Abstract
Despite the great success of vaccines that protect against RNA virus infections, and the development and clinical use of a limited number of RNA virus-specific drugs, there is still an urgent need for new classes of antiviral drugs against circulating or emerging RNA viruses. To date, it has proved difficult to efficiently suppress RNA virus replication by targeting host cell functions, and there are no approved drugs of this type. This opinion article discusses the recent discovery of a pronounced and sustained antiviral activity of the plant-derived natural compound thapsigargin against enveloped RNA viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), Middle East respiratory syndrome coronavirus (MERS-CoV), and influenza A virus. Based on its mechanisms of action, thapsigargin represents a new prototype of compounds with multimodal host-directed antiviral activity.
Keywords: thapsigargin, coronavirus, COVID-19, host-directed antivirals, ERAD, ER membranes, proteome
Activation of endoplasmic reticulum stress by the natural compound thapsigargin, an emerging new principle to combat coronavirus infections
The endoplasmic reticulum (ER) (see Glossary) is a large, perinuclear organelle that, in a dynamic manner, surveys the folding, assembly, transport, and degradation of membrane and secreted proteins [1]. The accumulation of unfolded or misfolded proteins in the ER lumen leads to the ER stress response and the unfolded protein response (UPR), a broad adaptive program that serves to increase the folding capacity of the ER and to restore protein homeostasis [2]. In general, ER stress and UPR act in concert and enable cells to detect and cope with misfolded proteins [3]. There is now strong evidence that this adaptive program is also activated in a broad range of cell types infected with intracellular pathogens [4]. In many cases, however, it remains unclear to what extent this cellular response is beneficial or detrimental to the pathogen (and/or host), and whether the interconnections of these processes can be pharmacologically tackled to help control the invading pathogen and thus ensure cell survival.
Both ER stress and the UPR can also be activated by chemicals, including thapsigargin, a natural compound that was isolated from the plant Thapsia garganica L. nearly 40 years ago [5]. Since then, thapsigargin has been used extensively to study its roles in (i) blocking calcium regulation, (ii) activating ER stress/UPR responses (by calcium depletion of the ER, see later), and (iii) mediating cytotoxicity [6].
In this opinion article, we highlight a newly discovered link between thapsigargin-mediated ER stress activation, UPR, and coronavirus (CoV) replication. As discussed later, there is now strong evidence that thapsigargin has antiviral effects and, based on a number of recent publications, it seems reasonable to suggest that this compound might pave the way for a new class of broad-spectrum antivirals against CoVs and, most likely, several other classes of enveloped RNA viruses.
CoV-associated changes in the ER structure and function as potential drug targets
CoVs are enveloped and contain four major structural proteins: the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins. The N protein encapsidates the large single-stranded, positive-sense RNA genome (+ssRNA) [7]. Cell entry is initiated by the interaction between the S protein and a cellular receptor. The subsequent fusion between viral and cellular membranes requires proteolytic cleavage by specific cellular proteases located at the plasma membrane or in endosomes, and leads to the release of the viral genome RNA, which is then translated into two large polyproteins (pp1a and pp1ab) which are proteolytically processed into individual nonstructural proteins (nsps) that together form the viral replication and transcription complex (vRTC). Specific components of the vRTC trigger a major reorganization of intracellular membrane structures, resulting, for example, in double-membrane vesicles (DMVs), zippered ER or convoluted membranes (CMs), and double-membrane spherules (DMSs). New 3D ultrastructural studies of SARS-CoV-2-infected cells provide strong evidence that viral RNA synthesis occurs inside ER-derived DMVs, which are considered as the major replicative organelles (ROs) [8., 9., 10., 11.]. In the course of infection, newly translated structural proteins translocate from ER membranes to the ER-to-Golgi intermediate compartment (ERGIC). Guided by interactions of newly formed nucleocapsids with other structural proteins, virus particles are formed by budding into the lumen of secretory vesicular compartments, and are released from the cell through the secretory exocytotic pathway [7]. This brief summary shows that key steps of the CoV replication cycle involve interactions with a large variety of cytoplasmic membranes, most prominently ER membranes [12., 13., 14.]. Although the molecular details of how viral and cellular factors (inter)act to trigger the profound membrane rearrangements in CoV-infected cells are not completely understood, the available information provides a solid basis to suggest that these processes may be attractive targets for antiviral therapies.
The discovery of the inhibitory effects of thapsigargin on CoV replication
In a recent study aimed at identifying commonalities and differences between mechanisms involved in canonical ER stress and CoV-mediated ER stress response, respectively, Shaban et al. used thapsigargin as a reference compound, as done before by many other groups working in this field; their study revealed that thapsigargin strongly inhibits the replication of human CoV 229E (HCoV-229E), MERS-CoV, SARS-CoV-2, and influenza A virus (IAV, strain KAN-1) in cell culture [15]. Altogether, thapsigargin was found to interfere with coronavirus replication at multiple levels, including viral RNA synthesis and genome replication, protein translation, and production of infectious virus progeny in different cell types [15].
The antiviral effect was observed with a half-maximal effective concentration (EC 50 ) in the lower nM range, well below the (known) half-maximal cytotoxic concentrations (CC 50 ) of thapsigargin [15]. The inhibitory effect was readily detectable, even when the compound was added as late as 8 h postinfection (p.i.). Moreover, the effect appeared to be remarkably long-lasting, with a single drug application being sufficient to suppress viral replication for up to 3 days [15]. Shortly after the paper by Shaban et al. was published as a preprint [16], Al-Beltagi et al. reported that thapsigargin has similar inhibitory effects on HCoV-OC43, respiratory syncytial virus (RSV), SARS-CoV-2, and various IAV H1N1 strains [17]. Both groups independently confirmed the suppression of SARS-CoV-2 replication in normal human differentiated bronchial epithelial (NHBE) cells grown at an air–liquid interface [15] or in undifferentiated NHBE cells [17,18], which were used as ex vivo models mimicking the natural site of infection in humans. In a follow-up study, Al-Beltagi et al. showed that thapsigargin also suppresses the SARS-CoV-2 Alpha, Beta, and Delta variants of concern [19]. The selectivity indices (CC50/EC50) reported in these studies ranged from >70 (for SARS-CoV-2) to >900 (for IAV, RSV, MERS-CoV, HCoV-229E, and HCoV-OC43) [15,17,18]. Taken together, these studies strongly support the idea that thapsigargin may be developed into a broad-spectrum anti-CoV drug for application in the upper respiratory tract, and warrants further preclinical and clinical studies.
How does thapsigargin work at the molecular level?
In the following sections we discuss recent observations that, mechanistically, the profound antiviral activity of thapsigargin is linked to the simultaneous interference with several interconnected cellular processes and pathways.
The differential effects of PERK inhibitors and thapsigargin on CoV replication
Shaban et al. went on to perform a detailed molecular study to obtain more insight into how thapsigargin inhibits viral replication. Both thapsigargin and CoV infection activate the PERK protein kinase, a major ER stress sensor that phosphorylates eukaryotic translation initiation factor (eIF) 2α to block protein translation [20]. A highly selective PERK inhibitor, GSK2656157, was shown to suppress viral replication of MERS-CoV and HCoV-229E by approximately one order of magnitude, but failed to abrogate the more than 100-fold suppression of replication resulting from treatment with thapsigargin [15]. These data show that PERK has a role in the replication of (at least some) CoVs. In contrast, thapsigargin appears to act downstream of PERK, and was shown to block the replication of all human CoVs investigated so far, as discussed earlier. It thus appears possible that PERK inhibitors might be further developed into strain-specific CoV antivirals, while thapsigargin appears to have a much more potent and broad-spectrum antiviral activity.
Thapsigargin relieves translational arrest and induces proteome-wide changes in metabolic pathways in CoV-infected cells
In previous transcriptome studies unrelated to viral infections, thapsigargin was reported to cause multiple changes in cellular mRNA expression, thereby significantly extending the list of changes that are typically involved in physiological adaptive UPR responses induced by an excess of folding-competent or folding-defective model protein substrates targeted to the ER [21,22]. These data suggested that additional mechanisms contribute to the phenotypes observed for thapsigargin-treated cells.
It was also unknown to what extent thapsigargin-induced mRNA changes correlate with changes in the proteome, in particular under conditions of the profound global translational shutdown that is typically observed in RNA virus-infected cells. Along with a strong activation of PERK, CoV infection was found to reduce cellular protein biosynthesis by about 90%. Thapsigargin treatment of infected cells was observed to improve translation rates significantly. Together with data from cell viability assays, these results suggest that the compound improves, but does not completely restore, the overall metabolic status of infected cells [15].
To address these points, Shaban et al. performed comparative proteomic and bioinformatics studies of cells infected with MERS-CoV and SARS-CoV-2, respectively. (Pathway) enrichment analysis revealed a thapsigargin-mediated activation of a broad spectrum of metabolic pathways which, probably, contribute to the protective response mounted in infected cells that were treated with this compound [15].
To further corroborate the significance of these observations, it will be important to define the enzymes and their metabolites that contribute to the thapsigargin-mediated metabolic changes in infected host cells, and to answer the question of whether metabolic reprogramming can be exploited as a general (cell-intrinsic) strategy to suppress RNA virus replication.
Thapsigargin changes expression levels of proteins that regulate membrane compartments
At the level of individual components, Shaban et al. identified a set of proteins that are specifically induced (120 proteins) or downregulated (63 proteins) in thapsigargin-treated and simultaneously infected cells, as shown in Figure 7 of their publication [15]. Many of the upregulated factors are annotated as being involved in the regulation of intracellular membrane compartments (ER, Golgi apparatus-associated vesicle biogenesis and transport, endocytosis) and membrane-associated processes such as ER stress and ER-associated protein degradation (ERAD, Box 1 ) [15]. As discussed above, membrane rearrangement and fusion events are major hallmarks of the coronavirus replication cycle and are critically involved in virus entry, replication, and budding [8., 9., 10.]. Based on the proteomic data, it is tempting to suggest that thapsigargin prevents, or activates, unknown membrane-associated processes that counteract efficient viral replication. To elucidate the underlying mechanisms, it will be important to study individually the (enzymatic) activities, interactomes (i.e., protein–protein interactions and networks), localizations, and functions of each of the specific factors identified by proteomics. Ideally, this approach should also include information from the plethora of proteomic, interactomic, and CRISPR/Cas9-based single-guide (sg) RNA genome-wide screens that have been performed recently for SARS-CoV-2 and other coronaviruses [23., 24., 25., 26., 27., 28., 29.]. We firmly believe that such studies could provide new avenues for antiviral approaches that target very specifically the profound structural and functional changes in intracellular membrane compartments in +RNA virus-infected cells.
Box 1. ERQC, ERAD, ER-phagy/autophagy and protein degradation.
One third of mammalian proteins are synthesized in the ER whose maturation is overseen by the ERQC system. ERQC employs lectin chaperones or the chaperone BiP (in concert with ER-localized cofactors) that, together with protein disulfide or peptidylprolyl isomerases, catalyze protein-folding reactions. Accurately folded and assembled proteins are released from chaperones, exit the ER, and are transported to the Golgi via coat complex II (COPII) vesicles to their final destinations. If ERQC fails, proteins will be disposed of by two degradative systems: ERAD or ER-phagy. ERAD encompasses the identification of misfolded proteins, their retrotranslocation to the cytosol, and degradation by the UPS. ERAD clients first bind to Sel1L which interacts directly with the E3 ligase HRD1. By oligomerization of its transmembrane domains, HRD1 forms the central, funnel-like structure of the retrotranslocon. Upon insertion into the HRD1 channel, the client starts to cross the ER membrane, reaches the cytosol, and becomes (poly)-ubiquitinylated by HRD1. At this point, the AAA-ATPase, p97, is recruited to the ER membrane via its association with valosin-containing protein-interacting membrane protein (VIMP), recognizes the polyubiquitin chains on the ERAD client, and provides the final energy to extract it fully from the ER membrane. The ERAD client is then delivered to the 26S proteasome and degraded. ERAD involves many additional (UPR-regulated) proteins as assessed by large-scale screens [73]. ER-phagy, a (macro)autophagy-related process, is used to remove larger proteins, protein aggregates, and damaged ER, and to reduce ER size after stress-induced expansion. During autophagy, multiple autophagy-related (ATG) proteins cooperate to elongate and bend pieces of membrane to engulf cytoplasmic components or organelles. The resulting structure, the autophagosome, is decorated with phosphatidylethanolamine (PE)-conjugated, lipidated LC3 (LC3-II) at the inner side of the lipid bilayer membrane. In case of selective autophagy, substrates often carry ubiquitin chains to tether them to specific autophagy receptors (e.g., p62/SQSTM1). Mammalian ER-phagy is promoted by six ER-resident receptors (FAM134B, RTN3L, CCPG1, SEC62, TEX264, and ATL3) that possess LC3-interacting regions (LIR). LIR domains help to recruit autophagy substrates to the interior of the autophagosome precursor, called isolation membrane or phagophore. To complete autophagy, the pore in the phagophore closes, and the outer autophagosomal membrane fuses with the lysosome, resulting in disintegration of the inner autophagosomal membrane and degradation of the sequestered materials (for excellent reviews on these topics see [30,74., 75., 76.]).
Alt-text: Box 1
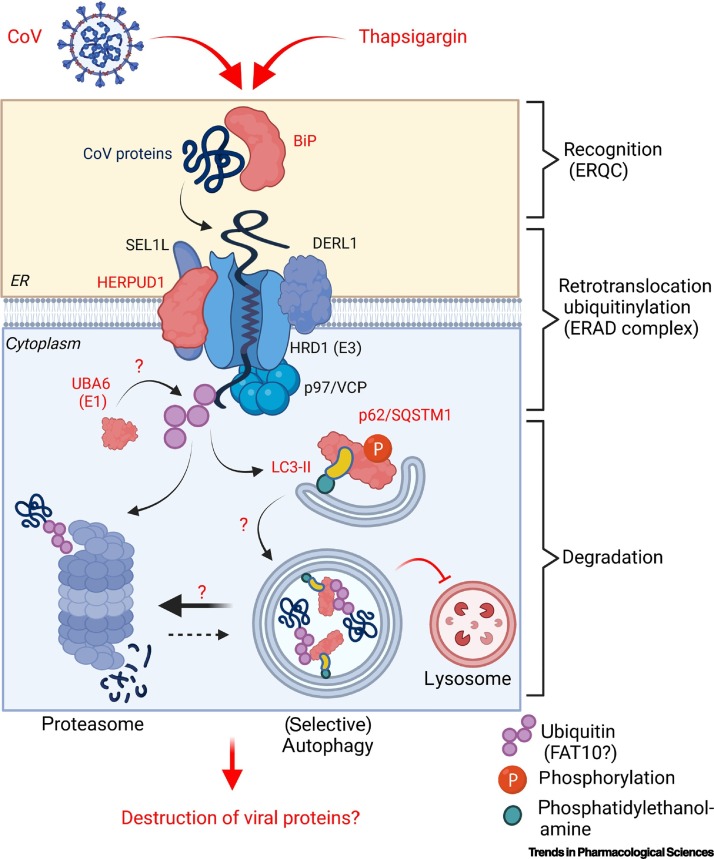
Thapsigargin induces multiple ER quality control (ERQC) and ERAD factors
There are multiple connections between ERQC, ERAD, the ubiquitin proteasome system (UPS) and autophagy pathways as outlined in Box 1. ERAD is a UPR-activated degradative mechanism that serves to retrotranslocate misfolded proteins from the lumen or membrane of the ER into the cytosol, whereupon they get ubiquitinylated and proteasomally hydrolyzed [30]. The regulation of ERAD and the related process called ER-phagy by viruses is incompletely understood [31,32]. The proteomic analyses mentioned earlier revealed that thapsigargin induces a strong upregulation of several ERAD factors, including homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 (HERPUD1), a central scaffold for ERAD complexes assembling in the ER membrane [33] (Figure 1 , Key figure). Thapsigargin also induced proteins involved in ubiquitinylation and ubiquitin-like post-translational modifications. Ubiquitinylation of target proteins proceeds in three steps through ubiquitin-activating (E1), -conjugating (E2) and -ligating (E3) enzymes [34]. Shaban et al. observed an upregulation of ubiquilins (UBQLN 1 and 4), the E2 enzyme ubiquitin-conjugating enzyme E2 G1 (UBE2G1), and the E1 enzyme ubiquitin-like modifier-activating enzyme 6 (UBA6) in cells infected with HCoV-229E, MERS-CoV, and SARS-CoV-2, respectively [15]. UBA6 and ubiquitin-activating enzyme E1 (UBE1, also called ubiquitin-like modifier-activating enzyme 1, UBA1) are the two main E1 enzymes that redirect proteins to the proteasome [35]. UBA6 can also activate the ubiquitin-like protein (UBL) FAT10 (also called ubiquitin D, UBD) and transfer FAT10 to its substrate proteins for rapid proteasomal destruction [35,36]. FAT10 is upregulated by the proinflammatory mediators interferon γ (IFN-γ), tumor necrosis factor α (TNF-α), or lipopolysaccharide (LPS), and has been found to modify ubiquitin-binding protein p62/sequestosome-1 (p62/SQSTM1), connecting this pathway to autophagy and suggesting a broader role for FAT10 in immune functions [37]. These results suggest a role for specific components of the UPS in viral protein turnover downstream of ERAD/autophagy. Therefore, it is an intriguing hypothesis that the thapsigargin-mediated autophagic block described below redirects CoV proteins to an increased ERAD, thereby contributing to their rapid clearance and, as a result, suppression of viral replication (Figure 1).
Figure 1.
Key figure. Hypothetical antiviral mechanism of thapsigargin involving endoplasmic reticulum (ER)-associated protein degradation (ERAD).
The scheme shows the organization of the ER membrane-associated ERAD complex as recently reviewed [30]. ERAD is intimately linked to the major proteasomal and autophagy-related protein degradation pathways [70., 71., 72.] (Box 1). Red colors highlight the proteins endoplasmic reticulum chaperone binding immunoglobulin protein (BiP; GRP-78, HSPA5), homocysteine-responsive endoplasmic reticulum-resident ubiquitin-like domain member 1 (HERPUD1), ubiquitin-like modifier-activating enzyme 6 (UBA6), autophagy-related protein LC3 (microtubule-associated proteins 1A/1B light chain 3A/B), and ubiquitin-binding protein p62/sequestosome-1 (p62/SQSTM1) that, by mass spectrometry and immunoblotting, have consistently been found to be induced by thapsigargin in coronavirus (CoV)-infected cells. Question marks indicate potential functional links between these factors. Because thapsigargin-treated infected cells show a strong reduction in CoV protein levels, it is tempting to suggest that thapsigargin enhances ERAD activity in favor of autophagy (which is blocked by the compound late in the autophagy pathway) and, thereby, causes disposal of viral proteins primarily through the proteasome pathway as part of its antiviral mechanism. Abbreviations: DERL1, Derlin-1; FAT10, a ubiquitin-like protein (ubiquitin D); HRD1, E3 ubiquitin-protein ligase synoviolin; SEL1L, protein sel-1 homolog 1; VCP, transitional endoplasmic reticulum ATPase, 5S Mg2+-ATPase p97 subunit, valosin-containing protein.
Thapsigargin blocks (selective) autophagy flux
Another factor that is consistently found to be regulated by thapsigargin is p62/SQSTM1 (Box 1, Figure 1), a multifunctional signaling protein and cargo receptor for selective autophagy clients [38]. The potential role of (macro)autophagy in CoV replication is under active debate, with evidence for both proviral and antiviral functions, depending on the virus strains or model systems used (reviewed in [39,40]). As shown in the study by Shaban et al., HCoV-229E, MERS-CoV, or SARS-CoV-2 infections downregulate the level of p62/SQSTM1, albeit to a different extent. All three viruses have comparably limited effects on LC3B-II, which is the lipidated (i.e., phosphatidylethanolamine-conjugated) form of autophagy-related protein LC3/microtubule-associated proteins 1A/1B light chain 3B (Atg8/LC3B), a ubiquitin-like molecule or modifier (ULM) that is essential for autophagosome formation early in the autophagy pathway [41]. Autophagy is a highly dynamic process that proceeds through several steps until the respective membrane-engulfed cargo is degraded by fusion of autophagosomes with lysosomes [42]. The activity of this pathway is commonly assessed by determining the turnover of autophagy proteins, which is generally referred to as the autophagic flux [43]. Application of bafilomycin A1, an inhibitor of the lysosomal enzyme V-ATPase, to CoV-infected cells resulted in (i) a strong increase in the p62 (and, to lesser extent, LC3B-II) protein levels, and (ii) a tenfold reduction in viral titers for some of the CoVs included in this study [15]. These results are in line with (more variable) antiviral effects observed for several other lysosomal inhibitors, including (hydroxy)chloroquine [44]. Taken together, the available data indicate that human CoVs stimulate selective autophagy at some point during viral replication, but clearly more evidence is needed to corroborate this hypothesis. In the presence of thapsigargin, strongly increased levels of p62/SQSTM1 and LC3B were observed in virus-infected cells by a bafilomycin A1-independent mechanism. Also, thapsigargin strongly inhibited the autophagic flux in these cells [15]. These results are best reconciled with the work by Ganley et al. who showed that, independently of ER stress signaling and the endosomal pathway, thapsigargin blocks the fusion of autophagosomes with lysosomes late in the autophagy pathway by an unknown mechanism [45,46]. It is therefore possible that such a mechanism (irreversibly) prevents the correct fusion of lysosomes with intracellular vesicles throughout the CoV replication cycle and, possibly, also interferes with membrane fusion events required for the formation of replicative organelles early in the infection cycle [7].
Calcium-related effects of thapsigargin
The best understood mechanism of the action of thapsigargin is related to intracellular calcium homeostasis [47]. Thapsigargin was first described as a modulator of the Ca2+-dependent histamine release from mast cells [48., 49., 50.]. Functional and structural evidence suggested that thapsigargin increases intracellular Ca2+ by specific and potent inhibition of the sarco-/endoplasmic reticulum (ER) Ca 2+ -ATPase (SERCA) (Box 2 ) [51., 52., 53.]. The depletion of ER Ca2+ and prolonged activation of the UPR are considered to cause the massive cell death seen with thapsigargin or its analogues in various cell types [54]. Furthermore, a recent report suggested that overexpression of SERCA may counteract the antiviral effects of thapsigargin against two RNA viruses infecting sheep and goats (Peste des petits ruminants virus, PPRV) or birds (Newcastle disease virus, NDV) [55]. In Drosophila melanogaster, the SERCA Ca2+ pump is important for membrane fusion events of lysosomes [56], suggesting that the block of autophagy and RNA virus replication caused by thapsigargin may be related, at least in part, to changes in subcellular Ca2+ gradients that support vesicle fusion events. This hypothesis may be an attractive avenue for further investigations into the antiviral effects of thapsigargin.
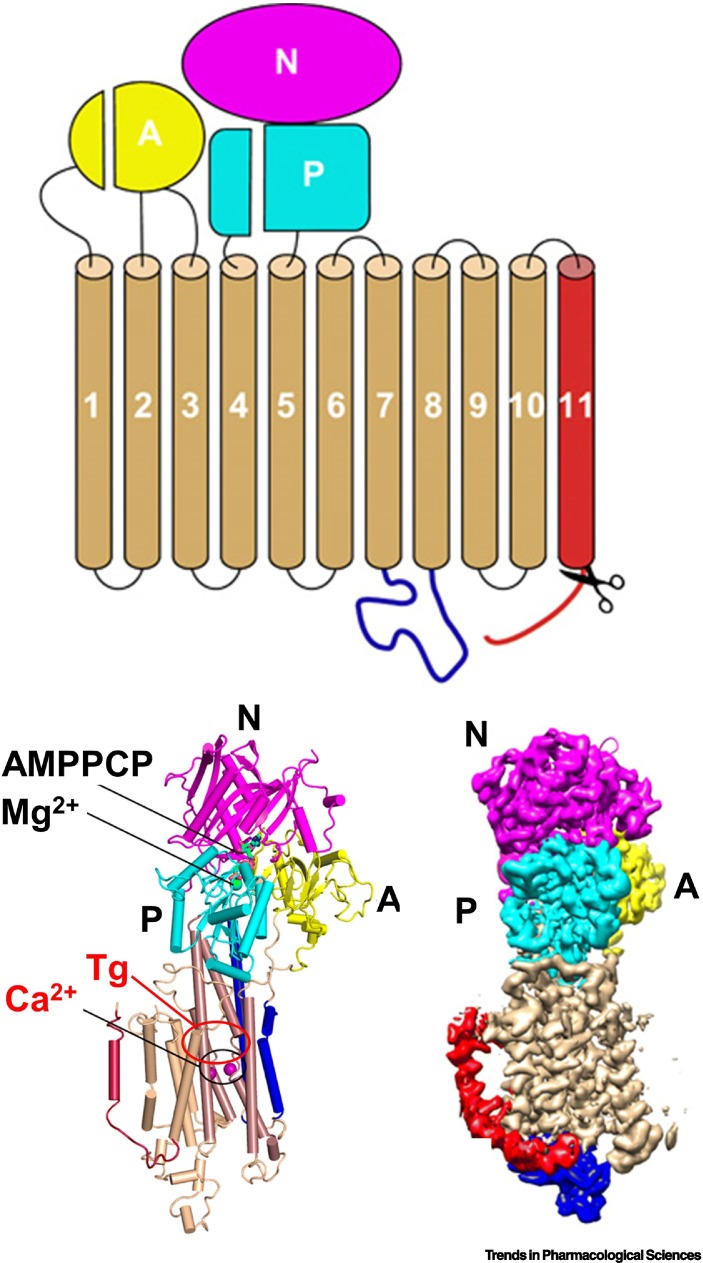
Box 2. Thapsigargin and Ca2+-transport.
In mammalian cells, there is a >1000-fold concentration gradient of Ca2+ between the lumen of the ER and the cytoplasm, which is maintained by SERCA ATPase calcium pumps. These enzymes constantly transfer Ca2+ from the cytoplasm into the ER, where the ion supports protein folding. The lipophilic thapsigargin (Tg), within minutes, enters the cell, fully (or irreversibly) inhibits SERCA, and depletes Ca2+ from the ER (reviewed in [66]). Several studies of the atomic structures revealed that thapsigargin allosterically inhibits Ca2+-ATPases as shown in Figure I. The conserved structure of SERCA proteins consists of three cytoplasmic domains – comprising an actuator domain (A), a nucleotide-binding domain (N), and a phosphorylation (P) domain – and ten transmembrane (TM) helices. Tg binds to a groove formed by TM helices TM3, TM4, TM5, and TM7, and this interaction is stabilized by hydrophobic interactions with residues F256, L260, V263, Ile761, V769, I829, F834, and M838 [77]. An interesting question is: whether the large pattern of gene expression changes observed in Tg-treated cells [21] is solely due to inhibition of SERCA, or whether Tg also has additional mechanisms of actions.
Figure I.
Scheme and structure models of SERCA with the Ca2+ and thapsigargin (Tg) binding sites indicated by circles.
Reproduced and adapted from [78] under a Creative Commons Attribution Non Commercial License 4.0 (CC BY-NC). Abbreviations: AMPPCP, adenylyl methylenediphosphonate; SERCA, sarco-/endoplasmic reticulum (ER) Ca2+-ATPase.
Alt-text: Box 2
The pharmacology of thapsigargin: can thapsigargin be developed into a (pan-)antiviral drug?
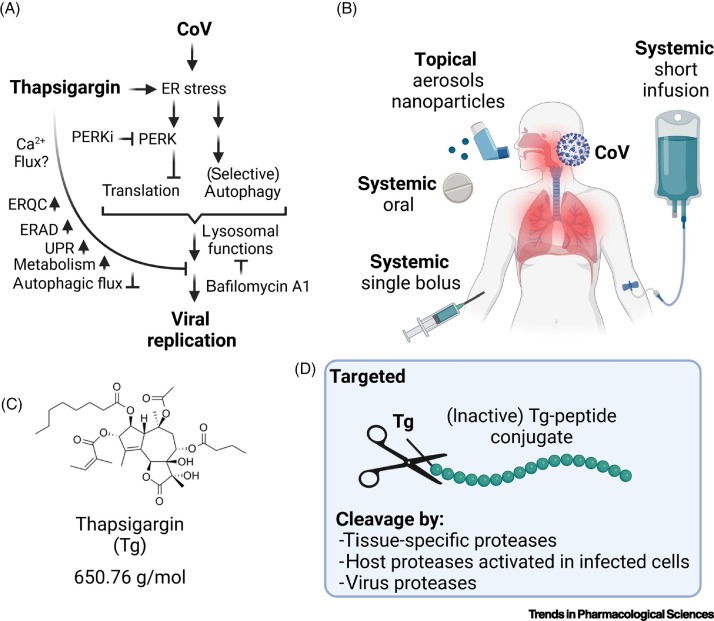
Taken together, the observations discussed earlier suggest that thapsigargin hits, in a unique manner, a combination of cellular processes that are essential for CoV replication (Figure 2A). It remains to be studied whether these processes can be targeted even more specifically using a combination of pathway-specific drugs, such as combinations of PERK and lysosomal inhibitors, with the aim of reducing potential side effects (Figure 2A).
Figure 2.
Summary of thapsigargin effects in coronavirus (CoV)-infected cells and possible modes of application in humans.
(A) The scheme shows the different levels of cellular processes that have been shown to be activated or suppressed by thapsigargin in cells infected with various CoVs. Also included are the effects of drugs blocking the lysosome (bafilomycin A1) or PERK kinase (PERKi, GSK2656157 or GSK2606414). At present, the role of thapsigargin-mediated calcium depletion in coronavirus replication is unclear. Adapted from [15]. Abbreviations: Tg, thapsigargin; ER, endoplasmic reticulum; ERQC, ER quality control; ERAD, ER-associated protein degradation; PERK, protein kinase RNA-activated (PRKR)-like endoplasmic reticulum kinase; UPR, unfolded protein response. (B) Possible routes and modes of application of thapsigargin in humans. While short-term oral administration was effective in mice, the bioavailability and pharmacokinetics of thapsigargin for the use as antiviral therapy in humans are currently unclear. (C) Structure of thapsigargin. (D) Potential protease-cleavable prodrugs in which thapsigargin is coupled to a peptide (in green) that will be cleaved off by cell type-specific proteases, by (inducible) proteases present only in infected cells, or by coronavirus proteases, in order to minimize side effects by restricting the active drug primarily or exclusively to infected cells.
The pharmacodynamics of thapsigargin in the context of depletion of Ca2+ from the ER and induction of cell death have been studied for four decades, primarily with the aim of using this drug in cancer therapy [6,49]. The synthesis and production of thapsigargin and derivatives thereof are well established, and formulations are available for application in humans [57., 58., 59.]. A major obstacle for its potential use in antiviral therapy relates to the toxicity expected to occur in vivo. In this context, two studies have recently been performed in animal models. Oral doses of thapsigargin applied to mice 12 h before or 12 h after infection with IAV significantly improved the survival rates [17,18]. Starting 1 day after a lethal challenge with IAV, a daily oral application of thapsigargin for 5 days was found to prevent infection-related death of mice infected with this virus [17]. In another study, thapsigargin was applied 15 h before a lethal dose of LPS or cecal ligation and puncture (CLP)-induced sepsis. Thapsigargin was shown to improve, in a dose-dependent manner, the survival rate of two mouse strains after 7 days [60]. Although these preclinical data on a short-term application of the drug are encouraging, more in vivo data are required to thoroughly assess the pharmacokinetics, efficacy, and toxicity of thapsigargin using animal models of infection and inflammation. This includes the application of thapsigargin in suitable SARS-CoV-2 animal models [61].
While systemic thapsigargin-mediated toxicity in humans may be appropriately addressed by using very low, single-bolus doses, or specific topical applications in the upper respiratory tract (Figure 2B), it may also be possible to design suitable prodrugs in which thapsigargin is coupled to peptides or other moieties to limit potential toxic side effects (Figure 2C,D) [62]. Thus, for example, mipsagargin, a protease-cleavable inactive prodrug of thapsigargin, has been applied in Phase I and Phase II clinical trials for prostate or liver cancer therapy, with acceptable toxicity [63., 64., 65., 66.]. By using a similar approach, it may be feasible to develop a cell-permeable thapsigargin derivative that is specifically cleaved by viral (or specific cellular) proteases, thereby restricting the release of the active drug to virus-infected cells (Figure 2C,D).
An additional clinical benefit of thapsigargin may arise from the compound-mediated suppression of inflammatory cytokines at the post-transcriptional level by interfering with their translation or secretion [67]. These effects have been observed in CoV-infected cultured cells and in the LPS/CLP animal models [15,60]. The suppression of inflammation-mediated lung injury by the anti-inflammatory glucocorticoid dexamethasone reduces COVID-19 mortality [68]. By analogy, it will be important to find out whether low-dose thapsigargin will not only suppress CoV replication but also tissue-specific or systemic immunopathologies relevant to COVID-19 disease.
Concluding remarks and future perspective
In this opinion article we reviewed the available evidence supporting a potent antiviral effect of thapsigargin against CoVs. This antiviral effect is linked to an improvement (but not complete rescue) of the cellular metabolic state and the survival of CoV-infected cells treated with this natural compound. There is also initial evidence that thapsigargin inhibits the replication of the flavivirus tick-borne encephalitis virus (TBEV), albeit by a different mechanism that involves the UPR-dependent priming of an interferon response [69]. Although many questions remain to be answered regarding virus-specific effects and the underlying antiviral actions of this compound at the molecular level (see Outstanding questions), particularly with respect to changes in intracellular membrane structures and functions in infected cells, we think it is worth pursuing further (preclinical and clinical) studies on thapsigargin and its derivatives (i) to establish topical, low-dosage, and short-term therapeutic applications, (ii) to restrict toxic effects to virus-infected cells, and (iii) to address the question of whether or not RNA viruses evolve drug-resistant variants that escape these multimodal antiviral mechanisms.
Outstanding questions.
Are SERCA and SERCA-dependent processes such as Ca2+-signaling, Ca2+-dependent regulation of cellular factors or enzymes, and subcellular changes in Ca2+ concentrations relevant for thapsigargin-mediated antiviral activities? What are the roles of local Ca2+-sensing or effector mechanisms inside organelles and at their interfaces?
How is cell morphology affected by thapsigargin early upon viral replication, and, related to this, can thapsigargin lead to the resolution of already established ROs? Can these questions be solved by 3D high-resolution imaging, including advanced electron microscopy?
Present evidence suggests that thapsigargin antiviral activity crucially relies on the inhibition/activation of several cellular processes, but it is unclear whether some of these are more important than others. Does thapsigargin hit a process or a specific molecular event that is common to all enveloped viruses, even though they have highly divergent life cycles (e.g., CoV or IAV)? Can these issues be addressed by meta-analyses of functional genome-wide genetic screens performed in various infection models?
Several hACE2-transgenic mice and also mouse-adapted SARS-CoV-strains are now available. Will thapsigargin suppress the replication of SARS-CoV-2 in animal models of COVID-19 disease? What are the longer-term consequences?
Multiple chemical analogues of thapsigargin have been synthesized recently and can be studied in RNA virus infection models with respect to replication versus cell survival. Can they be used to separate thapsigargin-mediated cytotoxicity from its antiviral effects?
Can thapsigargin analogues be designed in a way that they are cleaved, after internalization, by cell-specific or CoV main proteases?
Can thapsigargin be specifically targeted to sites of infection in vivo? For example, can (inactive) peptide–thapsigargin conjugates be targeted to cell-surface molecules only expressed on CoV-infected cells?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
This work was supported by the following grants from the Deutsche Forschungsgemeinschaſt (DFG, German Research Foundation): KR1143/9-2 and ZI618/6-2 [KFO309, P3 (to M.K. and J.Z.), project 284237345]; SFB1021/3 [A01 (to J.Z.), C02 (to M.K.), Z03 (to M.K., U.L.), project 197785619]; and GRK 2573 [RP4 (to M.L.S), RP5 (to M.K.), project 416910386]. The work was also supported by the German Ministry for Education and Research (RAPID, COVINET, and DZIF TTU 01.806, to J.Z.) and the LOEWE program of the state of Hesse (DRUID, B02 to J.Z.). Work in the laboratory of M.K. is also supported by the IMPRS program of the Max Planck Society and the Excellence Cluster CardioPulmonary Institute (EXC 2026: Cardio-Pulmonary Institute (CPI), project 390649896) and the DZL/UGMLC/ILH program. The work of J.Z. and M.K. is further supported by the Von-Behring-Roentgen-Stiftung and the Pandemics Network Hesse.
Author contributions
M.K. wrote the initial draft of the article and prepared the figures. M.S.S., C.M-B., J.M-S., B.V.A., J.Z. and M.L.S. helped to finalize the paper. All authors approved the submitted version of the paper.
Declaration of interests
The authors declare no competing interests.
Glossary
- Air–liquid interface
cell culture systems in which the apical site of a cell layer is exposed to air.
- Atg8/LC3
the autophagy-related protein LC3, also called microtubule-associated proteins 1A/1B light chain 3A/B; it is a major autophagy regulator.
- Autophagic flux
the turnover of components degraded by the autophagy pathway.
- BiP
endoplasmic reticulum chaperone BiP (GRP-78, HSPA5), a major ER factor supporting protein folding.
- Cecal ligation and puncture (CLP)-induced sepsis
a procedure for modeling sepsis in vivo, in which puncture of the cecum, which is full of bacteria, results in polymicrobial peritonitis, bacteremia, septic shock, multiorgan dysfunction, and death.
- Coronaviruses (CoVs)
a group of RNA viruses with very large, mRNA-like, +ssRNA genomes.
- Efficacy
the maximum effect or therapeutic response achievable from a pharmaceutical drug.
- Endoplasmic reticulum (ER)
an interconnected, intracellular network of flattened, membrane-enclosed sacs involved in multiple processes, including protein synthesis, folding, and transport.
- Endosomal pathway
a membrane transport pathway derived from the trans Golgi network to allow transport from the plasma membrane to the lysosome or back to the cell membrane.
- ERGIC
ER–Golgi intermediate compartment mediating the trafficking between the ER and the Golgi complex.
- ER stress response
a signaling network activated by ER stress sensors PERK, IRE1α, and ATF6.
- FAT10
a ubiquitin-like protein also called ubiquitin D.
- Global translational shutdown
inhibition of de novo protein synthesis by phosphorylation of the eukaryotic translation initiation factor 2α subunit (eIF2α) in response to infection, starvation, or ER stress.
- Half-maximal cytotoxic concentrations (CC50)
the concentration of a compound at which 50% of a drug’s cytotoxic effect is observed.
- Half-maximal effective concentration (EC50)
the concentration of a compound at which 50% of its maximal (antiviral) effect is observed. EC50 corresponds to the drug’s potency.
- (Pathway) enrichment analysis
a bioinformatics method that identifies biological pathways that are enriched in a gene list more than would be expected by chance.
- PERK
protein kinase RNA-activated (PRKR)-like endoplasmic reticulum kinase, or eukaryotic translation initiation factor 2-α kinase 3, is an ER-resident eIF2α kinase activated by ER stress.
- Replicative organelles (ROs)
modified endomembrane structures which are the sites of CoV genome replication.
- Sarco-/endoplasmic reticulum (ER) Ca2+-ATPase (SERCA)
a calcium ATPase type P-ATPase that transports calcium from the cytosol into the ER.
- Ubiquilins
a family of ubiquitin-binding proteins that function in both the proteasome and autophagy pathways.
- Ubiquitin
a small (8.6 kDa) regulatory protein that is covalently attached to a substrate by ubiquitinylation to regulate its decay or activity.
- Ubiquitin proteasome system (UPS)
ubiquitin-mediated proteolysis of unneeded/damaged proteins by cytosolic protein complexes called proteasomes.
- Unfolded protein response (UPR)
a transcriptional response program that induces multiple genes/corresponding proteins to compensate ER stress.
- Viral replication/transcription complexes (vRTCs)
several nonstructural CoV proteins (nsps) that replicate the CoV genome inside ROs.
References
- 1.Hetz C., et al. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020;21:421–438. doi: 10.1038/s41580-020-0250-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marciniak S.J., et al. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat. Rev. Drug Discov. 2022;21:115–140. doi: 10.1038/s41573-021-00320-3. [DOI] [PubMed] [Google Scholar]
- 3.Karagoz G.E., et al. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grootjans J., et al. The unfolded protein response in immunity and inflammation. Nat. Rev. Immunol. 2016;16:469–484. doi: 10.1038/nri.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rasmussen U., et al. Thapsigargine and thapsigargicine, two new histamine liberators from Thapsia garganica L. Acta Pharm. Suec. 1978;15:133–140. [PubMed] [Google Scholar]
- 6.Andersen T.B., et al. Thapsigargin – from Thapsia L. to mipsagargin. Molecules. 2015;20:6113–6127. doi: 10.3390/molecules20046113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.V’Kovski P., et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortese M., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866 e855. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein S., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snijder E.J., et al. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loconte V., et al. Using soft X-ray tomography for rapid whole-cell quantitative imaging of SARS-CoV-2-infected cells. Cell Rep. Methods. 2021;1 doi: 10.1016/j.crmeth.2021.100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romero-Brey I., Bartenschlager R. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses. 2016;8:160. doi: 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z., et al. The interplay between emerging human coronavirus infections and autophagy. Emerg. Microbes Infect. 2021;10:196–205. doi: 10.1080/22221751.2021.1872353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaban M.S., et al. Multi-level inhibition of coronavirus replication by chemical ER stress. Nat. Commun. 2021;12:5536. doi: 10.1038/s41467-021-25551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaban M.S., et al. Inhibiting coronavirus replication in cultured cells by chemical ER stress. bioRxiv. 2020 doi: 10.1101/2020.08.26.266304. Published online August 26, 2020. [DOI] [Google Scholar]
- 17.Al-Beltagi S., et al. Thapsigargin is a broad-spectrum inhibitor of major human respiratory viruses: coronavirus, respiratory syncytial virus and influenza A virus. Viruses. 2021;13:234. doi: 10.3390/v13020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulding L.V., et al. Thapsigargin at non-cytotoxic levels induces a potent host antiviral response that blocks influenza A virus replication. Viruses. 2020;12:1093. doi: 10.3390/v12101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Beltagi S., et al. Emergent SARS-CoV-2 variants: comparative replication dynamics and high sensitivity to thapsigargin. Virulence. 2021;12:2946–2956. doi: 10.1080/21505594.2021.2006960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urra H., Hetz C. Fine-tuning PERK signaling to control cell fate under stress. Nat. Struct. Mol. Biol. 2017;24:789–790. doi: 10.1038/nsmb.3478. [DOI] [PubMed] [Google Scholar]
- 21.Bergmann T.J., et al. Chemical stresses fail to mimic the unfolded protein response resulting from luminal load with unfolded polypeptides. J. Biol. Chem. 2018;293:5600–5612. doi: 10.1074/jbc.RA117.001484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergmann T.J., Molinari M. Three branches to rule them all? UPR signalling in response to chemically versus misfolded proteins-induced ER stress. Biol. Cell. 2018;110:197–204. doi: 10.1111/boc.201800029. [DOI] [PubMed] [Google Scholar]
- 23.Bailey A.L., Diamond M.S. A Crisp(r) new perspective on SARS-CoV-2 biology. Cell. 2021;184:15–17. doi: 10.1016/j.cell.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu Y., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun. 2021;12:961. doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baggen J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat. Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 26.Terracciano R., et al. Mapping the SARS-CoV-2–host protein–protein interactome by affinity purification mass spectrometry and proximity-dependent biotin labeling: a rational and straightforward route to discover host-directed anti-SARS-CoV-2 therapeutics. Int. J. Mol. Sci. 2021;22:532. doi: 10.3390/ijms22020532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stukalov A., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]
- 28.Samavarchi-Tehrani P., et al. A SARS-CoV-2-host proximity interactome. bioRxiv. 2020 doi: 10.1101/2020.09.03.282103. Published online September 4, 2020. [DOI] [Google Scholar]
- 29.McDougall W.M., et al. CRISPR genetic screens to discover host–virus interactions. Curr. Opin. Virol. 2018;29:87–100. doi: 10.1016/j.coviro.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Oikonomou C., Hendershot L.M. Disposing of misfolded ER proteins: a troubled substrate's way out of the ER. Mol. Cell. Endocrinol. 2020;500 doi: 10.1016/j.mce.2019.110630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Molinari M. ER-phagy: eating the factory. Mol. Cell. 2020;78:811–813. doi: 10.1016/j.molcel.2020.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Fregno I., Molinari M. Proteasomal and lysosomal clearance of faulty secretory proteins: ER-associated degradation (ERAD) and ER-to-lysosome-associated degradation (ERLAD) pathways. Crit. Rev. Biochem. Mol. Biol. 2019;54:153–163. doi: 10.1080/10409238.2019.1610351. [DOI] [PubMed] [Google Scholar]
- 33.Leitman J., et al. Herp coordinates compartmentalization and recruitment of HRD1 and misfolded proteins for ERAD. Mol. Biol. Cell. 2014;25:1050–1060. doi: 10.1091/mbc.E13-06-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grabbe C., et al. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Groettrup M., et al. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 2008;33:230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Wang F., Zhao B. UBA6 and its bispecific pathways for ubiquitin and FAT10. Int. J. Mol. Sci. 2019;20:2250. doi: 10.3390/ijms20092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aichem A., et al. The ubiquitin-like modifier FAT10 interferes with SUMO activation. Nat. Commun. 2019;10:4452. doi: 10.1038/s41467-019-12430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamark T., et al. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem. 2017;61:609–624. doi: 10.1042/EBC20170035. [DOI] [PubMed] [Google Scholar]
- 39.Miller K., et al. Coronavirus interactions with the cellular autophagy machinery. Autophagy. 2020;16:2131–2139. doi: 10.1080/15548627.2020.1817280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fecchi K., et al. Coronavirus interplay with lipid rafts and autophagy unveils promising therapeutic targets. Front. Microbiol. 2020;11:1821. doi: 10.3389/fmicb.2020.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pankiv S., et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 42.Dikic I., Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018;19:349–364. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 43.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 2017;18:1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorshkov K., et al. The SARS-CoV-2 cytopathic effect is blocked by lysosome alkalizing small molecules. ACS Infect Dis. 2021;7:1389–1408. doi: 10.1021/acsinfecdis.0c00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganley I.G., et al. Thapsigargin distinguishes membrane fusion in the late stages of endocytosis and autophagy. Autophagy. 2011;7:1397–1399. doi: 10.4161/auto.7.11.17651. [DOI] [PubMed] [Google Scholar]
- 46.Ganley I.G., et al. Distinct autophagosomal–lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol. Cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bagur R., Hajnoczky G. Intracellular Ca(2+) sensing: its role in calcium homeostasis and signaling. Mol. Cell. 2017;66:780–788. doi: 10.1016/j.molcel.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ali H., et al. The ability of thapsigargin and thapsigargicin to activate cells involved in the inflammatory response. Br. J. Pharmacol. 1985;85:705–712. doi: 10.1111/j.1476-5381.1985.tb10567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patkar S.A., et al. On the mechanism of histamine release induced by thapsigargin from Thapsia garganica L. Agents Actions. 1979;9:53–57. doi: 10.1007/BF02024109. [DOI] [PubMed] [Google Scholar]
- 50.Christensen S.B., et al. From plant to patient: thapsigargin, a tool for understanding natural product chemistry, total syntheses, biosynthesis, taxonomy, ATPases, cell death, and drug development. Prog. Chem. Org. Nat. Prod. 2021;115:59–114. doi: 10.1007/978-3-030-64853-4_2. [DOI] [PubMed] [Google Scholar]
- 51.Thastrup O., et al. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. U. S. A. 1990;87:2466–2470. doi: 10.1073/pnas.87.7.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sagara Y., Inesi G. Inhibition of the sarcoplasmic reticulum Ca2+ transport ATPase by thapsigargin at subnanomolar concentrations. J. Biol. Chem. 1991;266:13503–13506. [PubMed] [Google Scholar]
- 53.Toyoshima C., et al. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 54.Sehgal P., et al. Inhibition of the sarco/endoplasmic reticulum (ER) Ca(2+)-ATPase by thapsigargin analogs induces cell death via ER Ca(2+) depletion and the unfolded protein response. J. Biol. Chem. 2017;292:19656–19673. doi: 10.1074/jbc.M117.796920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar N., et al. Inhibitor of sarco/endoplasmic reticulum calcium-ATPase impairs multiple steps of paramyxovirus replication. Front. Microbiol. 2019;10:209. doi: 10.3389/fmicb.2019.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mauvezin C., et al. Autophagosome–lysosome fusion is independent of V-ATPase-mediated acidification. Nat. Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu H., et al. Divergent synthesis of thapsigargin analogs. Bioorg. Med. Chem. Lett. 2018;28:2705–2707. doi: 10.1016/j.bmcl.2018.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu H., et al. Scalable synthesis of (–)-thapsigargin. ACS Cent. Sci. 2017;3:47–51. doi: 10.1021/acscentsci.6b00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lopez C.Q., et al. Use of a temporary immersion bioreactor system for the sustainable production of thapsigargin in shoot cultures of Thapsia garganica. Plant Methods. 2018;14:79. doi: 10.1186/s13007-018-0346-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei Y., et al. Pharmacological preconditioning with the cellular stress inducer thapsigargin protects against experimental sepsis. Pharmacol. Res. 2019;141:114–122. doi: 10.1016/j.phrs.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 61.Lee C.Y., Lowen A.C. Animal models for SARS-CoV-2. Curr. Opin. Virol. 2021;48:73–81. doi: 10.1016/j.coviro.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao Z., et al. Targeting strategies for tissue-specific drug delivery. Cell. 2020;181:151–167. doi: 10.1016/j.cell.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Doan N.T., et al. Targeting thapsigargin towards tumors. Steroids. 2015;97:2–7. doi: 10.1016/j.steroids.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mahalingam D., et al. A phase II, multicenter, single-arm study of mipsagargin (G-202) as a second-line therapy following sorafenib for adult patients with progressive advanced hepatocellular carcinoma. Cancers (Basel) 2019;11:833. doi: 10.3390/cancers11060833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mahalingam D., et al. Mipsagargin, a novel thapsigargin-based PSMA-activated prodrug: results of a first-in-man phase I clinical trial in patients with refractory, advanced or metastatic solid tumours. Br. J. Cancer. 2016;114:986–994. doi: 10.1038/bjc.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Isaacs J.T., et al. Mipsagargin: The beginning-not the end-of thapsigargin prodrug-based cancer therapeutics. Molecules. 2021;26:7469. doi: 10.3390/molecules26247469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmitz M.L., et al. The crosstalk of endoplasmic reticulum (ER) stress pathways with NF-kappaB: complex mechanisms relevant for cancer, inflammation and infection. Biomedicines. 2018;6:58. doi: 10.3390/biomedicines6020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cain D.W., Cidlowski J.A. After 62 years of regulating immunity, dexamethasone meets COVID-19. Nat. Rev. Immunol. 2020;20:587–588. doi: 10.1038/s41577-020-00421-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carletti T., et al. Viral priming of cell intrinsic innate antiviral signaling by the unfolded protein response. Nat. Commun. 2019;10:3889. doi: 10.1038/s41467-019-11663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quiroga C., et al. Herp depletion protects from protein aggregation by up-regulating autophagy. Biochim. Biophys. Acta. 2013;1833:3295–3305. doi: 10.1016/j.bbamcr.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 71.Wilkinson S. Emerging principles of selective ER autophagy. J. Mol. Biol. 2020;432:185–205. doi: 10.1016/j.jmb.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bhattacharya A., Qi L. ER-associated degradation in health and disease – from substrate to organism. J. Cell Sci. 2019;132:jcs232850. doi: 10.1242/jcs.232850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Christianson J.C., et al. Defining human ERAD networks through an integrative mapping strategy. Nat. Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chino H., Mizushima N. ER-phagy: quality control and turnover of endoplasmic reticulum. Trends Cell Biol. 2020;30:384–398. doi: 10.1016/j.tcb.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 75.Ferro-Novick S., et al. ER-phagy, ER homeostasis, and ER quality control: implications for disease. Trends Biochem. Sci. 2021;46:630–639. doi: 10.1016/j.tibs.2020.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020;21:439–458. doi: 10.1038/s41580-020-0241-0. [DOI] [PubMed] [Google Scholar]
- 77.Aguayo-Ortiz R., Espinoza-Fonseca L.M. Linking biochemical and structural states of SERCA: achievements, challenges, and new opportunities. Int. J. Mol. Sci. Int. 2020;21:4146. doi: 10.3390/ijms21114146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Y., et al. Cryo-EM structures of SERCA2b reveal the mechanism of regulation by the luminal extension tail. Sci. Adv. 2020;6:eabb0147. doi: 10.1126/sciadv.abb0147. [DOI] [PMC free article] [PubMed] [Google Scholar]