Graphical abstract

Keywords: Type III polyketide synthase, Heterologous expression, SARS-CoV-2 main protease inhibition, ESKAPE pathogens inhibition, New adipostatins
Abstract
Antibiotic resistance and emerging viral pandemics have posed an urgent need for new anti-infective drugs. By screening our microbial extract library against the main protease of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the notorious ESKAPE pathogens, an active fraction was identified and purified, leading to an initial isolation of adipostatins A (1) and B (2). In order to diversify the chemical structures of adipostatins toward enhanced biological activities, a type III polyketide synthase was identified from the native producer, Streptomyces davawensis DSM101723, and was subsequently expressed in an E. coli host, resulting in the isolation of nine additional adipostatins 3–11, including two new analogs (9 and 11). The structures of 1–11 were established by HRMS, NMR, and chemical derivatization, including using a microgram-scale meta-chloroperoxybenzoic acid epoxidation-MS/MS analysis to unambiguously determine the double bond position in the alkyl chain. The present study discovered SARS-CoV-2 main protease inhibitory activity for the class of adipostatins for the first time. Several of the adipostatins isolated also exhibited antimicrobial activity against selected ESKAPE pathogens.
1. Introduction
Infectious diseases are among of the leading causes of global morbidity and mortality. There has been a resurgence of infectious diseases that once were treatable and controllable, such as infections caused by drug-resistant Gram-negative bacteria [1]. In addition, emerging viral infections have further imposed immense global health challenges [1], as represented by the pandemic of the coronavirus disease in 2019 (COVID-19). Together, these challenges have sounded an alarm on the spread of severe infectious diseases and posed an urgent need for new anti-infective drugs.
Microbial natural products have been a rich source of anti-infective agents and have inspired the development of a wealth of antibiotics used in clinical practice and veterinary medicine [2], [3]. As part of our efforts to discover antimicrobial compounds, especially antiviral agents to cope with the emerging coronavirus, we screened our microbial library comprising over 500 geographically and phylogenetically diverse bacterial and fungal strains. After cultivating these strains in 24-well plate microbioreactors, each culture broth was extracted in situ with 1-butanol and then dried to afford extracts. Two facile and convenient bioassays were selected to initiate the screening of these extracts. First, an inhibitory assay of the main protease, 3CLpro, of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was used, given the essential role of this protease in processing the coronavirus replicase polyproteins and the absence of this target in humans [4], [5]. Second, a broth microdilution antibacterial assay against the notorious ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumonia, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) was used, considering that these strains have the ability to escape or evade common antibiotics through antimicrobial resistance [6].
Through screening, we identified an extract prepared from a commercial soil actinomycete Streptomyces davawensis DSM101723 that decreased the activity of 3CLpro by 90% at the concentration of 20 µg/mL. This extract also inhibited the growth of methicillin-resistant Staphylococcus aureus USA300 at 8 µg/mL. To identify the compounds contributing to these observed activities, we performed a scale-up cultivation of S. davawensis DSM101723 in SPM broth (10 L), followed by a bioactivity-guided fractionation using the same bioassays. Two alkylresorcinols were isolated from the bioactive fraction and were subsequently identified as previously known adipostatins A (1) and B (2) [7], [8] based on HRMS and NMR analyses (Tables S5 and S6, Fig. 4, and Figures S8-S11).
Fig. 4.
Chemical structures of adipostatins 1–11 isolated in this study.
Alkylresorcinols are a widespread class of phenolic lipids predominantly discovered from plants and sometimes found in algae, insects, animals, bacteria, and fungi (Fig. 1 ) [9]. Structurally, they comprise a 1,3-dihydroxylated benzene ring (resorcinol core) and at least an alkane chain (alkyl tail) anchoring at C-5. The structural diversity of alkylresorcinols is attributed to the modifications in resorcinol core (e.g. substitution of phenolic protons) and the alkyl tail (e.g. dehydrogenation and hydroxylation). Due to their amphiphilic properties, alkylresorcinols are involved in certain biological processes (e.g. protecting cellular lipid components from oxidation), and also displayed diverse biological activities, such as antioxidant, antibacterial, cytotoxic, antiparasitic, genotoxic, and signalling properties [9].
Fig. 1.
Examples of representative alkylresorcinols from different sources and their biological activities.
As a subclass of alkylresorcinols, adipostatins have the common resorcinol core and various alkyl tails attached to C-5. Some adipostatins were found to inhibit eukaryotic glycerol-3-phosphate dehydrogenase [10], bacterial coenzyme A biosynthesis [8], and asparaginyl-tRNA synthetase of Brugia malayi parasites [7]. It is noted that the length of the branches and the degree of unsaturation of the alkyl tails have significant but various impacts on the biological activities of this class of compounds. For instance, the inhibitory activity of adipostatins with branched alkyl tails toward E. faecalis is 5 to 9-fold more potent than that of adipostatins with linear tails [8], whereas the trend is opposite for the killing effect toward adult B. malayi worms [7]. This observation inspired us to identify the biosynthetic gene(s) of the alkylresorcinols from S. davawensis DSM101723, aiming to manipulate the biosynthetic gene(s) for production of derivatives with diverse alkyl tails to improve anti-infective activities.
We first analyzed the biosynthesis of alkylresorcinols in S. davawensis DSM101723. Previous work has shown that bacterial adipostatins are assembled by iterative type III polyketide synthases (T3PKSs) that condense a long-chain acyl-CoA starter derived from fatty acid metabolism and three malonyl-CoA extenders, followed by an aldol intramolecular cyclization at C-2/C-7 (Figure S1) [11]. To identify the T3PKS encoding adipostatins A (1) and B (2), we analyzed the published genome sequence [12] of the producing strain S. davawensis DSM101723 by antiSMASH 5.0 [13]. Among the 50 predicted biosynthetic gene clusters, four enzymes, Adp, Pks9, Pks27 and Pks30, showed high sequence similarity of 70%, 51%, 94%, or 95% to the known T3PKSs, AgqA (WP_014447859) from Actinoplanes missouriensis NBRC 102,363 (Table S3) [14], LpmD (ADC96652) from Streptomyces sp. SN-1061 M [15], 1,3,6,8-tetrahydroxynaphthalene synthase (SCO1206) from Streptomyces coelicolor A3(2) [16], and 1,3,6,8-tetrahydroxynaphthalene synthase (BAB91443) from Streptomyces antibioticus IFO13271 [17], respectively, suggesting the potential role of Adp, Pks9, Pks27 and Pks30 as T3PKSs. Thus, we performed phylogenetic analysis of Adp, Pks9, Pks27 and Pks30 with other characterized bacterial T3PKSs which encode 1,3,6,8-tetrahydroxynaphthalene [16], [17], pyrones [15], alkylresorcinols [14], [18], and 3,5-dihydroxyphenylacetic acid [19]. As shown in Fig. 2 , only Adp was clustered with two resorcinol-producing T3PKSs, AgqA [14] and ArsB [18], while Pks9, Pks27 and Pks30 were clustered with T3PKSs known to encode products different from resorcinols. In fact, based on the scores of sequence alignment, Adp exhibited high sequence identity (57%) to AgqA which was known to encode an alkylresorcinol derivative, 6‐alkyl‐4‐O‐dihydrogeranyl‐2‐methoxyhydroquinone [14]. Together, these analyses suggested that adp is highly likely responsible for adipostatin biosynthesis in S. davawensis DSM101723.
Fig. 2.
Phylogenetic analysis of Adp, Pks9, Pks27 and Pks30 with characterized bacterial T3PKSs. Sequences were aligned using ClustalW, and the tree was constructed using the neighbour-joining method. The reliability of the tree was evaluated by bootstrap analysis with 1000 replicates. Scale: number of substitutions per nucleotide.
To test this hypothesis, we first cloned the adp gene from the genomic DNA of S. davawensis DSM101723, and subsequently expressed this gene in the engineered Streptomyces host S. coelicolor M1152, under the control of an ermE promoter. LC-MS data showed that S. coelicolor M1152/adp (Fig. 3 iii; Figure S2) readily produced adipostatins A (1) and B (2) in a higher yield compared to that of the native strain (Fig. 3i; Figure S2), indicating that the adp gene is indeed responsible for the biosynthesis of adipostatin.
Fig. 3.
Comparative LC-MS analysis of the EtOAc extracts from (i) the wild-type strain S. davawensis DSM101723, (ii) S. coelicolor M1152/pCAP01, (iii) S. coelicolor M1152/pLLH104, (iv) E. coli BL21 (DE3)/pACYCDuet-1, and (iv) E. coli BL21 (DE3)/pLLH102. The number labelled above each peak corresponds to individual compound identified in this study shown in Fig. 4. The HPLC chromatograms were monitored at 210 nm. The MS extracted ion chromatograms (EICs) of 1–11 were analyzed and shown in Figure S2.
Considering that different host strains may have distinct genetic capacity to provide different biosynthetic building blocks (e.g. different alkyl tails derived from diverse fatty acid metabolic steps) for adipostatin biosynthesis, we attempted to express the adp gene in engineered E. coli host as a facile and convenient way to diversify the structures of adipostatins toward enhancing their anti-infective activities. To this end, we amplified the adp gene from the genomic DNA of S. davawensis DSM101723 and cloned it into the pACYCDute-1 vector under the control of a T7 promoter to give pLLH102. After transforming the construct into E. coli BL21 (DE3), the resulting strain was cultured and induced by isopropyl-β-d-1-thiogalactopyranoside (IPTG). LC-MS data showed that E. coli BL21 (DE3)/pLLH102 produced nine additional alkylresorcinol analogs (3–11) in addition to adipostatins A (1) and B (2) (Fig. 3v; Figure S2), likely due to the incorporation of distinct biosynthetic building blocks from E. coli in contrast to those from Streptomyces.
To accumulate adequate amounts of alkylresorcinols for structure elucidation and anti-infective evaluation, E. coli BL21 (DE3)/pLLH102 harboring the adp gene was grown in a scaled-up cultivation (6 L). Fractionation of the EtOAc extract on C18 reversed-phase columns led to the purification of nine additional alkylresorcinols (3–11) besides 1 and 2 (Fig. 4 ), including two new adipostatin congeners, namely adipostatins K (9) and L (11), with a C16:1 cis-Δ9 and a 2′-OH C17:1-cis-Δ10 alkyl tails, respectively. The known alkylresorcinols were identified as 5-undecyl-1,3-benzenediol (3), grevillol (4), 5-tetradecyl-1,3-benzenediol (5), virenol B (6), (Z)-5-(trideca-4-enyl)resorcinol (7), bilobol (8), and 5-[(Z)-10-heptadecenyl] resorcinol (10) by detailed NMR analysis (see Supporting Information). Interestingly, compounds 3–8 and 10 had never been discovered from bacteria, although they were previously isolated from multiple plants in the genera of Ardisia, Grevillea, Parathesis, Dendrosenecio, Lithraea, and Ginkgo (Table S4) [20], [21], [22]. In addition, as a common challenge for structure identification of alkylresorcinols, the position and geometric configuration of the double bonds in the alkyl chains of these compounds had not been solidly established. Thus, in this study, the structures of 1–11, including the position and configuration of the double bonds, were unambiguously determined by HRMS, NMR, meta-chloroperoxybenzoic acid (m‐CPBA) epoxidation-MS/MS analysis, and Mosher ester analysis, as described below.
Comparison of the NMR (CD3OD) data for 3–11 (Tables 1 , S5 and S6, Figures S12-S35) with those for adipostatins A (1) and B (2) (Tables S5 and S6, Figures S8-S11) revealed resonances attributed to a 1,3-dihydroxylated benzene ring (resorcinol core) bearing an alkyl chain at C-5. This was further supported by the diagnostic HMBC correlations (Fig. 5 ). HRESI(–)MS analysis of compounds 3–6 returned [M – H]– ions indicative of molecular formulas C17H28O2 (ΔmDa 0.0), C19H32O2 (ΔmDa + 0.4), C20H34O2 (ΔmDa 0.0), and C21H36O3 (ΔmDa 0.0), respectively. All four double bond equivalents (DBE) are attributed to the resorcinol core, indicating that the C-5 substituted alkyl chains in 3–6 are all saturated. Analysis of 1D and 2D NMR data of 3–6 revealed linear alkyl chains as evidenced by the only resonance (δ H 0.90, t, J = 6.7–6.8; δ C 14.4) for one methyl group at the terminus of the alkyl tail. Particularly, HRMS analysis of compound 6 revealed that it possesses one additional oxygen compared to 3–5, suggesting hydroxylation of the alkyl chain, which is in agreement with the presence of a resonance for an oxygenated methine (δ H 3.71, δ C 73.6) in 6. The HMBC correlation from the oxygenated methine H-2′ to C-1′ (δ C 45.3), C-3′ (δ C 37.7) and C-5 (δ C 142.7) unambiguously positioned the hydroxyl group at C-2′. The absolute configuration of C-2′ in 6 was determined as R by esterification of 6 with Mosher’s reagents [(R)-(–)-MTPA-Cl or (S)-(+)-MTPA-Cl], followed by the calculation of 1H chemical shifts of both (S)- and (R)-Mosher esters (Fig. 6 , Figures S36, S38, S39 and Table S7). This conclusion was further validated by the comparison of specific optical rotation value ([α]24.3 D −4.5, c 0.12, CHCl3) of 6 with those of (R)-1-(2-bromo-3,5-dimethoxyphenyl)-2-pentanol ([α]21 D −24.4, c 1.10, CHCl3) [23] and virenol B ([α]25 D −3.7, c 0.52, MeOH) [24], both of which possess 2′-(R)–OH. Thus, the alkyl tails in compounds 3–6 were assigned as C11:0, C13:0, C14:0, and C15:0 2′-(R)–OH, respectively.
Table 1.
1H and 13C NMR (CD3OD) data for new adipostatins 9 and 11 in CD3OD.
| Pos. | Adipostatin K (9) |
Adipostatin L (11) |
||
|---|---|---|---|---|
| δC | δH (mult., J (Hz)) | δC | δH (mult., J (Hz)) | |
| 1 | 159.3 | − | 159.3 | − |
| 2 | 100.9 | 6.07 (t, 2.2) | 101.4 | 6.11 (t, 2.2) |
| 3 | 159.3 | − | 159.3 | − |
| 4 | 107.9 | 6.12 (d, 2.2) | 108.9 | 6.16 (d, 2.2) |
| 5 | 146.3 | − | 142.7 | − |
| 6 | 107.9 | 6.12 (d, 2.2) | 108.9 | 6.16 (d, 2.2) |
| 1′ | 37.0 | 2.44 (t, 7.7) | 45.3 | 2.53 (dd, 13.4, 6.4)2.59 (dd, 13.4,7.0) |
| 2′ | 32.5 | 1.58 (m) | 73.6 | 3.72 (m) |
| 3′ | 30.0–30.8 | 1.25–1.37 | 37.7 | 1.36 (m), 1.50 (m) |
| 4′ | 30.0–30.8 | 1.25–1.37 | 26.8 | 1.34 (m), 1.48 (m) |
| 5′ | 30.0–30.8 | 1.25–1.37 | 30.0–30.9 | 1.25–1.37 |
| 6′ | 30.0–30.8 | 1.25–1.37 | 30.0–30.9 | 1.25–1.37 |
| 7′ | 30.0–30.8 | 1.25–1.37 | 30.0–30.9 | 1.25–1.37 |
| 8′ | 28.1 | 2.03 (m) | 30.0–30.9 | 1.25–1.37 |
| 9′ | 130.8 | 5.34 (t, 4.9) | 28.1 | 2.03 (m) |
| 10′ | 130.8 | 5.34 (t, 4.9) | 130.8 | 5.34 (td, 4.6, 2.2) |
| 11′ | 28.1 | 2.03 (m) | 130.8 | 5.34 (td, 4.6, 2.2) |
| 12′ | 30.0–30.8 | 1.25–1.37 | 28.1 | 2.03 (m) |
| 13′ | 30.0–30.8 | 1.25–1.37 | 30.0–30.9 | 1.25–1.37 |
| 14′ | 32.9 | 1.25–1.37 | 30.0–30.9 | 1.25–1.37 |
| 15′ | 23.7 | 1.25–1.37 | 32.9 | 1.25–1.37 |
| 16′ | 14.4 | 0.91 (t, 6.7) | 23.7 | 1.25–1.37 |
| 17′ | 14.4 | 0.89 (t, 6.7) | ||
Fig. 5.
Diagnostic 2D NMR (CD3OD) correlations for 4, 9 and 11 as representative examples.
Fig. 6.
ΔδS−R [Δ(δS − δR)] values for (S)- and (R)-MTPA esters of 6 and 11. Values of ΔδS−R were presented in ppm. Regions (+and − ) were marked in blue and red, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
HRESI(–)MS analysis of compounds 7–11 returned [M – H]– ions suggestive of molecular formulas C19H30O2 (ΔmDa + 0.3), C21H34O2 (ΔmDa + 0.5), C22H36O2 (ΔmDa + 0.1), C23H38O2 (ΔmDa –0.2), and C23H38O3 (ΔmDa 0.0), requiring five DBEs. Four out of the five DBEs are attributed to the resorcinol core and the remaining one DBE is derived from the alkyl tail. A comparison of the 1H NMR (CD3OD) data for 7–11 (Tables 1 and S5) with those for 1–6 clearly revealed the presence of overlapping signals for two olefinic protons (δ H 5.34, δ C 130.8). Diagnostic 2D NMR correlations (Fig. 5) further confirmed the C-5 substitution of resorcinol moiety by monounsaturated alkyl chains. The configuration of each olefinic double bond in the alkyl chains of 7–11 was individually assigned as (Z) on the basis of the diagnostic chemical-shift values of the allylic 13C NMR signals observed around 28 ppm. In contrast, the (E) configuration of an olefin located at a long alkyl chain was reported to show allylic 13C NMR signals around 33 ppm [25], [26], [27]. In addition, this assignment is consistent with the biosynthesis of alkylresorcinols, as the C-5 alkyl chain is derived from fatty acids metabolism which predominantly produces (Z)-unsaturated fatty acids in E. coli. Another challenge was to determine the localization of the olefinic double bonds in 7–11. We recently developed an innovative analytical method for identification of double bond position in physiologically relevant lipids of primary metabolism [28]. This method uses meta-chloroperoxybenzoic acid (m‐CPBA) to epoxidize olefins at microgram scale, followed by fragmentation analysis in collision-induced dissociation (CID)-MS/MS. The diagnostic fragment ion pair with 16 Da mass difference indicates the localization of double bonds. Thus, we applied this method to the structure elucidation of 7–11 and successfully assigned the cis-double bonds at C-4′, C-8′, C-9′, C-10′, and C-10′ for compounds 7–11, respectively (Fig. 7 , Figures S3–S7). This was the first demonstration of using this novel method to determine the position of double bonds in secondary metabolites, suggesting its great potential as a facile and convenient method that is performed only at microgram scale, especially suitable for natural product research due to the low yield of these compounds. The last step to complete the structure elucidation of 7–11 was to determine the absolute configuration of the hydroxy group of 11. Similar to 6, the alkyl chain in compound 11 was hydroxylated at C-2′, as evidenced by its molecular formula (C23H38O3), downfield shift of δ C-2 ′ (Δ + 39.8 ppm), and HMBC correlations from H-1′ to C-2′ (δ C 73.6), C-3′ (δ C 37.7) and C-4/C-6 (δ C 108.9) (Fig. 5). The negative sign of the optical rotation of 11 ([α]24.3 D −11.5, CHCl3) suggested 2′-OH to be R configuration which is identical to 6. This assignment was further confirmed by Mosher ester analysis (Fig. 6, Figures S37, S40, S41 and Table S8). Altogether, the alkyl tails in compounds 7–11 were unambiguously determined to be C13:1 (Δ4′), C15:1 (Δ8′), C16:1 (Δ9′), C17:1 (Δ10′), and C17:1 [Δ10′, 2′-(R)–OH], respectively.
Fig. 7.
A microgram-scale meta-chloroperoxybenzoic acid epoxidation-MS/MS analysis for unambiguous determination of the double bond position for 7–11 (here uses 9 as an example).
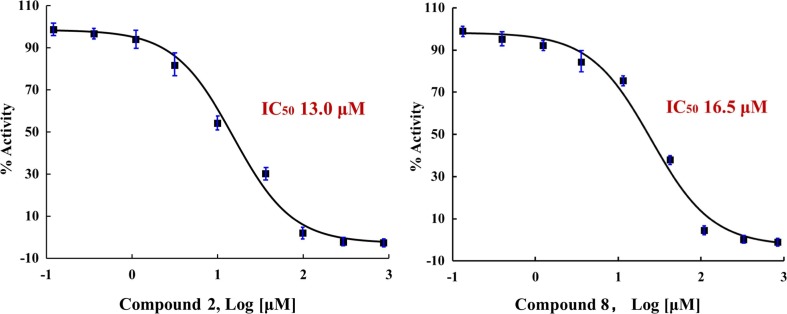
We next evaluated the anti-infective activities of compounds 1–11 using in vitro bioassays. In the SARS-CoV-2 3CLpro inhibition assay, 2 and 8 displayed dose-dependent inhibitory activity against this main protease of SARS-CoV-2, with IC50 values of 13.0 µM and 16.5 µM, respectively (Fig. 8 ). Neither 2 nor 8 showed any cytotoxicity using a HeLa cell line at a concentration of up to 200 µM. The lack of cytotoxicity warrants further investigation of these compounds as inhibitors of SARS-CoV-2 replication. Taking advantage of the recently reported crystal structure of SARS-CoV-2 3CLpro main protease, we performed a molecular docking to explore the potential binding of 2 and 8 to 3CLpro. A blind docking study was performed alongside a targeted docking to the literature reported inhibitor binding site [5], [29]. Interestingly, while 2 and 8 showed moderate affinity to the literature reported inhibitor binding site, these two compounds showed higher affinity to the central cavity formed between the two monomers of 3CLpro (Figure S42), suggesting that 2 and 8 may potentially exert 3CLpro inhibitory activity other than only binding to the literature reported inhibitor binding site. From the docking study, we also noticed that there was a binding difference in the hydrophobic tail region between 2 and 8, as the geometry of the C8′-C9′ double bond of 8 seemed to decrease hydrophobic interactions between 3CLpro and the region of C4′ to C10′ in 8, which may support the difference of the 3CLpro inhibitory activity between 2 and 8. Furthermore, we observed that the length, presence of an isopropyl group, and double bond geometry of the aliphatic chains of the compounds also affected the binding pose as a whole, which therefore affected the interactions of the 1,3-diphenol head with 3CLpro. For example, the C1 and C3 hydroxy groups of 2 were shown to interact with Ala285 of the 3CLpro chain A, and with Ala285, Gly283, Leu286, and Ser284 of the 3CLpro chain B. In contrast, these two hydroxy groups of 8 were shown to interact only with Lys5 of the 3CLpro chain A, and with Arg4 and Lys5 of the 3CLpro chain B. These differential interactions may also contribute to the observed differences of the 3CLpro inhibitory activity for 2 and 8. We also performed the same docking analysis for other adipostatins that did not show obvious 3CLpro inhibition in our assay. Although these compounds may possess certain binding affinity to the same pocket as that of 2 and 8, we also observed that these compounds were shown to interact with different amino acid residues of 3CLpro. For example, 3 exhibited a different binding pose compared to 2 and 8 (Figure S42), due to the significantly shorter aliphatic chain of 3. It is notable that while the docking study may help explain the observed different 3CLpro inhibitory activities of the compounds, further experiments are needed to validate the docking analysis.
Fig. 8.
Dose-dependent inhibition curves of 2 and 8 against 3CLpro of SARS-CoV-2.
Regarding antibacterial activity, intriguingly, most of our adipostatins inhibited the growth of Gram-positive ESKAPE pathogens in the broth microdilution assay, with the minimum inhibitory concentrations (MICs) ranging from 1 to 64 µg/mL (Table 2 ). Based on our results, adipostatins with shorter alkyl chains (e.g. 3, 4 and 7) generally displayed stronger inhibitory activities (MICs 1–8 µg/mL) against B. subtilis 168, methicillin-resistant S. aureus, and vancomycin-resistant E. faecium. However, 11 also exhibited moderate inhibitory activity against these three Gram-positive strains despite having the longest alkyl chain, suggesting that the activity is likely due to hydroxylation at its alkyl chain. None of the adipostatins showed significant inhibitory activity against Gram-negative pathogens, such as Klebsiella pneumoniae and Pseudomonas aeruginosa, with the exception of 3, which inhibited the growth of K. pneumoniae with an MIC value of 8 µg/mL. Structurally, 3 has the shortest alkyl chain among all the 11 adipostatins tested, suggesting that the length of the alkyl chain may play a role in the activity of adipostatins against K. pneumonia ATCC 10031. Altogether, these biological activity data suggested that the length, degree of unsaturation, and hydroxylation of the lipid chain of these adipostatins may have a variety of effects on the SARS-CoV-2 3CLpro inhibitory activity and antibacterial activity against ESKAPE pathogens. This provides both motivation and opportunity to perform follow-up studies on structure–activity relationship and carry out subsequent mechanism of action study for adipostatins.
Table 2.
SARS-CoV-2 3CLpro inhibitory activity and antibacterial activity of adipostatins 1–11.
| Compound | MIC (µg/mL) |
IC50 (µM) | |||||
|---|---|---|---|---|---|---|---|
| S. aureus USA300 | B. subtilis168 | E. faecium EF16 | S. epidermidis ATCC 35984 | P. aeruginosa PAO1 | K. pneumoniaATCC 10031 | 3CLpro | |
| 1 | > 64 | 64 | 64 | > 64 | > 64 | > 64 | > 100 |
| 2 | > 64 | 64 | 32 | > 64 | > 64 | > 64 | 13.0 |
| 3 | 4 | 1 | 4 | 2 | > 64 | 8 | > 100 |
| 4 | > 64 | 8 | 8 | > 64 | > 64 | > 64 | > 100 |
| 5 | > 64 | > 64 | 32 | > 64 | > 64 | > 64 | > 100 |
| 6 | 32 | 16 | 16 | 32 | > 64 | > 64 | > 100 |
| 7 | 4 | 1 | 4 | 4 | > 64 | > 64 | > 100 |
| 8 | 64 | 32 | 2 | 64 | > 64 | > 64 | 16.5 |
| 9 | > 64 | > 64 | > 64 | > 64 | > 64 | > 64 | > 100 |
| 10 | > 64 | > 64 | 64 | > 64 | > 64 | > 64 | > 100 |
| 11 | 8 | 8 | 8 | > 64 | > 64 | > 64 | > 100 |
| Daptomycina | 0.5 | 0.25 | 0.25 | 1 | > 64 | > 64 | NAb |
| GC376c | NAb | NAb | NAb | NAb | NAb | NAb | 0.68 |
Positive control for antibacterial assay.
Not tested.
Positive control for SARS-CoV-2 3CLpro inhibitory assay. The significant values were highlighted with light pink background.
In summary, to address drug-resistant bacterial infections and emerging viral pandemics, we screened a microbial library comprising over 500 diverse bacterial and fungal strains, aiming to discover anti-infective natural products as lead compounds. Through bioactivity-guided isolation and heterologous expression of a type III polyketide synthase, adipostatins 1–11 were purified and some of them exhibited in vitro activities against SARS-CoV-2 main protease (3CLpro) and drug-resistant ESKAPE strains. Notably, a recently developed method involving meta-chloroperoxybenzoic acid-induced epoxidation coupled with CID-MS/MS analysis was employed here at microgram scale, enabling unambiguous determination of the double bond position of the isolated adipostatins. Furthermore, it is notable that the wild-type strain affording the initial active extract, S. davawensis DSM101723, only produced two major adipostatins A (1) and B (2), while heterologous expression of the responsible biosynthetic gene in E. coli led to isolation of nine additional adipostatins 3–11, including two new analogs (9 and 11), which provided an economic and convenient means of diversifying the structures of adipostatins toward enhanced anti-infective activities.
2. Experimental section
2.1. General experimental procedures
UV–visible spectra were obtained on a Jasco V-730 UV–visible spectrophotometer with 1 cm quartz cells. Nuclear magnetic resonance (NMR) spectra were acquired on either a Bruker Avance III HD 400 MHz spectrometer with a 5 mm BBO 1H/19F-BB-Z-Gradient prodigy cryoprobe or a Bruker Avance III HD 500 MHz spectrometer with a PA BBO 500S2 BBF-H-D_05 Z SP probe. High-resolution ESIMS spectra were obtained on a Thermo Scientific Orbitrap Velos Pro hybrid ion trap-orbitrap mass spectrometer by direct injection. LC-DAD-MS analysis was performed on a Thermo Dionex Ultimate 3000 UHPLC system equipped with a diode array multiple wavelength detector and an LTQ XL linear ion trap mass spectrometer. Semi-preparative HPLC separation was carried out on a Thermo Dionex Ultimate 3000 HPLC system equipped with a PDA detector. Methanol‑d 4 used for NMR experiments was purchased from ACROS Organics. Other reagents and solvents used in this study were purchased from Fisher Scientific and Sigma-Aldrich.
2.2. Bioactivity screening procedures
A collection of taxonomically diverse bacterial and fungal strains (>500 isolates) was isolated from marine animals and sediments, medicinal plants, and rainforests soils collected from various places in the United States (e.g. San Diego Bay, Beaches at Charleston, Riverbanks Botanic Garden, and Congaree National Park) and other countries (e.g. Great Barrier Reef region in Australia). These strains were grown under various culture media (e.g. ISP-2, A1, DSM67, and PYG) using a high-throughput 24-well plate microbioreactor. For the strains that did not yield enough amounts of extracts due to the culture volume of the microbioreactor, a 50 mL scaled-up culture in 250 mL baffled flasks was performed. Each broth from the microbioreactor or the baffled flasks was extracted using butanol, transferred to 24-well deep well plates, and dried under vacuum to assemble a library with ~ 1,000 microbial extracts. Each extract was prepared as a stock solution in DMSO at a concentration of 10 mg/mL for screening. Subsequently, two facile and convenient bioassays were used for the screening of these extracts. First, a high throughput SARS-CoV-2 3CLpro inhibitory assay was performed (see 3CLpro Protease Inhibition Assay below for detailed information on the testing method). The final testing concentration of the extracts was 50 µg/mL with less than 1% DMSO, tested in triplicate using 384-well assay plates. The extracts with an activity of > 50% 3CLpro inhibition were considered as being active for further investigation. Second, the extracts that inhibited 3CLpro were further screened for their antibacterial activity against two indicator strains Staphylococcus aureus USA300 and P. aeruginosa PAO1, using a broth microdilution assay with 96-well microtiter plates. The final concentration of each extract in a well was 64 µg/mL. After incubation for overnight at 37 °C, the optical density of each well was measured at 600 nm and the percentage viability of bacterial cells was calculated. The extracts that inhibited > 50% bacterial growth were considered as hits.
2.3. Strains and plasmids
All strains and plasmids used in this study are listed in Table S1. Escherichia coli DH5α was used as the host for general subcloning. Escherichia coli BL21 (DE3) was used for adipostatin expression. Escherichia coli ET12567/pUZ8002 was used as the cosmid donor host for E. coli-Streptomyces intergeneric conjugation. Streptomyces davawensis DSM101723 was purchased from DSMZ. Streptomyces coelicolor M1152 were used as the host strain for heterologous expression. Plasmid extractions and DNA purifications were carried out using standardized commercial kits (OMEGA, Bio-Tek). Oligonucleotide synthesis and DNA sequencing were performed by Eton Bioscience (North Carolina, USA). PCR amplification was carried out with primers listed in Table S2 using PrimeSTAR HS DNA polymerase (Takara Bio). DNA assembly was conducted using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs).
2.4. Bacterial growth conditions
E. coli strains were routinely cultured in Luria–Bertani (LB; Becton, Dickinson and Company) liquid medium at 37 °C, 200 rpm, or on LB agar plate at 37 °C, with appropriate antibiotic selection. S. davawensis DSM101723 was recovered from 30% glycerol at 30 °C on R2YE medium (5% sucrose, 0.02% potassium sulfate, 1% magnesium chloride, 1% glucose, 0.5% yeast extract, and 0.01% Difco casamino acid), and cultured in Tryptic Soy Broth (TSB; Becton, Dickinson and Company) medium for genomic DNA preparation. S. coelicolor M1152 was grown at 30 °C on MS medium (3% soya flour, 2% mannitol, and 2% agar) for sporulation and conjugation. The S. coelicolor M1152 conjugants were fermented in SPM medium (1% soytone, 1% soluble starch, 2% d-maltose, and 5 mL of trace elements per liter) for compound production.
Bioinformatic Analysis of Type III PKS Gene adp. The type III PKS gene adp was identified from the S. davawensis DSM101723 genome using the antiSMASH 5.0 online program [13]. The evolutionary history was inferred using the Neighbour-Joining method [30]. Phylogenetic tree was built using MEGA5 [31].
Cloning of Type III PKS Gene adp. The primers used for cloning are summarized in Table S2. Briefly, the adp gene was PCR amplified from the genome of S. davawensis DSM101723 using the primers pACYCDute-1-adp-Fwd and pACYCDute-1-adp-Rev. After BamHI/AflII digestion of pACYCDute-1 vector, the PCR product was cloned into the linearized vector by Gibson assembly to give pLLH102. The pLLH102 construct was confirmed by Sanger sequencing and then transformed into E. coli BL21 (DE3) for overexpression. The construct pLLH104 used for gene expression in Streptomyces host was also constructed by Gibson assembly. Briefly, the adp gene and promoter PermE were amplified from the genome of S. davawensis DSM101723 and the plasmid pMXT19 [32] using primer pairs of pCAP01-Perm-Fwd/pCAP01-Perm-Rev and pCAP01-adp-Fwd/pCAP01-adp-Rev (Table S2), respectively. The PCR products, adp and PermE promotor, were cloned into SpeI/XhoI-digested pCAP01 vector by Gibson assembly to give pLLH104. Subsequently, pLLH104 was transformed into E. coli ET12567/PUZ8002 and then transferred to S. coelicolor M1152 via intergeneric conjugation. The empty vector pCAP01 was performed in the same way as the negative control.
Expression of Type III PKS Gene adp. S. coelicolor M1152/pLLH104 and pCAP01 conjugants were inoculated into 25 mL of SPM media (1% soytone, 1% soluble starch, 2% d-maltose, and 5 mL of trace elements per liter) in 125 mL flasks and cultivated at 30 °C, 220 rpm for 7 d. Cells were harvested and disrupted by sonication at room temperature. The lysates were extracted twice with an equal volume of EtOAc. The organic phase was evaporated and then resuspended in MeOH (100 µL) for LC-MS analysis (XBridge BEH-C18 column, 2.1 × 100 mm, 2.5 μm, 0.5 mL/min isocratic elution at 30% H2O/MeCN over 15 min, with 0.1% formic acid; UV 280 nm). In the meanwhile, E. coli BL21 (DE3) strain transformed with pLLH102 was cultivated at 37 °C in LB broth supplemented with 50 µg/mL chloramphenicol for 4–5 h as the seed culture. 250 µL of each seed culture was transferred into 25 mL of fresh LB supplemented with chloramphenicol (50 µg/mL). After cultivation at 37 °C to an OD600 of 0.4 to 0.6, 200 μM of IPTG was added into the culture broth and grown for additional 12–14 h at 23 °C, 220 rpm. Cells were extracted and prepared for LC-MS analysis, using the same method as described above for S. coelicolor M1152/pLLH104 and pCAP01.
Solvent Extraction and Purification of Adipostatins.S. davawensis DSM101723 culture (10 L) in SPM broth were harvested and extracted as above. The extract (3 g) was further fractionated on a reversed-phase C18 open column with a 10% stepwise gradient elution from 90% H2O/MeOH to MeOH. The bioactive fractions 9 and 10 were combined and subjected to Sephadex LH-20 fractionation eluting with MeOH to afford 20 subfractions. The combined bioactive subfractions 12–15 (41.4 mg) were further fractionated on semi-preparative HPLC (Waters XBridge BEH C18 OBD Prep column, 150 × 10 mm, 5 µm, 130 Å, 3.5 mL/min isocratic elution at 31% H2O/MeOH over 30 min with constant 0.1% formic acid, monitored at 280 nm) to afford adipostatins A (1) (t R = 9.0 min, 4.1 mg) and B (2) (t R = 7.9 min, 5.5 mg). The cells of E. coli BL21 (DE3)/pLLH102 in LB broth (6 L) were harvested and extracted using EtOAc to afford the extract (659.0 mg), which was fractionated on reversed-phase C18 open column and Sephadex LH-20 by following the same procedure. The combined subfractions (134.1 mg) were further fractionated on semi-preparative HPLC (Waters XBridge BEH C18 OBD Prep column, 150 × 10 mm, 5 µm, 130 Å, 3.5 mL/min isocratic elution at 31% H2O/MeOH over 30 min with constant 0.1% formic acid, monitored at 280 nm) to yield adipostatin A (1) (t R = 9.0 min, 5.7 mg), 5-undecyl-1,3-benzenediol (3) (t R = 2.3 min, 1.3 mg), grevillol (4) (t R = 4.3 min, 2.6 mg), 5-tetradecyl-1,3-benzenediol (5) (t R = 6.2 min, 0.7 mg), virenol B (6) (t R = 2.8 min, 1.6 mg), (Z)-5-(trideca-4-enyl)resorcinol (7) (t R = 2.6 min, 3.0 mg), bilobol (8) (t R = 5.0 min, 10.0 mg), adipostatin K (9) (t R = 8.2 min, 3.0 mg), 5-[(Z)-10-heptadecenyl] resorcinol (10) (t R = 10.3 min, 4.8 mg), and adipostatin L (11) (t R = 3.1 min, 3.0 mg).
Adipostatin K(9). White powder; UV–vis (CH3OH) λmax (log ε) 276 (2.98) nm; 1D and 2D NMR (500 MHz, CD3OD) data, see Table 1 and Figures S24-S28; HR-ESI (–) MS m/z 331.2642 [M – H]– (calcd for C22H35O2, 331.2643).
Adipostatin L(11). White powders; [α]24.3 D −11.5 (c 0.1, CHCl3); UV–vis (CH3OH) λmax (log ε) 276 (3.04) nm; 1D and 2D NMR (500 MHz, CD3OD) data, see Table 1 and Figures S31-S35; HR-ESI (–) MS m/z 361.2748 [M – H]– (calcd for C23H37O3, 361.2748).
m‐CPBA Epoxidation-MS/MS Analysis[28]. Each compound (10 µg) was mixed with 10 μL dichloromethane (DCM) containing 10 μg/μL meta-chloroperoxybenzoic acid (m-CPBA). The reaction was incubated at room temperature for 10 min. After incubation, 500 μL of 50% ACN was added to quench and dilute the reaction system. Diluted sample was used for direct infusion analysis by collision-induced dissociation (CID)-MS/MS.
Modified Mosher Ester Analysis. Compounds 6 and 11 (1 mg) were individually dissolved in pyridine (0.5 mL) in a 4 mL vial, and (R)-(–)-α-methoxy-α-(trifluoromethyl)-phenylacetyl chloride [(R)-MTPA-Cl] was added to the reaction vial. After 16 h at room temperature, the reaction mixture was dried under N2 and then redissolved in MeOH for the purification of (S)-MTPA ester using semi-preparative HPLC (Waters XBridge BEH C18 OBD Prep column, 150 × 10 mm, 5 µm, 130 Å, 3.5 mL/min isocratic elution at 31% H2O/MeOH over 30 min with constant 0.1% formic acid). Tris-(R)-MTPA esters of 6 and 11 were obtained by the entirely analogous method described above, using (S)-(+)-α-methoxy-α-(trifluoromethyl)-phenylacetyl chloride [(S)-MTPA-Cl]. The purified (S)- or (R)-MTPA esters of 6 and 11 were analyzed by 1H NMR, 1H/1H COSY and 1H/13C HSQC experiments in deuterated methanol.
3CLpro Protease Inhibition Assay[4], [5]. 3CLpro protease inhibition assay was measured by 3CL Protease (SARS-CoV-2) Assay Kit which was ordered from BPS Bioscience. Briefly, 3CL protease was diluted by assay buffer (with 1 mM DTT) at 10 ng/µL. The compounds to be tested were dissolved in DMSO to give a stock concentration of 10 mM. Then compounds were diluted with 3CL protease assay buffer (with 1 mM DTT) to give a series of concentration from 0.69 µM to 1.5 mM. Mixed 5 µL each dilution and 15 µL 3CL protease together in a 384-well microtiter plate and preincubated 30 min at room temperature with slow shaking. Then added 5 µL substrate solution to each well to start reaction. Incubated at room temperature for 4 h. Sealed the plate with the plate sealer. Measured the fluorescence intensity in a microtiter plate-reading fluorimeter capable of excitation at a wavelength of 360 nm and detection of emission at a wavelength of 460 nm. 3CL inhibitor GC376 was used as a positive control. The final concentration of DMSO in the assay did not exceed 1%. The GraphPad Prism 6.0 software (GraphPad) was used for the calculation of the IC50 values. All samples and controls were tested in triplicate and were presented as the mean ± SD.
Molecular Docking of Adipostatins against 3CLpro. Compounds identified in this work were subjected to molecular docking against the crystal structure of the SARS-CoV-2 3CLpro (PDB entry: 6Y2E) [31] retrieved from the RCSB Protein Data Bank (PDB) in .pdb format. To determine molecular binding modes, a blind docking study was used alongside a targeted docking to the literature reported inhibitor binding site [31], [32]. The compounds were converted to three dimensional ligands with Chem3D and saved in .mol2 format. Then, the AutoDockTools (ADT) suite [33] was used to generate .PDBQT files of the ligands and protein receptor. Docking studies were performed using AutoDock Vina (version 1.1.2) [34] with two sets of parameters. The blind study used exhaustiveness 100, number of binding modes 10, and energy range 5 with a docking box of 80 × 80 × 90 Å (x, y, z) centered at −4.0, −26.0, 22.0 (x, y, z). The targeted docking study used identical parameters with a docking box of 24 × 24 × 24 Å centered at 6.8, −25.5, 42.8. For each compound, we identified 10 binding poses. Binding poses were evaluated by docking score as determined by the highly effective scoring function of AutoDock Vina and the single best binding pose of each compound was selected for subsequent analysis. Docking results were visualized using PyMOL [35] and ligand receptor interactions were calculated and collected using a combination of PyMOL functions and custom python scripts.
Broth Microdilution Antibacterial Assay. Antibacterial activities were measured against Staphylococcus aureus USA300 (methicillin-resistant), Bacillus subtilis 168, Enterococcus faecium EF16 (vancomycin-resistant), Staphylococcus epidermidis ATCC 35984 (methicillin-resistant), Pseudomonas aeruginosa PAO1 (chloramphenicol-resistant), and Klebsiella pneumoniae ATCC 10031 by the broth microdilution method [36]. The bacterial strain to be tested was streaked onto a LB agar plate and was incubated at 37 °C for 24 h. Single colony was then transferred into fresh LB broth and incubated for 37 °C for 24 h. Then the cell was diluted 5000-fold using LB media. The compounds to be tested were dissolved in DMSO and diluted with sterile LB broth to give the concentrations ranging from 1.25 to 640 µg/mL. An aliquot (5 µL) of each dilution was transferred to a 96-well microtiter plate and freshly prepared microbial broth (45 µL) was added to each well. The plates were incubated at 37 °C overnight, and MICs were determined visually, as the lowest concentration showing no visible growth. Daptomycin was used as a positive control. All samples and controls were tested in triplicate.
3. Notes
The authors declare no competing financial interest.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work is partially funded by a National Institutes of Health (NIH) grant P20GM103641 and a National Science Foundation EPSCoR Program OIA-1655740. X. T. acknowledges a partial support from Shenzhen Bay Laboratory Startup Funds (21230051). L. L. acknowledges support by NIH grants R01DK071801 and S10RR029531. We thank Dr. Perry J. Pellechia and Miss Toni Johnson from UofSC NMR Facility for help with acquiring NMR data, as well as Dr. Michael D. Walla and Dr. William E. Cotham from UofSC Mass Spectrometry Facility for acquiring HRMS data. We thank Dr. Bradley S. Moore (University of California, San Diego) and Dr. Mervyn Bibb (John Innes Centre, UK) for providing pCAP01 and S. coelicolor M1152, respectively.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioorg.2021.104925.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Jones K.E., Patel N.G., Levy M.A., Storeygard A., Balk D., Gittleman J.L., Daszak P. Nature. 2008;451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lam K.S. Trends Microbiol. 2007;15:279–289. doi: 10.1016/j.tim.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Newman D.J., Cragg G.M. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 4.Morse J.S., Lalonde T., Xu S.Q., Liu W.R. ChemBioChem. 2020;21:730–738. doi: 10.1002/cbic.202000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang L.L., Lin D.Z., Sun X.Y.Y., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Oliveira D.M., Forde B.M., Kidd T.J., Harris P.N., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Clin. Microbiol. Rev. 2020;33:e00181–19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rateb M.E., Yang D., Vodanovic-Jankovic S., Yu Z.G., Kron M.A., Shen B. J. Antibiot. 2015;68:540–542. doi: 10.1038/ja.2015.22. [DOI] [PubMed] [Google Scholar]
- 8.Gómez-Rodríguez L., Schultz P.J., Tamayo-Castillo G., Dotson G.D., Sherman D.H., Tripathi A. Tetrahedron Lett. 2020;61 doi: 10.1016/j.tetlet.2019.151469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozubek A., Tyman J.H. Chem. Rev. 1999;99:1–26. doi: 10.1021/cr970464o. [DOI] [PubMed] [Google Scholar]
- 10.Tsuge N., Mizokami M., Imai S., Shimazu A., Seto H. J. Antibiot. 1992;45:886–891. doi: 10.7164/antibiotics.45.886. [DOI] [PubMed] [Google Scholar]
- 11.Martins T.P., Rouger C., Glasser N.R., Freitas S., de Fraissinette N.B., Balskus E.P., Tasdemir D., Leão P.N. Nat. Prod. Rep. 2019;36:1437–1461. doi: 10.1039/c8np00080h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jankowitsch F., Schwarz J., Ruckert C., Gust B., Szczepanowski R., Blom J., Pelzer S., Kalinowski J., Mack M. J. Bacteriol. 2012;194:6818–6827. doi: 10.1128/JB.01592-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blin K., Shaw S., Steinke K., Villebro R., Ziemert N., Lee S.Y., Medema M.H., Weber T. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Awakawa T., Fujita N., Hayakawa M., Ohnishi Y., Horinouchi S. ChemBioChem. 2011;12:439–448. doi: 10.1002/cbic.201000628. [DOI] [PubMed] [Google Scholar]
- 15.Kaysser L., Siebenberg S., Kammerer B., Gust B. ChemBioChem. 2010;11:191–196. doi: 10.1002/cbic.200900637. [DOI] [PubMed] [Google Scholar]
- 16.Izumikawa M., Shipley P.R., Hopke J.N., Thomas O., Xiang L.K., Noel J.P., Moore B.S. J. Ind. Microbiol. Biotechnol. 2003;30:510–515. doi: 10.1007/s10295-003-0075-8. [DOI] [PubMed] [Google Scholar]
- 17.Funa N., Ohnishi Y., Ebizuka Y., Horinouchi S. Biochem. J. 2002;367:781–789. doi: 10.1042/BJ20020953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funa N., Ozawa H., Hirata A., Horinouchi S. Proc. Natl. Acad. Sci. 2006;103:6356–6361. doi: 10.1073/pnas.0511227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfeifer V., Nicholson G.J., Ries J., Recktenwald J., Schefer A.B., Shawky R.M., Schröder J., Wohlleben W., Pelzer S. J. Biol. Chem. 2001;276:38370–38377. doi: 10.1074/jbc.M106580200. [DOI] [PubMed] [Google Scholar]
- 20.Valcic S., Wächter G.A., Eppler C.M., Timmermann B.N. J. Nat. Prod. 2002;65:1270–1273. doi: 10.1021/np020068n. [DOI] [PubMed] [Google Scholar]
- 21.Liu H.W., Zhao F., Yang R.Y., Wang M.Y., Zheng M.Q., Zhao Y.S., Zhang X., Qiu F., Wang H.S. Phytochemistry. 2009;70:773–778. doi: 10.1016/j.phytochem.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Lin B.B., Wang G.K., Wang Q., Ge C.Y., Qin M.J. Fitoterapia. 2011;82:1137–1139. doi: 10.1016/j.fitote.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 23.Uchida K., Watanabe H., Kitahara T. Tetrahedron Lett. 1998;54:8975–8984. [Google Scholar]
- 24.Chang H.S., Lin Y.J., Lee S.J., Yang C.W., Lin W.Y., Tsai I.L., Chen I.S. Phytochemistry. 2009;70:2064–2071. doi: 10.1016/j.phytochem.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 25.Wu Z.W., Li Y., Liu D., Ma M., Chen J.L., Lin W.H. Chem. Biodivers. 2017;14 doi: 10.1002/cbdv.201700059. [DOI] [PubMed] [Google Scholar]
- 26.De Haan J.W., Van de Ven L.J.M. Org. Magn. Reson. 1973;5:147–153. [Google Scholar]
- 27.T.A. Alvarenga, P.F. de Oliveira, J.M. de Souza, D.C. Tavares, M.L. Andrade e Silva, W.R. Cunha, M. Groppo, A.H. Januário, L.G. Magalhães, P.M. Pauletti, J. Agric. Food Chem. 64 (2016) 8821–8827. [DOI] [PubMed]
- 28.Feng Y., Chen B.M., Yu Q.Y., Li L.J. Anal. Chem. 2019;91:1791–1795. doi: 10.1021/acs.analchem.8b04905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosconati S., Forli S., Perryman A.L., Harris R., Goodsell D.S., Olson A.J. Expert Opin. Drug Discov. 2010;5:597–607. doi: 10.1517/17460441.2010.484460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saitou N., Nei M. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 31.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang X.Y., Li J., Millan-Aguinaga N., Zhang J.J., O'Neill E.C., Ugalde J.A., Jensen P.R., Mantovani S.M., Moore B.S. ACS Chem. Biol. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trott O., Olson A.J. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
- 36.Stalons D.R., Thornsberry C. Antimicrob. Agents Chemother. 1975;7:15–21. doi: 10.1128/aac.7.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.