Abstract
The coronavirus disease (COVID-19) first appeared at the end of December 2019 and is still spreading in most countries. To diagnose COVID-19 using reverse transcription - Polymerase chain reaction (RT-PCR), one has to go to a dedicated center, which requires significant cost and human resources. Hence, there is a requirement for a remote monitoring tool that can perform the preliminary screening of COVID-19. In this paper, we propose that a detailed audio texture analysis of COVID-19 sounds may help in performing the initial screening of COVID-19. The texture analysis is done on three different signal modalities of COVID-19, i.e. cough, breath, and speech signals. In this work, we have used 1141 samples of cough signals, 392 samples of breath signals, and 893 samples of speech signals. To analyze the audio textural behavior of COVID-19 sounds, the local binary patterns LBP) and Haralick’s features were extracted from the spectrogram of the signals. The textural analysis on cough and breath sounds was done on the following 5 classes for the first time: COVID-19 positive with cough, COVID-19 positive without cough, healthy person with cough, healthy person without cough, and an asthmatic cough. For speech sounds there were only two classes: COVID-19 positive, and COVID-19 negative. During experiments, 71.7% of the cough samples and 72.2% of breath samples were classified into 5 classes. Also, 79.7% of speech samples are classified into 2 classes. The highest accuracy rate of 98.9% was obtained when binary classification between COVID-19 cough and non-COVID-19 cough was done.
Keywords: COVID-19, Cough, Speech, Breath, Audio texture, Spectrogram, Local binary pattern, Haralick features
1. Introduction
The novel coronavirus disease (COVID-19) first appeared at the end of December 2019 and by December 2, 2021, close to 267 million cases have been reported in over 186 countries in the world [1]. The early symptoms of COVID-19 are fever, dry cough, loss of taste and smell, and fatigue. In severe cases, it might lead to shortness of breath, respiratory disorders, pneumonia, heart problems, and may even lead to death [2], [3], [4]. The spread of COVID-19 is mainly from the aerosols suspended through an infected person’s cough or sneeze.
To date, the RT-PCR test is recommended by the majority of countries. This test is providing high accuracy but has lots of limitations too [5]. Going to a dedicated test center, the need for trained medical practitioner, non-reusable testing kits, invasive nature, and time is taken to receive results are a few of its limitations. The home-based monitoring of COVID-19 is getting popular especially with the emergence of a new omicron variant. But because of its low performance, it shows no benefit in reducing the use of RT-PCR tests [6]. Hence, there is a strong need for additional screening or monitoring tools that could help to reduce the burden of the testing centers and could be used from remote locations, and is suitable for large-scale testing and monitoring [7], [8], [9]. The machine learning and artificial intelligence (AI)-based solutions could help smartphone data provide these additional tools.
To date, many AI-based algorithms have been implemented by researchers. But the main focus was on the screening of COVID-19 patients by using CT scans [10], [11], [12], [13] and X-ray images [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]. These methods are mainly based on convolutional neural networks (CNNs) and they use existing pre-trained networks to screen and classify COVID-19 patients from others [25], [26]. The main challenge among these methods is the unavailability of the large data sets and image acquisition process where the person needs to visit a clinic for getting a CT scan or an X-ray image.
We are living in the era of smartphones and biomedical wearables, which may provide an alternative, scalable, agile, and non-invasive way of screening COVID-19 patients [27], [28]. An AI-based preliminary diagnosis is done by using the cough samples that are recorded by a smartphone application [29]. This study is based on the recent analysis of the pathomorphological changes that are caused by COVID-19, and it states that the airway passage in COVID-19 remains dry and consistent because there is minimal movement of the phlegm. While for the other types of cough e.g. asthmatic or allergic cough, this airway passage is changing because of the movement of phlegm. In this work, the authors have used deep learning-based classifiers such as CNN for binary class and multi-class classification problems. In the same work, the Mel frequency cepstral coefficient feature is extracted from the cough samples and is classified by using a weighted K-NN classifier. They have claimed an overall accuracy of 92% [29]. In [30], the COVID-19 cough and respiratory sounds are taken for diagnosis of COVID-19, and an accuracy of 82% has been reported. In this work, researchers have extracted handcrafted features such as tempo, root mean square (RMS) energy, MFCCs, centroid, roll-off frequency, and used 256-dimensional VGG networks to classify COVID-19 and non-COVID-19 cough samples. For all the features that generate time-series such as MFCCs, authors have extracted several statistical parameters such as mean, median, skewness, kurtosis, and root mean square (RMS) from the original features. In [31], authors have proposed that COVID-19 and heart failure have some common symptoms and implemented a graph-based local feature generator (DNA pattern) method to diagnose these two diseases using cough sounds only. A high accuracy rate of up to 100% is reported in this work. Another similar work is proposed in [32], [33] where authors have implemented deep neural networks such as CNN, LSTM, and Resnet50. In this work, COVID-19 and non-COVID-19 sounds are classified and the highest ROC AUC of 0.98 is obtained for cough sounds, followed by 0.94 for breath, and 0.92 for speech sounds. In [34], the authors have used a deep rank-based average pooling network to diagnose COVID-19, pneumonia, pulmonary tuberculosis, and healthy users. Authors have reported the F1 score up to 95.94%.
In this paper, we will use a subset of the dataset recorded by the University of Cambridge, U.K which has the collection of COVID-19 sounds and non-COVID-19 sounds [30]. Also, cough when analyzed with other physiological parameters such as heart rate, respiration, and temperature can provide an enriched symptom tracker for monitoring disease progressions and any needed intervention or emergency medical attention.
The main contribution of the paper is the detailed analysis of audio textures present in the COVID-19 sounds. Because COVID-19 changes the airway passage in a different way than the other respiratory diseases, it is reasonable to hypothesize that COVID-19 sounds must have a distinct texture than the non-COVID-19 sounds. To our knowledge, this is the first time that the audio textural analysis on COVID-19 cough and breath sounds was done on following five different classes: COVID-19 positive with cough, COVID-19 positive without cough, healthy person with cough, healthy person without cough, and an asthmatic cough. The proposed work describes that spectrograms of the non-pitch related sounds such as cough and breath have some kind of texture present in it. The textural information is captured by employing local binary patterns and Haralick’s features [35]. These latent features are distinct for COVID-19 sounds than for non-COVID-19 sounds. During classification, high accuracy has been observed while classifying normal cough, breath, and speech samples from COVID-19 samples.
The rest of the sections are: Section 2 discusses the dataset used. Section 3 describes the proposed methodology, the next Section is about the experimental setup, and Section 5 describes the results obtained. Section 6 is about discussion, conclusion, and future work.
2. Dataset used
The COVID-19 sounds dataset has recordings of cough, breath, and speech sounds from COVID-19 and non-COVID-19 users. This dataset is collected by the University of Cambridge, UK [30]. The android and web-based applications were used to collect the samples. The dataset is well labeled and also has a list of other symptoms and medical history reported by the user. The dataset has samples from 4352 users from web-based applications and 2261 users from android-based smartphones. But, due to the sensitive nature of the database, the whole dataset is not released for research purposes, but only a portion of the dataset has been made accessible. Given the wide prevalence of smartphone users, in this work, we have used a subset of the dataset which is collected by the smartphone application only. The dataset has 1141 cough samples, 893 speech samples, and 392 breath samples [30]. Table 1 shows the distribution of audio samples in various classes. For speech signals, every user utters the following sentence three times: ”I hope my data can help manage the virus pandemic”. The speech utterances are sampled at 16 kHz.
Table 1.
Distribution of samples for cough, breath, and speech modality.
| Modality | Class | No. of samples |
|---|---|---|
| Cough | COVID-19 + ve with cough | 204 |
| COVID-19 + ve without cough | 64 | |
| COVID-19 -ve with cough | 631 | |
| COVID-19 -ve no symptom | 138 | |
| Asthma user with cough | 104 | |
| Breath | COVID-19 + ve with cough | 46 |
| COVID-19 + ve without cough | 51 | |
| COVID-19 -ve with cough | 64 | |
| COVID-19 -ve no symptom | 127 | |
| Asthma user with cough | 104 | |
| Speech | COVID-19 + ve | 308 |
| COVID-19 -ve | 585 |
3. Proposed methodology
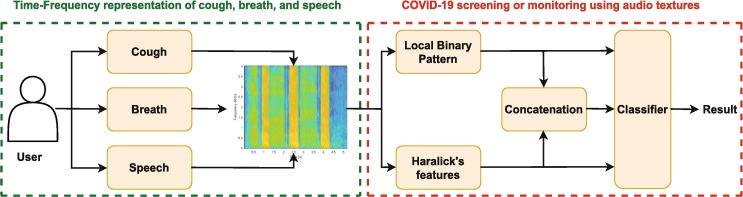
The proposed methodology to screen COVID-19 sounds from non-COVID-19 sounds is shown in Fig. 1 . The proposed system has two main blocks: First, time–frequency representation of COVID-19 sounds i.e. cough, breath, and speech signals. Second, audio texture analysis and classification of COVID-19 sounds. Both of the blocks are explained below in detail.
Fig. 1.
Proposed system architecture and flow diagram for texture analysis and classification of COVID-19 sounds.
3.1. Time–frequency representation of COVID-19 sounds
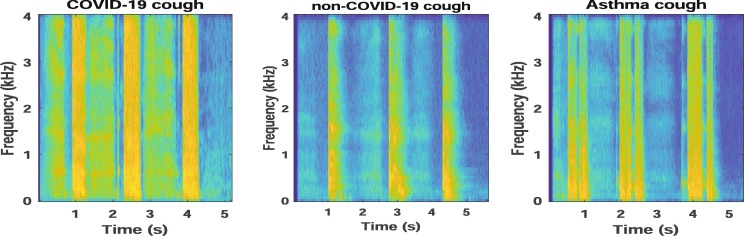
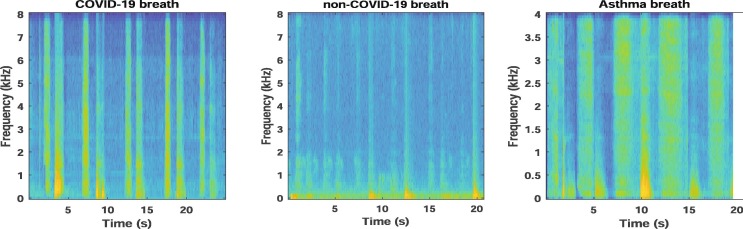
The COVID-19 and non-COVID-19 sounds are converted into their respective time-frequency representations (TFR). We have used spectrograms as the TFR of COVID-19 sounds. The spectrogram reflects the spectro-temporal correlations present in the signal in the form of an image. To get the spectrogram, a Hamming window is employed to divide the audio signal into the frames of length 128 samples and overlap of 120 samples. A 128-point FFT is performed on each frame to get the spectrogram. Fig. 2 and Fig. 3 show the spectrograms for cough and breath sounds for a COVID-19 positive user, non-COVID-19 (or healthy) user, and a user with asthma.
Fig. 2.
Spectrograms for COVID-19, non-COVID-19, and asthma cough sound.
Fig. 3.
Spectrograms for COVID-19, non-COVID-19, and asthma breath sound.
Fig. 2 shows that the COVID-19 cough is more consistent in nature and has a longer duration in comparison to the non-COVID-19 cough. The COVID-19 cough lasts for at least 1.2 s and has a noise-like texture that is uniform throughout the frequency bands, while the non-COVID-19 cough has a smaller stroke and is concentrated within a few frequency bands. The consistency of the COVID-19 cough leads to the coarse texture that is more uniform in nature just like in high textural sounds [29]. On the other hand, the non-COVID-19 sounds have a low texture. These initial observations open the avenues for audio texture analysis in COVID-19 and non-COVID-19 cough sounds, more rationale for breath and speech signals as well.
3.2. Audio texture analysis and classification of COVID-19 sounds
An audio sound could qualify as a texture if it is long-term, non-sinusoidal, stochastic but still homogeneous in nature, holds uniform energy throughout the signal, has poor harmonic content, and exhibits noise-like characteristics [36]. Recently audio textures have shown good machine learning results for pathological speech screening and analysis [35], [36]. COVID-19 sounds also exhibit texture-like properties and could be analyzed by textural features.
The spectrograms generated by the previous block are analyzed for texture extraction. In this work, we have analyzed the two textural features i.e. LBP and Haralick from the spectrogram of the COVID-19 cough, breath, and speech samples.
The audio texture could be described by two complementary measures: one is the local spatial pattern and another one is grayscale contrast [37]. The LBPs extract the local spatial patterns present in the spectrogram image here. Haralick’s features calculate the basic statistical parameters from the grayscale distribution.
LBP is one of the most explored image texture features [38]. It is primarily used for image processing based applications such as face recognition, pattern classification, and object detection. Hence if we are considering the spectrograms as an image, these LBPs could capture the textural patterns present in the audio. The basic version of LBP is called a uniform pattern LBP. In uniform patterns, the binary number is replaced by the number of transitions occurring between 0 and 1, and vice versa. There are a total of 58 uniform patterns present in the LBP. Rest all other patterns are non-uniform in nature and concatenated together to form a single pattern. Hence, the uniform LBP is a 59-dimensional feature vector. The basic version of LBP does not encode the rotation information of the image. In another version of LBP rotational invariant features are also encoded to generate a final set of features. The size of this rotational invariant LBP is not fixed and is dependent on the cell size of the mask taken. We have taken a cell size of , radius of 3 units, and got 120-dimensional LBP. Before using this feature vector for classification, we have normalized it by norm. The normalization makes the features vector invariant to the rotation of spectrograms. This makes the LBP features invariant to rotations [35], [39]. While extracting LBP, we have limited our experiments to 8 sampling points and a radius of 3 units, because it was observed by Ojala et al. that 94% of the local patterns could be captured by using 8 sampling points on the radius of 3 units [38].
Another textural feature that we have used in this work is Haralick’s feature. It describes the correlation between intensities of the neighboring pixels. The fourteen Haralick’s features are derived from the co-occurrence matrix [35]. It describes the texture present in the spectrogram images with the help of statistical parameters. The fourteen features are angular second moment, contrast, correlation, variance, inverse difference moment, sum average, sum variance, entropy, sum entropy, difference variance, difference entropy, information measure of correlation 1, information measure of correlation 2, and maximum correlation coefficient. In our work, we have considered all 14 Haralick’s features for screening of COVID-19 coughs [35]. These features are computed from the spectrogram image , denoted P, with dimension where is the number of gray levels in spectrogram image [35], [36]. Table 2 describes the 14 Haralaick’s features in detail.
Table 2.
Haralick’s features description [35].
| Haralick’s feature | Equation | Description |
|---|---|---|
| Angular second moment | measures homogeneity of local gray scale distribution | |
| Contrast | , describes quantity of local changes happening in an audio | |
| Correlation | , , , and are mean and standard deviations, value lies between to | |
| Variance | , measures spread of the signal | |
| Inverse difference moment | measures local homogeneity | |
| Sum average | measures mean of the gray level sum distribution | |
| Sum variance | calculates dispersion of the gray level sum distribution | |
| Sum entropy | measures disorder related to the gray level sum distribution | |
| Entropy | measure randomness | |
| Difference variance | describes heterogeneity | |
| Difference entropy | measures disorder related to the distribution of gray scale difference | |
| Information measure correlation 1 | and are the entropies of and respectively, and | |
| Information measure of correlation 2 | (i) | |
| Maximum correlation coefficient |
The experiments are performed for 3 sets of features. First, when only 120-dimensional LBP feature is fed to the classifier, second, when only 14-dimensional Haralick’s features are used, and third when we combine the LBP and Haralick’s features to make a single feature set of 134-dimensions. We have performed all our experiments on a weighted K-NN classifier with 10-fold cross-validation.
The weighted K-NN is employed with 10 number of neighbor points. For distance metric, euclidean distance is used, and to adjust distance weights, a squared inverse distance weighing function is used.
4. Experimental setup
In this work, we focus on the following clinically meaningful classification experiments. During experimentation, the classification models are trained for LBP only, and LBP + Haralick’s features.
-
•
Experiment 1: Here we do 3-class classifications from cough sounds to distinguish between COVID-19 positive users who have a cough as a symptom (COVID-19 cough), and COVID-19 negative users who have a cough (non-COVID-19 cough), asthma cough.
-
•
Experiment 2: Expanding our Experiment 1, in this, we perform 5-class cough classification with the following 5 classes: COVID-19 positive users who have a cough as a symptom (COVID-19 cough), COVID-19 positive users who don’t have a cough as a symptom (COVID-19 no cough), COVID-19 negative users who have a cough (non-COVID-19 cough), the COVID-19 negative users who don’t have a cough (non-COVID-19 no cough), and asthma cough.
-
•
Experiment 3: Using breath sounds to distinguish between users who have declared that they are COVID-19 positive (COVID-19 breath), healthy users (non-COVID-19 breath), and users having asthma.
-
•
Experiment 4: Using breath sounds to distinguish between all 5 classes i.e. breath sounds from COVID-19 positive users who have a cough as a symptom (COVID-19 breath), breath sounds from COVID-19 positive users who don’t have a cough as a symptom, breath sounds from COVID-19 negative users who have a cough, breath sounds from COVID-19 negative users who don’t have a cough, and breath sounds from users having asthmatic cough.
-
•
Experiment 5: Combine two modalities i.e. cough and breath to distinguish between all 5 classes.
-
•
Experiment 6: Distinguish between COVID-19 positive and COVID-19 negative users from their speech samples only.
-
•
Experiment 7: Here we do a simple binary classification between COVID-19 positive cough sounds and COVID-19 negative cough sounds.
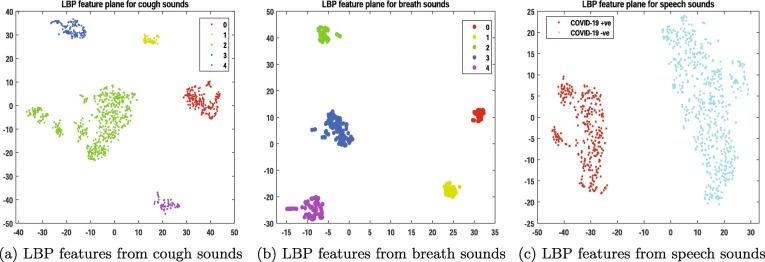
Feature visualization and cross validation As it has been proven that COVID-19 causes distinct pathomorphological changes in the respiratory system from those other non-COVID-19 respiratory diseases [29]. Hence, the latent features of COVID-19 must also be distinct from other classes. Fig. 4 shows the visualization of LBP features for the five classes using t-distributed stochastic neighborhood embedding (t-SNE) for cough, breath, and speech modality. All the experiments are performed with audio textural features fed into a weighted K-NN classifier with 10-fold cross-validation for ensuring that the model neither underfits nor overfits. To estimate the performance of the classifier, we performed several standard evaluation metrics such as Receiver operating characteristics - Area under the curve (ROC-AUC) and accuracy rate.
Fig. 4.
(a), (b): Visualization of LBP features from cough and breath modality for all 5 classes, 0: COVID-19 + ve with cough, 1: COVID-19 + ve without cough as a symptom, 2: COVID-19 -ve with cough, 3: COVID-19 -ve with no symptoms, 4: Asthma users with cough, (c): Visualization of LBP features from speech modality for 2 classes: COVID-19 + ve speech and COVID-19 -ve speech.
5. Results
Table 1 shows the set of samples and classes used for classifying COVID-19 cough, breath, and speech sounds. Table 3 reports the classification results for all 7 experiments performed. For each experiment, we report the best results for each set of texture features i.e. when using LBP only, Haralick feature only, and a combination of LBP and Haralick features. The first row reports classification for Experiment 1: the three-class classification between COVID-19 cough, non-COVID-19 cough, and asthma cough. The results reflect that almost 80% of the cough signals are well discriminated by the classifier when the combination of LBP and Haralick feature is used. Similarly, encouraging results are obtained for Experiment 2 as well, where the full 5 class classification task is done. The classification accuracy reduces a bit, which is expected as we are adding more classes to the data. Nevertheless, these results give us confidence that cough signals could be modeled by audio textures and there is a significant textural difference between various types of cough.
Table 3.
Classification results for all experiments.
| Experiment | Modality | Feature | ROC-AUC | Accuracy rate |
|---|---|---|---|---|
| Experiment 1 | cough | LBP only | 0.77 | 78.1 % |
| Haralick only | 0.69 | 72.9% | ||
| LBP + Haralick | 0.80 | 79.9% | ||
| Experiment 2 | cough | LBP only | 0.77 | 69.9% |
| Haralick only | 0.67 | 62.3% | ||
| LBP + Haralick | 0.79 | 71.7% | ||
| Experiment 3 | breath | LBP only | 0.92 | 82.7% |
| Haralick only | 0.90 | 80.8% | ||
| LBP + Haralick | 0.96 | 87.9% | ||
| Experiment 4 | breath | LBP only | 0.85 | 70.9% |
| Haralick only | 0.76 | 63.5% | ||
| LBP + Haralick | 0.86 | 72.2% | ||
| Experiment 5 | cough + breath | LBP only | 0.78 | 66.8 % |
| Haralick only | 0.69 | 60.5% | ||
| LBP + Haralick | 0.75 | 70.1% | ||
| Experiment 6 | Speech | LBP only | 0.83 | 79.5% |
| Haralick only | 0.73 | 73.1% | ||
| LBP + Haralick | 0.83 | 79.7% | ||
| Experiment 7 | cough | LBP only | 0.97 | 98.7 % |
| Haralick only | 0.90 | 92% | ||
| LBP + Haralick | 0.98 | 98.9% |
Experiment 3 and Experiment 4 also reflect good classification results for breath sounds. For 3-class classification we got an accuracy rate up to 87.9% and for 5-class classification, the accuracy rate is 72.2%. These experiments have better AUC than the previous Experiment 1 and Experiment 2. We performed Experiment 5 just to check if the combination of two modalities is giving any meaningful results or not. We found that the performance of the classifier dips when we combine two modalities.
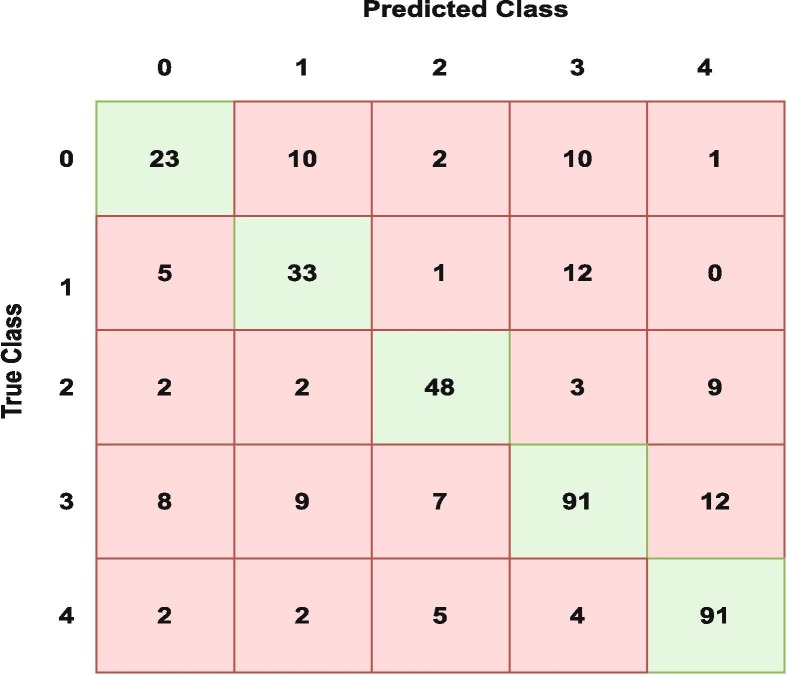
It is interesting to see that breath sounds have more discriminatory features than cough sounds, which adds value to the current state-of-the-art that suggests COVID-19 cough is one of its primary biomarkers. Fig. 5 shows the confusion matrix for the breath signals when all 5 classes are considered for LBP + Haralick textural features (Experiment 4).
Fig. 5.
Confusion matrix for breath modality for all 5 classes using LBP + Haralick (Experiment 4).
In Experiment 6, we have distinguished between speech samples collected from COVID-19 positive and COVID-19 negative users. We got an accuracy rate of 79.7% which means 79.7% of COVID-19 speech samples are correctly classified. The results achieved reflect the presence of discriminatory features in COVID-19 and non-COVID-19 cough, breath, and speech signals. Experiment 7, shows the accuracy rate up to 98.9% when a simple binary classification between COVID-19 positive cough sounds and COVID-19 negative cough sounds is done. To cross-validate the performance of our proposed system, we performed leave one subject out (LOSO) cross-validation on our dataset. We got encouraging results, for 5 class cough sound classification the average validation was 85.79%. For 5-class breath sound classification, the average validation accuracy is 98.5%. Similarly, for two-class speech sound classification, we got an accuracy rate of 68.89%.
5.1. Comparison with state-of-the-art works
In the last two years, many AI researchers have worked on the classification of COVID-19 pathological samples and non-COVID-19 pathological samples. A huge amount of work is done by using chest X-rays and CT scans. These imaging modalities are fed into some kind of deep neural networks such as CNN, LSTM, and RNN for classification purposes [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23]. A lot of work has been done on COVID-19 audio analysis also. In [30], binary classification of COVID-19 + ve users and non-COVID-19 users is done on cough, breath, and cough + breath samples. An accuracy of up to 82% is achieved when only cough samples are considered for binary classification. In [40], authors have classified cough sounds from non-cough sounds using a threshold of 0.8 and got an accuracy rate of almost 82% when acoustic features are fed to the binary classifier. Authors in [41] have used deep neural networks such as CNN to perform binary classification on COVID-19 cough sounds. They have reported an accuracy rate of 72%. In [32], authors have used CNN and Resnet50 to classify between COVID-19 cough and healthy cough sounds. They have achieved an accuracy of 95%. In [42], authors have implemented CNN, RNN, and LSTM to binary classify COVID-19 cough, breath, and voice sounds. The accuracy rate of up to 98.2% has been reported by using 240 samples only. Interestingly, they have also achieved higher accuracy with breath signals rather than cough signals. Table 4 compares the proposed work with the closest state-of-the-art works. The proposed system is performing very well when binary classification between cough of COVID-19 and healthy users is performed. For 5-class classification (refer to Section 4 and Table 3), we got encouraging results with an accuracy of 72.2%.
Table 4.
Comparison with the state-of-the-art works.
| Citation | Modality | No. of Classes | Accuracy rate | Features |
|---|---|---|---|---|
| Brown et al. | cough, breath | 2 | 82% | Acoustic |
| Orlandic et al. | cough | 2 | 82% | Acoustic |
| Bagad et al. | cough | 2 | 72% | Acoustic |
| Pahar et al. | cough | 2 | 95% | Acoustic |
| Pahar et al. | cough, breath, and speech | 2 | 98% | Deep networks |
| Hassan et al. | cough, breath, and speech | 2 | 98.2% | Acoustic |
| Proposed work | cough, breath, and speech | 2 | 98.9% | Textural |
| Proposed work | cough, breath, and speech | 5 | 72.2% | Textural |
6. Discussion, conclusion and future work
In this work, we have presented the scope of using audio textures to analyze and classify COVID-19 sounds. Based on the current COVID-19 knowledge, it has been proved that COVID-19 affects the respiratory airways in a different way than the other respiratory diseases. Hence, we hypothesize that COVID-19 sounds will also have distinct latent features to other respiratory diseases. Fig. 4 shows the distribution of LBP features for all 5 classes of cough and breath sounds. Our results in Fig. 4 and Table 3 support our hypothesis. Our proposed model can classify 5 types of cough sounds with an accuracy rate of 71.7%, 5 types of breath sounds with an accuracy rate of 72.2%, and 79.7% of speech sounds. The system offers the highest accuracy rate of 98.9% while performing binary classification on COVID-19 and non-COVID-19 cough sounds.
This work is one of its first kinds of work, where audio textural features are analyzed to screen COVID-19 using 3 different modalities i.e. cough, breath, and speech, and 5 different classes. The proposed method has a few advantages over the state-of-the-art works. Foremost, the method is computationally simple and explainable to some extent, unlike the complex deep neural networks. Unlike the existing AI-based work using cough as the only modality, we have extended our research to three modalities of COVID-19 sounds including cough, breath, and speech. This method builds the foundation that breath and speech sounds may be considered bio-markers along with cough sounds for COVID-19. Also, the audio texture analysis gives a new perspective to solving other audio signal-based problems. The proposed method has a few limitations as well, such as it could not be solely used to screen or diagnose COVID-19. But it opens new avenues for developing COVID-19 screening tools for telemedicine and remote monitoring [43]. Because cough is a symptom of more than 30 types of respiratory diseases, the proposed method could be improved if we have more data samples and more classes to analyze. Another limitation is that the manifestation of COVID-19 is changing with the emergence of new variants and the presence of asymptomatic cases that do not allow us to have a complete spectrum of COVID-19 sounds.
In the future, we would like to analyze and classify more types of cough caused by various other respiratory diseases and want to compare those with COVID-19 cough. In general, there is a treatment period of 14 days for COVID-19 positive persons. Hence, it would be interesting to investigate how COVID-19 sound’s textural behavior changes when a person undergoes treatment. We would also like to experiment further on breath and speech modalities for COVID-19 screening. To add more depth to the current work, we may introduce more advanced textural features to analyze and classify COVID-19 sounds. With the manifestation of new variants, the dataset is growing and as we start to get more samples we may use the current models and transfer learning-based models such as AlexNet, GoogleNet, ResNet, or DenseNet to analyze COVID-19 sounds. Hence, there is a lot of scope for the expansion of this work.
CRediT authorship contribution statement
Garima Sharma: Conceptualization, Methodology, Software, Writing - original draft. Karthikeyan Umapathy: Conceptualization, Supervision, Writing - review & editing, Project administration. Sri Krishnan: Conceptualization, Supervision, Writing - review & editing, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.[Online]. Available: URL: https://coronavirus.jhu.edu.
- 2.Atzrodt C.L., Maknojia I., McCarthy R.D., Oldfield T.M., Po J., Ta K.T., Stepp H.E., Clements T.P. A guide to covid-19: a global pandemic caused by the novel coronavirus sars-cov-2. FEBS J. 2020;287(17):3633–3650. doi: 10.1111/febs.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, china. Jama. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carfì A., Bernabei R., Landi F., et al. Persistent symptoms in patients after acute covid-19. Jama. 2020;324(6):603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.[Online]. Available: URL: https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/updates46-testing-strategies.pdf?sfvrsn=c9401268_6.
- 6.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for covid-19 diagnosis. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bong C.-L., Brasher C., Chikumba E., McDougall R., Mellin-Olsen J., Enright A. The covid-19 pandemic: effects on low-and middle-income countries. Anesthesia and analgesia. 2020 doi: 10.1213/ANE.0000000000004846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.M. Salathé, C.L. Althaus, R. Neher, S. Stringhini, E. Hodcroft, J. Fellay, M. Zwahlen, G. Senti, M. Battegay, A. Wilder-Smith, et al., Covid-19 epidemic in switzerland: on the importance of testing, contact tracing and isolation, Swiss Med. Weekly 150(1112) (2020). [DOI] [PubMed]
- 9.Sepulveda J., Westblade L.F., Whittier S., Satlin M.J., Greendyke W.G., Aaron J.G., Zucker J., Dietz D., Sobieszczyk M., Choi J.J., et al. Bacteremia and blood culture utilization during covid-19 surge in new york city. J. Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.00875-20. e00 875–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li K., Wu J., Wu F., Guo D., Chen L., Fang Z., Li C. The clinical and chest ct features associated with severe and critical covid-19 pneumonia. Investig. Radiol. 2020 doi: 10.1097/RLI.0000000000000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., et al. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology. 2020 doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Xia L. Coronavirus disease 2019 (covid-19): role of chest ct in diagnosis and management. Am. J. Roentgenol. 2020;214(6):1280–1286. doi: 10.2214/AJR.20.22954. [DOI] [PubMed] [Google Scholar]
- 13.J. Zhang, Y. Xie, Y. Li, C. Shen, Y. Xia, Covid-19 screening on chest x-ray images using deep learning based anomaly detection, arXiv preprint arXiv:2003.12338, vol. 27, 2020.
- 14.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput. Biol. Med. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahmud T., Rahman M.A., Fattah S.A. Covxnet: A multi-dilation convolutional neural network for automatic covid-19 and other pneumonia detection from chest x-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duran-Lopez L., Dominguez-Morales J.P., Corral-Jaime J., Vicente-Diaz S., Linares-Barranco A. Covid-xnet: a custom deep learning system to diagnose and locate covid-19 in chest x-ray images. Appl. Sci. 2020;10(16):5683. [Google Scholar]
- 17.Wang L., Lin Z.Q., Wong A. Covid-net: A tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain R., Gupta M., Taneja S., Hemanth D.J. Deep learning based detection and analysis of covid-19 on chest x-ray images. Appl. Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.L.O. Hall, R. Paul, D.B. Goldgof, G.M. Goldgof, Finding covid-19 from chest x-rays using deep learning on a small dataset, arXiv preprint arXiv:2004.02060, 2020.
- 20.Nayak S.R., Nayak D.R., Sinha U., Arora V., Pachori R.B. Application of deep learning techniques for detection of covid-19 cases using chest x-ray images: A comprehensive study. Biomed. Signal Process. Control. 2021;64 doi: 10.1016/j.bspc.2020.102365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadre R., Sundaram B., Majumdar S., Ushizima D. Validating deep learning inference during chest x-ray classification for covid-19 screening. Sci. Rep. 2021;11(1):1–10. doi: 10.1038/s41598-021-95561-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (covid-19) using x-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021:1–14. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.K. Ahammed, M.S. Satu, M.Z. Abedin, M.A. Rahaman, S.M.S. Islam, Early detection of coronavirus cases using chest x-ray images employing machine learning and deep learning approaches, medRxiv, 2020.
- 24.S.-H. Wang, Z. Zhu, Y.-D. Zhang, Pscnn: Patchshuffle convolutional neural network for covid-19 explainable diagnosis, Front. Public Health, vol. 9, 2021. [DOI] [PMC free article] [PubMed] [Retracted]
- 25.Bayoudh K., Hamdaoui F., Mtibaa A. Hybrid-covid: a novel hybrid 2d/3d cnn based on cross-domain adaptation approach for covid-19 screening from chest x-ray images. Phys. Eng. Sci. Med. 2020;43(4):1415–1431. doi: 10.1007/s13246-020-00957-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agarwal M., Saba L., Gupta S.K., Carriero A., Falaschi Z., Paschè A., Danna P., El-Baz A., Naidu S., Suri J.S. A novel block imaging technique using nine artificial intelligence models for covid-19 disease classification, characterization and severity measurement in lung computed tomography scans on an italian cohort. J. Med. Syst. 2021;45(3):1–30. doi: 10.1007/s10916-021-01707-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Mohammed, H. Syamsudin, S. Al-Zubaidi, R.R. AKS, E. Yusuf, Novel covid-19 detection and diagnosis system using iot based smart helmet, Int. J. Psychosocial Rehab., vol. 24, no. 7, pp. 2296–2303, 2020.
- 28.M. Mohammed, N.A. Hazairin, S. Al-Zubaidi, S. AK, S. Mustapha, E. Yusuf, Toward a novel design for coronavirus detection and diagnosis system using iot based drone technology, Int. J. Psychosoc. Rehabil., vol. 24, no. 7, pp. 2287–2295, 2020.
- 29.Imran A., Posokhova I., Qureshi H.N., Masood U., Riaz M.S., Ali K., John C.N., Hussain M.I., Nabeel M. Ai4covid-19: Ai enabled preliminary diagnosis for covid-19 from cough samples via an app. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown C., Chauhan J., Grammenos A., Han J., Hasthanasombat A., Spathis D., Xia T., Cicuta P., Mascolo C. Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. 2020. Exploring automatic diagnosis of covid-19 from crowdsourced respiratory sound data; pp. 3474–3484. [Google Scholar]
- 31.M.A. Kobat, T. Kivrak, P.D. Barua, T. Tuncer, S. Dogan, R.-S. Tan, E.J. Ciaccio, U.R. Acharya, Automated covid-19 and heart failure detection using dna pattern technique with cough sounds, Diagnostics, vol. 11, no. 11, 2021. [Online]. Available: URL: https://www.mdpi.com/2075-4418/11/11/1962. [DOI] [PMC free article] [PubMed]
- 32.M. Pahar, M. Klopper, R. Warren, T. Niesler, Covid-19 cough classification using machine learning and global smartphone recordings, Comput. Biol. Med. 2021, p. 104572. [DOI] [PMC free article] [PubMed]
- 33.M. Pahar, M. Klopper, R. Warren, T. Niesler, Covid-19 detection in cough, breath and speech using deep transfer learning and bottleneck features, Comput. Biol. Med., p. 105153, 2021. [DOI] [PMC free article] [PubMed]
- 34.Shui-Hua W., Khan M.A., Govindaraj V., Fernandes S.L., Zhu Z., Yu-Dong Z. Deep rank-based average pooling network for covid-19 recognition. Comput. Mater. Continua. 2022:2797–2813. [Google Scholar]
- 35.G. Sharma, D. Prasad, K. Umapathy, and S. Krishnan, “Screening and analysis of specific language impairment in young children by analyzing the textures of speech signal,” in 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC). IEEE, 2020, pp. 964–967. [DOI] [PubMed]
- 36.Sharma G., Zhang X.-P., Umapathy K., Krishnan S. Audio texture and age-wise analysis of disordered speech in children having specific language impairment. Biomed. Signal Process. Control. 2021;66 [Google Scholar]
- 37.P. Sidike, C. Chen, V. Asari, Y. Xu, and W. Li, “Classification of hyperspectral image using multiscale spatial texture features,” in 2016 8th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS). IEEE, 2016, pp. 1–4.
- 38.Ojala T., Pietikainen M., Maenpaa T. Multiresolution gray-scale and rotation invariant texture classification with local binary patterns. IEEE Trans. Pattern Anal. Mach. Intell. 2002;24(7):971–987. [Google Scholar]
- 39.Sharma G., Umapathy K., Krishnan S. Trends in audio signal feature extraction methods. Appl. Acoust. 2020;158 [Google Scholar]
- 40.Orlandic L., Teijeiro T., Atienza D. The coughvid crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Scientific Data. 2021;8(1):1–10. doi: 10.1038/s41597-021-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.P. Bagad, A. Dalmia, J. Doshi, A. Nagrani, P. Bhamare, A. Mahale, S. Rane, N. Agarwal, R. Panicker, Cough against covid: Evidence of covid-19 signature in cough sounds, arXiv preprint arXiv:2009.08790, 2020.
- 42.A. Hassan, I. Shahin, M.B. Alsabek, Covid-19 detection system using recurrent neural networks, in 2020 International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI). IEEE, 2020, pp. 1–5.
- 43.Krishnan S. Elsevier; 2021. Biomedical Signal Analysis for Connected Healthcare. [Google Scholar]