Antimicrobial Susceptibilities in CREC Isolates
The MIC values of the 11 tested antibiotics against the 3 isolates of blaNDM-1-carrying CREC are shown in Table 1. The 3 isolates were resistant to imipenem, meropenem, and doripenem with high MIC values ranging from 16 to 64 µg/mL. These isolates were resistant to all tested fluoroquinolones, ciprofloxacin (32 µg/mL), and levofloxacin (64 and 128 µg/mL), while sitafloxacin resistance (8 µg/mL) was found in 2 isolates. The low level of colistin resistance was also observed in all isolates. In addition, resistance to gentamicin and fosfomycin was detected in 2 (16 and 32 µg/mL) and 1 (256 µg/mL) isolates, respectively. However, all isolates were still susceptible to tigecycline and amikacin.
Table 1.
Minimum Inhibitory Concentration (MIC) Values of 11 Antibiotics and 1 β-Lactamase Inhibitor Against blaNDM-1-Harboring CREC Isolates
| Isolate Code | MIC Value (µg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOS | IPM | MEM | DOR | CIP | LVX | TIG | STFX | CT | GN | AMK | SUL | |
| CREC003 | 0.25 (S) | 32 (R) | 64 (R) | 16 (R) | 128 (R) | 32 (R) | 0.5 (S) | 8 (R) | 4 (R) | 32 (R) | 2 (S) | 128 |
| CREC004 | 0.5 (S) | 16 (R) | 32 (R) | 32 (R) | 64 (R) | 32 (R) | 0.5 (S) | 4 (I) | 4 (R) | 16 (R) | 2 (S) | 128 |
| CREC038 | 256 (R) | 32 (R) | 64 (R) | 32 (R) | 128 (R) | 32 (R) | 1 (S) | 8 (R) | 4 (R) | 1 (S) | 2 (S) | 128 |
Abbreviations: FOS, fosfomycin; IPM, imipenem; MEM, meropenem; CIP, ciprofloxacin; DOR, doripenem; LVX, levofloxacin; SUL, sulbactam; TIG, tigecycline; STFX, sitafloxacin; CT, colistin; GN, gentamicin; AMP, amikacin; R, resistance; I, intermediate resistance; S, susceptible.
Synergistic Effects Against CREC Isolates
The synergy study showed that synergistic effects of fosfomycin plus gentamicin, amikacin, ciprofloxacin, sitafloxacin, colistin, or tigecycline were observed in 1 isolate, while there was no difference in the combination of fosfomycin with imipenem, meropenem, doripenem, and levofloxacin in any isolates (Table 2). The antagonistic effect of fosfomycin in combination with sulbactam was seen in 1 isolate (Table 2).
Table 2.
The Synergistic Effects of Fosfomycin in Combination with Other Antimicrobial Agents Against blaNDM-1-Harboring CREC Isolates
| Combination | No. of Isolates | ||
|---|---|---|---|
| Synergy | Indifference | Antagonistic | |
| Fosfomycin + Imipenem | 0 | 3 | 0 |
| Fosfomycin + Meropenem | 0 | 3 | 0 |
| Fosfomycin + Doripenem | 0 | 3 | 0 |
| Fosfomycin + Gentamicin | 1 | 2 | 0 |
| Fosfomycin + Amikacin | 1 | 2 | 0 |
| Fosfomycin + Ciprofloxacin | 1 | 2 | 0 |
| Fosfomycin + Levofloxacin | 0 | 3 | 0 |
| Fosfomycin + Sitafloxacin | 1 | 2 | 0 |
| Fosfomycin + Sulbactam | 0 | 2 | 1 |
| Fosfomycin + Colistin | 1 | 2 | 0 |
| Fosfomycin + Tigecycline | 1 | 2 | 0 |
Sequence Types (STs) and Antimicrobial Resistance (AMR) Genes in CREC Isolates
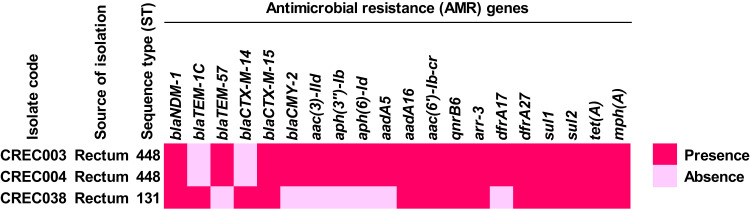
In the MLST analysis, CREC003 and CREC004 were identified in ST448, while CREC048 was identified in ST131, as shown in Table S1. The study found that the blaNDM-1 gene was present in the 3 isolates noted previously. Other β-lactam resistance genes, blaTEM-1C, blaTEM-57, blaCTX-M-14, blaCTX-M-15, and blaCMY-2, were also found in these CREC isolates. Additionally, the CREC isolates carried other AMR genes, conferring resistance to aminoglycosides (aac(3)-IId, aph(3”)-Ib, aph(6)-Id, aadA5, aadA16, and aac(6’)-Ib-cr), fluoroquinolone (qnrB6), rifampicin (ARR-3), trimethoprim (dfrA17 and dfrA27), sulfonamide (sul1 and sul2), tetracycline (tet(A)), and macrolide (mph(A)). The distribution of AMR genes is illustrated in Figure 1 and Table S2.
Figure 1.
Distribution of antimicrobial resistance (AMR) genes in blaNDM-1-harboring CREC isolates.
Mobile Genetic Elements (MGEs) in CREC Isolates
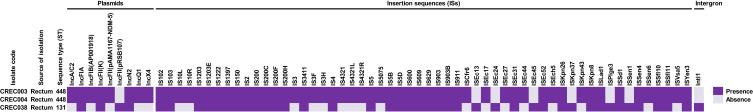
Various mobile genetic elements (MGEs) were identified in the blaNDM-1-harboring CREC isolates, as shown in Figure 2 and Tables S3–S5. For plasmids, we found IncFIA, IncFII(K), and IncN2 plasmids in all isolates. The IncA/C2, IncFIB (AP001918), IncFII (pAMA1167-NDM-5), IncQ1, and IncX4 plasmids were identified in only the ST448 isolates, while the IncFII (pRSB107) plasmid was only observed in the ST131 isolate.
Figure 2.
Distribution of mobile genetic elements (MGEs) in blaNDM-1-harboring CREC isolates.
Many insertion sequences (ISs), namely IS102, IS1222, IS2, IS200F, IS4, IS5, IS903, ISEc31, ISKpn26, ISLad1, ISSen4, and ISYen3, were also detected in all CREC isolates. ISEc17, ISEc27, ISEc44, ISEch5, and ISKpn43 were only found in the ST448 isolates, while ISEc13, ISEc24, ISEc45, ISKpn37, ISPlge3, and ISSen1 were only identified in the ST131 isolate. For integrons, a class 1 integron with the carriage of aac(6)-Ib gene responsible for aminoglycoside resistance was detected in the ST131 isolate.
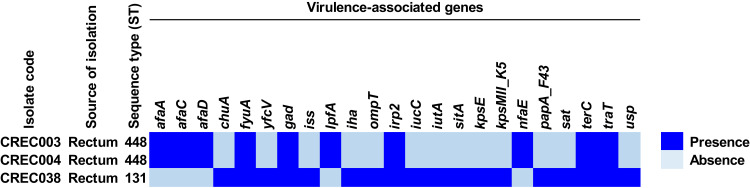
Virulence-Associated Genes in CREC Isolates
Besides the identification of AMR genes, we also looked for virulence-associated genes and the results are shown in Figure 3 and Table S6. We found the fyuA, gad, irp2, terC, and traT genes in all 3 blaNDM-1-harboring CREC isolates, while the afaA, afaC, afaD, lpfA, and nfaE genes were only detected in the ST448 isolates, and the chuA, yfcV, iss, iha, ompT, iucC, iutA, sitA, kpsE, kpsMII_K5, papA_F43, sat, and usp genes were only found in the ST131 isolate.
Figure 3.
Distribution of virulence-associated genes in blaNDM-1-harboring CREC isolates.
CRISPR-Cas Region in CREC Isolates
In the analysis of the CRISPR-Cas system, 5 to 7 CRISPR positions were found in 3 isolates of blaNDM-1-harboring CREC. The CRISPR positions contained 1 to 18 spacers. Additionally, 2 positions of cas locus was also identified in all ST448 isolates, which were further classified as CRISPR-Cas type-IF and type-IE. The details of the CRISPR-Cas region are provided in Tables 3 and S7.
Table 3.
CRISPR-Cas System in blaNDM-1-Harboring CREC Isolates
| Isolate Code | Source of Isolation | Sequence Type (ST) | CRISPR Region | Cas Region | ||||
|---|---|---|---|---|---|---|---|---|
| CRISPR Position | No. of Direct Repeat (DR) | No. of Spacer | Cas Position | cas Locus | Cas Type | |||
| CREC003 | Rectum | 448 | Position 1 | 2 | 1 | Position 1 | cas6_csy3_csy2_csy1_cas3-cas2_cas1 | Cas type-IF |
| Position 2 | 19 | 18 | Position 2 | cas2_cas1_cas6_cas5_cas7_cse2_cse1_cas3 | Cas type-IE | |||
| Position 3 | 14 | 13 | ||||||
| Position 4 | 2 | 1 | ||||||
| Position 5 | 12 | 11 | ||||||
| Position 6 | 10 | 9 | ||||||
| Position 7 | 2 | 1 | ||||||
| CREC004 | Rectum | 448 | Position 1 | 14 | 13 | Position 1 | cas1_cas3-cas2_csy1_csy2_csy3_cas6 | Cas type-IF |
| Position 2 | 19 | 18 | Position 2 | cas2_cas1_cas6_cas5_cas7_cse2_cse1_cas3 | Cas type-IE | |||
| Position 3 | 2 | 1 | ||||||
| Position 4 | 12 | 11 | ||||||
| Position 5 | 10 | 9 | ||||||
| Position 6 | 2 | 1 | ||||||
| Position 7 | 2 | 1 | ||||||
| CREC038 | Rectum | 131 | Position 1 | 2 | 1 | - | - | - |
| Position 2 | 2 | 1 | ||||||
| Position 3 | 5 | 4 | ||||||
| Position 4 | 7 | 6 | ||||||
| Position 5 | 8 | 7 | ||||||
Bacteriophage Genome in CREC Isolates
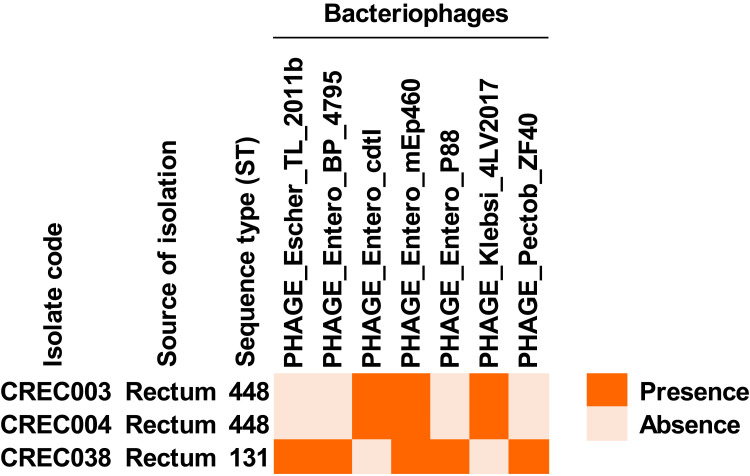
We found bacteriophage genomes in these isolates, as shown in Figure 4 and Table S8. PHAGE_Entero_mEp460 genome was found in all CREC isolates. Two bacteriophage genomes, PHAGE_Entero_cdtI and PHAGE_Klebsi_4LV2017, were only detected in the ST448 isolates, while 4 bacteriophage genomes, PHAGE_Escher_TL_2011b, PHAGE_Entero_BP_4795, PHAGE_Entero_P88, and PHAGE_Pectob_ZF40, were only found in the ST131 isolate.
Figure 4.
Presence of bacteriophage genomes in blaNDM-1-harboring CREC isolates.
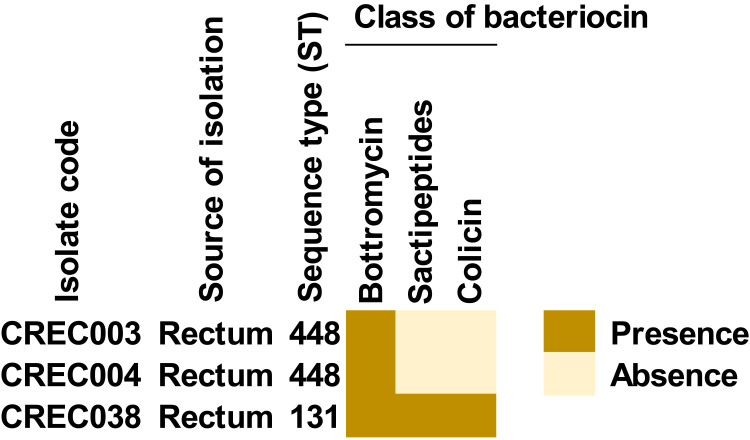
Bacteriocin Class in CREC Isolates
The results of bacteriocins are shown in Figure 5 and Table S9. We found that 3 genes, encoding bottromycin, sactipeptides, and colicin, were detected in the ST131 isolate, while only bottromycin was identified in the ST448 isolates.
Figure 5.
Presence of bacteriocin-encoding genes in blaNDM-1-harboring CREC isolates.
blaNDM-1-Bearing IncN Plasmid in CREC Isolates
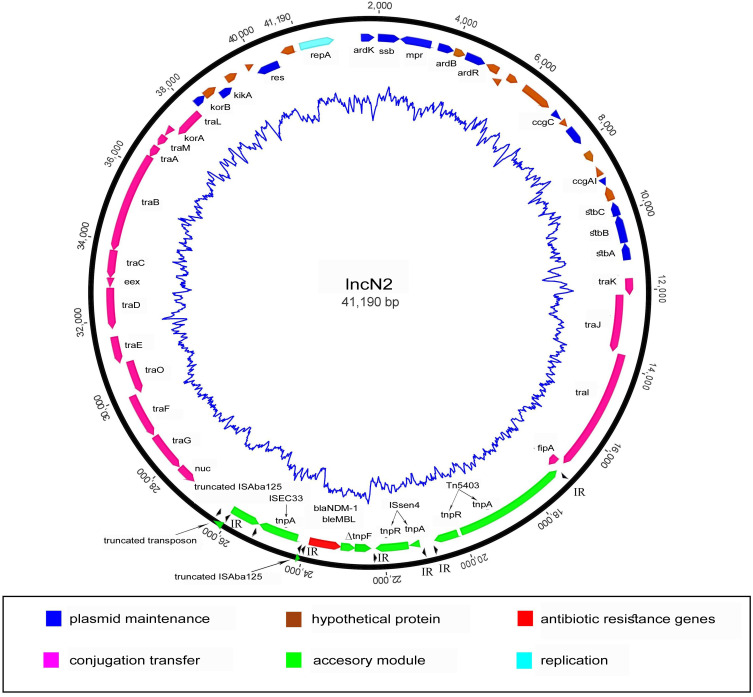
All 3 E. coli isolates carried the blaNDM-1 gene on a plasmid (pSK20-NDM1, pSK21-NDM1, and pSK22-NDM1) belonging to the IncN2 incompatibility group. The structure of the blaNDM-1-bearing IncN2 plasmid is shown in Figure 6. The pSK20-NDM1, pSK21-NDM1, and pSK22-NDM1 plasmids were closely similar, all having a length of 41,190 bp. All of the IncN2 plasmids had the same backbone, consisting of genes encoding for plasmid stability (stbA, stbB, and stbC), replication (repA), and conjugal transfer (tra). Only one antibiotic resistance gene was found, blaNDM-1. The genetic environment of blaNDM-1 shares a single conserved region, which was a Tn3-like structure, ISAba125, ISEc33, blaNDM-1, blaMBL, ΔtnpF, ISSen4, and Tn5403. The blaNDM−1 gene was bracketed by two insertion sequences (IS), ISEc33 and ISSen4, which belong to the IS630 and IS3 families, respectively. The upstream ISAba125 was interrupted by the insertion of ISEc33 but not the part of the promoter involved in the expression of blaNDM-1. The sequence analysis revealed that the accessory module was inserted into the IncN2 plasmid between the fipA and nuc genes.
Figure 6.
Structure of IncN2 plasmid in blaNDM-1-harboring CREC isolates.
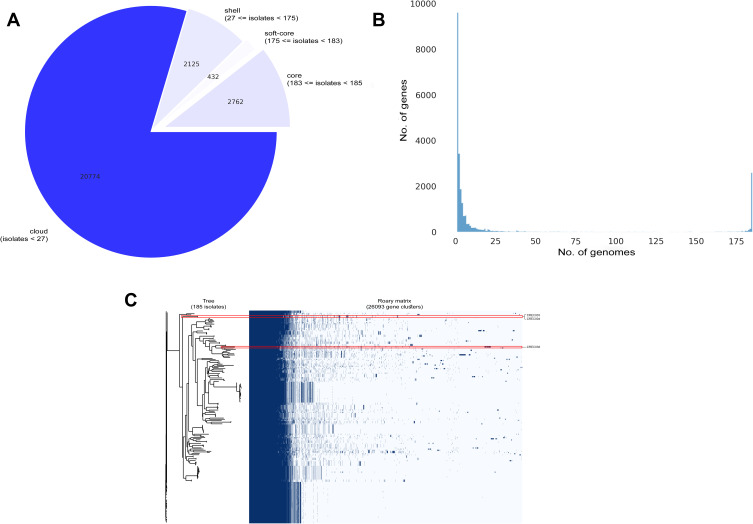
Pan-Genome and Phylogenetic Tree of CREC Isolates
The results of the pan-genome analysis are shown in Figure 7, while the phylogenetic tree is visualized in Figure S1. From 26,093 genes recorded in our study, 23,331 (89.41%) and 2,762 (10.59%) genes were identified as accessory and core genes, respectively. In comparison to previously published genomes, the CREC003 and CREC004 isolates (ST448) could be grouped in the same clade, while the ST131 isolate (CREC038) was located in a different clade (Figures 7 and S1). The ST448 isolates contained the ymfD, ymfE, and nfaA genes encoding the e14 prophage with SAM-dependent methyltransferase, the e14 prophage with inner membrane protein, and the non-fimbrial adhesin 1, respectively. The flhC_1, mntH_2, alba, moaA_2, and csoR genes encoding the flagellar transcriptional regulator FlhC, the divalent metal cation transporter MntH, the antilisterial bacteriocin subtilosin biosynthesis protein AlbA, the GTP 3’,8-cyclase, and the copper-sensing transcriptional repressor CsoR were present as important features in ST131 isolate.
Figure 7.
Genomic diversity among blaNDM-1-harboring CREC isolates compared to previously published genomes.
Notes: (A) A pie chart of the pan-genome, (B) a pan-genome frequency plot, and (C) a presence and absence matrix against a phylogenetic tree.
Discussion
In 2019, the Centers for Disease Control and Prevention (CDC) considered the infection caused by CRE as an urgent threat, due to the high level of estimated cases in hospitalized patients, mortality rates, and attributable health-care costs. CRE, particularly CREC, developed resistance mechanisms for many antibiotic classes such as carbapenems, aminoglycosides, and fluoroquinolones.33,34 Here, we report on the synergistic effects of fosfomycin in combination with other antimicrobial agents against blaNDM-1-harboring CREC isolates as well as the genomic characteristics of these pathogens. According to the MIC results, we found that all 3 blaNDM-1-harboring CREC isolates were resistant to more than 3 antibiotic classes, indicating multidrug resistance in these isolates. Although they were susceptible to tigecycline and amikacin, the use of these antibiotics as monotherapy still needs to be of concern since the target bacteria can develop resistance mechanisms during therapy.35 We then evaluated the synergistic activity of antibiotic combination for combating these pathogens. In the synergism study, we found that the use of fosfomycin combined with aminoglycosides, colistin, tigecycline, and sitafloxacin provided synergistic effects against the ST131 isolate of the blaNDM-1-harboring CREC. Similar to previous study, Cebrero-Cangueiro et al (2021) found that fosfomycin plus gentamicin showed a synergistic effect against carbapenemase-producing Enterobacteriaceae (CPE).36 The synergistic effects of fosfomycin plus colistin and tigecycline have been also reported in approximately 20% of the carbapenem-resistant K. pneumoniae (CRKP).10 The combination of fosfomycin with ciprofloxacin showed the synergistic effect against only 1 isolate of the ST448 in our study. The benefit of combination therapy is to suppress the emerging resistance mechanisms that probably occur during the treatment.37
In WGS analysis, 2 and 1 isolates of blaNDM-1-harboring CREC were identified in ST448 and ST131, respectively. These STs have been previously identified as an internationally spread clone.38,39 The E. coli ST131 clone has been reported as the major clone causing human infections and is predominantly found in Southeast Asia.40 The previous study has been demonstrated that the majority of this strain carried the blaKPC gene which commonly produces Klebsiella pneumoniae carbapenemase (KPC) and this gene is easily transferred to another bacteria through the MGEs. Thus, it raises the concern about the increase of carbapenem-resistant strains.41 In AMR detection, we found that the CREC isolates carried many AMR genes conferring resistance to carbapenems, third-generation cephalosporins, other β-lactams, aminoglycosides, fluoroquinolones, rifampicin, trimethoprim, sulfisoxazole, tetracycline, and macrolides. These isolates have also been classified as multidrug-resistant (MDR) isolates, which was in concordance with our MIC results. In Germany, Welker et al (2020) reported that E. coli ST131 carried both carbapenem and other β-lactam resistance genes.42 In South Korea, Shin et al (2021) also demonstrated that 23 AMR genes, especially the blaNDM gene, were present in E. coli isolates.43 Here, we also assessed the correlation among the antimicrobial susceptibility, synergistic activity of fosfomycin-based combination, and WGS results. The combinations of fosfomycin with aminoglycosides were synergy in the ST131 isolate having low MIC values of gentamicin and amikacin, which carried a lower number of aminoglycoside resistance genes than the ST448 isolates having a high MIC value of gentamicin (Tables 1 and 2, Figure 1).
In our study, MGEs were also investigated to assess the dissemination mechanisms of the AMR genes. A previous study reported the presence of 16 replicon types of blaNDM-1-bearing plasmids in Enterobacteriaceae, namely ColE, IncA/C, IncN2, IncFIA, IncFIB, IncFII, IncHI1, IncHI3, IncHIB, IncL/M, IncP, IncR, IncT, IncX1, IncX3, and IncY.3 In this study, IncFIA, IncF(II)K, and IncN2 plasmids were detected in all CREC isolates, while the blaNDM-1 gene was identified in the IncN2 plasmid. The blaNDM-1-bearing IncN2 plasmid shared >99% nucleotide identity and 100% query coverage with pJN24NDM1 (GenBank accession no. MK368725), pC2972-5-NDM (GenBank accession no. CP039806), pTR3 (GenBank accession no. JQ349086), and pNDM-ECS01 (GenBank accession no. KJ413946), according to the BLAST search. The pJN24NDM1 is an IncN2 plasmid first identified in an E. coli ST405 isolate from the abdominal fluid of a patient in China,44 while the pC2972-5-NDM and pTR3 plasmids have been reported in K. pneumoniae isolates from China45 and Singapore,46 respectively. These highly similar plasmids detected from different geographic areas and bacterial species can be assumed to spread by lateral dissemination.47 The first blaNDM-1-bearing IncN2 was identified in NDM-1-producing E. coli, which was isolated from a patient hospitalized in Bangladesh.48 In 2014, blaNDM-1-bearing IncN2 (pNDM-ECS01) was also detected in E. coli ST131 from Central Thailand,49 similar to our study. This could imply that the E. coli ST131 strain might be widely disseminated throughout Thailand. The IncN2 plasmid has the potential to horizontally transfer the blaNDM gene to other pathogenic bacteria, potential worldwide dissemination of the blaNDM gene. In addition, a previous study demonstrated that the blaNDM-1 gene located in transposon Tn125 was responsible for the blaNDM-1 spread among Acinetobacter spp.7 The structure of the Tn125 prototype includes ISAba125, blaNDM-1, bleMBL, ΔtrpF, dsbC, cutA, ΔgroES, groEL, ISCR27, and ISAba125. In a comparative analysis with the Tn125 prototype, the Tn125-like element in plasmids from this study lacked dsbC, cutA, ΔgroES, groEL, ISCR27, and downstream ISAba125.50 The sequences revealed that the accessory module was located between the fipA and nuc genes. We hypothesize that these two regions (between fipA and nuc genes) probably act as the main integration sites for the insertion of transposable elements within IncN plasmids. Same site insertion for the accessory module has also been reported in other IncN plasmids, pKT58A (GenBank accession no. JX065631) and pRSB206 (GenBank accession no. JN102344).51,52 Thus, this site might act as a significant integration site in IncN2 plasmids. Currently, IncN plasmids have been found all over the world with the highest prevalences in China and the United States.44
Furthermore, many ISs were present in these isolates, which are components of transposons providing a cut-and-paste mechanism.53 As shown in Figure 6, ISEc33 and ISSen4 bracketed the blaNDM-1 on IncN2 plasmid. This structure was the same as the previously reported p271A plasmid, which was isolated from E. coli ST101.54 Notably, we found a class 1 integron in the ST131 isolate, which also provides the ability to drive AMR genes to other bacteria.
To check the pathogenicity of these CREC isolates, the gene encoding siderophore receptor (fyuA), glutamate decarboxylase (gad), high molecular weight protein 2 non-ribosomal peptide synthetase (irp2), tellurium ion resistance protein (terC), and outer membrane protein complement resistance (traT) were identified in all CREC isolates. Most virulence genes can also be horizontally transferred to other bacteria through MGEs, similar to AMR genes.55
In the study of bacterial adaptive immunity system against foreign genetic elements, we found CRISPR-Cas systems in all CREC isolates. These CRISPR-Cas systems contained many spacers, indicating that these isolates may have been previously infected with bacteriophages or invaded by foreign DNA fragments. Bacteriophage genomes were also detected in all CREC isolates. The partial genome of Escherichia phage was identified in the ST131 isolate, while 4 partial genomes of Enterobacteria phage were present in the ST131 and/or ST448 isolates. Notably, we found the genomes of the Klebsiella phage in the ST448 isolates and Pectobacterium phage in the ST131 isolate. This may indicate that these CREC isolates had been priorly infected by bacteriophages with the lysogenic life cycle.56 Also, we hypothesize that these partial phage genomes might carry the AMR genes, as described in a previous study.57 To prove this phenomenon, long-read WGS is needed to elucidated further.
In the investigation of gene encoding bacteriocins, bottromycin was found in all isolates, while sactipeptides and colicin were only detected in the ST131 isolate. Bottromycin has been previously detected in E. coli isolated from the intestine of humans.58 It commonly provides a natural antimicrobial activity, particularly the modified bottromycin A2 which provides activity against methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE).59
In our study of the genomic diversity of our blaNDM-1-harboring CREC isolates compared to other published genomes, we found that the specific features of the ST448 and ST131 isolates were different from various STs. In ST488 isolates, the prophage-encoding proteins may be associated with bacterial adaptation to the bacteriophages,60,61 while non-fimbrial adhesin 1 is probably associated with cell attachment to abiotic surfaces and/or host cells, and the biofilm formation.62 For the ST131 isolate, the FlhC, MntH, AlbA, and CsoR proteins are probably related to the production of flagellar, the transportation of divalent metal cation, the production of antilisterial bacteriocin subtilosin, and the expression of a copper-sensing repressor, which might be beneficial for bacterial evolution and pathogenicity.63–66
Conclusion
The combinations of fosfomycin with aminoglycosides, colistin, tigecycline, and sitafloxacin might be considered as a treatment of infections caused by ST131 carrying the blaNDM-1 gene. Our study of the whole-genome and plasmid sequences of the blaNDM-1-harboring CREC isolates provides significant information, especially the presence of AMR genes, MGEs, virulence genes, genes involving bacterial defense mechanisms, and the structure of blaNDM-1-bearing IncN2 plasmid. These particular isolates contained specific features compared to some published genomes, which are probably associated with bacterial adaptation. The findings can be expected to be useful in the development of appropriate treatments and the control of these infections in the future.
Acknowledgments
The authors would like to acknowledge the Division of Infectious Diseases, Department of Internal Medicine, Faculty of Medicine, Prince of Songkla University for providing the bacterial isolates. We would also like to thank the Division of Biological Science, the Division of Computational Science, and the Molecular Evolution and Computational Biology (MECoB) Research Unit, Faculty of Science as well as the Department of Biomedical Science and Biomedical Engineering, Faculty of Medicine, Prince of Songkla University for use of their research facilities. Finally, we thank Mr. David Patterson from Department of International Affairs, Faculty of Medicine, Prince of Songkla University, for English language review in this study.
Funding Statement
This research was supported by the Human Research Ethics Committee (HREC), Faculty of Medicine, Prince of Songkla University (reference number: 59-352-14-1), and the National Research Council of Thailand (grant number: 1232545). In addition, the study was also supported by a Graduate Scholarship, Faculty of Medicine, Prince of Songkla University, Thailand and the Faculty of Science, Prince of Songkla University, Thailand (grant number: SCI64040135).
Ethical Approval
This study was approved by the Human Research Ethics Committee (HREC), Faculty of Medicine, Prince of Songkla University (Reference Number: 59-352-14-1). Also, the researchers were granted permission to extract the data from the database with waiver of consent because of the observational nature of the study. All data were fully anonymized before the researcher accessed and analyzed.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest.
References
- 1.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18(5):263–272. doi: 10.1016/j.molmed.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 2.Wailan AM, Paterson DL. The spread and acquisition of NDM-1: a multifactorial problem. Expert Rev Anti-Infect Ther. 2014;12(1):91–115. doi: 10.1586/14787210.2014.856756 [DOI] [PubMed] [Google Scholar]
- 3.Wu W, Feng Y, Tang G, et al. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2):e00115–18. doi: 10.1128/CMR.00115-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ripabelli G, Sammarco M, Salzo A, et al. New Delhi metallo-β-lactamase (NDM-1)-producing Klebsiella pneumoniae of sequence type ST11: first identification in a hospital of central Italy. Lett Appl Microbiol. 2020;71(6):652–659. doi: 10.1111/lam.13384 [DOI] [PubMed] [Google Scholar]
- 5.Snyder B, Montague B, Anandan S, et al. Risk factors and epidemiologic predictors of blood stream infections with New Delhi metallo-b-lactamase (NDM-1) producing Enterobacteriaceae. Epidemiol Infect. 2019;147:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chibabhai V, Nana T, Bosman N, et al. Were all carbapenemases created equal? Treatment of NDM-producing extensively drug-resistant Enterobacteriaceae: a case report and literature review. Infection. 2018;46(1):1–13. doi: 10.1007/s15010-017-1070-8 [DOI] [PubMed] [Google Scholar]
- 7.Bonnin RA, Poirel L, Nordmann P. New Delhi metallo-β-lactamase-producing Acinetobacter baumannii: a novel paradigm for spreading antibiotic resistance genes. Future Microbiol. 2014;9(1):33–41. doi: 10.2217/fmb.13.69 [DOI] [PubMed] [Google Scholar]
- 8.Jones LS, Toleman MA, Weeks JL, et al. Plasmid Carriage of bla NDM-1 in clinical Acinetobacter baumannii Isolates from India. Antimicrob Agents Chemother. 2014;58(7):4211–4213. doi: 10.1128/AAC.02500-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loose M, Link I, Naber KG, et al. Carbapenem-containing combination antibiotic therapy against carbapenem-resistant uropathogenic Enterobacteriaceae. Antimicrob Agents Chemother. 2019;64(1):e01839–19. doi: 10.1128/AAC.01839-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chukamnerd A, Pomwised R, Phoo MTP, et al. In vitro synergistic activity of fosfomycin in combination with other antimicrobial agents against carbapenem-resistant Klebsiella pneumoniae isolated from patients in a hospital in Thailand. J Infect Chemother. 2021;27(3):507–514. doi: 10.1016/j.jiac.2020.11.004 [DOI] [PubMed] [Google Scholar]
- 11.Samonis G, Maraki S, Karageorgopoulos D, et al. Synergy of fosfomycin with carbapenems, colistin, netilmicin, and tigecycline against multidrug-resistant Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa clinical isolates. Eur J Clin Microbiol Infect Dis. 2012;31(5):695–701. doi: 10.1007/s10096-011-1360-5 [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Vyas A, Mehra S. Utilization of MacConkey-meropenem screening agar for the detection of carbapenem resistant Enterobacteriaceae in a tertiary care hospital. SSRG int j med sci. 2018;2(4):e23. [Google Scholar]
- 13.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. approved standard M100. Wayne, PA: Clinical and Laboratory Standards Institute (CLSI); 2018. [Google Scholar]
- 14.Poirel L, Walsh TR, Cuvillier V, et al. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi: 10.1016/j.diagmicrobio.2010.12.002 [DOI] [PubMed] [Google Scholar]
- 15.Food and Drug Administration. Drug Safety Communication Increased Risk of Death with Tygacil (Tigecycline) Compared to Other Antibiotics Used to Treat Similar Infections. Washington, DC: FDA; 2010. [Google Scholar]
- 16.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. Version 9.0; 2019. Available from: http://www.eucast.org. Accessed April 5, 2022.
- 17.Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from. Escherichia Coli Anal Biochem. 1993;212(2):394–401. [DOI] [PubMed] [Google Scholar]
- 18.Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comp Biol. 2017;13(6):e1005595. doi: 10.1371/journal.pcbi.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 20.Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–3500. doi: 10.1093/jac/dkaa345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carattoli A, Zankari E, García-Fernández A, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi: 10.1128/AAC.02412-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malberg Tetzschner AM, Johnson JR, Johnston BD, et al. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol. 2020;58(10):e01269–20. doi: 10.1128/JCM.01269-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen MV, Cosentino S, Rasmussen S, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. 2012;50(4):1355–1361. doi: 10.1128/JCM.06094-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arndt D, Grant JR, Marcu A, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44(W1):W16–W21. doi: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Jong A, van Hijum SA, Bijlsma JJ, et al. BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 2006;34(suppl_2):W273–W279. doi: 10.1093/nar/gkl237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mbanga J, Amoako DG, Abia AL, et al. Genomic insights of multidrug-resistant Escherichia coli from wastewater sources and their association with clinical pathogens in South Africa. Front Vet Sci. 2021;8:137. doi: 10.3389/fvets.2021.636715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zingali T, Reid CJ, Chapman TA, et al. Whole genome sequencing analysis of porcine faecal commensal Escherichia coli carrying class 1 integrons from sows and their offspring. Microorganisms. 2020;8(6):843. doi: 10.3390/microorganisms8060843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid CJ, Blau K, Jechalke S, et al. Whole genome sequencing of Escherichia coli from store-bought produce. Front Microbiol. 2020;10:3050. doi: 10.3389/fmicb.2019.03050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paveenkittiporn W, Kamjumphol W, Ungcharoen R, et al. Whole-genome sequencing of clinically isolated carbapenem-resistant enterobacterales harboring mcr genes in Thailand, 2016–2019. Front Microbiol. 2021;11:3393. doi: 10.3389/fmicb.2020.586368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hadfield J, Croucher NJ, Goater RJ, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34(2):292–293. doi: 10.1093/bioinformatics/btx610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mediavilla JR, Patrawalla A, Chen L, et al. Colistin-and carbapenem-resistant Escherichia coli harboring mcr-1 and bla NDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4):e01191–e01216. doi: 10.1128/mBio.01191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482. doi: 10.3934/microbiol.2018.3.482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.León-Buitimea A, Garza-Cárdenas CR, Garza-Cervantes JA, et al. The demand for new antibiotics: antimicrobial peptides, nanoparticles, and combinatorial therapies as future strategies in antibacterial agent design. Front Microbiol. 2020;11:1669. doi: 10.3389/fmicb.2020.01669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cebrero-Cangueiro T, Labrador-Herrera G, Pascual Á, et al. Efficacy of fosfomycin and its combination with aminoglycosides in an experimental sepsis model by carbapenemase-producing Klebsiella pneumoniae clinical strains. Front Med. 2021;8:324. doi: 10.3389/fmed.2021.615540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrill HJ, Pogue JM, Kaye KS, et al. Treatment options for carbapenem-resistant Enterobacteriaceae infections. Open Forum Infect. Dis. 2015;2(2). doi: 10.1093/ofid/ofv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinal P, Nucleo E, Caltagirone M, et al. Genomics of Klebsiella pneumoniae ST16 producing NDM-1, CTX-M-15, and OXA-232. Clin Microbiol Infect. 2019;25(3):385.e1–385. e5. doi: 10.1016/j.cmi.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 39.Naseer U, Haldorsen B, Simonsen G, et al. Sporadic occurrence of CMY-2-producing multidrug-resistant Escherichia coli of ST-complexes 38 and 448, and ST131 in Norway. Clin Microbiol Infect. 2010;16(2):171–178. doi: 10.1111/j.1469-0691.2009.02861.x [DOI] [PubMed] [Google Scholar]
- 40.Chen SL, Ding Y, Apisarnthanarak A, et al. The higher prevalence of extended spectrum beta-lactamases among Escherichia coli ST131 in Southeast Asia is driven by expansion of a single, locally prevalent subclone. Sci Rep. 2019;9(1):1–14. doi: 10.1038/s41598-018-37186-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ripabelli G, Sammarco ML, Scutellà M, et al. Carbapenem-resistant KPC- and TEM-producing Escherichia coli ST131 isolated from a hospitalized patient with urinary tract infection: first isolation in Molise Region, Central Italy, July 2018. Microb Drug Resist. 2020;26(1):38–45. doi: 10.1089/mdr.2019.0085 [DOI] [PubMed] [Google Scholar]
- 42.Welker S, Boutin S, Miethke T, et al. Emergence of carbapenem-resistant ST131 Escherichia coli carrying blaOXA-244 in Germany, 2019 to 2020. Euro Surveill. 2020;25(46):2001815. doi: 10.2807/1560-7917.ES.2020.25.46.2001815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin H, Kim Y, Han D, et al. Emergence of high level carbapenem and extensively drug resistant Escherichia coli ST746 producing NDM-5 in influent of wastewater treatment plant, Seoul, South Korea. Front Microbiol. 2021;12. doi: 10.3389/fmicb.2021.645411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hao Y, Shao C, Geng X, et al. Genotypic and phenotypic characterization of clinical Escherichia coli sequence type 405 carrying IncN2 plasmid harboring blaNDM-1. Front. Microbiol. 2019;10:788. doi: 10.3389/fmicb.2019.00788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao H, Liu Y, Wang R, et al. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine. 2020;51:102599. doi: 10.1016/j.ebiom.2019.102599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y-T, Lin A-C, Siu LK, et al. Sequence of closely related plasmids encoding blaNDM-1 in two unrelated Klebsiella pneumoniae isolates in Singapore. PLoS One. 2012;7(11):e48737. doi: 10.1371/journal.pone.0048737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tijet N, Richardson D, MacMullin G, et al. Characterization of multiple NDM-1-producing Enterobacteriaceae isolates from the same patient. Antimicrob Agents Chemother. 2015;59(6):3648–3651. doi: 10.1128/AAC.04862-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poirel L, Lagrutta E, Taylor P, et al. Emergence of Metallo-β-Lactamase NDM-1-producing multidrug-resistant Escherichia coli in Australia. Antimicrob Agents Chemother. 2010;54(11):4914–4916. doi: 10.1128/AAC.00878-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Netikul T, Sidjabat HE, Paterson DL, et al. Characterization of an IncN2-type bla NDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother. 2014;69(11):3161–3163. doi: 10.1093/jac/dku275 [DOI] [PubMed] [Google Scholar]
- 50.Sun F, Yin Z, Feng J, et al. Production of plasmid-encoding NDM-1 in clinical Raoultella ornithinolytica and Leclercia adecarboxylata from China. Frontiers in Microbiology. 2015;6:458. doi: 10.3389/fmicb.2015.00458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolejska M, Villa L, Hasman H, et al. Characterization of IncN plasmids carrying bla CTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J Antimicrob Chemother. 2013;68(2):333–339. doi: 10.1093/jac/dks387 [DOI] [PubMed] [Google Scholar]
- 52.Eikmeyer F, Hadiati A, Szczepanowski R, et al. The complete genome sequences of four new IncN plasmids from wastewater treatment plant effluent provide new insights into IncN plasmid diversity and evolution. Plasmid. 2012;68(1):13–24. doi: 10.1016/j.plasmid.2012.01.011 [DOI] [PubMed] [Google Scholar]
- 53.Yoon E-J, Kim JO, Yang JW, et al. The bla OXA-23-associated transposons in the genome of Acinetobacter spp. represent an epidemiological situation of the species encountering carbapenems. J Antimicrob Chemother. 2017;72(10):2708–2714. doi: 10.1093/jac/dkx205 [DOI] [PubMed] [Google Scholar]
- 54.Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55(9):4224–4229. doi: 10.1128/AAC.00165-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grant MA, Weagant SD, Feng P. Glutamate decarboxylase genes as a prescreening marker for detection of pathogenic Escherichia coli groups. Appl Environ Microbiol. 2001;67(7):3110–3114. doi: 10.1128/AEM.67.7.3110-3114.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stone E, Campbell K, Grant I, et al. Understanding and exploiting phage–host interactions. Viruses. 2019;11(6):567. doi: 10.3390/v11060567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J, He X, Shen S, et al. Effects of the newly isolated T4-like phage on transmission of plasmid-borne antibiotic resistance genes via generalized transduction. Viruses. 2021;13(10):2070. doi: 10.3390/v13102070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yousaf S, Parvaiz H, Khan S, et al. Prediction of novel bacteriocin from human intestinal microbiome and their growth modeling. Appl Microbiol Biotechnol. 2020;104(9):3869–3884. doi: 10.1007/s00253-020-10493-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simons A, Alhanout K, Duval RE. Bacteriocins, antimicrobial peptides from bacterial origin: overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms. 2020;8(5):639. doi: 10.3390/microorganisms8050639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Casas V, Maloy S. The role of phage in the adaptation of bacteria to new environmental niches. In: Molecular Mechanisms of Microbial Evolution. Springer; 2018:267–306. [Google Scholar]
- 61.Campbell A. Phage evolution and speciation. In: The Bacteriophages. Springer; 1988:1–14. [Google Scholar]
- 62.Berne C, Ducret A, Hardy GG, et al. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria. Microbial Biofilms. 2015;3(4):163–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prüß BM, Liu X, Hendrickson W, et al. FlhD/FlhC-regulated promoters analyzed by gene array and lacZ gene fusions. FEMS Microbiol Lett. 2001;197(1):91–97. doi: 10.1016/S0378-1097(01)00092-1 [DOI] [PubMed] [Google Scholar]
- 64.Makui H, Roig E, Cole ST, et al. Identification of the Escherichia coli K-12 Nramp orthologue (MntH) as a selective divalent metal ion transporter. Mol Microbiol. 2000;35(5):1065–1078. doi: 10.1046/j.1365-2958.2000.01774.x [DOI] [PubMed] [Google Scholar]
- 65.Zheng G, Yan LZ, Vederas JC, et al. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol. 1999;181(23):7346–7355. doi: 10.1128/JB.181.23.7346-7355.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu T, Ramesh A, Ma Z, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat Chem Biol. 2007;3(1):60–68. doi: 10.1038/nchembio844 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The assembled genomes of the blaNDM-1-harboring CREC isolates from this study have been submitted to the NCBI GenBank under BioProject number PRJNA780210 with BioSample numbers SAMN23132882, SAMN23132883, and SAMN23132884.