Summary
Inter-organelle interactions are a vital part of normal cellular function; however, these have proven difficult to quantify due to the range of scales encountered in cell biology and the throughput limitations of traditional imaging approaches. Here, we demonstrate that Soft X-ray Tomography (SXT) can be used to rapidly map ultrastructural reorganization and inter-organelle interactions in intact cells. SXT takes advantage of the naturally occurring, differential X-ray absorption of the carbon-rich compounds in each organelle. Specifically, we use SXT to map the spatio-temporal evolution of insulin vesicles and their co-localization and interaction with mitochondria in pancreatic β-cells during insulin secretion and in response to different stimuli. We quantify changes in the morphology, biochemical composition, and relative position of mitochondria and insulin vesicles. These findings highlight the importance of a comprehensive and unbiased mapping at the mesoscale to characterize cell reorganization that would be difficult to detect with other existing methodologies.
Keywords: soft X-ray tomography, 3D cell mapping, mesoscale analysis, organelle interaction, pancreatic β-cell
eTOC blurb
Soft X-ray tomography combines rapid collection and high-resolution visualization to understand subcellular reorganization. Loconte et al. use the technique to quantify and describe changes in inter-organelle interactions within pancreatic β-cells, providing a unique approach for comprehensive 3D subcellular mapping.
Graphical Abstract
Introduction
Organelles are traditionally described as intracellular membrane compartments that separate biochemical processes in the cell (Valm et al., 2017; Cohen et al., 2018a). Each compartment is characterized by highly specialized functions that regulate cell homeostasis, division, and response to outside stimuli (Henne, 2021). Their functions are coordinated, and inter-organelle interactions facilitate the communication among different compartments. For example, the juxtaposition between mitochondria and endoplasmic reticulum (ER) is identified as the locus of lipid biosynthesis (Phillips and Voeltz, 2016); lipids are then transferred from mitochondria to lipid bodies for storage (Valm et al., 2017). Recently, many inter-organelle interactions have been demonstrated to take place at membrane contact sites, where interacting proteins from both organelles regulate the exchange of metabolites between compartments (Scorrano et al., 2019). For example, mitochondria-associated membranes are regions of ER that reversibly tether to mitochondria; determining their connectivity (i.e., contact numbers and localization) and temporal reorganization informs about cell calcium and lipid homeostasis (Vance, 2014). Highresolution imaging techniques provide an accurate identification of organelle interactions (Scorrano et al., 2019); and three-dimensional (3D) maps of entire cells are required to characterize the frequency of the interactions, to identify the number and location of inter-organelle contacts, and to determine whether they are altered during specific pathological conditions (Díaz et al., 2021).
Fluorescence microscopy largely contributed to the discovery of extensive communication between organelles. The rise of selective and synthetic fluorescence probes, together with the advancements in biotechnological methodologies, facilitated the discovery of subcellular mechanisms, such as the description of the role of the ER in mitochondrial fission and fusion (Cohen et al., 2018b; Guo et al., 2018; Valm et al., 2017). Similarly, transmission electron microscopy (TEM) (Graham et al., 2019) and focused ion beam-scanning electron microscopy (FIB-SEM) (Kizilyaprak et al., 2013; Kremer et al., 2015) have provided exquisite details of the 3D architecture of the cell at extremely high resolution, unraveling mechanisms related to the interaction between subcellular compartments and/or cytoskeleton (Müller et al., 2020). However, due to the limited penetration of the electron beam and specimen thickness, electron microscopy requires the samples to be fixed, stained, dehydrated, embedded in plastic, and sectioned (Noske et al., 2008). Cryo-electron tomography (cryo-ET) can image 200-nm thick sections of adherent cells in a near-native state with a resolution on the order of 30 Å (Lučić et al., 2013; Mahamid et al., 2016). However, due to the high magnification of the technique and the low penetration of the electron beam, the field of view is limited to a few hundred nanometers (Buckley et al., 2019). Moreover, scattering of the incident electron beam decreases the signal-to-noise ratio of the collected micrographs (Peet et al., 2019). Alternatively, imaging biological samples using soft X-ray tomography (SXT) enables imaging intact cells (Larabell and Nugent, 2010) with a spatial resolution of 35–60 nm (McDermott et al., 2012; Schneider et al., 2010) within the mesoscale size range, from 1 to 20 μm (Le Gros et al., 2012). Cells are imaged in the “water window” (284 – 583 eV photon energy), where the soft X-ray beam is attenuated almost entirely by bioorganic material. The density of the organic compounds in each voxel is quantified by measuring the X-ray attenuation (known as LAC – linear absorption coefficient), and its quantification adheres to Beer-Lambert’s law (Do et al., 2015; Ekman et al., 2017). Because each organelle has a distinct concentration of carbon-rich organic molecules, the LAC values can be used as a hallmark to identify individual cell substructures. SXT leverages the natural contrast of cells and does not require imaging probes or fluorescent dyes (Attwood, 2007). Compared to other 3D imaging approaches, SXT is a high-throughput technology with a data collection time of approximately 10 mins (Parkinson et al., 2012), which allows for comparison of larger numbers of cells under multiple biological conditions.
In this manuscript, we demonstrate the unique quantitative abilities of SXT. Our goal was to develop a more in-depth data analysis pipeline for mapping and quantifying inter-organelle interactions with SXT, using their natural contrast rather than stains. To do so, we build upon our initial observation of insulin vesicle-mitochondria contacts in pancreatic β-cells (White et al., 2020). We analyzed new datasets to explore interactions between insulin vesicles and mitochondria, adding a detailed analysis of the ultrastructural reorganization that occurs during a very early stage of the secretory process (~1 min) and a comparison with the later time points. In pancreatic βcells, mitochondria are involved in several mechanisms intended for the production and secretion of insulin. For example, ATP production triggers insulin secretion by closing the ATP-dependent potassium channels (Rorsman et al., 2000). Additionally, there is an association between mitochondrial metabolic products (i.e., Ca2+, NADH, pyruvate, and glutamate) and the activation of the ATP-independent insulin secretory pathways (Maechler et al., 2006; Wiederkehr and Wollheim, 2006). The interaction between insulin vesicles and mitochondria were examined previously (Maechler and Wollheim, 2001; Wollheim, 2000), but never mapped in whole cells.
To demonstrate the capability of SXT to map and quantify interactions between organelles, we examined the colocalization of insulin vesicles and mitochondria. We tracked these organelle interactions after stimulation with glucose and exendin-4, which is an incretin analog of the glucagon-like protein 1 receptor (GLP1R) agonist that potentiates the glucose effect on insulin stimulation (Eng et al., 1992; Gedulin et al., 2005; Kolterman et al., 2003). We captured organelle reorganization as a response to different types of stimulation and tracked alterations of mitochondria-vesicle interactions during the first and second phases of insulin secretion. We characterized the interactions that occur during the secretory process and evaluated the randomness of the interactions and drug stimulation on organelle association. Finally, we showed that vesicles in contact with the mitochondria network have similar LAC values, but different sizes, compared to vesicles farther from mitochondria.
Results
INS-1E architecture by SXT
We used SXT to investigate alterations in cell morphology and molecular density of INS-1E rat insulinoma cells at three different time points representing the early stage of the biphasic secretory process (1 min), the late stage of the first phase of secretion (5 min), and the early stage of the second phase of secretion (30 min) compared with unstimulated cells. Four experimental conditions were examined: no treatment, high glucose (25 mM), high glucose in combination with exendin-4 (Ex-4), and Ex-4 alone. This last condition was used as a control to verify the recapitulation of the features observed with co-stimulation with high glucose. The analysis was performed on 56 cells in total. The collective results in Table 1, though not all statistically significant, represent the type of large datasets that SXT can generate providing an informative and in-depth resource for exploring cellular structure.
Table 1: Quantitative analysis of the segmented organelles.
List of volume and LAC values of cells, nuclei, mitochondria, cytosol, and insulin vesicles for each condition and time point. The mean numbers of insulin 432 vesicles per condition and percentage of vesicles in contact with mitochondria are also reported. Each value is 433 reported with its standard deviation.
| Stimulation | 0 min (n=8) | 25 mM glucose | 25 mM glucose + 10 nM Ex-4 | 10 nM Ex-4 | |||||
|---|---|---|---|---|---|---|---|---|---|
| 1 min (n=4) | 5 min (n=8) | 30 min (n=8) | 1 min (n=4) | 5 min (n=8) | 30 min (n=8) | 5 min (n=4) | 30 min (n=4) | ||
| Cell volume (|jm3) | 715±148 | 636±137 | 752±125 | 873±261 | 633±138 | 1224±304 | 733±246 | 1040±232 | 801±157 |
| Nucleus volume (jm3) | 199±32 | 180±31 | 238±51 | 228±61 | 196±27 | 397±97 | 203±74 | 334±38 | 300±65 |
| Nucleus volume (%) | 28±4 | 31±23 | 32±5 | 27±38 | 29±3 | 33±2 | 28±5 | 33±5 | 37±4 |
| Nucleus LAC (jm−1) | 0.24±0.01 | 0.27±0.02 | 0.28±0.02 | 0.25±0.02 | 0.29±0.02 | 0.26±0.02 | 0.27±0.02 | 0.26±0.02 | 0.24±0.02 |
| Mitochondria volume (jm3) | 32±11 | 42±3 | 43±10 | 38±12 | 51±9 | 73±15 | 41±7 | 86±37 | 60±21 |
| Mitochondria volume (%) | 4.4±0.9 | 6.8±0.9 | 5.8±1.4 | 4.5±1.3 | 8.2±1.3 | 6.4±0.9 | 5.9±1.3 | 8.5±3.4 | 7.5±2.0 |
| Mitochondria LAC (jm−1) | 0.32±0.02 | 0.35±0.03 | 0.36±0.02 | 0.32±0.03 | 0.36±0.02 | 0.33±0.02 | 0.35±0.03 | 0.32±0.03 | 0.30±0.03 |
| Cytosol volume (jm3) | 516±130 | 441±110 | 514±96 | 644±205 | 453±113 | 827±210 | 530±181 | 706±197 | 501±103 |
| Cytosol volume (%) | 72±4 | 69±2 | 68±5 | 72±5 | 71±3 | 68±2 | 74±3 | 67±5 | 63±4 |
| Cytosol LAC (jm−1) | 0.25±0.01 | 0.28±0.02 | 0.29±0.02 | 0.28±0.01 | 0.28±0.03 | 0.27±0.03 | 0.28±0.03 | 0.26±0.01 | 0.25±0.02 |
| Insulin vesicle total number | 319±93 | 310±13 | 409±139 | 787±204 | 380±106 | 386±186 | 515±156 | 280±140 | 244±97 |
| Insulin vesicle total volume (jm3) | 4.0±1.9 | 3.1±1.3 | 4.0±2.6 | 7.5±2.3 | 3.2±0.8 | 3.8±2.0 | 5.4±2.8 | 4.7±3.9 | 2.1±0.9 |
| Insulin vesicle total volume (%) | 0.56±0.28 | 0.48±0.14 | 0.55±0.41 | 0.89±0.30 | 0.55±0.26 | 0.31±0.14 | 0.73±0.32 | 0.43±0.36 | 0.29±0.15 |
| Insulin vesicle LAC (jm−1) | 0.37±0.03 | 0.42±0.02 | 0.46±0.03 | 0.40±0.02 | 0.44±0.04 | 0.45±0.03 | 0.43±0.03 | 0.44±0.02 | 0.43±0.04 |
| Mitochondria-connected nsulin vesicles (%) | 9.7±6.1 | 14.7±6.2 | 21.1±9.8 | 10.3±2.9 | 23.8±15.7 | 20.1 ±11.7 | 15.3±5.7 | 16.9±8.9 | 28.8±10.1 |
Experimental manipulations were conducted using INS-1E cells in suspension. White et al. previously showed that there is no significant difference in the amount of insulin secretion between cells in suspension and adherent cells (White et al., 2020). In addition, the organization of the readily releasable and reserve pools is conserved (Bratanova-Tochkova et al., 2002). After manipulation, cells were placed in thin-walled glass capillaries, frozen, and imaged with SXT. The cells measured between 10–12 μm, which is easily penetrated by the soft X-ray beam (Duke et al., 2014). With a field of view of 16 μm x16 μm at 60 nm spatial resolution (Weinhardt et al., 2020), the whole-cell architecture can be collected in one single data acquisition. Data collection was performed by rotating the cells through 180º to ensure a near-isotropic reconstruction (Figure 1A); this eliminates loss of information caused by the “missing wedge” that is inherent to imaging cells grown on flat specimen mounts (Groen et al., 2019; Loconte et al., 2021).
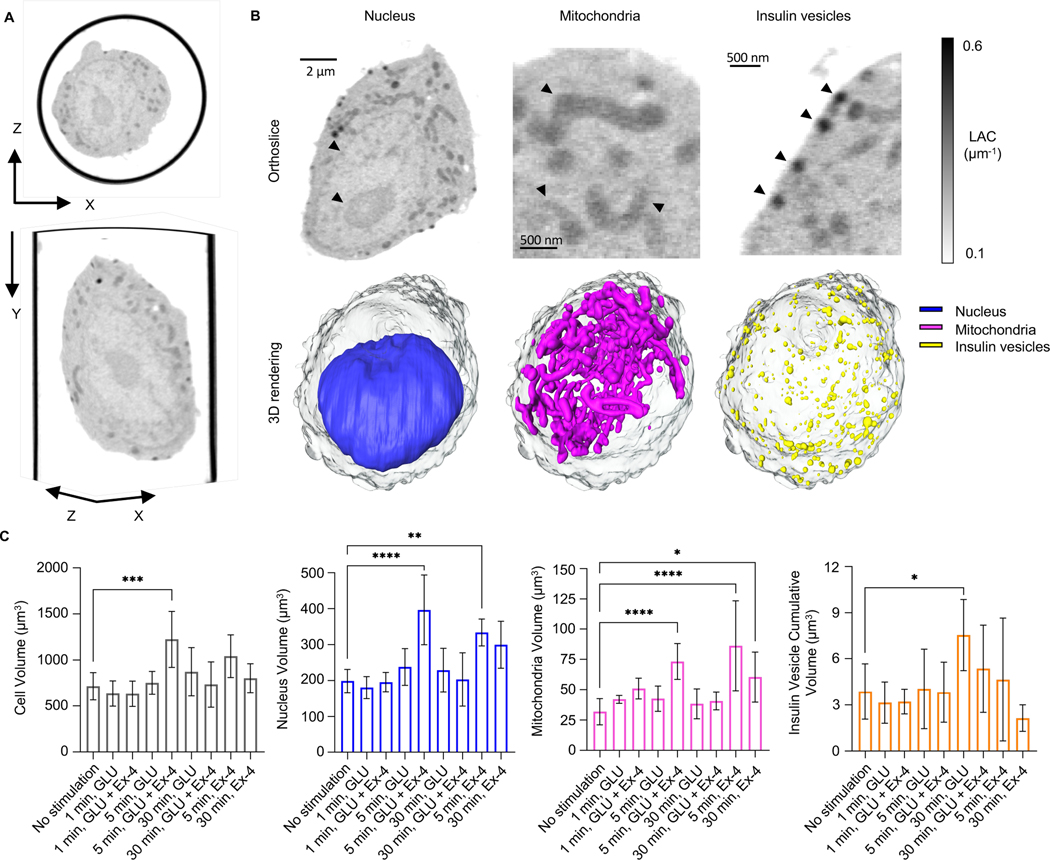
Figure 1: Isotropic reconstruction of a single pancreatic β-cell.
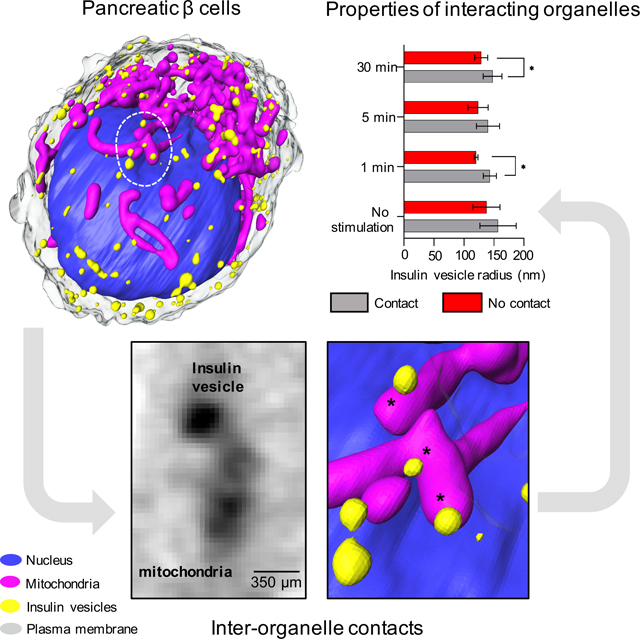
A) The whole cell volume is shown in the XY (bottom) and XZ plane (top). B) Organelles are imaged based on different levels of grey values, with a threshold between 0.1–0.6 μm−1. Single orthoslices (top) and relative 3D rendering (bottom) represent a single β-cell after 1 min of high glucose treatment (cell ID: 821_12). In detail, the rendering shows: nucleus (blue); mitochondrial network (magenta), and insulin vesicles (yellow) and their spatial distribution. All organelles in the orthoslices are highlighted by an arrowhead. C) Plots representing the overall volume of the organelle in the cells per condition and timepoint. We report alteration in the total volume of each type of organelles, in the following order: from left, changes in cell volume with signific difference between 5 min stimulated cells with high glucose+Ex-4 and no stimulated cells (***P=0.0002); changes in the nucleus volume with significant difference between no stimulated cells and 5 min stimulated cells with high glucose+Ex-4 (****P<0.0001), and 5 min stimulated cells with 10 nM Ex-4 (**P=0.0058); changes in the volume of the mitochondrial network with significant difference between no stimulated cells and 5 min stimulated cells with high glucose+Ex-4 (****P<0.0001), 5 min stimulated cells with Ex4 (****P<0.0001), and 30 min stimulated cells with Ex-4 (*P=0.0189); and finally, changes in the insulin vesicle cumulative volume with significant difference between no stimulated cells and 30 min stimulated cell with high glucose (*P=0.0170). Statical significance was validated using Dunnett’s multiple comparison test.
SXT generates 3D images of whole cells and subcellular organelles (Figure 1B), which makes it possible to determine the volumes of cells and organelles. We show that there was a broad range in cell size; the total volume of untreated INS-1E cells was 715±148 μm3 (Figure 1C and Table 1). To identify, label, and characterize specific organelles, we used a combination of LAC values and 3D morphology. We demarcated the cell nucleus by tracing the nuclear surface (Le Gros et al., 2016). The volume of the nucleus was 199±32 μm3, comprising an average of 30.7±3.4% of the total cell volume; the average LAC value was 0.26±0.02 μm-1. In the cytoplasm, we segmented the mitochondrial network and insulin vesicles (Figure 1B). Mitochondria, identified by their characteristic elongated shape, had an average LAC value of 0.32±0.02 μm−1, accounting for 4.37±0.86% of the whole cell volume in unstimulated cells. The spherical insulin vesicles had an average LAC value of 0.37±0.03 μm−1 and occupied just 0.56±0.28% of the total volume of unstimulated cells (Table 1).
Organelle properties and spatial distribution
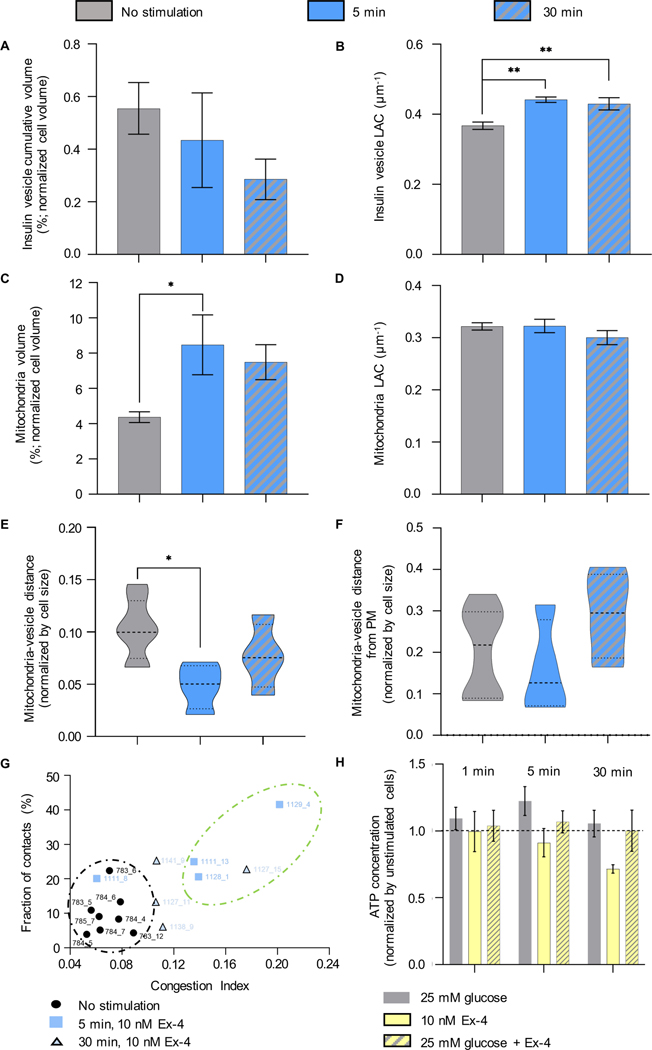
We first calculated the total volume of cells and organelles after stimulation with glucose alone or with Ex-4 at the three different time points (Figure 1C and Table 1). Glucose alone did not trigger any significant changes in the total volume of the cell, nucleus, or mitochondria after 1, 5, or 30-minutes; there was, however, a small but statistically significant increase in total insulin vesicle volume (from 4.0±.1.9 μm3 to 7.5±2.3 μm3) after 30 min. After 5 min stimulation with glucose+Ex-4, however, there was an increase in total cell volume (from 715±148 μm3 to 1224±304 μm3), which was accompanied by an increase in total nuclear volume (from 199±32 μm3 to 397±97 μm3) and total mitochondria volume (from 32±11 μm3 to 73±15 μm3). In cells treated with Ex-4 alone for 5 min, there was a statistically significant increase in total nuclear volume (from 199±32 μm3 to 334±38 μm3) and total mitochondrial volume (from 32±11 μm3 to 86±37 μm3), and an increase in total mitochondrial volume to 60±21 μm3 after 30-min treatment with Ex-4 alone (Figure 1C and Table 1).
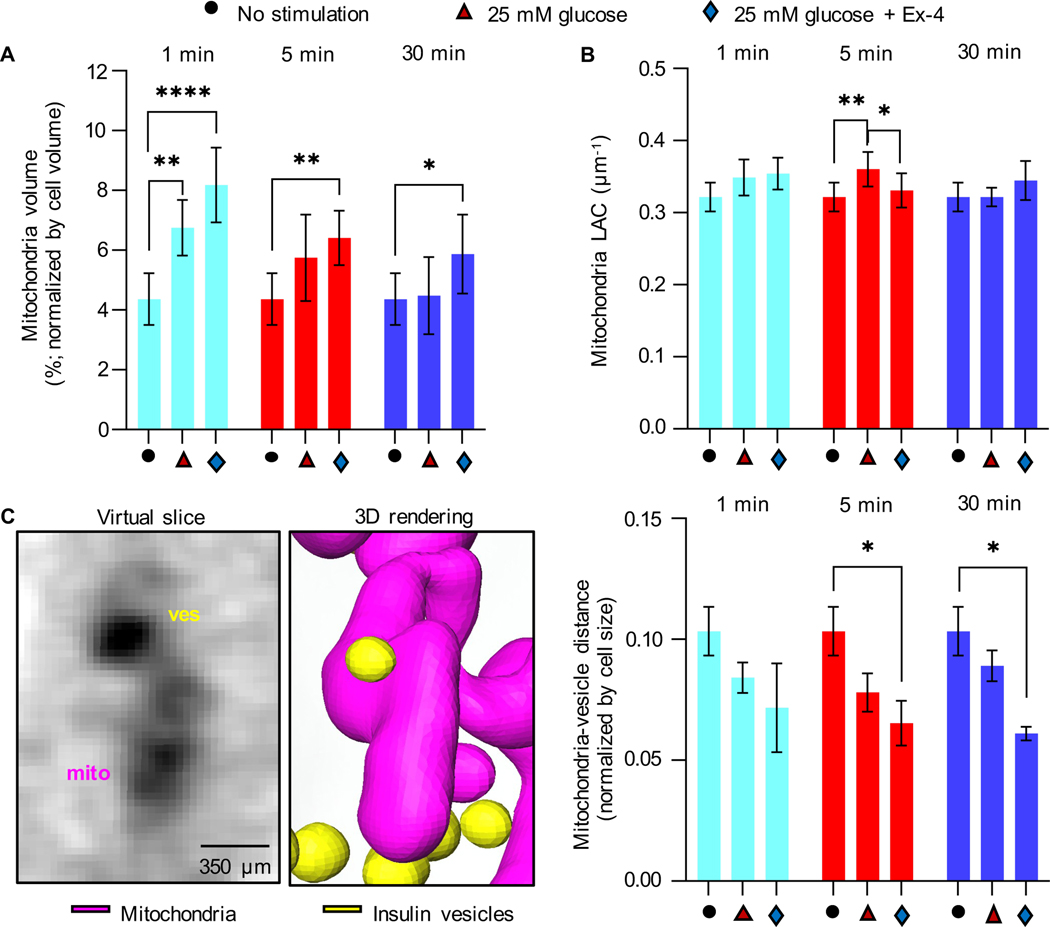
Given the heterogeneity seen in cultured cell lines, and the large variance in cell size, we analyzed changes in the volume of mitochondria and insulin vesicles normalized by cell volume. We observed an increase in the percent volume of mitochondria after 1 min of stimulation with high glucose, both without (6.76±0.93%) and with Ex-4 (8.18±1.25%), compared to unstimulated cells (4.37±0.86%). Mitochondrial volume began to decrease after 5 min of stimulation under both conditions, although more slowly in cells stimulated with glucose+Ex-4 (Figure 2A), suggesting there is an effect of Ex-4 on mitochondria biogenesis (Kang et al., 2015). SXT also provides quantitative information about the density of the biomolecules present in a subcellular structure and a specific LAC value for that organelle. The range of LAC values in mitochondria of untreated INS1E cells (0.32±0.02 μm1) was consistent with values we previously reported for other mammalian cells (https://ncxt.org/primer/). To determine whether the general biochemical composition of mitochondria was affected by each of the treatments, we calculated the average LAC values after treatment. As reported in Table 1 and Figure 2B, no significant changes were visible after treatment with glucose+Ex-4 at any timepoint compared to the unstimulated cells. After 5 min of stimulation with high glucose, however, the mitochondrial LAC value was significantly increased (0.36±0.02 μm−1) compared to both unstimulated (0.32±0.02 μm−1) and glucose+Ex-4 stimulated cells (0.33±0.02 μm−1) within the same timepoint. This indicates there was an increase in the concentration of carbon-rich components (i.e., proteins, lipids, and metabolites) per voxel in mitochondria after 5 min stimulation with glucose (Figure 2B).
Figure 2: Effect of glucose and Ex-4 stimulation on mitochondria properties and spatial distribution.
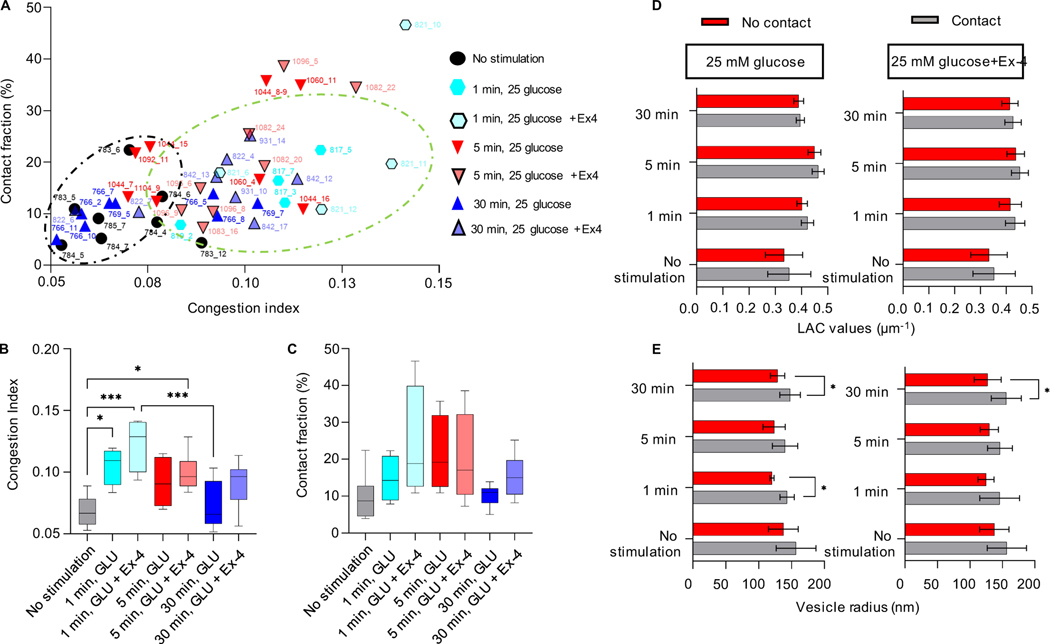
A) Percentage of cell volume occupied by mitochondria for each condition and time point. Compared to unstimulated cells, a significant difference is observed after 1 min (****P<0.001 and **P=0.042), 5 min (**P=0.0035), and 30 min (*P=0.0460) of stimulation with high glucose±Ex-4 (Tukey’s multiple comparison test). B) Mean mitochondria LAC value. Significant difference is observed between unstimulated cells and cells stimulated with 25 mM glucose for 5 min (**P=0.0071), and between 25 mM glucose vs. 25 mM glucose+10 nM Ex-4 stimulated cells for 5 min (*P=0.0426). Statistical significance was evaluated using Tukey’s multiple comparison test. C) Left: An orthoslice and 3D rendering of interacting insulin vesicles and mitochondria after 1 min of stimulation with high glucose. Right: mean distance between insulin vesicles and mitochondria normalized by cell size shows a significant decrease in the distance between unstimulated and glucose+Ex-4 stimulated cells after 5 min and 30 min (respectively, *P=0.0269 and *P=0.0121. Tukey’s multiple comparison test). Error bars represent SDs.
To identify and track the proximity between mitochondria and insulin vesicles under all conditions, we generated a 3D map of the segmented organelles (Figure 2C). We then calculated the minimal distances between vesicle centers and surfaces of the most proximal branch of the mitochondrial network and quantified the average distances. We found that the mean vesicle-mitochondrial distance decreased after stimulating the cells with high glucose+Ex-4. This was observed throughout the cells and is statistically significant after 5 and 30 min of stimulation compared to unstimulated cells, indicating a tighter association between the mitochondria and insulin vesicles under these conditions (Figure 2C).
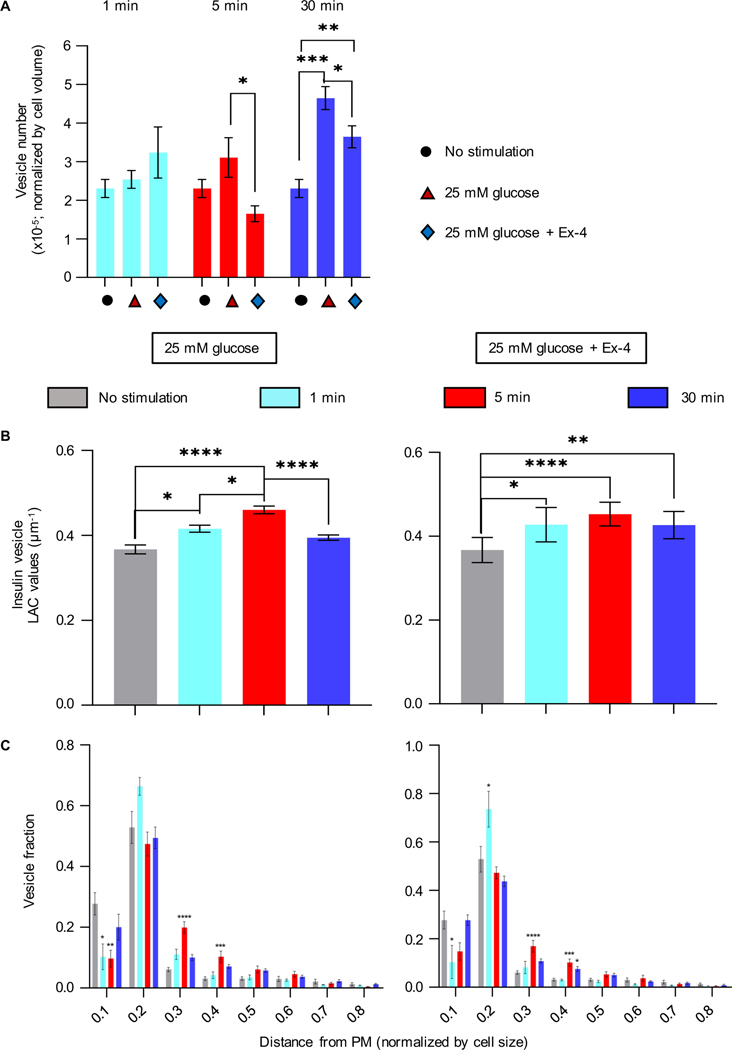
We then compared the number of insulin vesicles seen during the first minute of secretion with the later stages of the insulin secretory process (Figure 3A and Table 1). There was no significant change in the number of insulin vesicles at 1 min post stimulation with either glucose alone or in the presence of Ex-4. At 30 min post stimulation, there was a significant increase after stimulation with glucose alone (787±204) and with glucose+Ex-4 (515±156) compared with the number in unstimulated cells (319±93). At both 5 min and 30 min post stimulation there were significantly fewer vesicles after stimulation with glucose+Ex-4 than with glucose alone, suggesting that Ex-4 may alter the rate of insulin secretion during the second phase of secretion, as reported in White et al., 2020, but does not affect the first phase.
Figure 3: Effect of the high glucose stimulation with and without Ex-4 on insulin vesicle properties and spatial distribution.
A) Number of Insulin vesicles during the two phases of insulin secretion. There was no significant change in the number of vesicles after 1 minute stimulation with glucose alone or in the presence of Ex-4. A significant increase in vesicle numbers was seen at 30 min post stimulation with glucose alone (787±204, ****P<0.0001) and with glucose+Ex-4 (515±156, **P=0.0062), compared with unstimulated cells (319±93). Fewer vesicles were seen after stimulation with glucose+Ex-4 at both 5 (*P=0.0191) and 30 min (*P=0.0436) than when stimulated by glucose alone. B) Mean vesicle LAC values. Left: Treatment with 25 mM glucose. Insulin vesicle LAC values steadily increase after 1 min stimulation, from 0.37±0.03 μm−1 to 0.42±0.02 (*P=0.0138) and to 0.46±0.02 μm−1 after 5 min stimulation (****P<0.0001); there is also a significant difference in the increase between 1- and 5-min stimulation (*P=0.264). At 30 min post stimulation with glucose, there is a significant decrease in LAC value to 0.40±0.02 μm−1 compared with the LAC seen at 5 min post stimulation (****P<0.0001). Right: Treatment with 25 mM glucose+Ex-4. An increase in LAC values after stimulation is seen after 1 min glucose+Ex4 to 0.44±0.04 μm−1 (*P=0.0226), at 5 min to 0.45±0.03 μm−1 (****P<0.0001), and at 30 min to 0.43±0.03 μm−1 (**P=0.0051) (Tukey’s multiple comparison test). C) Plots representing the position of vesicles in the cell as normalized distance from the PM (0.0 is the closest distance to PM and 1.0 the farthest). With respect to the unstimulated cells, significant differences in vesicle population are found for 25 mM glucose-stimulated cells (left panel) within 0.1 from the PM (1 min stimulation: *P=0.0316, and 5 min: **P=0.0040), 0.2–0.3 (5 min stimulation: ****P<0.0001), and 0.3–0.4 (5 min stimulation: ***P=0.005); the same analysis is performed for 25 mM glucose+Ex-4 stimulated cells (right panel), with differences within 0.1 from the PM (1 min stimulated cells: *P=0.0337), 0.1–0.2 (1 min: *P=0.0129), 0.2–0.3 (5 min stimulated cells: ****P<0.0001), 0.3–0.4 (5 min: ***P=0.006 and *P=0.0488) (Dunnett’s multiple comparison test).
There was a significant increase in the molecular density (LAC value) of the vesicles stimulated with glucose and glucose+Ex-4 compared to unstimulated cells; the LAC values after 5 min glucose (0.46±0.02 μm−1) and glucose+Ex-4 (0.45±0.04 μm−1) stimulation are significantly higher than untreated cells (0.37±0.03 μm−1) (Figure 3B and Table 1). These data suggest there has been an alteration in vesicle maturation at the end of the first phase of secretion. Finally, measuring the distance of vesicles from the plasma membrane (PM) shows there is a decrease in the number of vesicles just beneath the PM (within 0.0–0.1, Figure 3C) after 1- and 5-min stimulation with glucose+Ex-4, and after 5 min stimulation with glucose alone, indicating a faster deployment to secretion of the vesicle population during the first phase of secretion (Figure 3C). At the same time there was an increase in the number of vesicles seen slightly farther from the PM (0.2–0.4) at 1 min after glucose+Ex-4 stimulation, and even deeper after 5 min glucose with and without Ex-4, indicating a re-population of the reserve pools (Rorsman et al., 2000).
Dynamics of mitochondria-vesicle interaction during insulin secretion
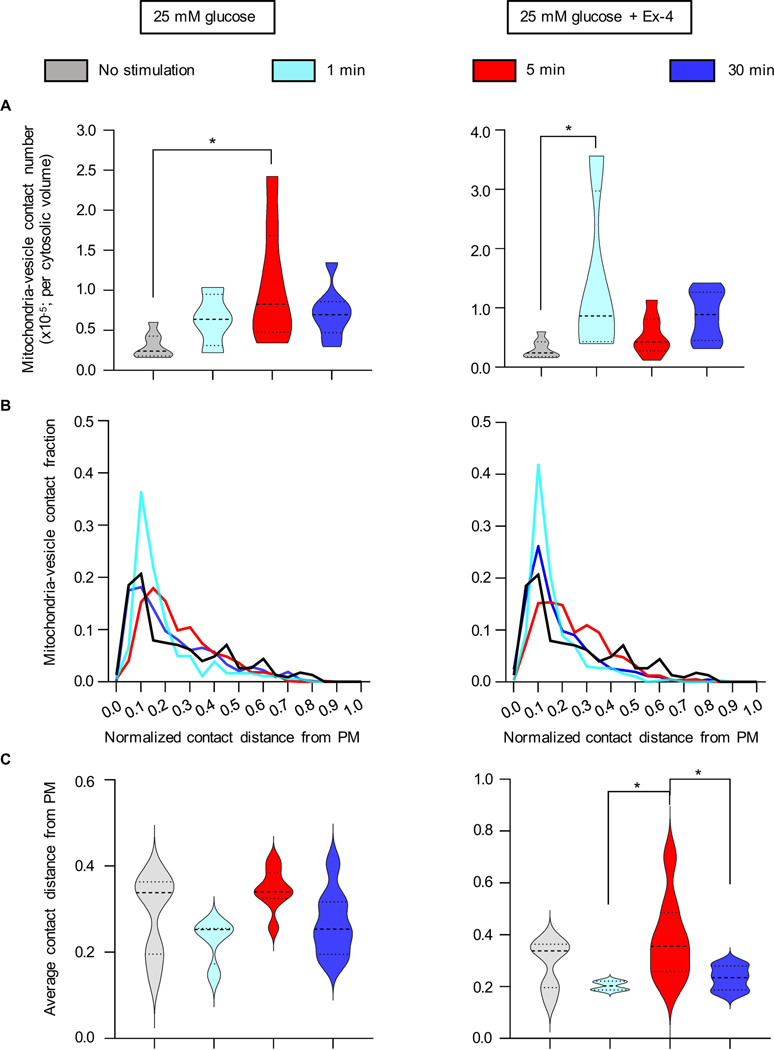
To investigate the frequency of the interaction between insulin vesicles and mitochondria, we quantified the number of mitochondria-vesicle contacts per cytosolic volume. We observed a statistically significant increase in mitochondria-vesicle contacts after 5 min treatment with high glucose without Ex-4 (Figure 4A, left panel) and after 1 min treatment with Ex-4 (Figure 4A, right panel) compared to unstimulated cells. We also analyzed the distribution of contacts with respect to the PM (Figure 4B) and found that there is an increase in contacts near the PM after 1-min treatment with high glucose±Ex-4. This was confirmed by determining the average distance of the contacts from the PM. The shortest distance of mitochondria-vesicle contact from the PM occurs after 1min and 30-min stimulations with high glucose±Ex-4 (Figure 4C). Interestingly, the contacts at 5 min cover a broad region beneath the PM after the stimulation with glucose+Ex-4, suggesting that this interaction in the cell might play a role in the active vesicle movement towards the PM and vesicle maturation.
Figure 4: Quantification of the spatial distribution of mitochondria-insulin vesicle contacts and localization with respect to PM.
A) Comparison of the number of mitochondria-insulin vesicle contacts per cell normalized by cytosolic volume. The mean value of contacts at each time shows a significant difference between unstimulated and 5-min stimulated cells with high glucose (*P=0.0126) (left panel), and 1-min stimulated cells with 25mM glucose+Ex-4 (*P=0.0267) (right panel) (Tukey’s multiple comparison test). B) Frequency distribution of mitochondria-vesicle contacts with respect to the PM; the distance is normalized by cell size. C) Average distance from the PM after 25 mM glucose of stimulation (left) and 25 mM glucose+Ex-4 (right). No significance difference was seen in cells treated with glucose alone. A significant difference in the distance was observed after 5-min stimulation with glucose+Ex-4 with respect to 1 min (*P=0.0453) and 30 min (*P=0.0397) (Tukey’s multiple comparison test).
Given the simultaneous increase in the mitochondria volume in cells treated with Ex-4 and the number of insulin vesicles after 30 min of stimulation with glucose±Ex-4, we aimed to evaluate the randomness of the interaction accounting for the crowdedness of the cell. We defined a congestion index (C.I., see Methods) and correlated its value to the fraction of insulin vesicles associated with mitochondria for each cell. Figure 5A shows that unstimulated cells and those stimulated with glucose for 30 min cluster at regions with a lower C.I. (black circle - below 0.08), with contacts between 3 and 13% of the total number of vesicles. On the other hand, those cells treated with glucose+Ex-4 for 1 and 5 min were randomly distributed in the higher C.I. range (greater than 0.08) with contacts between 4 and 25% of the vesicles in contact with mitochondria. There is a high degree of randomness in the populations of cells treated with glucose±Ex-4 for 1 and 5 min (Figure 5A–C), highlighting cell-dependent variability in vesicle-mitochondria interactions and cell-to-cell variability (White et al., 2020). Moreover, given the high variability of total cell volume among conditions and timepoints, the C.I. does not carry the accuracy required to determine the randomness of the interaction.
Figure 5: Analysis of the mitochondria-insulin vesicle interaction randomness.
A) The plot shows the correlation between the number of mitochondria-vesicle contacts and the congestion index (C.I.) per individual cell. The black circle shows clusters of cells with less crowding (lower C.I., 0.05–0.08) and percentage of contacts (3–12%); the green circle shows those with more crowding (higher C.I., 0.08–0.14) and percentage of contacts (425%) under different conditions. B) Mean value of the C.I. for each condition. Significant difference is reported between no stimulated cells and 1 min glucose stimulated cells (*P=0.0210), 1 min glucose+Ex-4 stimulated cells (***P=0.0002), and 5 min glucose+Ex-4 stimulated cells (*P=0.0153); significant difference is also reported between 1 min glucose+Ex-4 stimulated cells and 30 min glucose stimulated cells (***P=0.0007). C) Mean value of the faction of contacts per condition. D) Plot showing the LAC values of vesicles in contact (grey) and not in contact (red) with mitochondria, at each time point and stimulation, with no significant difference. E) Changes in the vesicle size whether in contact with the mitochondria network (grey) or not (red). The plot shows a significant difference between the size of vesicle after 1 min stimulation with glucose (143±11 nm contacts; 120±3.0 nm no contacts, *P=0.0195), 30 min stimulation with high glucose (148±16 nm contacts; 129±11 nm no contacts, *P=0.0147) and high glucose+Ex-4 (155±23 nm contacts; 127±21 nm no contacts, *P=0.0211). Error bars represent the SDs, and the statistical significance was evaluated using Welsh’s test.
To explore whether vesicle-mitochondria contacts are caused by crowdedness rather than biological function, we compared the properties of the vesicles associated with mitochondria to the vesicles farther away (Figure 5D–E). The vesicles in contact with the mitochondria did not show a significantly higher mean LAC value after the treatment with glucose and glucose+Ex-4 (Figure 5D). We did, however, observe an increase in the mean radius of the vesicles associated with mitochondria compared to vesicles that are not associated with mitochondria after treatment with glucose for 1 min (143±11 nm contacts; 120±3.0 nm no contacts) and 30 min (148±16 nm contacts; 129±11 nm no contacts). An increase in contacts was also seen after 30-min treatment with glucose+Ex-4 (155±23 nm contacts; 127±21 nm no contacts) (Figure 5E). The significant difference in the physical properties seen between those vesicles associated with mitochondria and those not associated with mitochondria suggests that the interactions may be of biological significance, potentially contributing to the vesicle maturation process.
Quantification of exendin-4 effect on the contact spatial distribution and organelle properties
It has been reported that Ex-4 enhances mitochondrial biogenesis and increases cell mass and function by restoring insulin secretion (Kalavalapalli et al., 2019; Kang et al., 2015). The role of Ex-4 has been explored primarily in combination with glucose co-stimulation, but the direct effect of Ex-4 on the ultrastructural organization and chemical composition of cells has not been investigated. We verified the direct effect of Ex-4 on cell ultrastructure by decoupling drug and glucose stimulation and treating the cells with 10 nM Ex-4 alone for 5- and 30-min. Ex-4 alone does not increase the cumulative insulin vesicle volume, unlike what we observed after co-stimulation with glucose for 30 minutes (Table 1). Thus, Ex-4 stimulation alone does not drive insulin vesicle biogenesis. On the other hand, Ex-4 affects the chemical composition of the vesicles both after 5 and 30 min from the stimulation (Figure 6B), increasing the LAC values (untreated: 0.37±0.03; 5 min: 0.44±0.02 μm−1; 30 min: 0.43±0.04 μm−1). Unlike the results shown in Figure 2A, Ex-4 increases the mitochondrial volume after 5 min, but this effect is not sustained during the second phase of secretion (30 min). Mitochondrial occupancy within the cell decreases during Ex-4 stimulation (unstimulated: 4.37±0.86%; 5 min: 8.47±3.40%; 30 min: 7.49±1.99%), indicating that the role of incretin in facilitating the mitochondrial biogenesis is dependent on glucose co-stimulation (Figure 6C). Like the treatment with high glucose+Ex-4, the LAC value of mitochondria is comparable between unstimulated cells (0.32±0.02 μm−1) and 5-min (0.32±0.03 μm−1) and 30-min Ex-4 stimulated cells (0.30±0.03 μm−1) (Figure 6D), confirming that the mitochondria molecular density is not altered by the treatment with Ex-4 alone. The distance between mitochondria and insulin vesicles is similarly affected by the treatment with Ex-4, inducing insulin vesicle and mitochondria association after 5 min of stimulation (Figure 6E). In terms of mitochondria-insulin vesicle interaction and contact distribution from the PM (Figure 6E–F), Ex-4 treatment is less effective after 30 min of stimulation compared to the high glucose+Ex-4 treatment (Figure 2C and Figure 4). Finally, we correlated the C.I. for each cell with the fraction of insulin vesicles in contact with the mitochondrial network. Also in this case, we found that the increase of the mitochondria network volume is not directly correlated to the number of contacts per cell; after 30 min of treatment, we observe different numbers of contacts in the cells 1138_9, 1127_11, and 1141_9, but similar C.I. (Figure 6G). Thus, we hypothesize that the vesicle-mitochondria association might be driven by cellular mechanisms that promote the enlargement of vesicle size (Figure 5E).
Figure 6: Effect of Ex-4 on the organelle properties.
A) Percentage of cell volume occupied by insulin vesicles in unstimulated, 5-min, and 30-min post stimulation with 10 nM Ex-4. B) LAC of insulin vesicles treated with Ex-4. Statistical significance was found between the unstimulated (0.37±0.03μm−1) and 5-min (0.44±0.02μm−1, **P=0.0026) and 30-min 0.43±0.04μm−1, **P=0.0089) post stimulation. C) Percentage of cell volume occupied by the mitochondrial network. Statistical significance was observed after stimulating the cells for 5 min with Ex-4 (8.47±3.40%, *P=0.0133), compared to unstimulated cells (4.37±0.86%). D) There were no significant differences in LAC values for mitochondria treated for 5 (0.32±0.03μm−1), and 30 min (0.30±0.03μm1) with Ex-4 compared with untreated cells (0.32±0.02μm−1). E) There was a significant difference in the mean distance between mitochondria and insulin vesicles in unstimulated cells and cells treated for 5-min (*P=0.0166). F) There was no significant difference in mitochondria-vesicle contacts at different distances from the PM (normalized by cell size) between unstimulated cells and after 5, and 30 min of stimulation. G) Correlation between C.I. and fraction of vesicles in contact with mitochondria in unstimulated cells and cells after 5-, and 30-min of stimulation with Ex-4. Two clusters are highlighted: the black circle shows cells with low C.I. (0.04–0.08) and fraction of contact; the green circle shows cells with high C.I. (0.12–0.20) and high fraction of contacts (20–42%). H) ATP concentration of cells treated with high glucose±Ex-4 and Ex-4 alone. The ATP concentration for each condition is normalized by the ATP concentration in unstimulated cells. The black line shows the distance between the ATP value in the unstimulated cells and cells treated with glucose and Ex-4. In all panels, statistical significance was evaluated using Tukey’s multiple comparison test.
Finally, we conducted an ATP quantification assay to rationalize the relationship between mitochondria biogenesis and the effect of Ex-4. Although we observed an increase in the mitochondria volume after the stimulation with high glucose and Ex-4, we did not detect a significant difference in the amount of ATP produced within the first 30 min of stimulation with either high glucose or high glucose+Ex-4, and a lower level of ATP was measured within 30 min of stimulation with Ex-4 alone (Figure 6H). Thus, we hypothesize that despite the increase in the mitochondrial volume, the production of ATP was not enhanced by the treatment with Ex-4.
Discussion
This study demonstrates the feasibility of quantifying rearrangements in the cellular ultrastructure at high resolution (~60 nm) using soft X-ray tomography. SXT can be used to quantify changes in organelle morphology and spatial distribution and to track the effects of cell stimulation over time. We detected rearrangements in mitochondria and insulin vesicle localization and their association during the first and second phase of the insulin secretion, consistent with previous observations reported in the literature (Maechler and Wollheim, 2001; White et al., 2020; Wollheim, 2000). Our data provide unprecedented insights into the cell-wide interactions between vesicles and mitochondria, showing that most of the interactions during the first and second phases of secretion take place in close proximity to the PM, in agreement with the hypothesis that the interaction is functional for vesicle docking and fast release (Maechler and Wollheim, 2001). Similar evidence in the insulin vesicle radius variability in different cellular neighborhoods was observed by Zhang et al. (Zhang et al., 2020) who suggested that the interaction with mitochondria may contribute to the larger radius of vesicles localized in close proximity to the PM, where the interaction is more frequent. Future use of cryo-electron tomography may help to capture the local mitochondria-insulin vesicle interaction; however, this would require correlation with fluorescence microscopy to target the sites of interaction. Ex-4 enhances the interaction of vesicles and mitochondria, contributing to a fast rearrangement of cellular ultrastructure. Ultrastructural rearrangements on this scale and at high resolution are difficult to investigate using conventional microscopy, since, for instance, the use of classical TEM reveals only local rearrangements in the cellular ultrastructure (Fava et al., 2012; Wollheim, 2000). More recently, developments of FIB-SEM have shown distinct interactions between insulin vesicles and microtubules in primary β-cells, allowing quantification of the distribution of insulin vesicles in contact with microtubules from the nucleus and PM and defining the role of the cytoskeleton in the process of vesicle transfer from the Golgi (Müller et al., 2020). Despite the high resolution achieved by electron microscopy over the past decade, the requirement of sample fixation, staining, dehydrating and slicing adds time-consuming steps that limit the numbers of cells that can be investigated. However, the combination of electron microscopy with SXT will be a valuable tool to identify the role of organelles not seen in SXT due to limitations of resolution and contrast (e.g., cytoskeleton) (Chichón et al., 2012).
A unique feature of SXT is the quantitative nature of the LAC value, which allows detection of alterations in the molecular density of subcellular compartments (McDermott et al., 2012). We observed a significant increase in the mitochondria LAC value after treating the cells for 5 min with 25 mM glucose, revealing the possible activation of mechanisms that involve the respiratory cycle and glucose metabolism (Maechler et al., 2006); on the contrary, we could not identify changes in the mitochondria LAC after treatment with only glucose or Ex-4 after 1- and 30-min of stimulation. Also, the average LAC values for cellular components in different stages allows us to locally track the effect of interactions between different organelles. In this specific case, we did not observe a statistically significant variation in the LAC as a response to association of insulin vesicles with mitochondria; however, we could see an increase in vesicle size, as detected with mature vesicles in proximity to the PM (Suckale and Solimena, 2010; Zhang et al., 2020). Our study paves the way for investigation of the metabolic and protein signaling processes that enhance the interaction between mitochondria and insulin vesicles to explore the dynamics of organelle cross-talk in the cell. We provide evidence for the spatial proximity of vesicles to mitochondria, suggesting possible intra-organelle communication. Similarly, such interaction is corroborated by the enlargement of the vesicles in contact with the mitochondrial network at different time points during the insulin secretion process. Altogether, these data can contribute to the design of live cell, proteomic, and metabolomic experiments, focusing on the enrichment of vesicle and mitochondria components. Moreover, to further investigate the role of this interaction, the use of fluorescent probes targeting putative proteins involved in the interaction will provide an additional tool to explore the cell dynamics via live-cell or pulse-chase experiments, informed by the timepoints observed via SXT.
A shortfall of SXT imaging is the lack of details at the molecular level, which would inform about physical interactions among organelles. This lack of information is readily overcome by correlating SXT reconstructions with fluorescence data acquired from the same specimen. Individual molecules can be spatially localized based on the overlay of the fluorescence signal with the X-ray absorption (Smith et al., 2014). Moreover, the development of cryogenic fluorescence microscopes has significantly improved this multi-scale description of the cell (Elgass et al., 2015). For instance, the use of fluorescently labeled insulin vesicles will help in elucidating the role of vesicle maturation in association with mitochondria, confirming that the association may activate the maturation process. Moreover, our results will inform future live cell imaging experiments by providing a general timescale, as well as the specific regions of the cell in which to observe vesicle-mitochondria contacts. Another limitation is the time-consuming segmentation process; the lack of an automatic pipeline for the organelle identification and annotation increases the time to obtain a segmented 3D reconstruction for analysis. However, recent efforts in the development of machine learning-based segmentation tools will facilitate the analysis and interpretation of whole-cell SXT data in a shorter time (Francis et al., 2020).
To conclude, SXT combines the short time of data collection (<10 min) with the quantitative description of subcellular features, opening the way to the spatiotemporal mapping of cell reorganization. This approach can, in fact, be applied to any organelle-organelle interaction within any cell type and can inform about intracellular systemic interactions with minimal cell manipulation. Therefore, SXT can be considered a very useful tool to augment data-driven models (Raveh et al., 2021; Singla et al., 2018; Singla and White, 2021) and to provide details about 3D subcellular reorganization in healthy and pathological conditions.
STAR Methods
Resource availability
Lead contact
Further information and requests for resources and reagents could be addressed to and fulfilled by the lead contact: Carolyn A. Larabell (carolyn.larabell@ucsf.edu).
Materials availability
This study did not generate new reagents.
Data and code availability
All original code and reconstructed data are available from the corresponding contacts upon request.
Any additional information required to reanalyze the data reported in this paper is available from the corresponding contacts upon request.
Experimental model and subject details
Cell line: Rat insulinoma INS-1E cell line was obtained from ATCC. Cells were cultured in T75 flasks with Optimized RPMI media supplemented with 10% FBS, 1× Pen/Strep, and 0.05 mM β-mercaptoethanol and seeded at a density of 4.0×105 cells/cm2. All cells were grown at 37°C, in a 5% CO2 atmosphere.
Method details
Cell stimulation
Before glucose treatment, the cells were incubated in 4 ml of Krebs-Ringer bicarbonate buffer (KREBs) (115 mM NaCl, 24 mM NaHCO3, 5 mM KCl, 1 mM MgCl2, and 1 mM CaCl2) supplemented with 10 mM Hepes (pH 7.4), 0.1% bovine serum albumin, and 1.1 mM glucose for 30 min at 37°C. The glucose starvation buffer was then removed and replaced with 2 ml of buffer containing either 25 mM glucose, 25 mM glucose+10 nM Ex-4, or 10 nM Ex-4 and incubated for 1, 5, or 30 min at 37°C, in 5% CO2 atmosphere.
SXT sample preparation and data collection
Cells were removed from the T75 flask and treated in suspension as reported in the previous paragraph. Cell were harvested by centrifugation at 300g for 6 min, resuspended in Krebs buffer, and loaded in thin-walled glass capillaries (1.5 μl per loading). Each capillary was then rapidly plunged-frozen in nitrogen-cooled liquid propane (Parkinson et al., 2013).
Projection images were collected at 517 eV using XM-2 (Le Gros et al., 2014), the National Center for X-ray Tomography soft X-ray microscope at the Advanced Light Source of Lawrence Berkeley National Laboratory; the microscope was equipped with a 60-nm resolution objective lens. During data collection, cells were maintained in a stream of helium gas cooled to liquid nitrogen temperatures (Parkinson et al., 2013), which allows the collection of projection images while reducing the effects of exposure to radiation. Projection images were collected sequentially around a rotation axis of 180°, with 2° increments. Depending on the synchrotron ring current, an exposure time of 140 and 350 ms was used. The image 3D reconstruction was achieved using an iterative reconstruction method in the software package AREC3D (Parkinson et al., 2012). For comparison, all data were normalized and transferred to linear absorption coefficient by accounting for pixel size.
ATP quantification assay
Cell ATP level was measured from cells grown in a 96-well plate, with 4 repeats for each condition. Cells were treated with 25 mM glucose, 25 mM glucose+Ex-4, and Ex-4 alone. The experiment was performed on unstimulated cells and cells stimulated for 1, 5, and 30 min. 50 μl of detergent (ATP luminescence detection kit, Abcam, ab113849) were added to each well to lyse the cells and stabilize the ATP, followed by 50 μl of substrate. Cell lysate was then incubated in the dark for 15 min. Luminescence was read for 2s using a plate reader. Substrate solution was used as blank.
Segmentation and LAC quantification
All cells were manually segmented using the software Amira 2019.1 (Thermo Scientific™). The identification of each type of organelles was based on previously published data (McDermott et al., 2009; White et al., 2020). The nucleus was manually segmented at first in every 10 ortho slices; each orthoslice was consequently interpolated to reconstruct the 3D label field. Mitochondria, lipid droplets, and insulin vesicles were both manually and semiautomatically segmented, setting the threshold of X-ray absorption to values reported in literature (McDermott et al., 2009; White et al., 2020). Lipid droplets were identified setting the threshold between 0.48 and 0.90 μm−1, with maximum peak per droplet set at about 0.66 μm-1. An average insulin vesicle LAC of 0.42±0.03−1 μm was used to label these organelles with a range of LAC values that spans from 0.28 to 0.62 μm−1 was used along with morphology and size.
Insulin vesicle distance from PM and mitochondria and vesicle-mitochondria contact distribution
The distance of each insulin vesicle from the PM and mitochondria was calculated using Euclidean distance transform (EDT). For distribution from the PM, the shortest distance between the vesicle center and the PM was considered, and for the distribution of the vesicles from the mitochondria we considered the shortest distance between vesicle center and the mitochondria mask. To also account for the change in the cell size across all conditions, the observed vesicle distance from both PM and mitochondria was normalized by the maximum EDT distance of the respective cell. The insulin vesicle-mitochondria contact-point distribution was generated applying the same strategy to the vesicle-mitochondria contact.
Identification of LAC and size of insulin vesicles associated to mitochondria
In each cell, the vesicle was considered in contact with mitochondria if the shortest distance between the vesicle surface and mitochondria mask was less than 1.5 voxels. The shortest distance was computed using EDT, similar to the distance of insulin vesicles from PM. Finally, the LAC and the volume of vesicles in contact or not in contact with the mitochondria network were extracted for each single instance. The mean values for each group were compared for each condition.
Congestion Index
The congestion index (C.I.) describes the crowdedness of a single cell under a well-defined treatment and is defined as:
where, and are the insulin vesicles, mitochondria, whole cell and nucleus volume in the single cell, respectively. The index identifies the fraction of the non-nuclear volume in the cell occupied by insulin vesicles and mitochondria. The higher value of the C.I. corresponds to the crowded ultrastructure in the cell.
Quantification and statistical analysis
Data statistical analysis was performed using Prism (version 9.2, GraphPad Software, La Jolla, CA). The analysis of variance test (ANOVA) was used to determine statistical difference in the mean of three or more data populations. Each test was followed up by a post hoc test, reported in the main body of the text for each analysis. Welsh’s test was used to determine statistical difference between the mean values of the LAC and the radius of insulin vesicles in contact and not in contact with mitochondria.
Key resource table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, Peptides and Recombinant proteins | ||
| Addex Bio Optimized RPMI media | Addex Bio | Cat# C0004-02 |
| Heat inactivated FBS | Fisher Scientific | Cat# 35-011-CV |
| PBS | Thermo Scientific | Cat# 10010023 |
| 1xPen/Strep | Invitrogen | Cat# 15140-148 |
| β-mercaptoethanol | Invitrogen | Cat# 21985-023 |
| Glucose | Sigma | Cat# 67021 |
| NaCl | Sigma | Cat# S9888 |
| NaHCO3 | Sigma | Cat# S5761 |
| KCl | Sigma | Cat# 746436 |
| MgCl2 | Sigma | Cat# M2670 |
| CaCl2 × 2H2O | Sigma | Cat# C7902 |
| BSA | Sigma | Cat# A3912 |
| HEPES | Sigma | Cat#H4034 |
| Exendin-4 | Med Chem Express | Cat# HY-13443 |
| Critical Commercial Assays | ||
| Luminescent ATP Detection Assay Kit | Abcam | Cat# Ab113849 |
| Deposited Data | ||
| Raw and analyzed data (no stimulated cells, 5 min and 30 min 25 mM glucose stimulated cells and 25 mM glucose+Ex-4 stimulated cells) | (White et al., 2020) | N/A |
| Raw and analyzed data (1 min 25 mM glucose stimulated cells and 25 mM glucose+Ex-4 stimulated cells; 5 min and 30 min 10 nM Ex-4 stimulated cells) | This paper | N/A |
| Experimental Models: Cell Lines | ||
| INS-1E | Addex Bio | Cat# C0018009 |
| Software and Algorithms | ||
| ImageJ - Fiji 1.53 c | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| GraphPad Prism 9.2 | GraphPad | https://www.graphpad.com/scientificsoftware/prism/ |
| Amira 2019.1 | Thermo Scientific | https://thermofisher.com/amira-avizo |
| IMOD - 4.10.42 | (Kremer et al., 1996) | https://bio3d.colorado.edu/imod/ |
| AREC reconstruction | (Parkinson et al., 2012) | N/A |
| Organelle distribution pipeline | (White et al., 2020) | N/A |
| Contact distribution pipeline | This paper | N/A |
| Identification of LAC and size of in vesicles associated to mitochondria | This paper | N/A |
Highlights.
Soft X-ray tomography can be used to quantify intracellular contacts
Organelles are identified by differential X-ray absorption
Soft X-ray tomography enables comprehensive subcellular mapping
Soft X-ray tomography identifies variations in the organelle molecular density
Acknowledgments
The authors would like to thank Prof. Raymond C. Stevens, Dr. Liping Sun, and the members of the Pancreatic Beta Cell Consortium for the insightful discussions. The authors also thank Dr. Bieke Vanslembrouck for the support during the data collection and analysis, and Dr. Claire Cato for her feedback on the manuscript. The authors would like to thank Dr. Michal Hammel at LBNL for the use of the plate reader.
Funding: The National Center for X-ray Tomography was supported by NIH (P41GM103445), DOE’s Office of Biological and Environmental Research (DE-AC02–5CH11231). K.L.W. was supported by the Bridge Institute and USC and the Burroughs Wellcome Fund. A.S. was supported by NIH/NIGMS R01GM083960 and P41GM109824. V.L. was supported by NSFC (3191101219).
Footnotes
Declaration of interest
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Attwood D. (2007). Soft x-rays and extreme ultraviolet radiation: principles and applications (Cambridge University Press; ). [Google Scholar]
- Bratanova-Tochkova TK, Cheng H, Daniel S, Gunawardana S, Liu Y-J, Mulvaney-Musa J, Schermerhorn T, Straub SG, Yajima H, and Sharp GWG (2002). Triggering and Augmentation Mechanisms, Granule Pools, and Biphasic Insulin Secretion. Diabetes 51, S83. [DOI] [PubMed] [Google Scholar]
- Buckley G, Gervinskas G, Taveneau C, Venugopal H, Whisstock JC, and de Marco A. (2019). Automated cryo-lamella preparation for high-throughput in-situ structural biology. bioRxiv. [DOI] [PubMed] [Google Scholar]
- Carlessi R, Chen Y, Rowlands J, Cruzat VF, Keane KN, Egan L, Mamotte C, Stokes R, Gunton JE, Bittencourt P.I.H.d., et al. (2017). GLP-1 receptor signalling promotes β-cell glucose metabolism via mTOR-dependent HIF-1α activation. Scientific Reports 7, 2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichón FJ, Rodríguez MJ, Pereiro E, Chiappi M, Perdiguero B, Guttmann P, Werner S, Rehbein S, Schneider G, Esteban M, et al. (2012). Cryo X-ray nano-tomography of vaccinia virus infected cells. Journal of Structural Biology 177, 202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Valm AM, and Lippincott-Schwartz J. (2018a). Interacting organelles. Current Opinion in Cell Biology 53, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Valm AM, and Lippincott-Schwartz J. (2018b). Multispectral Live-Cell Imaging. Current Protocols in Cell Biology 79, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Díaz P, Sandoval-Bórquez A, Bravo-Sagua R, Quest AFG, and Lavandero S. (2021). Perspectives on Organelle Interaction, Protein Dysregulation, and Cancer Disease. Frontiers in Cell and Developmental Biology 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do M, Isaacson SA, McDermott G, Le Gros MA, and Larabell CA (2015). Imaging and characterizing cells using tomography. Arch Biochem Biophys 581, 111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke E, Dent K, Razi M, and Collinson LM (2014). Biological applications of cryo-soft X-ray tomography. Journal of Microscopy 255, 65–70. [DOI] [PubMed] [Google Scholar]
- Ekman AA, Chen JH, Guo J, McDermott G, Le Gros MA, and Larabell CA (2017). Mesoscale imaging with cryo-light and X-rays: Larger than molecular machines, smaller than a cell. Biology of the Cell 109, 24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgass KD, Smith EA, LeGros MA, Larabell CA, and Ryan MT (2015). Analysis of ERmitochondria contacts using correlative fluorescence microscopy and soft X-ray tomography of mammalian cells. J Cell Sci 128, 2795–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J, Kleinman WA, Singh L, Singh G, and Raufman JP (1992). Isolation and characterization of exendin-4, an exendin-3 analogue, from Heloderma suspectum venom. Further evidence for an exendin receptor on dispersed acini from guinea pig pancreas. Journal of Biological Chemistry 267, 7402–7405. [PubMed] [Google Scholar]
- Fava E, Dehghany J, Ouwendijk J, Müller A, Niederlein A, Verkade P, Meyer-Hermann M, and Solimena M. (2012). Novel standards in the measurement of rat insulin granules combining electron microscopy, high-content image analysis and in silico modelling. Diabetologia 55, 1013–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis JP, Wang H, White K, Syeda-Mahmood T, and Stevens R. (2020). Neural network segmentation of cell ultrastructure using incomplete annotation. arXiv, 1183–1187. [Google Scholar]
- Gedulin BR, Nikoulina SE, Smith PA, Gedulin G, Nielsen LL, Baron AD, Parkes DG, and Young AA (2005). Exenatide (Exendin-4) Improves Insulin Sensitivity and β-Cell Mass in Insulin-Resistant Obese fa/fa Zucker Rats Independent of Glycemia and Body Weight. Endocrinology 146, 2069–2076. [DOI] [PubMed] [Google Scholar]
- Graham BJ, Hildebrand DGC, Kuan AT, Maniates-Selvin JT, Thomas LA, Shanny BL, and Lee WCA (2019). High-throughput transmission electron microscopy with automated serial sectioning. bioRxiv, 1–15. [Google Scholar]
- Groen J, Conesa JJ, Valcárcel R, and Pereiro E. (2019). The cellular landscape by cryo soft X-ray tomography. Biophysical Reviews 11, 611–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li D, Zhang S, Yang Y, Liu JJ, Wang X, Liu C, Milkie DE, Moore RP, Tulu US, et al. (2018). Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 175, 1430–1442.e1417. [DOI] [PubMed] [Google Scholar]
- Henne WM (2021). Organelle homeostasis principles: How organelle quality control and inter-organelle crosstalk promote cell survival. Developmental Cell 56, 878–880. [DOI] [PubMed] [Google Scholar]
- Kalavalapalli S, Bril F, Guingab J, Vergara A, Garrett TJ, Sunny NE, and Cusi K. (2019). Impact of exenatide on mitochondrial lipid metabolism in mice with nonalcoholic steatohepatitis. Journal of Endocrinology 241, 293–305. [DOI] [PubMed] [Google Scholar]
- Kang MY, Oh TJ, and Cho YM (2015). Glucagon-Like Peptide-1 Increases Mitochondrial Biogenesis and Function in INS-1 Rat Insulinoma Cells. Endocrinol Metab 30, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kizilyaprak C, Bittermann AG, Daraspe J, and Humbel BM (2013). FIB-SEM Tomography in Biology. In Electron Microscopy Methods in Molecular Biology (Methods and Protocols) (Humana Press, Totowa, NJ: ), pp. 541–558. [DOI] [PubMed] [Google Scholar]
- Kolterman OG., Buse JB., Fineman MS., Gaines E., Heintz S., Bicsak TA., Taylor K., Kim D., Aisporna M., Wang Y., et al. (2003). Synthetic Exendin-4 (Exenatide) Significantly Reduces Postprandial and Fasting Plasma Glucose in Subjects with Type 2 Diabetes. The Journal of Clinical Endocrinology & Metabolism 88, 3082–3089. [DOI] [PubMed] [Google Scholar]
- Kremer A, Lippens S, Bartunkova S, Asselbergh B, Blanpain C, Fendrych M, Goossens A, Holt M, Janssens S, Krols M, et al. (2015). Developing 3D SEM in a broad biological context. Journal of Microscopy 259, 80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN & McIntosh JR (1996) Computer Visualization of Three-Dimensional Image Data Using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Laedtke T, Kjems L, Pørksen N, Schmitz O, Veldhuis J, Kao PC, and Butler PC (2000). Overnight inhibition of insulin secretion restores pulsatility and proinsulin/insulin ratio in type 2 diabetes. American Journal of Physiology-Endocrinology and Metabolism 279, E520–E528. [DOI] [PubMed] [Google Scholar]
- Larabell CA, and Nugent KA (2010). Imaging cellular architecture with X-rays. Curr Opin Struc Biol 20, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros MA, Knoechel CG, Uchida M, Parkinson DY, McDermott G, and Larabell CA (2012). 2.6 Visualizing Sub-cellular Organization Using Soft X-ray Tomography. In Comprehensive Biophysics, Edward HE, ed. (Amsterdam: Elsevier; ), pp. 90–110. [Google Scholar]
- Le Gros MA, McDermott G, Cinquin BP, Smith EA, Do M, Chao WL, Naulleau PP, and Larabell CA (2014). Biological soft X-ray tomography on beamline 2.1 at the Advanced Light Source. Journal of Synchrotron Radiation 21, 1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Gros Mark A., Clowney EJ, Magklara A, Yen A, Markenscoff-Papadimitriou E, Colquitt B, Myllys M, Kellis M, Lomvardas S, and Larabell Carolyn A. (2016). Soft X-Ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell Reports 17, 2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loconte V, Chen J-H, Cortese M, Ekman A, Le Gros MA, Larabell C, Bartenschlager R, and Weinhardt V. (2021). Using soft X-ray tomography for rapid whole-cell quantitative imaging of SARS-CoV2-infected cells. Cell Reports Methods 1, 100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučić V, Rigort A, and Baumeister W. (2013). Cryo-electron tomography: The challenge of doing structural biology in situ. Journal of Cell Biology 202, 407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler P, Carobbio S, and Rubi B. (2006). In beta-cells, mitochondria integrate and generate metabolic signals controlling insulin secretion. The International Journal of Biochemistry & Cell Biology 38, 696–709. [DOI] [PubMed] [Google Scholar]
- Maechler P, and Wollheim CB (2001). Mitochondrial function in normal and diabetic β-cells. Nature 414, 807–812. [DOI] [PubMed] [Google Scholar]
- Mahamid J, Pfeffer S, Schaffer M, Villa E, Danev R, Cuellar LK, Förster F, Hyman AA, Plitzko JM, and Baumeister W. (2016). Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972. [DOI] [PubMed] [Google Scholar]
- McDermott G, Fox DM, Epperly L, Wetzler M, Barron AE, Le Gros MA, and Larabell CA (2012). Visualizing and quantifying cell phenotype using soft X-ray tomography. BioEssays 34, 320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott G, Le Gros MA, Knoechel CG, Uchida M, and Larabell CA (2009). Soft X-ray tomography and cryogenic light microscopy: the cool combination in cellular imaging. Trends in Cell Biology 19, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merglen A, Theander S, Rubi B, Chaffard G, Wollheim CB, and Maechler P. (2004). Glucose Sensitivity and Metabolism-Secretion Coupling Studied during Two-Year Continuous Culture in INS-1E Insulinoma Cells. Endocrinology 145, 667–678. [DOI] [PubMed] [Google Scholar]
- Müller A, Schmidt D, Xu CS, Pang S, D’Costa JV, Kretschmar S, Münster C, Kurth T, Jug F, Weigert M, et al. (2020). 3D FIB-SEM reconstruction of microtubule–organelle interaction in whole primary mouse β cells. Journal of Cell Biology 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S, Martin S, and Parton RG (2009). Lipid droplet-organelle interactions; sharing the fats. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1791, 441–447. [DOI] [PubMed] [Google Scholar]
- Noske AB, Costin AJ, Morgan GP, and Marsh BJ (2008). Expedited approaches to whole cell electron tomography and organelle mark-up in situ in high-pressure frozen pancreatic islets. Journal of Structural Biology 161, 298–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson DY, Epperly LR, McDermott G, Le Gros MA, Boudreau RM, and Larabell CA (2013). Nanoimaging cells using soft X-ray tomography. Methods in molecular biology 950, 457–481. [DOI] [PubMed] [Google Scholar]
- Parkinson DY, Knoechel C, Yang C, Larabell CA, and Le Gros MA (2012). Automatic alignment and reconstruction of images for soft X-ray tomography. J Struct Biol 177, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet MJ, Henderson R, Russo CJ (2019). The energy dependence of contrast and damage in electron cryomicroscopy of biological molecules. Ultramicroscopy 203, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips MJ, and Voeltz GK (2016). Structure and function of ER membrane contact sites with other organelles. Nature Reviews Molecular Cell Biology 17, 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh B., Sun L., White KL., Sanyal T., Tempkin J., Zheng D., Bharat K., Singla J., Wang C., Zhao J., et al. (2021). Bayesian metamodeling of complex biological systems across varying representations. 1, 0–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorsman P, Eliasson L, Renström E, Gromada J, Barg S, and Göpel S. (2000). The Cell Physiology of Biphasic Insulin Secretion. Physiology 15, 72–77. [DOI] [PubMed] [Google Scholar]
- Schneider G, Guttmann P, Heim S, Rehbein S, Mueller F, Nagashima K, Heymann JB, Müller WG, and McNally JG (2010). Three-dimensional cellular ultrastructure resolved by X-ray microscopy. Nature Methods 7, 985–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, et al. , (2012) Fiji: an open-source platform for biological-image analysis. Nature Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M, Kamoshita M, and Islinger M. (2020). Organelle interplay—peroxisome interactions in health and disease. Journal of Inherited Metabolic Disease 43, 71–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorrano L, De Matteis MA, Emr S, Giordano F, Hajnóczky G, Kornmann B, Lackner LL, Levine TP, Pellegrini L, Reinisch K, et al. (2019). Coming together to define membrane contact sites. Nature Communications 10, 1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva BSC, DiGiovanni L, Kumar R, Carmichael RE, Kim PK, and Schrader M. (2020). Maintaining social contacts: The physiological relevance of organelle interactions. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1867, 118800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla J, McClary KM, White KL, Alber F, Sali A, and Stevens RC (2018). Opportunities and Challenges in Building a Spatiotemporal Multi-scale Model of the Human Pancreatic β Cell. Cell 173, 1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla J, and White KL (2021). A community approach to whole-cell modeling. Current Opinion in Systems Biology 26, 33–38. [Google Scholar]
- Smith EA, McDermott G, Do M, Leung K, Panning B, Le Gros MA, and Larabell CA (2014b). Quantitatively imaging chromosomes by correlated cryo-fluorescence and soft x-ray tomographies. Biophysical Journal 107, 1988–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suckale J, and Solimena M. (2010). The insulin secretory granule as a signaling hub. Trends in Endocrinology & Metabolism 21, 599–609. [DOI] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, and Lippincott-Schwartz J. (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE (2014). MAM (mitochondria-associated membranes) in mammalian cells: Lipids and beyond. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids 1841, 595–609. [DOI] [PubMed] [Google Scholar]
- Weinhardt V, Chen JH, Ekman AA, Guo J, Remesh SG, Hammel M, McDermott G, Chao W, Oh S, Gros MAL, et al. (2020). Switchable resolution in soft x-ray tomography of single cells. PLoS ONE 15, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KL, Singla J, Loconte V, Chen JH, Ekman A, Sun L, Zhang X, Francis JP, Li A, Lin W, et al. (2020). Visualizing subcellular rearrangements in intact β cells using soft x-ray tomography. Science Advances 6, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederkehr A., and Wollheim CB. (2006). Minireview: Implication of Mitochondria in Insulin Secretion and Action. Endocrinology 147, 2643–2649. [DOI] [PubMed] [Google Scholar]
- Wollheim CB (2000). Beta-cell mitochondria in the regulation of insulin secretion: a new culprit in Type II diabetes. Diabetologia 43, 265–277. [DOI] [PubMed] [Google Scholar]
- Zhang X, Carter SD, Singla J, White KL, Butler PC, Stevens RC, and Jensen GJ (2020). Visualizing insulin vesicle neighborhoods in β cells by cryo-electron tomography. Science Advances 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All original code and reconstructed data are available from the corresponding contacts upon request.
Any additional information required to reanalyze the data reported in this paper is available from the corresponding contacts upon request.