Abstract
Twenty Acinetobacter baumannii strains resistant to various antibiotics were analyzed for integron content and sequences of the amplification products. Sixteen clinical isolates had a class 1 integron, 2 contained an additional class 1 or class 2 integron, but no class 3 integron was detected. Thirteen strains had integrons with a single cassette: aac(3)-Ia (9 strains), ant(2")-Ia (2 strains), or aac(6′)-Ib (2 strains); 1 had aac(6′)-Ib and oxa20 cassettes and an unknown gene; and 1 had an integron containing ant(2")-Ia and an oxa3 cassette truncated by IS6100. The remaining strains harbored class 1 integrons with gene cassettes previously found in Enterobacteriaceae. One integron had a hybrid structure composed of intI2 and the 3′ conserved segment of class 1 integrons. These data indicate that integrons play a major role in multidrug resistance in Acinetobacter.
Integrons are genetic elements that can integrate, by site-specific recombination, gene cassettes, usually antibiotic resistance genes (5). Three main classes of integrons have been described (22). All have a 5′ conserved segment, including an intI gene encoding an integrase and an attI recombination site, but have distinct 3′ conserved segments. In the class 1 integrons, the 3′ conserved segment includes three open reading frames (ORFs)—qacEΔ1, a deletion derivative of the antiseptic resistance gene qacE; the sul1 sulfonamide resistance gene; and ORF5, of unknown function—or tni genes, as in Tn402 (18). The second class of integrons was found in transposon Tn7 and its derivatives, and its 3′ conserved segment contains five tns genes involved in the movements of the transposon. A single class 3 integron has been reported to date, but its 3′ conserved segment has not been characterized (2). Cassettes usually include a single ORF and, downstream, an attC site which is an imperfect inverted repeat sequence related to a 60-bp consensus sequence (4). The movements of cassettes are catalyzed by the integrase, which can excise or integrate cassettes by site-specific recombination between two specific sequences, either attI and attC or two attC sites. Cassette mobility results in the dissemination of resistance genes, and more than 50 cassettes have been described for gram-negative bacteria (5). This genetic flexibility allows numerous cassette rearrangements under antibiotic selective pressure, and study of these various assortments can lead to a better understanding of integron evolution. Acinetobacter, in particular Acinetobacter baumannii, is increasingly responsible for nosocomial infections in intensive care units and is often resistant to multiple antibiotics (27). Integrons have already been found in this species, but their cassette content has not been fully characterized (3, 25). The aim of our study was to search for the presence of the three classes of integrons in multidrug-resistant A. baumannii clinical isolates and to characterize their gene cassette assortments.
MATERIALS AND METHODS
Bacterial strains.
Twenty clinical A. baumannii strains isolated between 1992 and 1998 were collected because of their multiresistance to antibiotics, in particular to aminoglycosides, sulfonamides, chloramphenicol, and β-lactams (Table 1). Seventeen strains were isolated from French hospitals, 1 was from Greece, and 2 were from the Czech Republic. Bacteria were grown in brain heart infusion broth (Biomerieux, Marcy l'Etoile, France) or on Mueller-Hinton agar (Biomerieux). Susceptibility to ampicillin, ticarcillin, ceftazidime, gentamicin, isepamicin, amikacin, tobramycin, kanamycin, streptomycin, spectinomycin, rifampin, chloramphenicol, and sulfonamide was tested by disk diffusion on Mueller-Hinton agar. The susceptibility tests were interpreted according to Comité de l'Antibiogramme de la Société Française de Microbiologie recommendations (1). Incubations were done at 37°C.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Acinetobacter baumannii | ||
| BM4420 | Ak Cm Gm Ip Km Su Tc | Clinical strain from France |
| BM4421 | Ak Cm Gm Ip Km Rif Sp Su Tc Tm | Clinical strain from France |
| BM4422 | Ak Cm Cz Gm Ip Km Sm Sp Su Tc Tm | Clinical strain from France |
| BM4423 | Ak Cm Gm Is Km Sm Sp Su Tc Tm | Clinical strain from France |
| BM4424 | Ak Cm Gm Ip Km Su Tc | Clinical strain from France |
| BM4425 | Cm Gm Km Sm Sp Su Tc | Clinical strain from France |
| BM4426 | Ak Cm Cz Ip Km Sm Sp Su Tc Tm | Clinical strain from France |
| BM4427 | Ak Cm Ip Km Sm Sp Tc | Clinical strain from France |
| BM4428 | Ak Cm Ip Km Gm Sm Sp Su | Clinical strain from France |
| BM4429 | Cm Cz Km Su Tc Tm | Clinical strain from France |
| BM4430 | Ak Cm Gm Ip Km Sm Sp Su Tm | Clinical strain from France |
| BM4431 | Ak Gm Ip Km Sm Sp Su Tc | Clinical strain from France |
| BM4432 | Ak Cm Cz Gm Ip Km Sm Sp Su Tm | Clinical strain from France |
| BM4433 | Cm Sm Sp | Clinical strain from France |
| BM4434 | Cm Gm Km Sm Sp Su Tc Tm | Clinical strain from France |
| BM4435 | Cm Gm Km Sm Sp Su Tc | Clinical strain from France |
| BM4436 | Cm Gm Km Sm Sp Su Tc Tm | Clinical strain from France |
| BM4437 | Ak Gm Ip Km Sm Su Tc | Clinical strain from France |
| BM4438 | Ak Cm Cz Gm Ip Km Sm Sp Su Tc | Clinical strain from Czech Republic |
| BM4439 | Cm Cz Gm Km Rif Tm Tc | Clinical strain from Czech Republic |
| Escherichia coli JM83 | araΦ (lac-proAB) rpsL [Φ80Δ(lacZ)M15] | 28 |
| Plasmids | ||
| pUC18 | Tra− Mob− Ap | 28 |
| pAT719 | Tra− Mob− Ap Tm Gm | 3.8-kb BamHI fragment of BM4430 cloned in pUC18 |
Abbreviations: Tra−, non-self-transferable; Mob−, nonmobilizable; Ak, amikacin resistance; Cm, chloramphenicol resistance; Cz, ceftazidime resistance; Gm, gentamicin resistance; Ip, isepamicin resistance; Km, kanamycin resistance; Rif, rifampin resistance; Sm, streptomycin resistance; Sp, spectinomycin resistance; Su, sulfonamide resistance; Tc, ticarcillin resistance; Tm, tobramycin resistance.
Preparation and analysis of DNA.
Total DNA was prepared by using a Qiamp tissue kit (Qiagen, Inc., Chatworth, Calif.). Small- and large-scale plasmid DNAs were prepared as described previously (24). DNA for PCR was obtained by boiling as described previously (16). Electrophoresis was performed with an 0.8% agarose gel (GIBCO BRL, Cergy Pontoise, France) and a Tris-borate-EDTA buffer system.
Genetic techniques.
Escherichia coli JM83 was transformed as described previously (24). Selective antibiotic concentrations were as follows: ampicillin, 100 μg/ml; and tobramycin, 10 μg/ml.
DNA techniques.
Integrons were screened for by PCR with three sets of primers specific for the intI1, intI2, and intI3 genes and mapped with primers complementary to conserved segments (Table 2). When the 3′ conserved segment was missing, the cassettes were cloned with selection for antibiotic resistance. Amplification was performed with 50-μl reaction mixtures consisting of Taq or Pfu DNA polymerase buffer, 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates, 50 pmol of each primer (Isoprim, Toulouse, France), 1 U of Taq or 2 U of Pfu DNA polymerase, and 25 ng of DNA by use of model 9600 DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). The PCR products were purified using a Qiaquick PCR purification kit (Qiagen) as recommended by the manufacturer. For strains with two PCR products, the two fragments were separated by agarose gel electrophoresis and purified using a Qiaquick gel extraction kit (Qiagen).
TABLE 2.
Oligodeoxyribonucleotides used for amplification
| Primer | Sequence | Strand | Location |
|---|---|---|---|
| intI1L | 5′-ACATGTGATGGCGACGCACGA-3′ | + | intI1 |
| intI1R | 5′-ATTTCTGTCCTGGCTGGCGA-3′ | − | intI1 |
| intI2L | 5′-CACGGATATGCGACAAAAAGGT-3′ | + | intI2 |
| intI2R | 5′-GTAGCAAACGAGTGACGAAATG-3′ | − | intI2 |
| intI3L | 5′-GCCTCCGGCAGCGACTTTCAG-3′ | + | intI3 |
| intI3R | 5′-ACGGATCTGCCAAACCTGACT-3′ | − | intI3 |
| 5′-CS | 5′-GGCATCCAAGCAGCAAG-3′ | + | 5′ conserved segment of class 1 integrons |
| 3′-CS | 5′-AAAGCAGACTTGACCTGA-3′ | − | 3′ conserved segment of class 1 integrons |
| int2S | 5′-ACCTTTTTGTCGCATATCCGTG-3′ | + | 5′ conserved segment of class 2 integrons |
| intCS2 | 5′-TACCTGTTCTGCCCGTATCT-3′ | − | tnsE (3′ conserved segment of class 2 integrons) |
| ORFL | 5′-GTCCGACATCCACGACGTCTGATC-3′ | + | qacEΔ1 |
| SulR | 5′-CGAACCTGCTAACTAGGTA-3′ | − | sul1 |
| aadAL | 5′-GTTGTGCACGACGACATCATTCC-3′ | + | ant(3")-Ia |
| ORFR | 5′-GTCGCTGCAACTCGCGACT-3′ | − | ORF5 |
| tnpL | 5′-ACACCCTCGGCTACCACCTC-3′ | − | tnpM |
The 3.8-kb BamHI fragment of BM4430 was cloned in pUC18 and transformed into E. coli JM83 with selection on tobramycin (28).
Random amplified polymorphic DNA (RAPD) analysis was performed with the A5 10-mer primer (5′GCCGGGGCCT3′; Bioprobe Systems, Montreuil-sous-Bois, France) as described previously (17). RAPD fingerprints were arbitrarily designated by letters.
DNA sequencing.
The PCR-amplified and cloned fragments were sequenced by using the ABI PRISM dRhodamine terminator protocol as recommended by the manufacturer (Perkin-Elmer Applied Biosystems, Les Ulis, France). Products were analyzed with an ABI PRISM 310 automated DNA sequencing apparatus (Perkin-Elmer Applied Biosystems).
Computer analysis of sequence data.
The nucleotide sequence analysis procedure was obtained online over the Internet at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov).
Enzymes and chemicals.
T4 DNA ligase (Pharmacia, St. Quentin-en-Yvelines, France) and restriction endonuclease BamHI (Boehringer, Meylan, France) were used according to the recommendations of the manufacturers. Tobramycin and ampicillin were obtained from Sigma Chemical Co. (St. Louis, Mo.). Deoxynucleoside triphosphates were obtained from Boehringer. Taq DNA polymerase was purchased from GIBCO BRL, and Pfu DNA polymerase was obtained from Stratagene Cloning Systems (La Jolla, Calif.). Antibiotic disks were obtained from Sanofi Diagnostics Pasteur (Marnes-la-Coquette, France).
Nucleotide sequence accession numbers.
The 403-bp ORFO cassette from BM4426 (see Results and Discussion), the class 1 integron from BM4430, and the class 2 hybrid integron from BM4431 have been deposited in the GenBank data library (Los Alamos, N.Mex.) under accession no. AJ251519, AJ289190, and AJ289189, respectively.
RESULTS AND DISCUSSION
Detection of intI genes.
The presence of integrons was detected by amplification of an internal fragment of the integrase genes in 80% (16 of 20) of A. baumannii isolates (Table 3), indicating that these elements are widely spread among multiresistant isolates of this species. Among the 20 clinical isolates, 16 contained an intI1 gene. The intI2 gene of class 2 integrons was detected in strain BM4431, which also contained a class 1 integron. The intI3 gene was not found. Previous studies have reported a high frequency of multiresistant gram-negative isolates containing integrons (8, 9, 12). Class 1 integrons were found in 68% of worldwide nosocomial isolates of A. baumannii (25). A study of 100 clinical isolates of A. baumannii from Chilean hospitals indicated that more than 50% of the isolates harbored an intI1 or an intI2 gene (3).
TABLE 3.
RAPD types, PCR amplification products, and cassette contents of A. baumannii clinical isolates
| Strain | RAPD type | intI gene | Class 1 integron 3′ conserved segment | Size (bp) of the PCR product obtained with primers 5′-CS and 3′-CS | Gene content and order (accession no.)b |
|---|---|---|---|---|---|
| BM4420 | A | 1 | + | 500 | aac(3)-Ia [aacC1] (X15852) |
| BM4421 | B | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4422 | C | 1 | + | 700 | ant(2") [aadB] (L06418) |
| BM4423 | C | 1 | + | 700 | ant(2") (L06418) |
| BM4424 | D | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4425 | B | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4426 | B | 1 | + | 2,400 | aac(6′)-Ib [aacA4]-ORFO-oxa20 (X60321-AJ251519-AF024602) |
| BM4427 | E | None | − | ||
| BM4428 | B | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4429 | D | 1 | + | 800 | aac(6′)-Ib (X60321) |
| BM4430 | F | 1 | − | None | ant(2")-oxa3Δ-IS6100-IS26-tnpMΔ (AJ289190) |
| BM4431 | G | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4431 | G | 2 | − | dfrA1-sat-ant(3")-Ia [aadA1] (AJ289189) | |
| BM4432 | H | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4433 | D | None | − | ||
| BM4434 | D | 1 | + | 800 | aac(6′)-Ib (X60321) |
| BM4435 | B | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4436 | I | 1 | + | 4,000 | ant(2")-cmlA1-oxa10-ant(3")-Ia (L06418-U12338-U37105-X12870) |
| BM4436 | I | 1 | + | 1,300 | dfrB1-aadA6 (U36276-AF140629) |
| BM4437 | J | None | − | ||
| BM4438 | B | 1 | + | 500 | aac(3)-Ia (X15852) |
| BM4439 | K | None | − |
+, yes; −, no.
Alternative nomenclature for aminoglycoside-modifying enzymes is shown in brackets.
Mapping of class 1 integrons.
The cassette assortments in all the strains were characterized by PCR with the 5′-CS and 3′-CS primers and by sequencing of the amplification products (Table 3). Thirteen strains had integrons containing a single cassette which encoded an AAC(3)-Ia, an AAC(6′)-Ib, or an ANT(2") aminoglycoside-modifying enzyme. Previous studies reported integrons with a single cassette, often an ant(3")-Ia gene, in various gram-negative bacteria (8, 13, 23, 25). In this study, the ant(3")-Ia gene cassette was not detected alone, but nine strains had an integron, not reported so far, carrying only aac(3)-Ia. Most strains were resistant to both streptomycin and spectinomycin, suggesting an aminoglycoside adenylyltransferase activity. An ant(3")-Ia gene cassette (GenBank accession no. X12870) was detected only in strains BM4431 and BM4436. Although most ant(3")-Ia genes are known to be part of integron cassettes, these genes have also been found outside integrons, such as in R538-1 (6). Another explanation is that an ant(3")-Ia gene cassette was included in a large integron which could not be detected by our PCR protocol.
Total DNA from A. baumannii BM4436 yielded two PCR products. The first fragment, 1.3 kb, contained two known cassettes, dfrB1 and aadA6; the second fragment, approximately 4 kb, contained a new assortment of four known cassettes, ant(2"), cmlA1, oxa10, and ant(3")-Ia.
The integron in BM4426 was found to contain three cassettes (Table 3). The upstream one, adjacent to the 5′ conserved segment, was aac(6′)-Ib. The most downstream one was the oxa20 gene, recently found in an integron from Pseudomonas aeruginosa (15). These two cassettes were separated by a 466-bp sequence containing an ORF, designated ORFO, of 362 bp and displaying no homology with known genes. ORFO was followed by an attC site, indicating that it was part of a cassette. Other ORFs of unknown functions have also been found in integrons (19). A. baumannii BM4426 and BM4436 contained, respectively, oxa20 and aadA6 cassettes, already described for P. aeruginosa (15), confirming that integrons may be transferred via plasmids and/or transposons between these two species.
The integron in BM4430.
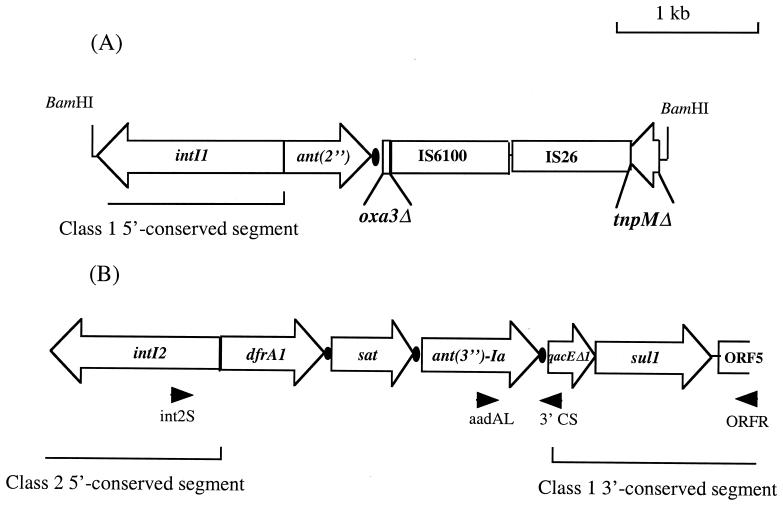
Class 1 integrons lacking a 3′ conserved segment have been reported (21). In our study, the 3′ conserved segment was strongly linked to class 1 integrons, since it was detected by PCR with primers ORFL and SulR in 15 out of 16 strains. The deleted integron in the remaining strain, BM4430, was characterized by cloning with selection for tobramycin resistance. Analysis of the sequence of the 3.8-kb BamHI insert of the recombinant plasmid obtained, pAT719, showed that the integron had an unusual organization, with an ant(2") gene cassette and, downstream, the first 51 bases of an oxa3 cassette (Fig. 1A). The oxa3 cassette was truncated by a copy of IS6100, an insertion sequence found in Mycobacterium fortuitum (11). In BM4430, IS6100 was followed by a 14-bp sequence and a copy of IS26 (14) and a portion of the tnpM gene, proposed to encode a modulator protein which enhances Tn21 transposition and suppresses the resolution of cointegrate replicons in vivo (7). The location of the tnpM gene in BM4430 is unusual compared to the structure of the integron In2 of Tn21, in which this gene is located between the intI1 and tnpR genes, upstream from the 5′ conserved segment (10). The lack of an amplification product of PCR with primers intI1R and tnpL suggests that the sequence at the right end of the truncated tnpM gene is not the beginning of a second integron.
FIG. 1.
Physical maps of the class 1 integron of BM4430 (A) and of the hybrid integron of BM4431 (B). Filled circle, attC site; open arrows, coding sequences; arrowheads, primers.
Mapping of class 2 integrons.
A. baumannii BM4431 yielded two PCR products with primers specific for the intI1 and intI2 genes (Table 3). To characterize these integrons, multiplex PCR with primers 5′-CS and 3′-CS, specific for the class 1 integrons, and primers int2S and intCS2, specific for the class 2 integrons, was performed and yielded two amplification products. The first one, 500 bp, contained a class 1 integron with a single aac(3)-Ia cassette (Table 3). The second one, 2.5 kb, contained the intI2 gene and the dfrA1 and sat cassettes, which are usually part of class 2 integrons (26). The ant(3")-Ia (also called aadA1) cassette, located downstream from the sat cassette, is identical to those previously reported for class 1 integrons (GenBank accession no. X12870) and differed from the ant(3")-Ia cassette found in class 2 integrons (GenBank accession no. X03043). This gene contains the codon AGA instead of AGG at position 684, the codon GTC instead of GTT at position 750, and the additional triplet GAA at position 706. Downstream from these cassettes, we did not find the tns genes usually described for the 3′ conserved segment of class 2 integrons, but we did find the first 56 bp of the 3′ conserved segment of class 1 integrons.
To characterize the entire 3′ segment in BM4431, amplification with primers aadAL and ORFR (Fig. 1B) was performed and yielded a 1.6-kb fragment. Sequence determination of this fragment showed that the 3′ end of the integron in BM4431 contained the same ORFs as the 3′ conserved segment of class 1 integrons, i.e., qacEΔ1, sul1, and ORF5 (Fig. 1B). This first report of a chimeric integron indicates that recombination between class 1 and class 2 integrons may occur in nature and is of interest in view of the evolution of integrons and their implication in the natural engineering of bacterial genomes. This hybrid structure may have resulted from cointegrate formation catalyzed by the IntI1 integrase between a class 2 integron and a class 1 integron with the ant(3")-Ia cassette at the left end of the 3′ conserved segment. Another possibility involves RecA-dependent homologous recombination between two copies of the ant(3")-Ia cassette, which has been found in both classes of integrons. Class 2 integrons have already been detected in Acinetobacter spp., but their structure has not been characterized (3, 25).
Distribution of integrons among RAPD types.
Eleven RAPD profiles were observed among the 20 clinical isolates (Table 3). Integrons containing the same organization of cassettes were found in various RAPD genotypes (Table 3), suggesting a horizontal transfer of integrons, also suspected in other studies (23, 25). Martinez-Freijo et al. (12) also found similar integrons irrespective of host species or geographic origin. In addition, the promoter sequences were mostly conserved, even in isolates from different countries with distinct selective pressure, suggesting that acquisition of resistance likely is due to transfer of entire integrons via plasmids and/or transposons rather than of individual cassettes. In contrast, certain strains with similar RAPD types (B and D) contained distinct integrons.
Conclusions.
Many studies have reported the high prevalence of integrons in human clinical isolates (9, 12, 13, 23, 26, 27) as well as in strains of animal origin (20) or from aquatic ecosystems (21), suggesting the important role of integrons in the dissemination of antibiotic resistance genes in the environment. Detection of integrons was often based on PCR or hybridization experiments (3, 8, 9, 12). Systematic sequencing of the integrons allows complete description of the cassettes and the detection of unusual rearrangements. Of special interest is the hybrid integron resulting from a recombination event between class 1 and class 2 integrons.
ACKNOWLEDGMENTS
We thank H. Giamarellou and M. Kettner for the gift of Acinetobacter strains.
REFERENCES
- 1.Acar J, Carret G, Cavallo J D, Chardon H, Choutet P, Courvalin P, Dabernat H, Drugeon H, Duval J, Dubreuil L, Goldstein F, Jarlier V, Leclercq R, Nicolas-Chanoine M H, Philippon A, Rouveix B, Sirot J, Soussy C J. Communiqué 1999. Paris, France: Comité de l'Antibiogramme de la Société Française de Microbiologie; 1999. [PubMed] [Google Scholar]
- 2.Arakawa Y, Murakami M, Suzuki K, Ito H, Wacharotayankun R, Ohsuka S, Kato N, Ohta M. A novel integron-like element carrying the metallo-β-lactamase gene blaIMP. Antimicrob Agents Chemother. 1995;39:1612–1615. doi: 10.1128/aac.39.7.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gonzalez G, Sossa K, Bello H, Dominguez M, Mella S, Zemelman R. Presence of integrons in isolates of different biotypes of Acinetobacter baumannii from Chilean hospitals. FEMS Microbiol Lett. 1998;161:125–128. doi: 10.1111/j.1574-6968.1998.tb12937.x. [DOI] [PubMed] [Google Scholar]
- 4.Hall R M, Brookes D E, Stokes H W. Site-specific insertion genes into integrons: role of the 59-base element and determination of the recombination cross-over point. Mol Microbiol. 1991;5:1941–1959. doi: 10.1111/j.1365-2958.1991.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 5.Hall R M, Collis C M. Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist Updates. 1998;1:109–119. doi: 10.1016/s1368-7646(98)80026-5. [DOI] [PubMed] [Google Scholar]
- 6.Hollingshead S, Vapnek D. Nucleotide sequence analysis of a gene encoding a streptomycin/spectinomycin adenyltransferase. Plasmid. 1985;13:17–30. doi: 10.1016/0147-619x(85)90052-6. [DOI] [PubMed] [Google Scholar]
- 7.Hyde D R, Tu C P D. tnpM: a novel regulatory gene that enhances Tn21 transposition and suppresses cointegrate resolution. Cell. 1985;42:629–638. doi: 10.1016/0092-8674(85)90120-5. [DOI] [PubMed] [Google Scholar]
- 8.Jones M E, Peters E, Weersink A M, Fluit A, Verhoef J. Widespread occurrence of integrons causing multiple antibiotic resistance in bacteria. Lancet. 1997;349:1742–1743. doi: 10.1016/S0140-6736(05)62954-6. [DOI] [PubMed] [Google Scholar]
- 9.Lévesque C, Piché L, Larose C, Roy P H. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39:185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liebert C A, Hall R M, Summers A O. Transposon Tn21, flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–522. doi: 10.1128/mmbr.63.3.507-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, Gicquel B. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 12.Martinez-Freijo P, Fluit A C, Schmitz F J, Grek V S C, Verhoef J, Jones M E. Class I integrons in gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple-antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Freijo P, Fluit A C, Schmitz F J, Verhoef J, Jones M E. Many class I integrons comprise distinct stable structures occurring in different species of Enterobacteriaceae isolated from widespread geographic regions in Europe. Antimicrob Agents Chemother. 1999;43:686–689. doi: 10.1128/aac.43.3.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mollet B, Iida S, Shepherd J, Arber W. Nucleotide sequence of IS26, a new prokaryotic mobile genetic element. Nucleic Acids Res. 1983;11:6319–6330. doi: 10.1093/nar/11.18.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naas T, Sougakoff W, Casetta A, Nordmann P. Molecular characterization of OXA-20, a novel class D β-lactamase, and its integron from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;42:2074–2083. doi: 10.1128/aac.42.8.2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ploy M C, Giamarellou H, Bourlioux P, Courvalin P, Lambert T. Detection of aac(6′)-I genes in amikacin-resistant Acinetobacter spp. by PCR. Antimicrob Agents Chemother. 1994;38:2925–2928. doi: 10.1128/aac.38.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ploy M C, Grélaud C, Martin C, Denis F. Emergence at the Limoges teaching hospital of Enterobacter aerogenes strains producing a derepressed cephalosporinase: molecular epidemiology using the RAPD technique. Pathol Biol. 1997;45:404–408. [PubMed] [Google Scholar]
- 18.Radström P, Sköld O, Swedberg G, Flensburg J, Roy P H, Sundström L. Transposon Tn5090 of plasmid R751, which carries an integron, is related to Tn7, Mu, and the retroelements. J Bacteriol. 1994;176:3257–3268. doi: 10.1128/jb.176.11.3257-3268.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Recchia G D, Hall R M. Gene cassettes: a new class of mobile element. Microbiology. 1995;141:3015–3027. doi: 10.1099/13500872-141-12-3015. [DOI] [PubMed] [Google Scholar]
- 20.Ridley A, Threlfall E J. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. FEMS Microbiol Lett. 1998;4:113–118. doi: 10.1089/mdr.1998.4.113. [DOI] [PubMed] [Google Scholar]
- 21.Rosser S J, Young H K. Identification and characterization of class 1 integrons in bacteria from an aquatic environment. J Antimicrob Chemother. 1999;44:11–18. doi: 10.1093/jac/44.1.11. [DOI] [PubMed] [Google Scholar]
- 22.Rowe-Magnus D A, Mazel D. Resistance gene capture. Curr Opin Microbiol. 1999;2:481–486. doi: 10.1016/s1369-5274(99)00004-1. [DOI] [PubMed] [Google Scholar]
- 23.Sallen B, Rajoharison A, Desvarenne S, Mabilat C. Molecular epidemiology of integron-associated antibiotic resistance genes in clinical isolates of Enterobacteriaceae. Microb Drug Resist. 1995;1:91–98. doi: 10.1089/mdr.1995.1.195. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Seward R J, Towner K J. Detection of integrons in worldwide nosocomial isolates of Acinetobacter spp. Clin Microbiol Infect. 1999;5:308–318. doi: 10.1111/j.1469-0691.1999.tb00149.x. [DOI] [PubMed] [Google Scholar]
- 26.Sundstrom L, Roy P H, Skold O. Site-specific insertion of three structural gene cassettes in transposon Tn7. J Bacteriol. 1991;173:3025–3028. doi: 10.1128/jb.173.9.3025-3028.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Villers D, Espaze E, Coste-Burel M, Giauffret F, Ninin E, Nicolas F, Richet H. Nosocomial Acinetobacter baumannii infections: microbiological and clinical epidemiology. Ann Intern Med. 1998;129:182–189. doi: 10.7326/0003-4819-129-3-199808010-00003. [DOI] [PubMed] [Google Scholar]
- 28.Yanish-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]