Abstract
Background and objectives
Successful clinical integration of genomic sequencing (GS) requires evidence of its utility. While GS potentially has benefits (utilities) or harms (disutilities) across multiple domains of life for both patients and their families, there is yet no empirically informed conceptual model of these effects. Our objective was to develop an empirically informed conceptual model of perceived utility of GS that captures utilities and disutilities for patients and their families across diverse backgrounds.
Methods
We took a patient-centered approach in which we began with a review of existing literature followed by collection of primary interview data. We conducted semi-structured interviews to explore types of utility in a clinically and sociopolitically diverse sample of 60 adults from seven Clinical Sequencing Evidence-Generating Research (CSER) consortium projects. Interviewees had either personally received, or were parents of a child who had received, GS results. Qualitative data was analysed using thematic analysis. Findings from interviews were integrated with existing literature on clinical and personal utility to form the basis of an initial conceptual model that was refined based on expert review and feedback.
Results
Five key utility types that have been previously identified in qualitative literature held up as primary domains of utility and disutility in our diverse sample. Interview data were used to specify and organize subdomains of an initial conceptual model. After expert refinement, the five primary domains included in the final model are: clinical, emotional, behavioral, cognitive, and social, and several sub-domains are specified within each.
Conclusion
We present an empirically-informed conceptual model of perceived utility of GS. This model can be used to guide development of instruments for patient-centered outcome measurement that capture the range of relevant utilities and disutilities and inform clinical implementation of GS.
1. Introduction
Genomic sequencing (GS) is central to the advancement of genomic medicine, which aims to deliver care to patients based on their genetic makeup. While GS is increasingly applied in clinical care, data on the utility of GS remain limited [1–6]. Lack of consensus on what constitutes GS utility and, relatedly, how to best measure it from a patient perspective, hinder the development of a robust evidence base.
GS utility has been conceptually described as a “composite construct” made up of multiple domains, consisting of utilities (benefits) and disutilities (harms) [7] that impact not only individual patients, but also their families and society [8]. However, assessments of GS utility have traditionally been much more limited, focused on one of two well-described types of utility: clinical utility or personal utility. Assessments of clinical utility reflect a standard, objective approach for evaluating novel clinical diagnostics by measuring whether the test changes clinical management and improves patient health outcomes [9,10]. The concept of personal utility recognizes that, because of the nature of information returned from GS, patients may experience important non-health outcomes [11–14]. These non-health effects may be in the form of positive utility, or positive effects on informed decision making about future reproduction, improved self-knowledge, or interpersonal connection with others facing similar clinical circumstances [15,16]. Non-health effects may also be experienced as a disutility in the form of discrimination or negative psychosocial effects [15,17]. Limiting patient-reported assessments to the realm of personal utility alone, however, may not adequately capture the full range of health and non-health related impacts that patients and their families experience from GS. To adequately capture the full range of relevant domains for GS utility assessment, best practices require research on patient experience to inform conceptual models, especially for models that will support development of outcome measures [18,19].
Several qualitative studies have explored patients’ and parents’ perspectives on the impact of GS and understandings of its utility, with findings suggesting that patients and their parents experience utilities and disutilities across multiple domains of life. Malek and colleagues categorize the impacts of GS reported by parents of pediatric patients into clinical, psychological, and pragmatic domains [11]. In a systematic review of studies exploring personal utility, Kohler and colleagues identify multiple elements of personal utility (e.g., self-knowledge, understanding of health condition, altruism, and coping) that can be categorized into affective, cognitive, and behavioral domains; these investigators also acknowledge that there are also social utilities of GS that deserve attention [15,16].
These studies provide an important first step in identifying the range of utilities and disutilities of GS, but they are limited by a lack of participant diversity in sociopolitical characteristics such as race and ethnicity, as well as institutional setting and clinical context [11,20–22]. Furthermore, specific sub-domains to support future measure development have not yet been identified. In-depth qualitative exploration in more diverse patient populations, settings, and clinical contexts is therefore needed to develop a robust conceptual model of patient-centered utility that could inform the development of future measurement instruments with wide applications for GS evaluation.
In this paper, we report primary findings from qualitative interviews conducted with a sociopolitically and clinically diverse group of participants who received a variety of GS results through the Clinical Sequencing Evidence-Generating Research (CSER) consortium. Informed by those findings, we present a conceptual model of patient-perceived utility of clinical GS. The proposed model can be used to guide future approaches to the evaluation of GS, including the development of patient-centered outcome measures [18,23], that reflect the relevant range of patient and parent experiences.
2. Method
This paper reports on the initial phase of measure development (i.e., identification of domains and subdomains) following best practices for developing psychometric scales for use in health and social sciences [18,19]. We took a patient-centered approach in which we began with a review of existing literature followed by collection of primary interview data and development of a conceptual model, which is intended to support the development of quantitative measurement tools in future work. We follow guidelines for reporting of formative qualitative research for health preference measures [24].
2.1. Sample Selection and Recruitment
The CSER consortium, described in detail elsewhere, is comprised of seven clinical sequencing projects, reflecting diverse study populations, medical conditions, and US geographic locations [25]. We recruited participants via email or phone from all seven CSER projects using purposive sampling, prioritizing individuals who both (1) were representative of the respective project by gender and race and ethnicity, and (2) represented a diversity of GS findings, including positive primary or secondary findings, heterozygote (carrier) status, negative findings, and variants of uncertain significance. Participants had previously received GS results and were either adults receiving their own results or parents of a child who received their child’s GS results. The projects included six extramural CSER projects: Baylor College of Medicine (KidsCanSeq), HudsonAlpha Institute for Biotechnology (SouthSeq), Icahn School of Medicine at Mt. Sinai (NYCKidSeq), Kaiser Permanente Northwest (CHARM Study), University of California San Francisco (P3EGS), University of North Carolina at Chapel Hill (NCGENES2), and one NHGRI intramural project (ClinSeq). While most projects recruited participants receiving results from their ongoing CSER project, the KidsCanSeq project also recruited participants from their CSER-phase one study (BASIC3) [26]. Similarly, the NCGENES2 project recruited participants from a GS study of children and infants (NC NEXUS) from the Newborn Sequencing In Genomic medicine and public HealTh (NSIGHT) consortium [27]. The Institutional Review Board (IRB) of Baylor College of Medicine approved this research (#H-44856, initial approval 03/19/2019), and the study procedures were approved by each project’s IRB.
2.2. Data Collection
We performed structured database searches (see Online Resource) to update and expand the search protocol employed in a previous systematic literature review published by a study team member (BB) [15]. In addition to three domains of personal utility (affective, cognitive, and behavioral) and one social utility domain previously described, we also identified a clinical utility domain (as perceived by patients) for a total of five primary domains of utility and disutility across the studies reviewed. This literature review was used to develop a semi-structured interview guide (Online Resource), which was pilot tested and refined. The interviews focused on participants’ own experiences of receiving results and the impacts the results had on them, rather than on hypothetical outcomes. Interviews began with open-ended questions about what participants hoped to learn from the GS study and the impact of receiving GS, followed by probes related to the key utility domains extracted from our literature review [11,16,28]. Interviews concluded with another open-ended probe to identify additional experienced impacts.
One-on-one telephone interviews were conducted between March and August 2019. Study team members at each CSER project conducted interviews, as they were most familiar with the participants in their own project. All interviewers (BB, SM, MW, CG, CR, SO, SA, BA, KS, BW) were graduate-level trained researchers or had extensive experience (>10 years) conducting qualitative research. One interviewer identifies as male/man and the rest as female/woman. Participants were compensated $20–50 for participation, consistent with project standards. All interviews were audio-recorded, professionally transcribed, and reviewed for accuracy (but not returned to participants for comment). Identifiable information was removed from transcripts. No additional interviews were conducted when the research team agreed that informational redundancy had been reached, determined by the team’s assessment that additional data did not lead to any new emergent themes [29,30].
2.3. Qualitative data analysis
Using a content analysis orientation, we took an integrated, iterative approach to development of the coding structure for interview data [31]. The qualitative data analysis was designed and led by research team members with expertise in qualitative methods and psychometrics. Transcripts were coded by researchers with considerable experience with clinical GS research projects. Broad a priori codes, based on our literature-informed interview guide, served as the organizing analytical framework. These codes were applied to transcripts by one study team member from each project and then reviewed by a second team member (IC, JOR, SM). Next, data within each a priori code were reviewed by a subset of the team to inductively identify secondary and tertiary codes representing the range of impacts experienced as a result of sequencing (SM, JM, KBB, CR). Qualitative analysis was facilitated using Dedoose Version 8.0.35 (2018; Los Angeles CA: SocioCultural Research Consultants, LLC).
2.4. Model development
A subset of the study team (HSS, SRM, JOR, IC, JM, CKR, CSB, ALM) drafted an initial conceptual model based on the qualitative analysis. Deductive, a priori codes (Online Resource) reflected the major themes found and formed the basis of the primary domains of the model; inductive codes (secondary and tertiary codes) formed the basis of the sub-domains of the model. The draft model was then refined through a process of expert review by the full study team, which is comprised of researchers with deep experience and expertise in bioethics, genomics, medicine, public health, psychology, and health economics. Feedback on model structure and its domains and sub-domains was elicited in written form, followed by a videoconference for group discussion. Final decisions were made by consensus.
3. Results
3.1. Interview participants
Sixty participants across all seven CSER projects participated in interviews (Table 1). The majority (67%) were parents whose children had undergone GS because they had either cancer, a suspected genetic condition, or a severe developmental disorder. The remainder of participants were adults, evenly split between those at risk of hereditary cancer and those who were ostensibly healthy. Forty-two percent of participants self-identified as non-Hispanic White (Table 2). Interview participants reflected recipients of various, and sometimes multiple, GS result types (Table 3), including: primary diagnostic results (32%), secondary results (30%), heterozygote status (63%), variants of uncertain significance or inconclusive results (35%), and negative results (48%). Median time between disclosure of GS results and interview participation was 5 months (range: <1 month to 5 years). Interviews lasted 25–40 minutes.
Table 1.
Clinical Sequencing Evidence-generating Research (CSER) consortium project descriptions
| Project Name (number of interviews conducted) |
Population Description | Type of results discloseda |
|---|---|---|
| BASIC3/KidsCanSeq (n=10) | Children with cancer | Primary, Secondary, Heterozygote Status, Pharmacogenomic, VUS, Negative |
| CHARM Study (n=10) | Adults at risk for hereditary cancer | Primary, Secondary, Heterozygote Status, VUS, Negative |
| ClinSeq (n=10) | Adults, no specific phenotype | Primary, Secondary, VUS, Negative |
| NCGENES2 through NC NEXUS (n=10) | Healthy newborns and children with metabolic diseases or hearing loss | Primary, Secondary, Heterozygote Status |
| NYCKidSeq (n=3) | Children with suspected neurologic, immunologic and cardiac genetic conditions | Primary, Secondary, VUS, Negative |
| P3EGS (n=10) | Infants and children with severe developmental disorders, with or without congenital anomalies; parents whose fetus has a structural anomaly | Primary, Secondary, VUS, Negative |
| SouthSeq (n=7) | Newborns with suspected genetic conditions | Primary |
VUS, variant of uncertain significance; Primary results are defined as known or likely pathogenic results relevant to clinical phenotype or diagnosis; Secondary results are medically actionable, consistent with American College of Medical Genetics and Genomics guidelines.
Table 2.
Interview participant characteristicsa
| n (%) | |
|---|---|
| Relationship to patient (n=60) | |
| Self - adult patient | 20 (33%) |
| Parent of pediatric patient | 40 (67%) |
| Sex (n=60) | |
| Female | 49 (82%) |
| Male | 11 (18%) |
| Race and ethnicity (n=60) | |
| American Indian | 2 (3%) |
| Asian | 7 (12%) |
| Black or African American | 13 (22%) |
| Hispanic or Latinx | 7 (12%) |
| Multiracial or multiethnic | 3 (5%) |
| Non-Hispanic White | 28 (47%) |
| Age in years (n=43) | |
| Mean (SD) | 36 (9.5) |
| Min - Max | 18 – 54 |
| Education (n=42) | |
| Some high school | 2 (5%) |
| High school graduate, GED or equivalent | 6 (14%) |
| Some college | 10 (24%) |
| Associates degree | 2 (5%) |
| Bachelors degree | 10 (24%) |
| Graduate or professional degree | 12 (29%) |
No enrolled participants dropped out of the study; based on enrollment data from 6 projects, 8 invited individuals declined to participate.
Table 3.
Sequencing results among interview participants
| Sequencing Result Typea | |||||
|---|---|---|---|---|---|
| Project | Positive Primary |
Positive Secondary |
Heterozygote Status |
VUS or Inconclusive | Negative |
| BASIC3/KidsCanSeqb (n=10) | 2 | 3 | 8 | 8 | 5 |
| CHARM Study (n=10) | 2 | 3 | 4 | 1 | 2 |
| ClinSeq (n=10) | NA | NA | NA | NA | 10 |
| NCGENES2 through NC NEXUS (n=10)c | -- | -- | 7 | -- | -- |
| NYCKidSeq (n=3) | NA | NA | NA | 1 | 2 |
| P3EGS (n=10) | 4 | NA | NA | 3 | 3 |
| SouthSeq (n=7) | 4 | NA | NA | 1 | 2 |
| Total | 12/37 (32%) | 6/20 (n=30%) | 19/30 (63%) | 14/40 (35%) | 24/50 (48%) |
VUS, variant of uncertain significance.
NA = not applicable for sites that did not return that type of result to participants included in this study. Total column subtracts NA and missing data from the denominator.
BASIC3/KidsCanSeq participants also received pharmacogenomics results.
Missing data for NC NEXUS results returned.
3.2. Conceptual Model Structure
Interview data were generally consistent with the types of GS utility and disutility identified in the conceptual and empirical literature. Seven major themes emerged as meaningful patterns identified through analysis of the interview data: patient clinical care, family clinical care, emotions, future impact, future conditions, social impact, and scientific/media awareness. These themes were consistent with the a priori codes used as our organizing framework for analysis. In the first iteration of the conceptual model, these major themes were mapped to four utility domains according to their relevance to aspects of life for patients and parents: Health and Healthcare, Affective, Social, and Decisional and Pragmatic. Through the process of expert review, the model domain names were simplified and the Decisional and Pragmatic domain was divided into two domains, labeled Behavioral and Cognitive.
Conceptual model development – both structure and domain nomenclature – was an iterative process, guided by interview data, expert opinion, and published studies (many of which involved members of the study team). The final proposed model consists of five primary domains of perceived utility of GS (Figure 1): Clinical, Emotional, Behavioral, Cognitive, and Social. Sub-domains, which were also informed through a combination of interview data, literature, and expert input, were mapped within this model. Each domain includes 2–7 sub-domains, appropriate to support the development of an item pool for psychometric instrument development [18]. Subdomains are considered to be the important dimensions along which a continuum of impact from utility (benefit) to disutility (harm) exists. Each domain is described below, with sub-domain labels identified in italics. Domains, sub-domains, and exemplary quotes are presented in Table 4. We describe qualitative findings related to domains and sub-domains below.
Figure 1. Utility Domains.
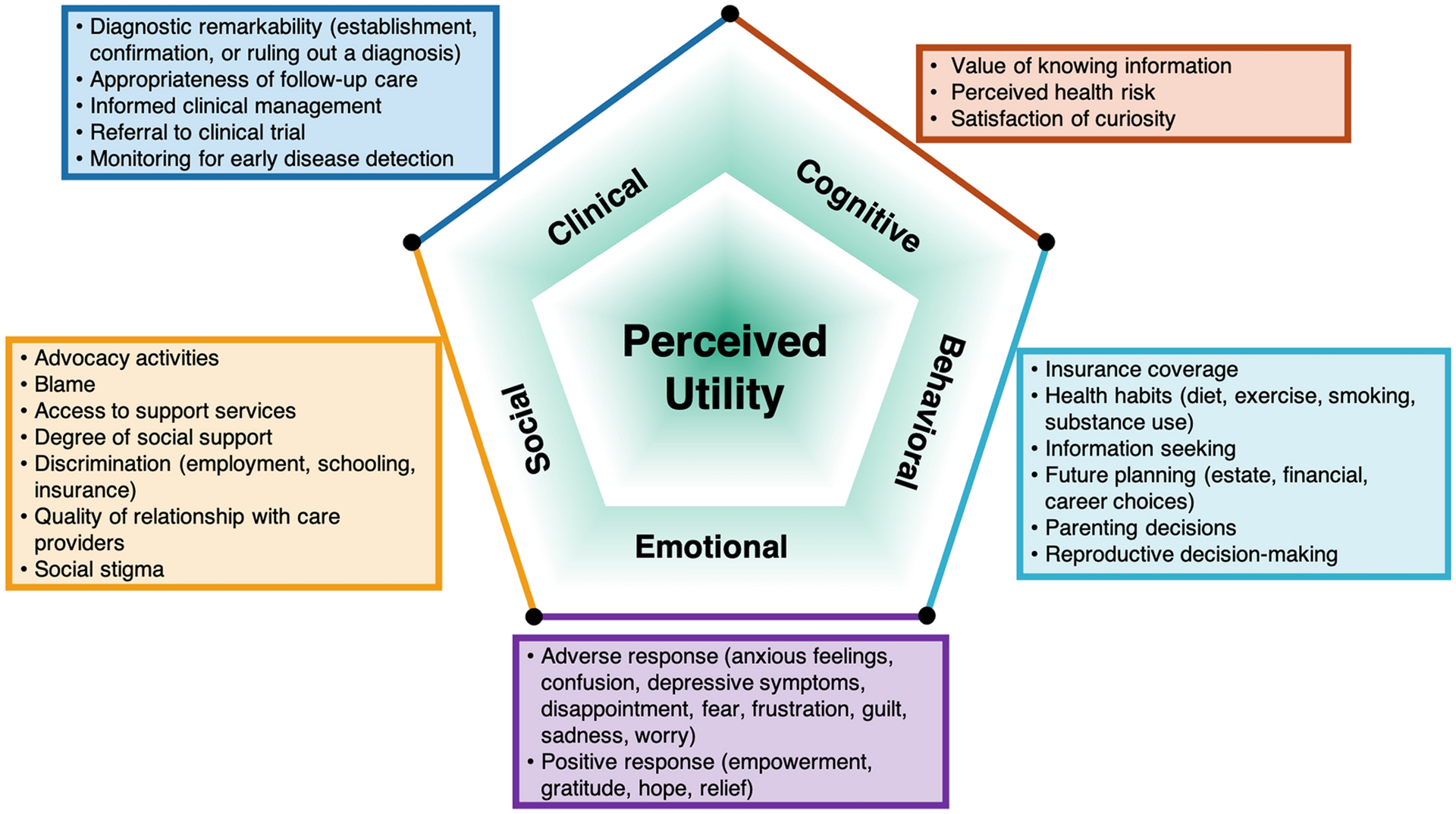
Conceptual model of patient perceived utility of genomic sequencing
Table 4.
Domains, subdomains, and exemplary quotes
| Domain | Subdomain | Exemplar Quote | Project, Study ID |
|---|---|---|---|
| Clinical | Diagnostic remarkability (establishment, confirmation, or ruling out a diagnosis) |
|
Site 2, ID 116 |
| Appropriateness of follow-up care |
|
Site 1, ID 131 | |
| Informed clinical management |
|
Site 4, ID 153 | |
| Referral to a clinical trial | None coded in interview data | ||
| Monitoring for early disease detection |
|
Site 3, ID 113 | |
| Emotional | Adverse response (anxious feelings, confusion, depressive symptoms, disappointment, fear, frustration, guilt, sadness, worry) |
|
Site 2, ID 109 |
| Positive response (empowerment, gratitude, hope, relief) |
|
Site 4, ID 118 | |
| Behavioral | Insurance coverage |
|
Site 3, ID 120 |
| Health habits (diet, exercise, smoking, substance use) |
|
Site 5, ID 117 | |
| Information seeking |
|
Site 5, ID 156 | |
| Future planning (estate, financial, career choices) |
|
Site 3, ID 113 |
|
| Parenting decisions |
|
Site 3, ID 126 | |
| Reproductive decision-making |
|
Site 4, ID 124 | |
| Cognitive | Value of knowing information |
|
Site 3, ID 147 |
| Perceived health risk |
|
Site 5, ID 156 | |
| Satisfaction of curiosity |
|
Site 5, ID 117 | |
| Social | Advocacy activities | None coded in interview data | |
| Blame |
|
Site 3, ID 138 | |
| Access to support services |
|
Site 3, ID 143 | |
| Degree of social support |
|
Site 1, ID 125 | |
| Discrimination (employment, schooling, insurance) |
|
Site 3, ID 120 | |
| Quality of relationship with care providers |
|
Site 3, ID 157 | |
| Social stigma | None coded in interview data |
3.3. Utility Domains
3.3.1. Clinical utility
Some participants described GS as clinically useful for either the patient or the patient’s family by, for example, enabling, confirming, or ruling out a diagnosis (diagnostic remarkability). As one participant explained, sequencing “ruled out a lot of diagnoses…it helped rule out what other possibilities there were…” (Project 2, ID 116). Although a few participants reported that GS impacted their clinical care (informed clinical management), the majority described GS as having little to no impact on managing clinical care, regardless of the type of result they received. As one parent explained, “…There wasn’t really any change, there was nothing that they could do with genetic testing that would’ve affected his treatment.” (Project 3, ID 120). Some of those who reported GS informed their clinical management explained that GS led their clinicians to recommend additional screening or testing, including additional genetic testing and earlier initiation of preventive screenings, such as colonoscopies (monitoring for early disease detection).
3.3.2. Emotional utility
Participants described a range of emotions associated with receiving results, with positive responses more commonly described than adverse ones. Some participants for whom GS established or confirmed a diagnosis described feeling appreciation for having an explanation (positive response). As one offered, “Honestly, I think I just feel more at peace, knowing that there is a reason for all of these little things…” (Project 4, ID 118). Others described GS as providing hope, including reassurance that there was no elevated risk for future disease. As one parent explained, “knowing that her genes were no different from any other normal kid really gave us hope that she might be ok [after cancer remission]” (Project 3, ID 126). Participants also described GS as providing relief, including alleviating worries about future health risks.
Some participants described an adverse response, such as frustration related to unrealized expectations that they would gain knowledge from GS, or disappointment that it did not yield a definitive “reason” for illness (adverse response): “there was some frustration because I was hoping to get a definitive answer from those tests.” (Project 2, ID 116). Others described similar reactions, characterizing their feelings about not receiving positive findings as leading them to feel confused as to the underlying cause of their/their child’s illness. As one parent explained, “after they gave me results that I didn’t have no genes that provoked my son’s cancer, I was more confused…where was the disease coming from?” (Project 3, ID 106). Another described feeling helpless, stating “I felt like I was at a dead end, at a loss, because that testing revealed nothing, and I had put so much hope into it revealing something, positive or negative, and because it revealed nothing, it was just a tremendous let down.” (Project 2, ID 109).
Some participants described feeling fearful after receiving results, either from learning about elevated risks of future conditions or related to downstream implications of a diagnosed illness. Notably, however, participants generally described the increased knowledge as being a worthwhile trade-off for this fear. One participant said: “…obviously it’s a little bit scary knowing that [result] for sure, but it’s also kind of helpful to know that this is something I have to watch out for because it’s better to know….than just having this hovering over you…”(Project 5, ID 156). Others expressed worry or sadness related to receiving a diagnosis. As described by one parent: “…at least the first couple months, it was hard to deal with. I mean, we both cried a lot…it was hard to talk about.” (Project 6, ID 128).
3.3.3. Behavioral utility
While most participants did not think that the results had utility for either their future planning or their intention to modify lifestyle behaviors or prepare for onset of conditions in the future, some participants described substantial importance of GS results for these purposes. Among those who described health behavior effects, the majority of comments related to general dietary or lifestyle improvements, including efforts to eat healthier or to increase physical activity (health habits). Participants commented that GS motivated them to undertake healthier lifestyle behaviors, regardless of whether results indicated elevated disease risk. As one participant with no prior health history described after receiving negative results: “…it made me want to be more proactive about staying healthy. You feel like you get kind of a clean sheet genetically when you receive a good test result like that. So it’s very motivating…it’s like, ‘Okay. The genetic part’s good. I don’t want to screw up the lifestyle part now.’” (Project 5, ID 117). However, two participants described health behavior changes related to a specific genetic finding, including the use of dietary supplements related to their specific diagnosis and actions to help preserve vision for a child diagnosed with a condition related to vision loss.
A few participants explained how GS impacted their future planning in terms of decision-making about selecting health insurance or long-term care coverage, career choices, and parenting decisions (future planning), even in the case of negative results. As one parent of a patient with negative results explained: “…given what the results were, it has helped us stay the course and be hopeful, and commit to giving our daughter a normal upbringing. She’s allowed to do everything.” (Project 3, ID 126) The most commonly described impact on future planning related to reproductive decision-making, for the participant or their children (reproductive decision-making). Some participants described results as providing reassurance regarding having additional children, which one described as giving both “confidence” and “relief.” For others, results discouraged having (additional) children. As one explained, “I always wanted three kids, but since I know…the other kid might also be affected by this, I would definitely put off my plan from three to two…” (Project 4, ID 124).
3.3.4. Cognitive utility
In addition to behaviorial effects of sequencing results, participants acknowledged that the information held value apart from its connection to any specific follow-up actions. Several participants reported utility related to having information from GS itself (value of knowing information). As one participant with negative results explained: “there’s absolutely value [in having information from sequencing]…there’s value in knowledge, there’s value in the answer ‘no.’ No, you don’t have diabetes, no you don’t have hypertension… there’s value in ‘no.’” (Project 7, ID 149). Others described value in the certainty of confirmed disease etiology. As one parent explained, “I feel better about knowing what was causing his problems.” (Project 1, ID 112) According to another parent “I think there is [value]…. Me as a person, if I know why, I feel more in control….Ok, I understand the research behind this…I feel more confident explaining to people, ‘Well, this is what’s going on’….and being able to take care of him and kind of see things before they happen, if that makes sense.” (Project 1, ID 119)
Others identified the utility of increased accuracy of health risks (perceived health risk), such as by helping to fill gaps in family health history, as one parent described: “as a parent, I want to be able to provide my kids the history…what you need to look for and what kind of testing need[s] to be done.” (Project 5, ID 151) Another dimension of cognitive utility related to satisfying curiosity for the results and their health implications (satisfaction of curiosity): “I just thought it was really cool. I was actually pretty excited to sign up and just to learn as much as I could. That’s just kind of my nature, though. I just want to know what I’m working with.” (Project 5, ID 117)
3.3.5. Social utility
Participants described a range of social utilities and disutilities from GS, shaping their interactions with family, schools, involvement in advocacy or support groups, and clinical care. Impact on families, both positive and adverse, was the most commonly mentioned type of social utility. Several participants described GS as facilitating enhanced social support from and understanding by family members about the genetic condition (degree of social support).
Social utilities related to a child’s education were also described, some of which were positive, including individualized educational planning or resources (access to support services), being able to “give [information from sequencing studies] to the teachers so that they can work with him on a personal level much better than just, you, ‘here’s a kid and you figure it out from there.’” (Project 3, ID 143). However, another parent described the adverse effect of perceiving that the school viewed the genetic diagnosis as a “reason” for why the child should be educated elsewhere (discrimination).
Participants described GS as facilitating their engagement in both individual and group activities related to advocacy (advocacy activities) and support (access to support services), such as linking them into in-person or online support groups for rare diseases. One parent of a child for whom sequencing diagnosed a rare disease described connecting weekly with another parent via Facebook messenger, an exchange she described as “reassuring,” and that it was “just nice to have someone else that know[s] where I’m coming from.” (Project 1, ID 125). Finally, one participant described participating in a research study involving GS as enhancing her family’s relationship with their child’s clinical care team (quality of relationship with care providers).
4. Discussion
We developed a conceptual model of patient-perceived utility that was both informed by primary qualitative data from a diverse group of patients and parents who had received GS results and grounded in prior conceptual and empirical literature. Interview participants were recruited from seven clinical GS research studies and were diverse in terms of race and ethnicity, geographic locations in the US, and medical conditions. After expert review and feedback on the empirical findings, the resulting conceptual model (Figure 1) consists of five domains, each with sub-domains: clinical, emotional, behavioral, cognitive, and social (Table 4).
The types of personal utility (affective, cognitive, and behavioral) and social utility identified in the review of literature on personal utility of genetics and genomics by Kohler et al. are represented in our model [15], along with clinical utility, which Malek et al. identified as a type of utility that patients and parents also find important [11]. Our findings suggest that both personal and clinical features of GS are important to patients, indicating that both are relevant for inclusion in a conceptual model and a subsequent patient-reported outcome measure of the perceived utility of GS. From a patient-centered perspective, our model supports a definition of utility that extends beyond traditional objective clinical outcome measurement to include features of clinical care that patients and parents value, as well as non-clinical domains of emotional, behavioral, cognitive, and social utility [8].
Given the lack of a widely accepted, specific, and systematically developed model of patient-perceived utility of GS and the importance of robust measurement to generate evidence for decisions about clinical use and reimbursement [16,32,33], our proposed model aims to fill an important gap. Our work builds upon and extends previous research through the addition of empirical evidence from a more diverse set of participants, organization of domains and identification of sub-domains, and synthesis of findings into a conceptual model. This proposed conceptual model of patient-perceived utility (Figure 1) can guide the future development of measurement tools for administration as part of clinical research studies and routine practice that capture the full range of perceived benefits and harms from GS and better reflect the experience of patients and their families, a central component to guide clinical implementation and evaluation of GS [23]. Existing measures of clinical utility, designed for administration to clinicians [32], and personal utility designed for administration to patients [16], might not capture the full range of benefits and harms from the patient’s perspective. Based on qualitative findings relevant to each domain across types of interview participants, we expect that our model will be appropriate to support the development of psychometric instruments for both adult patients and parents or caregivers of pediatric patients. Our findings also suggest that both positive and negative effects are plausible within each domain. While some scholars have positioned both clinical utility and personal utility along a spectrum of quality of life [17], traditional measures of health-related quality of life are not designed to accurately measure the effects of sequencing. Thus, we aim to support development of an instrument specifically intended for that purpose.
Among interview participants, the time between GS results disclosure and interview participation ranged from less than one month to five years, making the resultant conceptual model applicable to exploration of both shorter- and longer-term effects. Additionally, the variability in which themes arose across interview participants is a positive feature from a measurement perspective. However, our findings should be interpreted in light of several limitations. First, while we took several steps to elicit the full range of utilities from GS, including developing our interview guide from a literature review of previously described impacts and prompting participants to describe any additional impacts, there may be impacts we failed to uncover, and participants did not provide feedback on findings. While our sample was diverse in important ways, it did not reflect all possible clinical conditions for which GS might be sought and we did not conduct interviews with adolescents and young adults. Finally, selection bias could have diminished the extent to which we uncovered adverse impacts of sequencing if patients who were more adversely affected were unwilling to participate in interviews. Further investigation regarding the impact of result received on perceptions of utility is needed, and can be facilitated with measures developed based on this model.
5. Conclusions
Overall, our findings suggest that to comprehensively understand the effect of clinical GS and inform clinical integration and reimbursement policy, a broad lens should be used to capture the domains of GS utility experienced by patients or parents, including both clinical and non-clinical domains. Our proposed model can support such measurement development in the future.
Supplementary Material
Key Points.
Genomic sequencing is being rapidly integrated into clinical care, and there is a need for patient-centered outcome measures to assess key outcomes such as utility.
We integrated primary interview data with findings from the literature to organize and refine a conceptual model of patient-perceived utility of genomic sequencing.
Our proposed model is appropriate to support the future development of a patient-centered measure that captures the wide range of outcomes from genomic sequencing that are important to adult patients and parents of pediatric patients.
Acknowledgements
We thank Katie Lewis, Carla Rich, and Beatriz Anguiano for their contributions to this project. We are especially grateful for the participants and staff of all CSER studies for their important contributions.
Funding
The Clinical Sequencing Evidence-Generating Research (CSER) consortium is funded by the National Human Genome Research Institute (NHGRI) with co-funding from the National Institute on Minority Health and Health Disparities (NIMHD) and the National Cancer Institute (NCI), supported by U01HG006487 (UNC), U01HG007292 (KPNW), U01HG009610 (Mt Sinai), U01HG006485 (Baylor), U01HG009599 (UCSF), U01HG007301 (HudsonAlpha), and U24HG007307 (Coordinating Center). The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the NIH. More information about CSER can be found at https://cser-consortium.org/. CKR was supported by a San Diego State University Graduate Fellowship. CSB supported by NHGRI R01HG008753 (Bloss PI).
Footnotes
Conflicts of interest/Competing interests
The authors declare that they have no conflicts of interest.
Ethics approval
This research was approved by the Institutional Review Board (IRB) of Baylor College of Medicine, and the interview study procedures were approved by each individual project’s IRB. Informed consent was obtained from all participants.
Availability of data and material
Data available upon request.
References
- 1.Ginsburg GS, Phillips KA. Precision Medicine: From Science To Value. Health Affairs. 2018;37:694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Payne K, Gavan SP, Wright SJ, Thompson AJ. Cost-effectiveness analyses of genetic and genomic diagnostic tests. Nat Rev Genet. 2018;19:235–46. [DOI] [PubMed] [Google Scholar]
- 3.Smith HS, Swint JM, Lalani SR, Yamal J-M, de Oliveira Otto MC, Castellanos S, et al. Clinical Application of Genome and Exome Sequencing as a Diagnostic Tool for Pediatric Patients: a Scoping Review of the Literature. Genet Med. 2019;21:3–16. [DOI] [PubMed] [Google Scholar]
- 4.Parsons DW, Roy A, Yang Y, Wang T, Scollon S, Bergstrom K, et al. Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol. 2016;2:616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts JS, Robinson JO, Diamond PM, Bharadwaj A, Christensen KD, et al. Patient understanding of, satisfaction with, and perceived utility of whole-genome sequencing: findings from the MedSeq Project. Genet Med. 2018;20:1069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart MR, Biesecker BB, Blout CL, Christensen KD, Amendola LM, Bergstrom KL, et al. Secondary findings from clinical genomic sequencing: prevalence, patient perspectives, family history assessment, and health-care costs from a multisite study. Genet Med. 2019;21:1100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foster MW, Mulvihill JJ, Sharp RR. Evaluating the utility of personal genomic information. Genet Med. 2009;11:570–4. [DOI] [PubMed] [Google Scholar]
- 8.ACMG Board of Directors. Clinical utility of genetic and genomic services: a position statement of the American College of Medical Genetics and Genomics. Genet Med. 2015;17:505–7. [DOI] [PubMed] [Google Scholar]
- 9.Burke W Genetic Test Evaluation: Information Needs of Clinicians, Policy Makers, and the Public. American Journal of Epidemiology. 2002;156:311–8. [DOI] [PubMed] [Google Scholar]
- 10.Teutsch SM, Bradley LA, Palomaki GE, Haddow JE, Piper M, Calonge N, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malek J, Slashinski MJ, Robinson JO, Gutierrez AM, Parsons DW, Plon SE, et al. Parental Perspectives on Whole-Exome Sequencing in Pediatric Cancer: A Typology of Perceived Utility. JCO Precision Oncology. 2017;1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson JO, Wynn J, Biesecker B, Biesecker LG, Bernhardt B, Brothers KB, et al. Psychological outcomes related to exome and genome sequencing result disclosure: a meta-analysis of seven Clinical Sequencing Exploratory Research (CSER) Consortium studies. Genet Med. 2019;21:2781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lupo PJ, Robinson JO, Diamond PM, Jamal L, Danysh HE, Blumenthal-Barby J, et al. Patients’ perceived utility of whole-genome sequencing for their healthcare: findings from the MedSeq project. Personalized Medicine. 2016;13:13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravitsky V, Wilfond BS. Disclosing Individual Genetic Results to Research Participants. The American Journal of Bioethics. 2006;6:8–17. [DOI] [PubMed] [Google Scholar]
- 15.Kohler JN, Turbitt E, Biesecker BB. Personal utility in genomic testing: a systematic literature review. Eur J Hum Genet. 2017;25:662–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler JN, Turbitt E, Lewis KL, Wilfond BS, Jamal L, Peay HL, et al. Defining personal utility in genomics: A Delphi study. Clin Genet. 2017;92:290–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mighton C, Carlsson L, Clausen M, Casalino S, Shickh S, et al. Quality of life drives patients’ preferences for secondary findings from genomic sequencing. Eur J Hum Genet. 2020;28:1178–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boateng GO, Neilands TB, Frongillo EA, Melgar-Quiñonez HR, Young SL. Best Practices for Developing and Validating Scales for Health, Social, and Behavioral Research: A Primer. Front Public Health. 2018;6:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeVellis RF. Scale development: theory and applications. Fourth edition. Los Angeles: SAGE; 2017. [Google Scholar]
- 20.Mollison L, O’Daniel JM, Henderson GE, Berg JS, Skinner D. Parents’ perceptions of personal utility of exome sequencing results. Genet Med. 2020;22:752–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosell AM, Pena LDM, Schoch K, Spillmann R, Sullivan J, Hooper SR, et al. Not the End of the Odyssey: Parental Perceptions of Whole Exome Sequencing (WES) in Pediatric Undiagnosed Disorders. J Genet Counsel. 2016;25:1019–31. [DOI] [PubMed] [Google Scholar]
- 22.Sanderson SC, Linderman MD, Suckiel SA, Zinberg R, Wasserstein M, Kasarskis A, et al. Psychological and behavioural impact of returning personal results from whole-genome sequencing: the HealthSeq project. Eur J Hum Genet. 2017;25:280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horowitz CR, Orlando LA, Slavotinek AM, Peterson J, Angelo F, Biesecker B, et al. The Genomic Medicine Integrative Research Framework: A Conceptual Framework for Conducting Genomic Medicine Research. The American Journal of Human Genetics. 2019;104:1088–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollin IL, Craig BM, Coast J, Beusterien K, Vass C, DiSantostefano R, et al. Reporting Formative Qualitative Research to Support the Development of Quantitative Preference Study Protocols and Corresponding Survey Instruments: Guidelines for Authors and Reviewers. Patient. 2020;13:121–36. [DOI] [PubMed] [Google Scholar]
- 25.Amendola LM, Berg JS, Horowitz CR, Angelo F, Bensen JT, Biesecker BB, et al. The Clinical Sequencing Evidence-Generating Research Consortium: Integrating Genomic Sequencing in Diverse and Medically Underserved Populations. The American Journal of Human Genetics. 2018;103:319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green RC, Goddard KAB, Jarvik GP, Amendola LM, Appelbaum PS, Berg JS, et al. Clinical Sequencing Exploratory Research Consortium: Accelerating Evidence-Based Practice of Genomic Medicine. The American Journal of Human Genetics. 2016;98:1051–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg JS, Agrawal PB, Bailey DB, Beggs AH, Brenner SE, Brower AM, et al. Newborn Sequencing in Genomic Medicine and Public Health. Pediatrics. 2017;139:e20162252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosse SD, Khoury MJ. What is the clinical utility of genetic testing? Genet Med. 2006;8:448–50. [DOI] [PubMed] [Google Scholar]
- 29.Given LM. 100 questions (and answers) about qualitative research. Los Angeles, California: SAGE; 2016. [Google Scholar]
- 30.Saunders B, Sim J, Kingstone T, Baker S, Waterfield J, Bartlam B, et al. Saturation in qualitative research: exploring its conceptualization and operationalization. Qual Quant. 2018;52:1893–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley EH, Curry LA, Devers KJ. Qualitative Data Analysis for Health Services Research: Developing Taxonomy, Themes, and Theory. Health Serv Res. 2007;42:1758–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hayeems RZ, Luca S, Ungar WJ, Bhatt A, Chad L, Pullenayegum E, et al. The development of the Clinician-reported Genetic testing Utility InDEx (C-GUIDE): a novel strategy for measuring the clinical utility of genetic testing. Genet Med. 2020;22:95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens Smith H, Russell HV, Lee BH, Morain SR. Using the Delphi method to identify clinicians’ perceived importance of pediatric exome sequencing results. Genet Med. 2020;22:69–76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available upon request.


