Abstract
Infections due to Candida albicans are usually treated with azole antifungals such as fluconazole, but treatment failure is not uncommon especially in immunocompromised individuals. Relatedly, in vitro studies demonstrate that azoles are nonfungicidal, with continued growth at strain-dependent rates even at high azole concentrations. We hypothesized that upregulation of ERG11, which encodes the azole target enzyme lanosterol demethylase, contributes to this azole tolerance in Candida species. RNA analysis revealed that ERG11 expression in C. albicans is maximal during logarithmic-phase growth and decreases as the cells approach stationary phase. Incubation with fluconazole, however, resulted in a two- to fivefold increase in ERG11 RNA levels within 2 to 3 h, and this increase was followed by resumption of culture growth. ERG11 upregulation also occurred following treatment with other azoles (itraconazole, ketoconazole, clotrimazole, and miconazole) and was not dependent on the specific medium or pH. Within 1 h of drug removal ERG11 upregulation was reversed. Azole-dependent upregulation was not limited to ERG11: five of five ERG genes tested whose products function upstream and downstream of lanosterol demethylase in the sterol biosynthetic pathway were also upregulated. Similarly, ERG11 upregulation occurred following treatment of C. albicans cultures with terbinafine and fenpropimorph, which target other enzymes in the pathway. These data suggest a common mechanism for global ERG upregulation, e.g., in response to ergosterol depletion. Finally, azole-dependent ERG11 upregulation was demonstrated in three additional Candida species (C. tropicalis, C. glabrata, and C. krusei), indicating a conserved response to sterol biosynthesis inhibitors in opportunistic yeasts.
The dimorphic yeast Candida albicans is a common cause of vaginitis and, in immunocompromised individuals, of oropharyngeal and systemic infections. Antifungal azoles such as fluconazole (oral and intravenous) and miconazole (topical) are used for treatment or prophylaxis of most C. albicans infections. While azoles have little or no toxicity they generally lack fungicidal activity. Consequently, in immunocompromised individuals azoles must be administered for extended periods of time. This practice, combined with the increased use of azoles in recent years, is most likely responsible for the increased isolation of azole-resistant strains of C. albicans and of intrinsically resistant Candida species such as C. glabrata and C. krusei (2, 17, 28, 40). Even in the absence of resistance, treatment failures or recurrent infections are not uncommon, especially in immunocompromised individuals (28). These clinical limitations associated with azole use have in vitro correlates. In susceptibility assays, significant “trailing” growth occurs even at high fluconazole concentrations with many Candida isolates (23, 26, 30). In agar diffusion assays, trailing is visualized as background growth which may obscure the zone of inhibition (33).
Azoles inhibit the enzyme lanosterol demethylase in the sterol biosynthetic pathway. This pathway is conserved in eukaryotes, leading to cholesterol in mammals and ergosterol in fungi (Fig. 1). In Saccharomyces cerevisiae and C. albicans sterols have been shown to be important in membrane fluidity, membrane permeability, cell morphology, enzyme activity, and cell cycle progression (5, 13, 18, 19, 20). Sterol precursors are also involved in heme and glycolipid biosynthesis as well as protein prenylation (20). In addition to azoles, other classes of antifungals target ergosterol synthesis, including allylamines (e.g., terbinafine) and morpholines (e.g., fenpropimorph).
FIG. 1.
Ergosterol biosynthesis pathway. Only selected substrates or products are shown. Genes (in italics) that encode enzymes in the pathway are labeled according to the convention for S. cerevisiae; those whose expression was monitored in this study are shown in bold. The three groups of sterol biosynthesis inhibitors examined in this study are underlined and are shown to the right of the gene encoding the targeted enzyme. CoA, coenzyme A.
Lanosterol demethylase is encoded by the gene ERG11 (also known as CYP51). Multiple mechanisms for azole resistance in C. albicans clinical isolates have been identified, including increased expression of multidrug transporters (encoded by CDR1, CDR2, and MDR1), mutations in lanosterol demethylase that reduce azole binding, and increased expression of ERG11 (7, 22, 34, 36, 41, 42). Genetic studies with S. cerevisiae confirm that each of these mechanisms can operate alone to confer various degrees of azole resistance (1, 16, 36). Specifically, GAL1 promoter-mediated overexpression of ERG11 in S. cerevisiae was recently shown to confer high-level fluconazole resistance, further implicating ERG gene upregulation as a factor in azole resistance (14).
Regulation of the ergosterol biosynthetic pathway has been studied in some detail in S. cerevisiae. Changes in the activities of selected enzymes or in the levels of expression of selected ERG genes in response to sterol availability, genetic lesions in the pathway, or treatment with specific inhibitors have been described (3, 6, 13, 20, 24, 31, 37). More recently, microarray techniques in combination with the S. cerevisiae genome sequence have permitted investigation of global changes in gene expression associated with ERG mutations and inhibitor treatment (4, 32).
In light of its potential role in modulating azole susceptibility, we have examined the effects of azole treatment on ERG expression in C. albicans. We report that azole exposure leads to ERG11 upregulation, which was followed by resumed culture growth. In addition, we demonstrate that (i) other ERG genes are upregulated by azole treatment, (ii) other inhibitors of ergosterol biosynthesis upregulate ERG11, and (iii) azole treatment upregulates ERG11 expression in other Candida species.
(This work was presented in part at the 39th Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, Calif., 26 to 29 September 1999 [K. W. Henry and T. D. Edlind, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 455, p. 553, 1999].)
MATERIALS AND METHODS
Strains, media, and drugs.
The strains used in this study are listed in Table 1, along with their azole susceptibilities. Candida species were maintained on liquid or agar YPD medium (1% yeast extract, 2% peptone, 2% dextrose). Susceptibility assays were performed in RPMI 1640 medium with l-glutamate and without NaHCO3 (Sigma, St. Louis, Mo.), buffered to pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS). DOB medium (0.17% yeast nitrogen base, 2% dextrose, 0.5% ammonium sulfate) was prepared as recommended by the manufacturer (Bio 101, Inc., Vista, Cal.). RNA expression studies were done in YPD or, where indicated, RPMI 1640 medium or DOB medium. The following drugs were purchased from the indicated supplier: fluconazole, Pfizer (Groton, Conn.); terbinafine, Novartis (East Hanover, N.J.); itraconazole and ketoconazole, Jannsen (Titusville, N.J.); fenpropimorph, Cresent Chemical (Hauppauge, N.Y.); and miconazole and clotrimazole, Sigma. The concentrations of the fluconazole stocks were 2 mg/ml in saline, and the stocks were stored at 4°C; the concentrations of all others were 10 mg/ml in dimethyl sulfoxide and the stocks were stored at −20°C.
TABLE 1.
Strains used in this study and azole susceptibilities
| Candida species | Strain | MIC (μg/ml)
|
|||||
|---|---|---|---|---|---|---|---|
| Fluconazole
|
Itraconazole
|
Ketoconazole
|
|||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | ||
| C. albicans | ATCC 24433 | 0.25 | 1.0 | 0.016 | 0.031 | 0.016 | 0.031 |
| ATCC 90028 | 0.25 | 2.0 | 0.016 | 0.031 | 0.016 | 0.062 | |
| 630-15.3a | 0.25 | 0.50 | 0.016 | 0.016 | 0.016 | 0.016 | |
| 707.15a | 0.50 | >256 | 0.031 | >8.0 | 0.016 | >8.0 | |
| C. glabrata | ATCC 66032 | 16 | 128 | 0.062 | 2.0 | 0.031 | 2.0 |
| C. tropicalis | ATCC 750 | 0.50 | >256 | <0.031 | 0.12 | <0.031 | 0.50 |
| C. krusei | ATCC 14243 | 32 | 64 | 0.031 | 0.062 | 0.031 | 0.25 |
Obtained from J. Rex (30).
Susceptibility assays.
Susceptibility testing of Candida species was done according to the guidelines in document M27-A (26) in flat-bottom 96-well polystyrene microtiter plates. Briefly, a 100-μl volume of RPMI 1640 medium was added to a series of wells, except for the initial well, to which 195 μl was added. Drugs (5 μl) were added to the initial well at twice the maximum concentration to be tested (16 μg/ml for itraconazole and ketoconazole), and twofold serial dilutions were made by transferring 100 μl of this solution to subsequent wells. When testing for fluconazole susceptibility, only 174.4 μl of RPMI 1640 medium was added to the initial well, as 25.6 μl of the drug stock was required to achieve a final concentration of 256 μg/ml in 200 μl. The final well in each series received no azole and served as a growth control. Control series received RPMI 1640 medium alone or RPMI 1640 medium plus 0.5% DMSO depending upon the drug vehicle used. Logarithmic-phase cultures of yeast were diluted in RPMI 1640 medium, and 100 μl was added to each well to give a final density of 104 cells/ml. Plates were incubated at 35°C, and the absorbance at 630 nm was read at 24 and 48 h with a microplate reader (Bio-Tek Instruments, Winooski, Vt.). Dilutions (fivefold) of additional drug-free wells were prepared and served as references for readings of the MIC (80% reduction in turbidity).
RNA isolation.
Logarithmic-phase cultures (3 × 107 to 5 × 107 cells/ml) were exposed to drugs at the indicated concentrations and times at 30°C (or 35°C where indicated) with shaking. Controls received an equivalent amount of DMSO or were untreated. RNA was extracted as described previously (9). Briefly, at the indicated times, 108 cells from each culture were transferred to microcentrifuge tubes, pelleted, and washed twice in ice-cold 50 mM sodium acetate–10 mM EDTA buffer (pH 4.5). The pellet was resuspended in 200 μl of ice-cold sodium acetate–EDTA buffer, followed by the addition of 200 μl of glass beads, 20 μl of 10% sodium dodecyl sulfate, and 200 μl of prewarmed buffer-saturated phenol. The cells were disrupted by periodic vigorous vortexing and incubation at 65°C for 5 to 10 min. The samples were cooled on ice and centrifuged, and the aqueous phase was reextracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by extraction with chloroform-isoamyl alcohol (24:1). (The reextractions were omitted when RNA was isolated for slot blot analysis.) RNA was precipitated overnight with 2.5 volumes of ethanol and 0.1 volume of 3 M sodium acetate and was collected by centrifugation.
RNA hybridization analysis.
For slot blot analysis, RNA was dissolved in 500 μl of H2O and denatured by the addition of 200 μl of 37% formaldehyde and 300 μl of 20× SSPE (20× SSPE is 3.6 M sodium chloride, 0.2 M sodium phosphate, and 20 mM EDTA [pH 7.0]) with incubation at 65°C for 15 min. Either 100 μl (for the ACT1 probe) or 250 μl (for the other probes) of denatured RNA was applied to a positively charged nylon membrane (Boehringer Mannheim, Indianapolis, Ind.) with a Bio-Dot SF apparatus (Bio-Rad, Richmond, Calif.). The membranes were rinsed in 2× SSPE and UV cross-linked. Slot blots were hybridized overnight to C. albicans ACT1-, ERG11-, CDR1-, or CDR2-specific PCR products (see Table 2) that were random primer labeled with digoxigenin-dUTP, as recommended by the manufacturer (Boehringer Mannheim). The blots were washed twice under high-stringency conditions (0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 0.1% sodium dodecyl sulfate at 68°C) and prepared for chemiluminescence detection with CSPD substrate, as recommended. After 15 min of incubation at 37°C, the blots were exposed to Kodak X-OMAT LS film for 1 to 15 min. RNA levels were quantified with a densitometer (Bio-Rad GS-670 imaging densitometer with Molecular Analyst software), with ERG11, CDR1, and CDR2 levels normalized to the levels of the ACT1 controls.
TABLE 2.
Primers used in this study
| Gene | Protein | GenBank accession no. | Primer | Locationa | Sequence (5′ to 3′)b |
|---|---|---|---|---|---|
| ERG9 | Squalene synthase | D89610 | Forward | 448 to 467 | AAAATGGGTAATGGTATGGC |
| Reverse | 895 to 914 | ACTTGGGGAATGGCACAAAA | |||
| ERG1 | Squalene epoxidase | U69674 | Forward | 583 to 605 | TTGACAWTTAKTTGTGATGGTAT |
| Reverse | 1243 to 1265 | CTTTGGAAATATTTGAAACAACC | |||
| ERG7 | Squalene cyclase | L04305 | Forward | 1584 to 1603 | TATCCATACGTGGAATGTAC |
| Reverse | 2133 to 2154 | TGTATAWACCTAATGCCTTAAT | |||
| ERG11 | Lanosterol 14α-demethylase | X13296 | Forward | 903 to 931 | ATTGGTATTCTTATGGGTGGTCAACATAC |
| Reverse | 1390 to 1416 | CCCAATACATCTATGTCTACCACCACC | |||
| ERG25 | C-4 Methyl sterol oxidase | AF051914 | Forward | 235 to 257 | ATTCCWTATTTTAGAAAATGGAA |
| Reverse | 805 to 825 | GWAATCCCACCATCTAAAAGA | |||
| ERG3 | Δ5,6-Desaturase | AF069752 | Forward | 459 to 482 | CCWMTTTGAAAAACCAAATG |
| Reverse | 961 to 984 | GAATTGACCGTAGTTGTAGTTGAA | |||
| ACT1 | Actin | X16377 | Forward | 975 to 1001 | ACCGAAGCTCCAATGAATCCAAAATCC |
| Reverse | 1465 to 1491 | GTTTGGTCAATACCAGCAGCTTCCAAA | |||
| CgACT1 | C. glabrata actin | AF069746 | Forward | 45 to 64 | ATGTGTAAGGCKGGKTTTGC |
| Reverse | 1050 to 1069 | ATCCACATTTGTTGGAAKGT | |||
| CDR | ABC transporters CDR1 and CDR2 | X77589 and U63812 | Forward | 901 to 924 | GTGGTGTTTCCGGTGGTGAAAGAAA |
| Reverse | 1379 to 1403 | CCTGGTGTTGGATCGTTCACATTCA | |||
| CDR1 | ABC transporter CDR1 specific | X77589 | Forward | −184 to −163 | TTTTTTTTTTTAGTTCATCATC |
| Reverse | 242 to 262 | GGTCATTATTTATTTCTTCAT | |||
| CDR2 | ABC transporter CDR2 specific | U63812 | Forward | −187 to −167 | CTCTATTATGAATACTAGTAG |
| Reverse | 239 to 258 | TTGTTCATGACTATTGTTGC | |||
| MDR1 | Major facilitator superfamily transporter | X53823 | Forward | 451 to 476 | GAGTCGTAGCTACATTGCCATTAACA |
| Reverse | 1014 to 1040 | GGTGATTTCTAATGGTCTCCATAATGT |
Location refers to the nucleotide sequence within the C. albicans gene (except CgACT1), with the “A” of the start codon (ATG) representing +1.
Code for mixed bases within the primer sequence: W = A + T, K = G + T, and M = A + C.
Reverse transcription-PCR (RT-PCR) analysis.
RNA pellets were washed in 70% ethanol and resuspended in 50 μl of RNase-free water. Aliquots (1 μg) were treated with RNase-free DNase I as recommended by the manufacturer (Promega, Madison, Wis.). cDNAs were prepared with reverse transcriptase (Moloney murine leukemia virus reverse transcriptase; New England Biolabs, Beverly, Mass.), as recommended, by using the reverse primers listed in Table 2. Each reaction had a parallel reaction that lacked reverse transcriptase to confirm that the PCR product was derived from cDNA rather than genomic DNA contamination. PCR was performed with the primer sets listed in Table 2. Initially, genes were amplified by PCR for 23 cycles to determine levels of expression. Genes expressed at medium to high levels (genes for which PCR products were observed at 23 cycles; ACT1, ERG11, ERG3, and CDR) were subjected to three independent amplifications of 20, 23, and 25 cycles to ensure that amplification during the logarithmic phase was obtained. Genes expressed at low levels (little or no PCR product at 23 cycles; ERG9, ERG1, ERG7, and ERG25) were amplified for 25, 28, and 30 cycles. Cycling conditions for ACT1, CDR, and ERG11 were 94°C for 1 min, 57°C for 1 min, and 72°C for 1 min. All other genes were amplified with an annealing temperature of 52°C due to differences in primer melting temperatures. The PCR products were subjected to gel electrophoresis and ethidium bromide staining, and relative intensities were determined with a densitometer, with normalization to ACT1 levels.
RESULTS
ERG11 expression varies with growth phase.
In initial experiments, C. albicans gene expression was monitored by RNA slot blot hybridization. The probes used were specific for their targeted RNAs, as determined by preliminary Northern and Southern blotting and, in the case of the CDR1 and CDR2 probes, by slot blot hybridization to RNAs from strains from which those genes had been deleted (35). In subsequent experiments, gene expression was monitored by RT-PCR. While RNA hybridization is potentially more quantitative, RT-PCR facilitates the analysis of larger numbers of samples and of multiple genes. In selected experiments in which both assays were used, comparable results were obtained (compare Fig. 2A and 5).
FIG. 2.
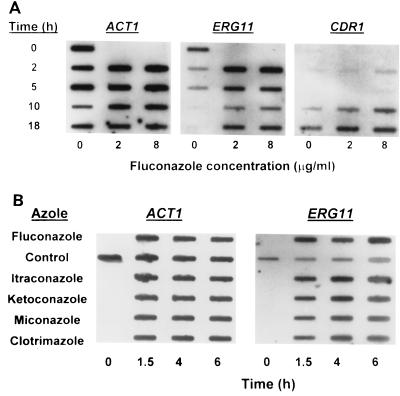
Effect of azole treatment on ERG11 expression in C. albicans. (A) RNA slot blot analysis of ACT1, ERG11, and CDR1 expression in C. albicans 24433 following fluconazole treatment (0, 2, or 8 μg/ml) for the indicated times. Similar results were observed in four independent experiments and with a second C. albicans strain (strain ATCC 90028). (B) RNA slot blot analysis of ACT1 and ERG11 expression in C. albicans 24433 following treatment with the indicated azoles for the indicated times. Azole concentrations were 9 μg/ml (fluconazole) and 0.5 μg/ml (all others). The control received DMSO vehicle (final concentration, 0.25%). Similar results were observed in a second independent experiment.
FIG. 5.
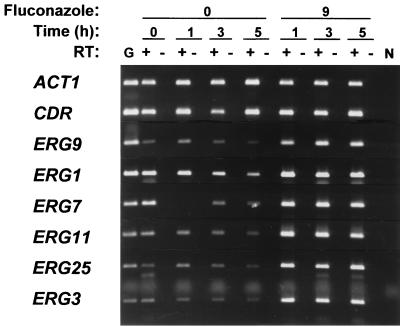
Global upregulation of C. albicans ERG genes following fluconazole treatment. RT-PCR was used to analyze the relative expression of ACT1, CDR (CDR1 and CDR2), and the indicated ERG genes in cultures (strain 24433) treated with fluconazole at 9 μg/ml (labeled 9) or no drug (labeled 0) for 0, 1, 3, or 5 h, as indicated. Equal volumes from cDNA reactions with (+) or without (−) reverse transcriptase (RT) were amplified in parallel to detect genomic DNA contamination. Lane G, PCR positive control containing genomic DNA; lane N, PCR negative control without added template. The data shown are representative of two independent experiments, and similar data were obtained with an additional C. albicans isolate (isolate 630-15.3). Due to differences in basal levels of expression, PCR cycle numbers were optimized for each gene to ensure logarithmic-phase amplification. The cycle numbers used were 23 (ERG11 and ACT1), 25 (CDR), 28 (ERG3, ERG7, ERG9), and 30 (ERG1 and ERG25). On the basis of previous slot blot data, CDR expression is primarily due to CDR1.
Both slot blot (Fig. 2A) and RT-PCR (see Fig. 5) analyses revealed that the level of ERG11 expression in C. albicans strain 24433 varies dramatically with the growth phase. ERG11 RNA levels were maximal during logarithmic-phase growth (0 h; 3 × 107 to 5 × 107 cells/ml) and then decreased 3- to ≥10-fold as the cells approached the stationary phase (5 to 10 h; ≥1 × 108 cells/ml). The levels of ACT1 RNA were relatively constant over this interval. In contrast, expression of the multidrug transporter gene CDR1 increased as cells approached stationary phase (Fig. 2A), in agreement with previous studies (10, 15). CDR2 expression was not detected in untreated logarithmic- or stationary-phase cultures (data not shown).
Azole treatment of C. albicans leads to ERG11 upregulation.
Treatment of C. albicans cultures with the triazole fluconazole at a concentration of 2 to 9 μg/ml resulted in four- to fivefold increases in ERG11 RNA levels within 1.5 to 2 h of incubation (Fig. 2A and B). This increase was in marked contrast to the late-logarithmic-phase-associated reduction observed in the untreated controls. Lower but reproducible upregulation was also detected in cultures treated with as little as 0.5 μg of fluconazole per ml (data not shown). ERG11 expression remained elevated for up to 18 h (Fig. 2A). Treatment with four other azoles (the triazole itraconazole and the imidazoles ketoconazole, clotrimazole, and miconazole, all at 0.5 μg/ml) had an effect comparable to that of fluconazole (Fig. 2B). Furthermore, comparable fluconazole-dependent ERG11 upregulation was observed in three of three additional C. albicans strains tested, including a strain (strain 707.15) which exhibits high-level trailing (Table 1; data not shown).
Fluconazole (2 or 8 μg/ml) treatment of C. albicans cultures also resulted in increased CDR1 expression. However, this was observed only after 10 to 18 h of azole treatment (Fig. 2A). CDR2 expression was not detected (data not shown).
Correlation of ERG11 upregulation and C. albicans growth in fluconazole-treated cultures.
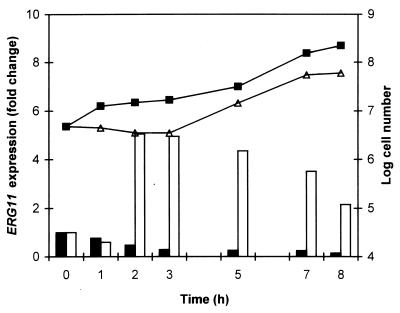
Under the conditions used, fluconazole at 9 μg/ml completely inhibited the growth of a C. albicans culture during the initial 3 h of treatment, whereupon growth resumed (Fig. 3). In the same experiment, fluconazole treatment resulted in a fivefold increase in ERG11 RNA levels (relative to those for the control at 0 h) after 2 h. Thus, ERG11 upregulation directly preceded and presumably contributed to the resumption of C. albicans growth.
FIG. 3.
Correlation of ERG11 expression and C. albicans 24433 growth in control and fluconazole-treated cultures. RNA levels (determined from slot blot analysis and normalized to ACT1 levels) are represented as fold increase or decrease relative to the level for the control at 0 h; solid bars, 0 μg of fluconazole per ml; open bars, 9 μg of fluconazole per ml. C. albicans growth is represented as cell number (determined in a hemocytometer) at the indicated times and treatments: no drug (■) and 9 μg of fluconazole per ml (▵). The data shown represent the averages of three independent experiments.
The experiment described above was repeated, except that the fluconazole was removed at the 2-h time point by careful washing. The 2.6-fold increase in ERG11 RNA levels after 2 h of fluconazole treatment was reduced to 1.1-fold after 0.5 h and to 0.5-fold (comparable to that for the untreated control) after 1 h of further incubation in drug-free medium (data not shown). Thus, fluconazole-dependent ERG11 upregulation is reversible.
ERG11 upregulation is independent of medium and pH.
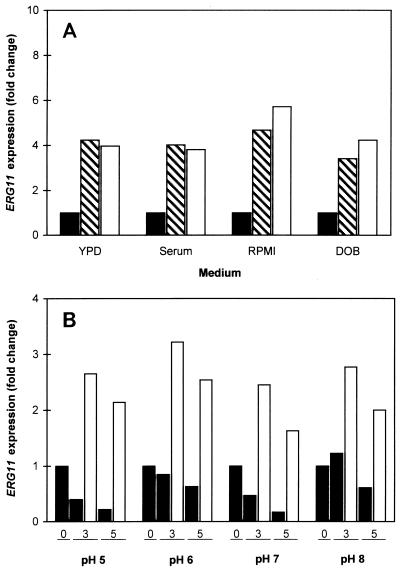
The activity of fluconazole against C. albicans is affected by changes in medium and pH (23, 29, 33). Therefore, the effects of these variables on fluconazole-dependent upregulation of ERG11 were examined. ERG11 expression after 2 h of fluconazole treatment was compared in YPD (used in all experiments described above), YPD with 20% fetal bovine serum (which induces germ tubes), a defined RPMI 1640 medium (according to the guidelines in document M27-A [26] for susceptibility testing), and a defined minimal medium (supplemented yeast nitrogen base [DOB medium]). Comparable levels of upregulation were observed in all four media (Fig. 4A), although there were different basal levels of ERG11 expression that positively correlated with the medium-dependent growth rate (data not shown).
FIG. 4.
Effects of medium and pH on fluconazole-dependent ERG11 upregulation in C. albicans 24433. (A) Media were YPD, YPD supplemented with 20% fetal bovine serum (Serum), RPMI 1640 medium prepared according to the susceptibility testing guidelines in document M27-A (26) (RPMI), and supplemented yeast nitrogen base (DOB). Effects on ERG11 expression were analyzed by slot blot analysis with normalization to ACT1 RNA levels and are represented as the fold change relative to the level for the untreated controls (solid bars). Treatment was for 2 h with fluconazole at 1 (hatched bars) or 9 (open bars) μg/ml. The data shown are the averages of two independent experiments. (B) RPMI 1640 medium was adjusted to the indicated pH, and ERG11 expression was analyzed by RT-PCR. Incubation was at 35°C instead of 30°C to conform with the guidelines in document M27-A (26). Treatment was for 0, 3, or 5 h, as indicated, with no drug (solid bars) or fluconazole at 9 μg/ml (open bars). The data represent the averages of four RT-PCRs from two independent experiments.
Fluconazole-induced upregulation was further examined in RPMI 1640 medium buffered to pH 5, 6, 7, and 8. Alteration of the pH did not significantly alter the ability of C. albicans to upregulate ERG11 in response to azole treatment (Fig. 4B). Alteration of the pH also had no detectable effect on the expression of the multidrug transporter genes CDR1, CDR2, and MDR1 (data not shown).
Azole treatment induces a global increase in sterol biosynthesis gene expression.
The data presented above demonstrate that azole treatment upregulates expression of ERG11, which encodes the azole target lanosterol demethylase. The effects of azole treatment on the expression of other ERG genes was examined next. Three of the genes examined (ERG9, ERG1, and ERG7) encode enzymes that act upstream, and two (ERG25 and ERG3) encode enzymes that act downstream of lanosterol demethylase in the ergosterol biosynthesis pathway (Fig. 1). As with ERG11 (see above), the expression of these genes was maximal in logarithmic-phase cells (0 h) and decreased as the cultures approached the stationary phase (3 to 5 h) (Fig. 5). Similarly, fluconazole (9 μg/ml) treatment of these cultures resulted in increased levels of expression of all five ERG genes, in addition to ERG11. This global ERG upregulation was also observed in a second C. albicans strain (strain 630-15.3; data not shown).
Inhibitors targeting other sterol biosynthesis enzymes upregulate ERG11.
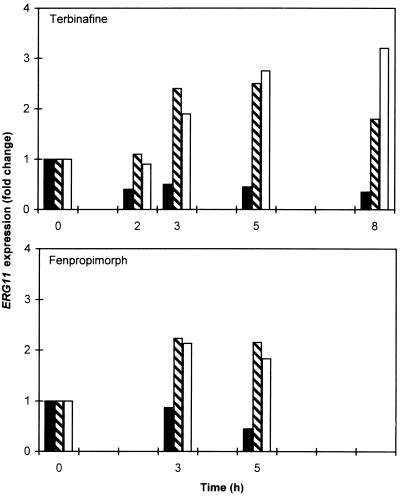
Since fluconazole upregulated the expression of multiple ERG genes, it seemed likely that inhibitors acting at other steps in the sterol biosynthesis pathway might upregulate ERG11. The allylamine terbinafine inhibits the ERG1 product squalene epoxidase, which acts upstream of the ERG11 product lanosterol demethylase (Fig. 1). Nevertheless, slot blot analysis showed that treatment with terbinafine (1 and 9 μg/ml) for 3 to 5 h upregulated ERG11 RNA two- to threefold (relative to that at 0 h) (Fig. 6). The morpholine fenpropimorph inhibits the ERG24 and ERG2 products, which act downstream of ERG11 (Fig. 1). Similarly, RT-PCR revealed that treatment with fenpropimorph (1 and 9 μg/ml) upregulated ERG11 1.8- to 2.2-fold (Fig. 6).
FIG. 6.
Upregulation of C. albicans ERG11 following treatment with terbinafine or fenpropimorph. ERG11 RNA levels were examined by slot blot hybridization (terbinafine) or RT-PCR (fenpropimorph), normalized to ACT1 RNA levels, and represented as the fold change relative to the RNA levels at 0 h. Cultures were treated with no drug (solid bars) or the indicated drug at 1 (hatched bars) and 9 (open bars) μg/ml for the indicated times.
Azole-dependent ERG11 upregulation is conserved in other Candida species.
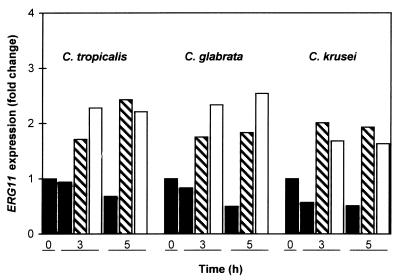
The ability of fluconazole to upregulate ERG11 in C. tropicalis, C. glabrata, and C. krusei was examined. As indicated in Table 1, these three species are either moderately (C. glabrata) or highly (C. krusei) resistant to fluconazole or exhibit high-level trailing (C. tropicalis; the MIC at 48 h was substantially greater than the MIC at 24 h). RT-PCR was used with a single primer pair that represented conserved ERG11 sequences present in all Candida species tested. As for C. albicans, fluconazole (9 μg/ml) treatment for 3 to 5 h resulted in ERG11 upregulation in C. tropicalis (1.7- to 2.4-fold), C. glabrata (1.8-fold), and C. krusei (2.0-fold) (Fig. 7). Itraconazole has a relatively high level of activity against several fluconazole-resistant fungi, including C. krusei (Table 1). C. tropicalis, C. glabrata, and C. krusei upregulated ERG11 1.8- to 2.5-fold in response to itraconazole treatment (0.1 μg/ml for 3 to 5 h) (Fig. 7).
FIG. 7.
Upregulation of ERG11 in three additional Candida species following azole treatment. RNA levels were examined by RT-PCR after 0, 3, and 5 h of treatment, as indicated, and are represented as the fold change relative to the levels at 0 h. Cultures were treated with no drug (solid bars), fluconazole at 9 μg/ml (hatched bars), or itraconazole at 0.1 μg/ml (open bars).
DISCUSSION
Azoles are the most widely used group of antifungals. Fluconazole, in particular, has negligible toxicity, excellent bioavailability, and a moderately high level of activity against commonly encountered yeasts such as C. albicans. Other azoles such as itraconazole have potent and broader-spectrum activity that extends to fluconazole-resistant Candida species and many molds. However, most azole treatments only suppress and do not eliminate fungal infections. In immunocompromised individuals, this leads to a requirement for long-term treatment, which in turn leads to a selection for azole-resistant fungal strains. In vitro, the nonfungicidal activity of azoles is readily apparent in the trailing observed in broth dilution assays (23, 30) and the background growth observed in agar diffusion assays such as the E-test (33). Indeed, these assays indicate that fluconazole and potentially other azoles lack even fungistatic activity toward common fungal pathogens. Understanding the cellular responses to these drugs may enhance our understanding of antifungal resistance and facilitate the development of new antifungal agents.
We hypothesized that the ability of C. albicans to tolerate azole treatment was at least partly due to the upregulation of the ERG11 gene, which encodes the azole target lanosterol demethylase. This hypothesis derived from previous reports that ERG11 was constitutively upregulated in certain fluconazole-resistant clinical isolates (7, 22, 41; A. M. Alarco, I. Balan, F. Comte, G. St-Germain, D. Falconer, T. Parkinson, C. A. Hitchcock, and M. Raymond, Abstr. ASM Conf. Candida and Candidiasis, abstr. C22, p. 54, 1999). Also, in S. cerevisiae ERG11 overexpression was directly shown to confer fluconazole resistance (14; K. W. Henry and T. D. Edlind, unpublished data). In support of the hypothesis, our data demonstrate C. albicans ERG11 upregulation in response to treatment with any of five different azoles. Upregulation is maximal 2 to 3 h after treatment and is most readily visualized in late-logarithmic-phase cultures in which ERG11 expression is normally declining. The effect was observed in four different media over a pH range of 5 to 8 and was reversed following drug removal.
Fluconazole treatment of C. albicans upregulated not only ERG11 encoding the azole target but also five of five other ERG genes tested. Consistent with this global upregulation, treatment with compounds that inhibit enzymes upstream (terbinafine) and downstream (fenpropimorph) of lanosterol demethylase also led to ERG11 upregulation. Finally, ERG11 upregulation in response to azole treatment was demonstrated in three other Candida species with various fluconazole susceptibilities: moderately resistant C. glabrata, highly resistant C. krusei, and C. tropicalis which exhibits substantial trailing.
Our studies were limited to the examination of ERG11 RNA levels rather than protein levels or enzyme activity. This was unavoidable due to the lack of specific antibodies or staining procedures and the inability to reliably measure lanosterol demethylase activity in the presence of azole. However, related studies with S. cerevisiae demonstrate that ERG mRNA levels correlate with the corresponding enzyme activities (6, 20, 24).
The results that we observed with Candida are similar to those previously reported for S. cerevisiae. For example, S. cerevisiae ERG9 expression increases following treatment with lovastatin (hydroxymethylglutaryl coenzyme A reductase inhibitor), zaragosic acid (squalene synthase inhibitor), or ketoconazole (13, 31). Genetic lesions in ergosterol biosynthesis result in both increased levels of ERG9 expression and increased ERG9 activity (13, 24). Enzyme inhibitors and genetic lesions in ergosterol biosynthesis also cause an increase in the levels of ERG3 mRNA (3, 37). Similarly, in C. glabrata deletion of ERG3 was associated with upregulation of ERG11 and deletion of ERG11 was associated with ERG3 upregulation (8). Recent studies with S. cerevisiae and microarrays for investigation of genome-wide expression in response to antifungal treatment or mutations in the ergosterol pathway support earlier studies and more clearly demonstrate global changes in gene expression. Specifically, Bammert and Fostel (4) and unpublished data cited by Rosamond and Allsop (32) document global ERG gene upregulation in response to azole, terbinafine, and amorolfine treatment or mutations in ERG1, ERG11, ERG6, ERG2, and ERG5. Since a complete C. albicans genome sequence is not yet publicly available, genome-wide microarray analysis with this organism in response to antifungal treatment has not been reported. It would no doubt further enhance our understanding of the antifungal response in this opportunistic pathogen.
Although ERG upregulation is likely to contribute significantly to the survival of azole-treated cells, our data do not support a specific connection between ERG upregulation and the trailing phenotype that is variably expressed in Candida isolates. Previous work has shown that lowering the pH reduces trailing growth in several C. albicans isolates (23; T. D. Edlind, K. W. Henry, and S. K. Katiyar, Abstr. 39th Intersci. Conf. Antimicrob. Agents Chemother., abstr. 297, p. 549, 1999), but pH had little effect on azole-dependent upregulation of C. albicans ERG11 (Fig. 4B). Additionally, azole-dependent upregulation of ERG11 did not vary in magnitude or kinetics between low-trailing-level strain C. albicans 630.15-3 and high-trailing-level strain 707.15 (30) (Table 1 and data not shown). Thus, the basis for trailing and its variation among Candida isolates remains to be defined.
The molecular mechanism behind the global upregulation of ERG genes in response to azoles and other sterol biosynthesis inhibitors is unknown. It is clearly initiated by either the depletion of a late product (e.g., ergosterol) of the pathway or the accumulation of an early substrate or toxic sterol by-product. Current evidence argues against accumulation of a specific substrate or by-product as the initiator of global ERG upregulation. Specifically, several different inhibitors that act on different enzymes in the ergosterol biosynthesis pathway cause upregulation of ERG genes in S. cerevisiae (4, 6, 13, 37) or, as shown here, Candida species. The substrates or by-products that accumulate in these treated cells would presumably vary depending upon the inhibitor used. These and other data have led to the suggestion that in S. cerevisiae the levels of ergosterol or other sterol formed late in the pathway regulate ERG expression (3, 27), and this may be the case in Candida species as well. As in yeasts, the inhibition of sterol biosynthesis in mammalian cells results in increased levels of sterol biosynthesis gene expression and enzyme activity (11, 12, 21, 25, 38, 39). Thus, while the mechanism remains unknown, it appears to be evolutionarily conserved.
ACKNOWLEDGMENTS
We thank J. Rex for providing C. albicans strains.
This work was supported by Public Health Service grants AI32433 and AI46768.
REFERENCES
- 1.Alarco A M, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 2.Ampel N M. Emerging disease issues and fungal pathogens associated with HIV infection. Emerg Infect Dis. 1996;2:109–116. doi: 10.3201/eid0202.960205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthington-Skaggs B A, Crowell D N, Yang H, Sturley S L, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 4.Bammert G F, Fostel J M. Genome-wide expression patterns in Saccharomyces cerevisiae: comparison of drug treatments and genetic alterations affecting biosynthesis of ergosterol. Antimicrob Agents Chemother. 2000;44:1255–1265. doi: 10.1128/aac.44.5.1255-1265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon R D, Kerridge D. Correlation between the sterol composition of membranes and morphology in Candida albicans. J Med Vet Mycol. 1988;26:57–65. doi: 10.1080/02681218880000071. [DOI] [PubMed] [Google Scholar]
- 6.Dimster-Dink D, Rine J. Transcriptional regulation of a sterol-biosynthetic enzyme by sterol levels in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3981–3989. doi: 10.1128/mcb.16.8.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franz R, Kelly S L, Lamb D C, Kelly D E, Ruhnke M, Morschhauser J. Multiple mechanisms contribute to a stepwise development of fluconazole resistance in clinical Candida albicans strains. Antimicrob Agents Chemother. 1998;42:3065–3072. doi: 10.1128/aac.42.12.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geber A, Hitchcock C A, Swartz J E, Pullen F S, Marsden K E, Kwon-Chung K J, Bennett J E. Deletion of the Candida glabrata ERG3 and ERG11 genes: effect on cell viability, cell growth, sterol composition, and antifungal susceptibility. Antimicrob Agents Chemother. 1995;39:2708–2717. doi: 10.1128/aac.39.12.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry K W, Cruz M C, Katiyar S K, Edlind T D. Antagonism of azole activity against Candida albicans following induction of multidrug resistance genes by selected antimicrobial agents. Antimicrob Agents Chemother. 1999;42:1968–1974. doi: 10.1128/aac.43.8.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernaez M L, Gil C, Pla J, Nombela C. Induced expression of the Candida albicans multidrug resistance gene CDR1 in response to fluconazole and other antifungals. Yeast. 1998;14:517–526. doi: 10.1002/(SICI)1097-0061(19980430)14:6<517::AID-YEA250>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 11.Hidaka Y, Hotta H, Nagata Y, Iwasawa Y, Horie M, Kamei T. Effect of a novel squalene epoxidase inhibitor, NB-598, on the regulation of cholesterol metabolism in HepG2 cells. J Biol Chem. 1991;266:13171–13177. [PubMed] [Google Scholar]
- 12.Kempen H J, von Son K, Cohen L H, Griffioen M, Verboom H, Havekes L. Effect of ketoconazole on cholesterol synthesis and on HMG-CoA reductase and LDL-receptor activities in Hep G2 cells. Biochem Pharmacol. 1987;36:1245–1249. doi: 10.1016/0006-2952(87)90077-3. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy M A, Barbuch R, Bard M. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 1999;1445:110–122. doi: 10.1016/s0167-4781(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 14.Kontoyiannis D P, Sagar N, Hirschi K D. Overexpression of Erg11p by the regulatable GAL1 promoter confers fluconazole resistance in Saccharomyces cerevisiae. Antimicrob Agents Chemother. 1999;43:2798–2800. doi: 10.1128/aac.43.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy S, Gupta V, Prasad R, Panwar S L, Prasad R. Expression of CDR1, a multidrug resistance gene of Candida albicans: transcriptional activation by heat shock, drugs and human steroid hormones. FEMS Microbiol Lett. 1998;160:191–197. doi: 10.1111/j.1574-6968.1998.tb12910.x. [DOI] [PubMed] [Google Scholar]
- 16.Lamb D C, Kelly D E, Schunck W H, Shyadehi A Z, Akhtar M, Lowe D J, Baldwin B C, Kelly S L. The mutation T315A in Candida albicans sterol 14α-demethylase causes reduced enzyme activity and fluconazole resistance through reduced affinity. J Biol Chem. 1997;272:5682–5688. doi: 10.1074/jbc.272.9.5682. [DOI] [PubMed] [Google Scholar]
- 17.Law D, Moore C B, Wardle H M, Ganguli L A, Keaney M G L, Denning D W. High prevalence of antifungal resistance in Candida spp. from patients with AIDS. J Antimicrob Chemother. 1994;34:659–668. doi: 10.1093/jac/34.5.659. [DOI] [PubMed] [Google Scholar]
- 18.Lees N D, Broughton M C, Sanglard D, Bard M. Azole susceptibility and hyphal formation in a cytochrome P-450-deficient mutant of Candida albicans. Antimicrob Agents Chemother. 1990;34:831–836. doi: 10.1128/aac.34.5.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lees N D, Skaggs B, Kirsch D R, Bard M. Cloning of the late genes in the ergosterol biosynthesis pathway of Saccharomyces cerevisiae—a review. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- 20.Lees N D, Bard M, Kirsch D R. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- 21.Lopez D, Chambers C M, Keller R K, Ness G C. Compensatory responses to inhibition of hepatic squalene synthase. Arch Biochem Biophys. 1998;351:159–166. doi: 10.1006/abbi.1997.0556. [DOI] [PubMed] [Google Scholar]
- 22.Lopez-Ribot J L, McAtee R K, Lee L N, Kirkpatrick W R, White T C, Sanglard D, Patterson T F. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob Agents Chemother. 1998;42:2932–2937. doi: 10.1128/aac.42.11.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marr K A, Rustad T R, Rex J H, White T C. The trailing end point phenotype in antifungal susceptibility testing is pH dependent. Antimicrob Agents Chemother. 1999;43:1383–1386. doi: 10.1128/aac.43.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M'baya B, Fegueur M, Servouse M, Karst F. Regulation of squalene synthetase and squalene epoxidase activities in Saccharomyces cerevisiae. Lipids. 1989;24:1020–1023. doi: 10.1007/BF02544072. [DOI] [PubMed] [Google Scholar]
- 25.Molowa D T, Cimis G M. Co-ordinate regulation of low-density-lipoprotein receptor and 3-hydroxy-3-methylglutaryl-CoA reductase and synthase gene expression in HepG2 cells. Biochem J. 1989;260:731–736. doi: 10.1042/bj2600731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. NCCLS document M27-A. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Pinto W J, Lozano R, Nes W R. Inhibition of sterol biosynthesis by ergosterol and cholesterol in Saccharomyces cerevisiae. Biochim Biophys Acta. 1985;836:89–95. doi: 10.1016/0005-2760(85)90224-3. [DOI] [PubMed] [Google Scholar]
- 28.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rex J H, Pfaller M A, Galgiani J N, Bartlett M S, Espinel-Ingroff A, Ghannoum M A, Lancaster M, Odds F C, Rinaldi M G, Walsh T J, Barry A L. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin Infect Dis. 1997;24:235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 30.Rex J H, Nelson P W, Paetznick V L, Lozano-Chiu M, Espinel-Ingroff A, Anaissie E J. Optimizing the correlation between results of testing in vitro and therapeutic outcome in vivo for fluconazole by testing critical isolates in a murine model of invasive candidiasis. Antimicrob Agents Chemother. 1998;42:129–134. doi: 10.1128/aac.42.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson G W, Tsay Y H, Kienzle B K, Smith-Monroy C A, Bishop R W. Conservation between human and fungal squalene synthetases: similarities in structure, function, and regulation. Mol Cell Biol. 1993;13:2706–2717. doi: 10.1128/mcb.13.5.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosamond J, Allsop A. Harnessing the power of the genome in the search for new antibiotics. Science. 2000;287:1973–1976. doi: 10.1126/science.287.5460.1973. [DOI] [PubMed] [Google Scholar]
- 33.Ruhnke M, Schmidt-Westhausen A, Engelmann E, Trautmann M. Comparative evaluation of three antifungal susceptibility test methods for Candida albicans isolates and correlation with response to fluconazole therapy. J Clin Microbiol. 1996;34:3208–3211. doi: 10.1128/jcm.34.12.3208-3211.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard D, Kuchler K, Ischer F, Pagani J-L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 36.Sanglard D, Ischer F, Koymans L, Bille J. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungals. Antimicrob Agents Chemother. 1998;42:241–253. doi: 10.1128/aac.42.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith S J, Crowley J H, Parks L W. Transcriptional regulation by ergosterol in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:5427–5432. doi: 10.1128/mcb.16.10.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamasawa N, Hayakari M, Murakami H, Matsui J, Suda T. Reduction of oxysterol levels up-regulates HMG-CoA reductase activity in rat liver. Atherosclerosis. 1997;131:237–242. doi: 10.1016/s0021-9150(97)00055-5. [DOI] [PubMed] [Google Scholar]
- 39.Qin W, Infante J, Wang S R, Infante R. Regulation of HMG-CoA reductase, apoprotein-B and LDL receptor gene expression by the hypocholesterolemic drugs simvastatin and ciprofibrate in Hep G2, human and rat hepatocytes. Biochim Biophys Acta. 1992;1127:57–66. doi: 10.1016/0005-2760(92)90201-6. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez J A, Sobel J D, Peng G, Steele-Moore L, Schuman P, Holloway W, Neaton J D. Evolution of vaginal Candida species recovered from human immunodeficiency virus-infected women receiving fluconazole prophylaxis: the emergence of Candida glabrata? Terry Beirn Community Programs for Clinical Research in AIDS (CPCRA) Clin Infect Dis. 1999;28:1025–1031. doi: 10.1086/514746. [DOI] [PubMed] [Google Scholar]
- 41.White T C. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob Agents Chemother. 1997;41:1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White T C. The presence of an R467K amino acid substitution and loss of allelic variation correlate with an azole-resistant lanosterol 14α-demethylase in Candida albicans. Antimicrob Agents Chemother. 1997;42:1488–1494. doi: 10.1128/aac.41.7.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]