Abstract
Secreted proteins are critical for the coordination of potent immune defenses, such as in engineered T cell therapies, however, there are few widely accessible approaches to accurately analyze and sort large numbers of cells based on their secretory functions. We report a workflow for the rapid screening and sorting of single individual T cells based on IL-2 secretion accumulated at high concentrations in nanoliter droplets and encoded back onto the secreting cell’s surface. In our method, droplets are used solely to partition cells, enabling rapid accumulation of signals onto cell surfaces, and eliminating diffusive crosstalk between neighbors. All downstream sorting leverages conventional high-throughput and readily accessible flow cytometry after the emulsion is disrupted. We achieve monodisperse droplet generation (CV<10%) at flow rates up to 200 μL/min using step emulsification, enabling processing of entire libraries of cells within tens of minutes without significant secretion crosstalk. In comparison to our approach, strong mitogenic activation overwhelmed the conventional bulk on-cell cytokine assay, rendering labeled, non-activated cells indistinguishable from actively secreting neighbors within one hour. Processing of identical cell mixtures following droplet encapsulation yielded no apparent crosstalk even after three hours. Instead, IL-2 production spanning several orders of magnitude was observed from roughly 20% of analyzed activated lymphocytes, representing an at least 10-fold increase in dynamic range compared to unencapsulated cells. Secreting cells could also be sorted using fluorescence activated cell sorting (FACS). The approach can ultimately enable sorting of cells based on functional properties with higher accuracy in a more accessible format to life science researchers.
Introduction
Direct modulation of T-lymphocyte biology, either genetically through the expression of chimeric receptors, or physically via the addition of antibodies against inhibitory checkpoint proteins, has demonstrated profound efficacy towards the treatment of systemic hematological malignancies1. These novel treatments, termed immunotherapies, refine the immune system’s ability to differentiate between self and nonself, enabling natural biological defense mechanisms to identify and eradicate otherwise intractable tumors. Unfortunately, broad translation of such approaches is currently limited, resulting in tremendous variation in clinical outcomes both between patients and amongst sub-types of cancers treated2–7. One barrier to the design of more effective personalized immunotherapies, stems from an incomplete characterization of the phenotypic traits which enable T cells to excel at policing the body, including factors that affect proliferative potential, cell stemness, and aggressiveness upon stimulation by their cognate antigen8–11.
Currently immunophenotyping approaches primarily focus on analyzing cells through labeled surface antigens. While these methods yield insights into the primary role of cells (e.g. Helper T cells, Cytotoxic T cells etc.) they reveal little about their metabolic activity or propensity to respond to immunogenic challenges12,13. A complimentary set of phenotypic information can be found by examining the diverse array of proteins secreted by T cells. These cytokines and chemokines directly coordinate the complex set of behaviors necessary for proper immune responses, including recruiting cells to the area of interest, stimulating them to induce clearance of pathogens, and restricting activity once the targets have been cleared. Cytokine production is also directly translatable to a variety of immunotherapeutic approaches. High concentrations of interferon-α and interleukin-2 (IL-2) have been administered directly to patients to respectively suppress tumor proliferation or induce T cell expansion14,15. Armored CAR T-Cells have been designed which constitutively express pro-inflammatory cytokines that can counteract immunosuppressive microenvironments typically associated with solid tumors16,17. Even more recently, engineered synthetic-notch receptors have enabled construction of user defined custom T cell secretion programs, facilitating targeted production of therapeutic molecules which may be too toxic to apply systemically18. While these synthetic biology approaches to cytokine control are beneficial, they add significant complexity to the manufacture of therapeutic T cell products, which is not ideal.
Interestingly, single-cell studies have alluded to the presence of a small fraction of otherwise indistinguishable polyfunctional T cells which secrete many cytokines simultaneously and in excess, and whose presence in pre-infusion products correlates well with clinical outcomes12,13,19. Therefore the development of profiling technologies which can screen and sort T cells based on functional behavior directly, by quantifying protein production from individual clones, could aid therapeutic strategies by enabling downstream analysis of the genetic or epigenetic mutations which improve clonal fitness20, and subsequently enriching pre-infusion cell populations with desirable phenotypes.
Several approaches to measure secreted proteins from individual cells have been introduced over the past few years. The most widely used of these, intracellular cytokine staining, is rapid and compatible with standard flow cytometry, but requires fixation and permeabilization, leaving cells non-viable21. Well-based micro-confinement approaches have also been developed and used to detect up to 42 different cytokines simultaneously using a combination of fluorescent and spatial barcoding, but throughput is limited to ~1,000 cells per experiment and recovery of interesting clones is not straightforward22,23. Microfluidic workflows leveraging water-in-oil emulsions partially circumvent these limitations by enabling cell recovery through emulsion destabilization24,25. However, most of these approaches rely on co-encapsulation of cells with cytokine capture beads, an inefficient process which necessitates the use of custom sorting tools that can process intact emulsions, to preserve the linkage between cells and their associated bead readout. Finally, new highly automated advanced analysis systems are being introduced to the market, but adoption of these technologies is limited in most research settings due to large capital costs and limited throughput.26,27
One particularly intriguing secretion profiling methodology makes use of bispecific antibodies to capture cytokines directly onto the surface of secreting cells, rendering them compatible with traditional immunostaining and flow cytometry workflows28. Unfortunately, these tools are standardly used to interrogate cells in bulk solution, leaving them susceptible to crosstalk between neighboring cells and to loss of signal from cytokines that diffuse away from the local microenvironment29.
In this work, we expand on the use of cell-surface based capture assays by combining bispecific antibody technology with high-throughput microfluidic droplet generation using step-emulsification (Fig 1). We demonstrate that assessment of single-cell cytokine production in partitioned aqueous volumes yields detectable secretion signals in as little as a few hours, while simultaneously increasing assay accuracy, and improving signal intensity on actively secreting cell clones over bulk. Moreover, the highly parallelized droplet generation scheme we employ, increases cell encapsulation throughputs by over an order of magnitude compared to previously reported microfluidic secretion screens24,25,29,30, preventing bulk background accumulation of secreted proteins during longer encapsulation workflows, and enabling screening of larger (e.g. hundreds of thousands) cell populations. The continued development of such easy-to-use functional profiling tools will remove fundamental limitations to immune cell screening and can directly yield large populations of phenotypically enriched, viable T-cells for subsequent molecular analysis and therapeutic use.
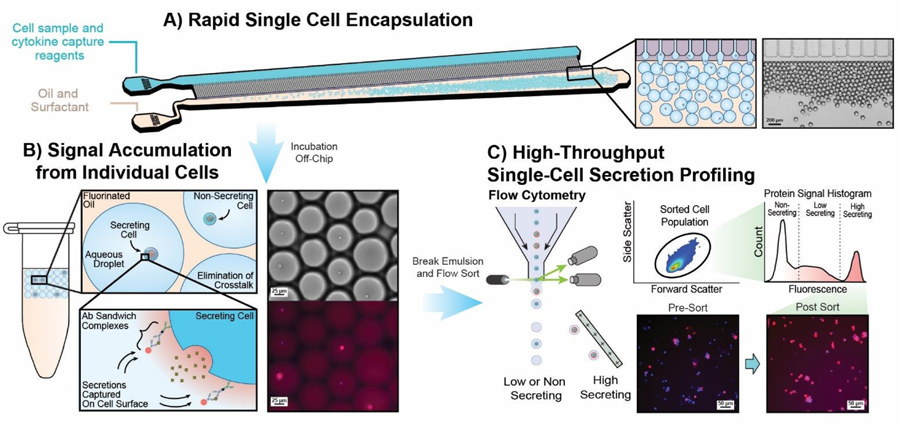
Figure 1:
Droplet enhanced secretion encoding workflow. A) Cells are co-encapsulated with anti-cytokine capture and detection probes within uniformly sized microdroplets using a highly parallelized step emulsification device. B) Droplets are incubated off chip, enabling accumulation of secreted proteins without crosstalk to neighbouring cells. The presence of secreted cytokines in each droplet is encoded back onto the secreting cell’s surface as a fluorescent sandwich immunocomplex. C) The emulsion is then broken and recovered cells can be analysed using standard flow cytometry. Analysed cells remain viable throughout the entire process and can be subsequently expanded in culture.
Results:
Highly Parallelized Single Cell Encapsulation
Analysis of secreted products from individual cells is inherently difficult due to diffusion of proteins away from their production source. Droplet microfluidic technology has emerged as a method of choice in single-cell secretion screening, because of its ability to partition cells into micron scale droplets which both trap and concentrate biomolecules. Unfortunately, most microfluidic designs generate droplets in a serial manner, rapidly reaching an upper processing limit. This restricts the number of cells that can be assayed before upstream accumulation of secreted products in wells or syringes reduces assay integrity. Such throughput limitations serve as a fundamental barrier to adoption of functional cellular profiling approaches in pre-clinical research, where large amounts of cells often must be screened simultaneously to identify and sort rare phenotypes.
To overcome these challenges, we adapted a highly parallelized step-emulsification device for our functional screening workflow (Fig. 2a)31,32. Step-emulsification exploits rapid pressure drops experienced by fluids emerging from confined channels into taller collection reservoirs to induce droplet pinch off 33. This primary advantage of this technique is that droplet generation is highly flow invariant, with channel geometric features dictating droplet size rather than individual fluid flow rates. The offset in height between the taller aqueous inlet and feed channel and the confined parallel drop generating channels serves to reduce the hydraulic pressure drop in the feed channel, and along with the taller height of the collection channel, is critical to maintaining a relatively uniform pressure drop across all drop generating channels. In our design a single aqueous inlet flow is evenly distributed between 200 parallel nozzles, each of which can generate droplets at a rate of ~4000 droplets/minute. While we have reported the use of step-emulsification previously to generate hydrogel microparticles, to the best of our knowledge it has not been leveraged previously to encapsulate objects into individual droplets.
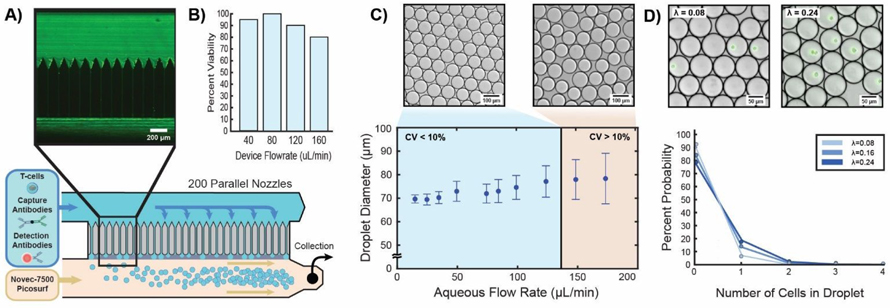
Figure 2:
Microfluidic droplet generator characterization. A) Step emulsification generates monodisperse droplets from 200 channels in parallel. Cells are loaded into the aqueous inlet along with capture and detection antibodies, pass uniformly between all channels, and are encapsulated into ~70 μm diameter droplets as the aqueous phase passes into a surfactant stabilized fluorinated oil. High exposure fluorescent images show that cells travel through the device uniformly, ensuring that loading is not biased to a specific region of the device (top). B) Cell viability after encapsulation at various flow rates. Cells remain viable over all conditions tested indicating that flow induced shear stressed are not a limiting factor in our cytokine screening assay. C) Droplet size and uniformity is minimally affected by flow rates. As we increased aqueous flow rates from 15 to 175 μL/min the average droplet diameter varied from 70 to 78 μm. When droplets are generated too rapidly they begin to crowd around the outlet of downstream nozzles, allowing more fluid to enter each droplet before pinch off and increasing sample CV. At our chosen flow rate of 125 μL/min roughly 12,000 droplets per second were generated. D) Known concentrations of primary human T-cells were encapsulated using our microfluidic step emulsifier and imaged to determine the number of cells in each droplet. The experimental cell distribution (dots) followed the expected Poisson distribution (lines) demonstrating that parallelized encapsulation followed predictable loading trends.
We determined the optimal operating conditions for our device by simultaneously assessing cell viability, and droplet uniformity as a function of flow rate, with the goal of maximizing viable cell throughput without sacrificing assay consistency. Primary human T cells flowing through our device remained viable at all tested flow rates, indicating that shear stresses encountered during passage through the narrow (35 μm) channel nozzles were not a significant assay limitation, and implying that cells would remain active for subsequent functional screens (Fig. 2a, 2b). The average droplet diameter also remained relatively stable over a large range of flow rates, ranging from 70–78 μm across all tested conditions (Fig 2c, Video S1). Interestingly, we found that the major limitation in device operation stemmed from accumulation of previously generated droplets around downstream nozzles at high flow rates, physically precluding the expansion of newer droplets over the channel step (Video S2), and resulting in the production of larger (>100 μm) diameter droplets from several nozzles towards the end of the device. Thus, we restricted device operation to 125 μL/min which was the highest tested flow rate preserving stringent droplet monodispersity (CV <10%). Importantly, at this throughput we are already capable of producing ~12,000 droplets/second, resulting in cell processing rates of thousands of cells per second when considering a target loading fraction of 10% following Poisson statistics (λ ~ 0.1).
Due to the parallelized nature of our device, we also verified that cell loading proceeded uniformly rather than biasing towards specific channel regions. To test this, we labeled T cells with CellTracker™ Green CMFDA dye and reconstituted them at known cell concentrations ranging from 400,000–1.4 million cells/mL. The resulting samples were then encapsulated and imaged using fluorescence microscopy to identify the number of cells within each droplet. The average cell occupancy for each condition closely followed expected Poisson distributions for all three tested conditions (λ = 0.08, 0.16, 0.24) (Fig 2d), indicating that single cell partitioning could be stringently enforced through control of initial cell concentrations. Thus, our step emulsification platform can be utilized as an efficient, high-throughput alternative to traditional serial encapsulation approaches, enabling compartmentalization of over 103 viable cells/second with less than 0.5% of droplets containing multiple cells.
Droplet Mediated On-Cell Cytokine Screening
Solid phase substrates are critical components of all secretion capture workflows due to their ability to concentrate diffuse biomolecules and translate their presence into detectable signals. However, assay schemes necessitating the co-delivery of both cells and substrates into a single droplet lead to subsequent non-idealities in both screening and sorting, as not all cells will have capture substrates present in their droplets, and traditional flow cytometers are generally incapable of screening intact oil droplets.
Our assay design relies on the use of commercially available capture and detection probes (Miltenyi Biotec) which instead transduce the presence of cytokines from the cellular microenvironment into a membrane localized fluorescence signal, allowing each secreting cell to function as its own solid phase capture substrate (Fig 3a). Typically, these assays are performed on bulk cell populations where cytokines can diffuse from secreting cells onto their neighbors yielding false positive signals. We hypothesized that translation of these on-cell molecular biosensing workflows into a droplet-based assay format would improve reliability by locally concentrating secreted molecules around producer cells, increasing signal, while simultaneously eliminating interaction between screened cells to reduce background accumulated from neighboring secreting cells.
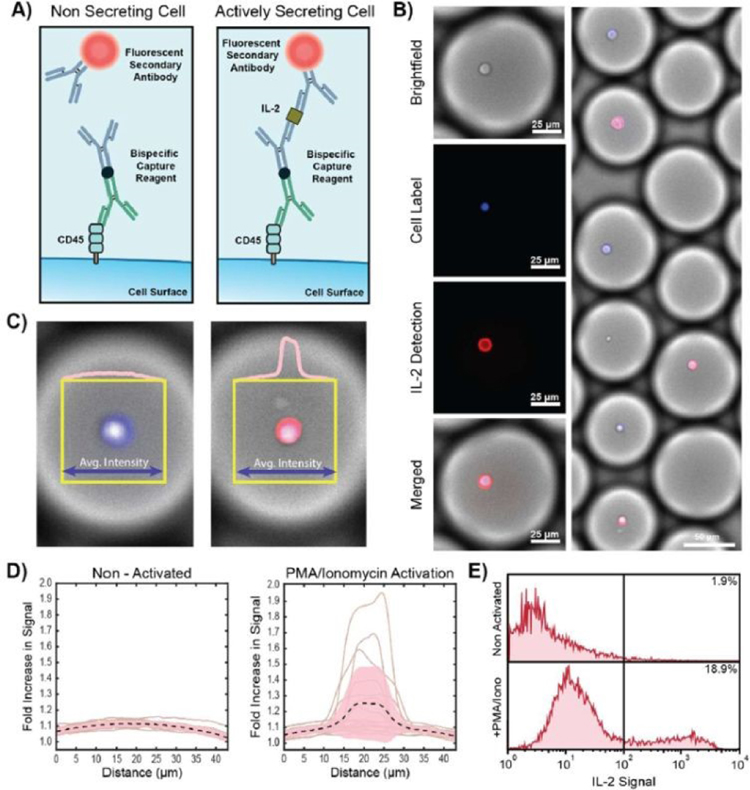
Figure 3:
Secretion encoding on the cell surface leads to amplified signal upon stimulation. A) Schematic of the assay for capturing and reporting on IL-2 secretion using bi-functional antibodies and an IL-2 sandwich assay. Stimulation of production of IL-2 leads to localized signal on the cell surface. B) Microscopy images in brightfield and two fluorescence channels showing detection of on-cell signal for a subset of T-cells that were stimulated with PMA/ionomycin. C) The average intensity within a box at the centre of the droplet is integrated over the box height and reported as an intensity profile. D) Intensity profiles for droplets with T-cells without stimulation (Non-Activated) and with PMA/ionomycin stimulation show distinct fluorescence profiles. E) After emulsion breaking, T-cells with IL-2 specific signal are analyzed by flow cytometry. Approximately 20% of stimulated T-cells showed IL-2 secretion signal compared to 2% of non-stimulated cells.
To test our screening approach, we compared the secretion signal obtained from T cells stimulated by the small molecule activators phorbol 12-myristate 13-acetate (PMA) and ionomycin versus control cells incubated in media without activation. We selected the T cell growth cytokine IL-2 as our analyte of interest because of its broad utility as a functional quality marker in cell therapy development pipelines, and due to its strong upregulation upon T cell activation, especially in CD4+ cells.
Because addition of reagents into formed droplets is nontrivial, we chose to pre-mix both anti-IL2 capture and phycoerethrin (PE) conjugated anti-IL2 detection probes with cells prior to encapsulation. Although this type of one pot assay may be prone to high background due to cell-cell and cell-reagent interactions in the feed syringe, care was taken to ensure that cells were thoroughly washed and encapsulated in under ten minutes after mixing with the capture and detection probes. Preliminary fluorescence visualization of newly formed droplets revealed a homogeneous fluorescence signal across each droplet volume (Fig. S1a). This indicated that although capture reagents were free to coat cell surfaces there was no detectable IL-2 within any droplet, leaving fluorescent secondary antibodies free floating. After emulsification, samples were incubated at 37°C for 1–12 hours to provide sufficient time for IL-2 accumulation in each cell-loaded droplet. Upon subsequent re-visualization notable differences were observed between the activated and non-activated cell populations (Fig. 3b, Fig. S1b), with a subset of activated T cells possessing a clear fluorescent signal localized along their periphery on average ~1.24 fold above background but up to 1.94 fold in some cells, while none of the control population was discernable over the fluorescent droplet background (Fig. 3c,d). This was indicative of sequestration of IL-2 by both capture and detection reagents, encoding individual secretion histories back onto the cell surface through the formation of fluorescent sandwich immunocomplexes.
Finally, after confirmation that our one pot droplet assay successfully translated IL-2 production into a detectable signal we destabilized the formed emulsions, recovered cells into an excess of fresh media, and analyzed the full population using flow cytometry (Fig. 3e). As expected, activation significantly increased IL-2 production, with 18.9% of cells staining positive for IL-2 after just one hour of incubation compared to 1.9% for the unstimulated control. Interestingly however, although activation increased the fraction of IL-2+ cells 10-fold over control only about 20% of T cells produced detectable levels of IL-2 even after strong stimulation and overnight incubation, alluding to an underlying heterogeneity in phenotypic polarization of the T cell population assayed.
Encapsulation Reduces Crosstalk
We next compared the crosstalk in interleukin-2 (IL-2) secretion between un-activated and activated populations mixed together in either a bulk or droplet encapsulated format using the full flow cytometry workflow. We prepared non-activated and activated peripheral blood mononuclear cell (PBMC) populations. Activated PBMCs had cytokine production induced by culturing for 6 hours in RPMI-1640 media supplemented with PMA/ionomycin. Nonactivated samples were prepared similarly but without addition of PMA/ionomycin. After incubation, samples were collected, washed to remove background secretions, and mixed at a 1:1 ratio of activated:non-activated. This mixture was resuspended in fresh media containing IL-2 capture and detection reagents, before being partitioned into individual droplets via step emulsification or left in a well plate for the standard bulk assay. Care was taken to ensure rapid sample processing, with all samples being completely emulsified.
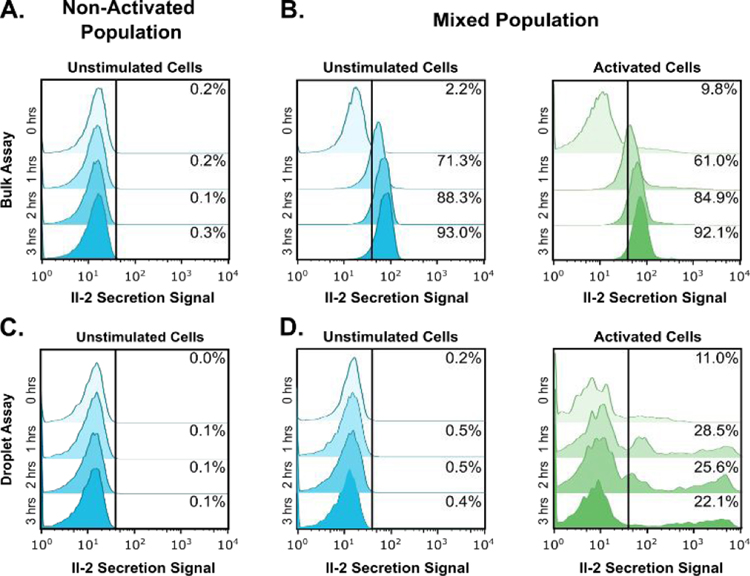
When non-activated cells were analyzed alone, no IL-2 production was detected in either bulk or emulsion assays at any timepoint (Fig. 4a, 4c). In contrast, when the two samples were mixed in the bulk assay, both activated and non-activated PBMC populations rapidly acquired fluorescent signal. The mean fluorescence of the non-activated cells was slightly higher than that of the activated population due to a small but noticeable amount of bleed through of the cell tracker fluorescence into the secretion fluorescence channel, however this does not account for the time-dependent increase in fluorescence for the unstimulated cells mixed with stimulated cells. Within one hour, both cell populations increased in fluorescence due to cytokine accumulation in the bulk solution, rendering the two populations indistinguishable (Fig. 4b). In contrast, partitioning through microfluidic step emulsification preserved clear variations in signal between the two populations, with less than 0.5% of non-activated PBMCs, and 22–29% of PMA/ionomycin activated PBMCs appearing as IL-2 positive after flow cytometric analysis (Fig. 4d). Additionally, in the droplet-mediated assay samples, IL-2 fluorescent secretion signals spanned over two orders of magnitude, with the highest secretors appearing over ten times brighter than the highest recorded signals in bulk phase analysis. This increase in dynamic range is critical for isolating cells with different secretion phenotypes which are not distinguishable without emulsification, because secreted proteins diffuse away and do not accumulate to reach higher concentrations.
Figure 4:
Encapsulation of primary T cells into individual droplets eliminates crosstalk in secretion assays. T cells were either chemically stimulated using PMA/ionomycin or fluorescently labelled and left unstimulated. The two populations were then mixed and left either in a single aqueous phase or partitioned into microdroplets. At the indicated timepoints cells were recovered and evaluated for IL-2 secretion using flow cytometry. A, C) When T cells were not activated no secreted IL-2 was detected in bulk or droplets at any time point. B) When the same population of unstimulated T cells was mixed with stimulated, actively secreting T cells both populations rapidly acquired fluorescent signal, rendering the two populations indifferentiable in as little as 1 hour. D) When mixtures of non-stimulated and chemically stimulated T cells were instead incubated as individual cells in microdroplets almost no signal was detected on non-stimulated cells at any timepoint. Instead activated T cells rapidly acquired a bright secretion signal spanning two orders of magnitude.
Enrichment of Functionally Active T Cell Clones through Standard FACS
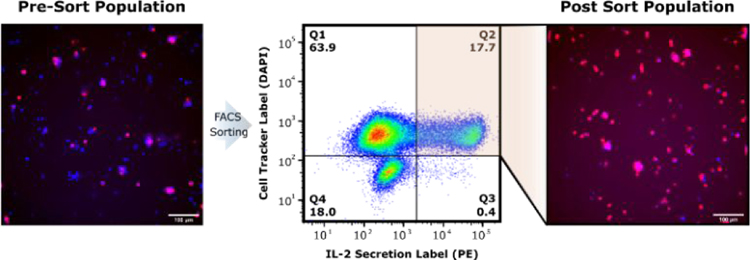
We also directly sorted cells based on IL-2 secretion signal following emulsification and de-emulsification using standard FACS technology. This enables the screening and enrichment of cells at rates of up to 104 cells/second, exceeding sorting throughputs using microfluidic droplet sorters. IL-2+ T cells were gated and sorted following recovery from droplets in our on-cell cytokine secretion screen (Fig. 5). Again, cytometric analysis revealed ~20% of viable T cells were active IL-2 producers. Sorting on this gate (upper right gate, Fig. 5) we were able to enrich IL-2 secreting cells to >95% of the final population.
Figure 5:
FACS sorting of IL-2 secreting cells following droplet encapsulation and on-cell cytokine capture. CellTracker Blue label is shown in blue and indicates viable cells with higher intensity. IL-2 (PE) secretion signal is show in magenta and shows the pre-sort population that comprises ~20% of the population with IL-2 secretion signal. Scatter plot showing the analyzed populations and upper right gate of IL-2+ and CellTracker+ cells that are sorted. Microscopy images of the sorted populations are shown on the right indicating significant enrichment of the IL-2 secreting cells.
Conclusions
We demonstrated that by using a high-throughput step emulsification microfluidic device we can improve the performance of an on-cell cytokine capture assay to analyze and select IL-2 secreting cells with higher signal and reduced crosstalk to nonsecreting cells. We found that the incubation of cells in droplets during on-cell secretion assay workflows significantly aids in the elimination of false positive signals stemming from proteins diffusing away from their producing cells, while simultaneously enhancing the concentration of secreted products around the cellular microenvironment leading to improved resolution between low and high secreting cell clones. Our results show that with this increased precision one observes ~20% of human T cells activated with PMA/ionomycin secreted IL-2, which differs from results using the on-cell cytokine capture assay in bulk, in which the entire population shifts (>90% of cells) over time. This increased signal is expected to be independent of the seeding density of cells, since it is driven by secretions being lost to the surrounding environment when cells are not encapsulated. Although crosstalk can be avoided with previous work where it is suggested that fewer than ~1×106 cells/mL in bulk should be used with fewer than 1% of actively secreting cells, the concentration effect and increased dynamic range of encapsulation are not achieved with these protocols. Altogether, our results suggest that care should be taken in interpreting the results of bulk cytokine capture assays when no method of compartmentalization is used.
The high-throughput processing (< 10 min) with the step emulsifier was enabling for this assay since cytokines released prior to cells reaching the droplet generator were reduced. Longer processing times with serial droplet generators could lead to crosstalk being initiated in the wells or syringes used to feed the device with cell samples, prior to encapsulation. Finally, the compatibility with standard flow cytometers and FACS systems and simpler operation of step emulsifiers makes this approach suited for life science researchers without microfluidics experience.
One outstanding issue with this phenotyping workflow stems from a reliance on surface molecule expression for the formation of capture sites. While CD45 is typically accepted to be broadly expressed across lymphocytes, our method of analyzing cells could feasibly omit the detection of high secreting cell clones with low CD45 expression. If such concerns are critical for the analysis of a specific cell type alternative strategies including recently reported lab on a particle approaches may be suitable alternatives for secretion assays.34–35
Ultimately, increased ability to adopt single-cell secretion assays in research labs can expand the investigation and use of functionally defined populations of cells for applications spanning cell therapy and basic science. While the current study focused on detecting secretions from a model system of cells stimulated with broadly activating chemical stimuli, it is not difficult to envision future studies building on these efforts to explore cell clones with potent responses to target antigens. The continued development of improved single-cell phenotyping tools will be imperative for informing the future of biotherapeutic design.
Materials and Methods
Culture of Primary T Cells
Trima filters were purchased from the UCLA virology core and eluted to recover concentrated volumes of whole blood. T cells were isolated from the trima filter eluent using the RosetteSep™ Human T Cell Enrichment Cocktail (STEMCELL Technologies) according to manufacturer instructions, and immediately frozen at a concentration 1×107 cells/mL of freezing media. Before analysis, each aliquot of T cells was rapidly thawed and transferred into pre-warmed ImmunoCult™-XF T Cell Expansion Media supplemented with 25 μL/mL ImmunoCult™ Human CD3/CD28 T cell activator, and 50 IU/mL IL-2 at a final concentration of 1×106 cells/mL. Cells were passaged twice a week. Culture media was replenished with 50 IU/mL of IL-2 every 2 days. Cells were cultured in a sterile incubator at 37°C and 5% CO2 for up to 3 weeks before thawing a fresh vial of cells.
Device Fabrication
Step emulsification devices were fabricated using master molds previously utilized for the high-throughput production of particle microgels. Detailed descriptions of wafer fabrication can be found in the report by de Rutte et al.32 Devices were formed from molds using a PDMS Sylgard 184 kit (Dow Corning). The polymer base and crosslinker were combined at a 10:1 ratio, poured into the mold, degassed, and cured in a 70°C oven overnight. The following morning devices were cut from the mold and entry/exit ports were formed by biopsy punching through the polymer at either end of the device. Cut PDMS slabs were then activated alongside glass microscope slides (VWR) in an air-based plasma cleaner (Harrick Plasma) and subsequently bound together. Each formed device was incubated for another 30 min in a 70°C oven to strengthen the bond between the PDMS and glass substrate. Finally, the device was filled with Aquapel, incubated for 1 minute, and flushed with Novec-7500 fluorinated oil to render the channels hydrophobic.
Droplet Based On-Cell Cytokine Capture Assay
T cells were taken from bulk culture, washed into fresh media and added into a single well of a 12-well plate at a concentration of 5×106 cells/mL. Wells were supplemented with 100 ng/mL phorbol 12-myristate 13-acetate (PMA) and 2.5 μM ionomycin, and cells were incubated in this activating mixture for a total of six hours. During the final hour of incubation CellTracker™ Blue CMAC dye was added into the activated cell mixture at a final concentration of 20 μM to label the cells. After incubation, cells were washed thoroughly a total of 3 times to remove any secreted IL-2 backgound that had accumulated within the solution over the course of the incubation. 1×106 cells were removed from the activated cell population and rapidly reconstituted into fresh media containing 50 μL/mL miltenyi IL-2 capture reagent and 50 μL/mL miltenyi phycoerythrin (PE) conjugated anti-IL-2 detection antibody within a 1 mL syringe. Simultaneously 2 mL of a 2% mixture of Pico-Surf™ fluorinated surfactant (3M) diluted into Novec-7500 engineered fluid (3M) was prepared and placed in a separate syringe. The two solutions were injected into our prefabricated step-emulsification droplet generator at flow rates of 125 μL/min and 250 μL/min for the aqueous cell mixture and fluorinated oil phases respectively. The tubing used to couple the feed syringes to the droplet generator was mechanically agitated every few minutes to prevent cells from settling before reaching the device, aiding the uniform distribution of cells between the 200 parallel droplet generating channels. Formed droplets were collected in 15 mL conical tubes, covered with 500 μL of light mineral oil (Sigma-Aldrich) to prevent evaporation, and incubated at 37°C and 5% CO2 for 1–6 hours depending on experimental need.
After sufficient incubation time, the emulsions were destabilized and the cells from each sample were recovered. Briefly, the bottom fraction of clear, un-emulsified Novec oil was first removed from the conical tube. Next, 5 mL of fresh media was added to the emulsified samples to serve as a diluent for uncaptured IL-2 secretions immediately after disruption of the droplets. Subsequently 500 μL of 1H,1H,2H,2H-Perfluoro-1-octanol (PFO) was added to the tube and samples were stored at room temperature for ~5 minutes while the emulsion destabilized and the cells were released. The top phase of the sample was then removed, leaving the bottom PFO phase behind. The recovered cell solution was spun down at 300g for 5 minutes and reconstituted into fresh media a total of 3 times to wash cells. Cell samples were then analyzed via flow cytometry.
Secretion Cross Talk Assay
T cells were split into two populations, one population was activated with PMA and ionomycin as described above but remained unlabeled, the other was not activated and instead maintained in fresh media at an equal concentration and time duration as the activated sample and labeled with 20 μM of a CellTracker™ DeepRed dye. The activated and non-activated cell populations were then analyzed either independently or in a 1:1 mixture in bulk culture or droplet assay formats.
Bulk Assay Format:
Cells were divided into three samples (non-activated, activated, and 1:1 mixture) and were washed 3X in cold media before analysis. 4×106 cells from each condition were obtained and resuspended into 80 μL of cold media. 20 μL of IL-2 capture reagent were then added to each sample and the cell/antibody mixture was incubated on ice for 5 minutes. Following this incubation, cells were quickly resuspended into 10 mL of warm media and incubated with vigorous agitation at 37°C. At each tested time point, 2.5 mL of sample was removed from the bulk solution, washed 3X, and resuspended into a final volume of 80 μL of cold media. Samples were then labeled with 20 μL of PE anti-IL-2 reporter antibodies on ice over a period of 10 minutes. Samples were washed 3X once more and analyzed via flow cytometry.
Droplet Assay Format:
Cells were analyzed as described in the droplet-based on-cell cytokine analysis section above with the notable exception that the three samples (non-activated, activated, and 1:1 mixture) were analyzed every hour between 0–4 hours. Nonactivated cells were labeled with 20 μM of a CellTracker™ DeepRed dye to differentiate from activated cells.
Flow Cytometry
All non-sorting experiments were conducted on a 2-laser BD FACS Canto II. PE fluorophores were excited using a 488 nm laser and filtered using 556LP and 585/42 filters. Cell tracker labels were excited using a 633 nm laser and filtered through a 686/20 filter.
All sorting experiments were performed at the UCLA flow cytometry core facility, using a 4-laser BD FACS Aria II. PE flurophores were excited using a 561 nm laser and filtered using a 585/15 filter. Blue cell tracker labels were excited using a 355 nm laser and filtered through a 450/50 filter.
Supplementary Material
Acknowledgements
Flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center (JCCC) and Center for AIDS Research Flow Cytometry Core Facility that is supported by National Institutes of Health awards P30 CA016042 and 5P30 AI028697, and by the JCCC, the UCLA AIDS Institute, the David Geffen School of Medicine at UCLA, the UCLA Chancellor’s Office, and the UCLA Vice Chancellor’s Office of Research. The authors acknowledge support from NCI IMAT grant 1R21CA256084.
Footnotes
Conflicts of interest
There are no conflicts to declare.
Notes and references
- 1.Lim WA and June CH, Cell, 2017, 168, 724–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vareki SM, Garrigos C and Duran I, Crit Rev Oncol Hemat, 2017, 116, 116–124. [DOI] [PubMed] [Google Scholar]
- 3.Chowell D, et al. , Science, 2018, 359, 582-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fraietta JA, et al. , Nat Med, 2018, 24, 563-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garfall AL, et al. , Blood Adv, 2019, 3, 2812–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Majzner RG and Mackall CL, Nat Med, 2019, 25, 1341–1355. [DOI] [PubMed] [Google Scholar]
- 7.Sharma P, Hu-Lieskovan S, Wargo JA and Ribas A, Cell, 2017, 168, 707–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu Y, et al. , Blood, 2014, 123, 3750–3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommermeyer D, et al. , Leukemia, 2016, 30, 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishna S, et al. , Science, 2020, 370, 1328–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, et al. , Nature Communications, 2020, 11. [Google Scholar]
- 12.Ma C, et al. , Cancer Discov, 2013, 3, 418–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, et al. , Nat Med, 2011, 17, 738–U133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Belardelli F, Ferrantini M, Proietti E and Kirkwood JM, Cytokine Growth F R, 2002, 13, 119–134. [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg SA, J Immunol, 2014, 192, 5451–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeku OO, et al. , Sci Rep-Uk, 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avanzi MP, et al. , Cell Rep, 2018, 23, 2130–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roybal KT, et al. , Cell, 2016, 167, 419-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossi J, et al. , Blood, 2018, 132, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Di Carlo D, Slas Technol, 2019, 24, 359–372. [DOI] [PubMed] [Google Scholar]
- 21.Sander B, Andersson J and Andersson U, Immunol Rev, 1991, 119, 65–93. [DOI] [PubMed] [Google Scholar]
- 22.Bradshaw EM, et al. , Clin Immunol, 2008, 129, 10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu Y, et al. , P Natl Acad Sci USA, 2015, 112, E607–E615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konry T, et al. , Biosensors and Bioelectronics, 2011, 26, 2707–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chokkalingam V, et al. , Lab on a Chip, 2013, 13, 4740–4740. [DOI] [PubMed] [Google Scholar]
- 26.Le K, et al. , Biotechnol Progr, 2018, 34, 1438–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, et al. , Nat Chem Biol, 2016, 12, 76-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell JDM, et al. , Clin Exp Immunol, 2011, 163, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, et al. , Lab on a Chip, 2020, 20, 1513–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazutis L, et al. , Nat Protoc, 2013, 8, 870–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhar M, et al. , P Natl Acad Sci USA, 2018, 115, 9986–9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de Rutte JM, Koh J and Di Carlo D, Adv Funct Mater, 2019, 29. [Google Scholar]
- 33.Montessori A, et al. , Phys Rev Fluids, 2018, 3. [Google Scholar]
- 34.de Rutte, et al. , SLAS Tech, 2021. (in press). [Google Scholar]
- 35.Lee, et al. , ACS Nano, 2021. (in press). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.