Abstract
We report the cloning and characterization of the gyrA gene of the Mycoplasma hominis DNA gyrase, which was previously shown to be associated with quinolone resistance in this organism. The 2,733-bp gyrA gene encodes a protein of 911 amino acids with a calculated molecular mass of 102.5 kDa. As expected, M. hominis GyrA exhibits higher homology with the GyrA subunits of the gram-positive bacteria Clostridium acetobutylicum, Bacillus subtilis, Streptococcus pneumoniae, and Staphylococcus aureus than with its Escherichia coli counterpart. Knowing the entire sequence of the gyrA gene of M. hominis could be very useful for confirming the role of the GyrA subunit in fluoroquinolone resistance. Twenty-nine mutants of M. hominis were selected stepwise for resistance to trovafloxacin, a new potent fluoroquinolone, and their gyrA, gyrB, parC, and parE quinolone resistance-determining regions were characterized. Three rounds of selection yielded 3 first-step, 12 second-step, and 14 third-step mutants. The first-step mutants harbored a single substitution, Glu460→Lys (E. coli coordinates), in ParE. GyrA changes, Ser83→Leu, Glu87→Lys, and Ala119→Glu or Val, were found only in the second round of selection. At the third step, additional substitutions, at ParC Ser80, Ser81, and Glu84 and ParE Leu440, associated with high-level resistance to fluoroquinolones, appeared. Thus, high-level resistance to trovafloxacin required three steps and was associated with alterations in both fluoroquinolone targets. According to these genetic data, in M. hominis, as in Staphylococcus aureus and Streptococcus pneumoniae, topoisomerase IV seems to be the primary target of trovafloxacin.
The intracellular targets of fluoroquinolones in bacteria are considered to be the type II topoisomerases, DNA gyrase and topoisomerase IV (23). DNA gyrase is composed of two A and two B subunits, encoded by the gyrA and the gyrB genes, respectively. This tetrameric enzyme catalyzes ATP-dependent negative supercoiling of DNA. Topoisomerase IV, a C2E2 tetramer encoded by the parC and parE genes, is essential for chromosome partitioning. Mutations in the quinolone resistance-determining regions (QRDRs) of GyrA and ParC mainly and GyrB and ParE less frequently have been described as the major mechanism for quinolone resistance (10, 23).
Mycoplasma hominis is a cause of urogenital tract infections and has been implicated in extragenital infections as well, especially in immunocompromised patients (46). We recently reported in vitro and in vivo fluoroquinolone-resistant mutants of M. hominis associated with alterations in GyrA, ParC, and ParE QRDRs (3, 5, 7). Furthermore, previous genetic studies showed that topoisomerase IV was the primary target of pefloxacin, ofloxacin, and ciprofloxacin, whereas DNA gyrase was the primary target of sparfloxacin (5, 25).
Concerning the target genes of fluoroquinolones in M. hominis, the gyrB, parC, and parE genes and only the QRDR sequence of gyrA have been cloned and sequenced (3, 4, 28). Here we report the cloning, sequencing, and organization of the complete M. hominis gyrA gene, as well as a detailed analysis of the gyrA, gyrB, parC, and parE QRDRs from M. hominis mutants selected in a stepwise-manner for resistance to trovafloxacin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and DNA manipulations.
The M. hominis reference strain PG21 (ATCC 23114) was grown in Hayflick modified broth medium supplemented with arginine (17). The Escherichia coli strain JM109 and the vector pGEM3zf(+) (Promega) were used to construct libraries and to subclone DNA inserts. Chromosomal DNA from M. hominis PG21 was obtained as previously described (47). Manipulations of DNA, including electrophoresis, Southern blotting, and in situ colony hybridization, were carried out by standard procedures (40). For Southern and colony hybridization, DNA was radiolabeled with 50 μCi of [α-32P]dCTP (3,000 Ci/mmol) using a NonaPrimer kit from Appligene. Plasmid DNA was amplified in E. coli by using a QIAprep spin miniprep kit (Qiagen).
Restriction mapping and cloning procedures for the gyrA locus.
Genomic DNA was singly or doubly digested with various restriction enzymes. Restriction fragments were separated by electrophoresis, blotted to nylon membranes, and hybridized to α-32P-labeled probes under standard stringent conditions. Two DNA probes, MH3-MH4 and 321-322, corresponding to the 5′ and 3′ regions of gyrA, respectively, were generated by PCR amplification of the M. hominis genomic DNA with primers MH3 and MH4 (3) and with primers 321 and 322 (this study; see below), respectively. A restriction map was constructed from the hybridization patterns of genomic DNA obtained with each of the two probes.
M. hominis PG21 genomic DNA was then digested with EcoRI, BglII, or BglII-HindIII. The fragments were ligated to the linearized vector dephosphorylated and digested with EcoRI, BamHI, or BamHI-HindIII, respectively. After transformation of E. coli by ligation mixtures, recombinant clones containing the gyrA sequences were selected by colony hybridization with the [α-32P]dCTP-labeled MH3-MH4 or 321-322 DNA fragments. Hybridization-positive clones were selected, and their plasmid content was determined. Three recombinant plasmids, pMH3.1, pMH14.1, and pMH2.20, containing M. hominis DNA inserts of 4.2, 3.7, and 0.95 kbp, respectively, were selected for sequencing studies.
PCR amplification.
PCRs were carried out with a Perkin-Elmer Cetus thermal cycler with 100 ng of template DNA for M. hominis PG21 or with 2 μl of a broth culture for trovafloxacin-resistant mutants and 1 μM each primer as described elsewhere (5). Primer sets MH3 and MH4, MH6 and MH7, MH11 and MH13, and MH27 and MH28, previously described (5), were used to amplify the gyrA, gyrB, parC, and parE QRDRs, respectively. The gyrA 3′-end probe from M. hominis PG21 was generated with primers 321 (5′-TTAACAAGCGATGGTGTTGC-3′) and 322 (5′-GATAATTTTCTGTCATTGTCTTC-3′). Amplification was achieved with an initial denaturation step of 10 min at 94°C; 40 cycles of 1 min at 94°C, 1 min at 55°C, and 1 min at 72°C; and a final 10-min extension step at 72°C.
DNA sequence analysis.
Double-stranded DNA was sequenced on both strands by using an ABI PRISM dRhodamine terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS and an ABI PRISM 377 sequencer (Perkin-Elmer Applied Biosystems) according to the manufacturer's instructions. Forward and reverse primers, flanking the multiple-cloning-site polylinker of the pGEM3zf(+) vector, as well as internal primers were used to obtain the complete sequences of the DNA inserts of the three recombinant plasmids. PCR products of the quinolone-resistant strains were directly sequenced after purification with a Wizard PCR Preps DNA purification system (Promega). Pairwise and multiple sequence alignments were done with ALIGNp, CLUSTAL W (Infobiogen), and BLAST (National Center for Biotechnology Information) software.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was performed as previously described by using restriction endonucleases BamHI, SalI, SmaI, and XhoI (4, 28). Fragments containing the gyrA gene were identified by using the [α-32P]dCTP-labeled MH3-MH4 DNA fragment as a probe.
Antimicrobial agents and determination of MICs.
Antibiotics were purchased from the following manufacturers: norfloxacin, Merck Sharp & Dohme, Roma, Italy; pefloxacin and sparfloxacin, Rhône-Poulenc-Rorer, Vitry-sur-Seine, France; ofloxacin, Hoechst Marion Roussel, Romainville, France; ciprofloxacin, Bayer-Pharma, Puteaux, France; and trovafloxacin, Pfizer, Orsay, France. The MICs of the fluoroquinolones were determined by the agar dilution method as previously described (2).
In vitro selection of trovafloxacin-resistant mutants.
Stepwise selection of trovafloxacin-resistant mutants was performed by plating approximately 2 × 107 color-changing units of strain PG21 onto Hayflick modified agar medium containing increasing inhibitory concentrations of trovafloxacin. After 48 h of incubation at 37°C, resistant colonies were grown in broth medium without antibiotic and used for the next round of selection. The frequency of mutation was determined as the number of colonies appearing on the plate with antibiotic divided by the number of colonies in the inoculum.
Nucleotide sequence accession number.
The DNA sequence corresponding to the gyrA-encompassing fragment has been assigned GenBank accession no. AF242654.
RESULTS AND DISCUSSION
Cloning and organization of the gyrA locus.
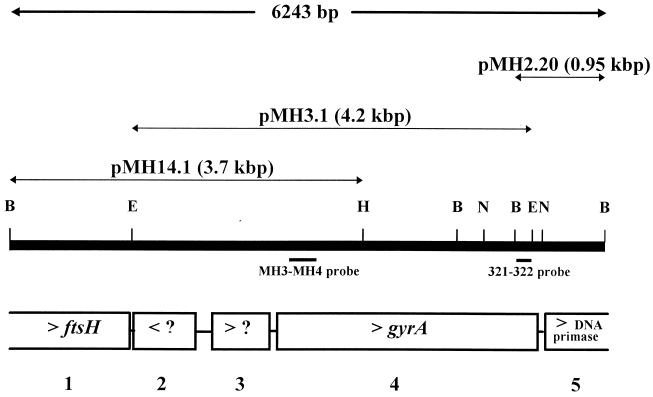
Southern blot hybridization of M. hominis genomic DNA with probes MH3-MH4 and 321-322 revealed that most of the gyrA gene was contained in a 4.2-kbp EcoRI fragment. By using combinations of single and double digests of the DNA with the enzymes BglII, EcoRI, HindIII, and NsiI, we established the restriction map of the gyrA gene region. As indicated in Fig. 1, the 3.7-kbp BglII-HindIII fragment overlapping the 4.2-kbp EcoRI fragment was found to contain the 5′ end of gyrA. These two fragments were recovered from genomic libraries of M. hominis by in situ colony hybridization, and the respective recombinant plasmids, pMH3.1 and pMH14.1, obtained were sequenced (Fig. 1). DNA probe 321-322, corresponding to the 3′ end of the M. hominis sequenced fragment, was chosen to clone the 3′ end of the gyrA gene and its flanking regions. The 0.95-kbp BglII fragment containing the gyrA 3′ end was cloned in E. coli, and recombinant plasmid pMH2.20 (Fig. 1) was sequenced.
FIG. 1.
Restriction map and organization of the M. hominis PG21 gyrA locus. E, EcoRI; H, HindIII; B, BglII; N, NsiI. ?, hypothetical gene. ORFs are numbered 1 to 5. Arrowheads indicate the transcription sense of the ORFs. Sizes of DNA inserts are indicated in parentheses.
Sequencing of the inserts of the three recombinant plasmids allowed the characterization of a 6,243-bp genomic DNA fragment of M. hominis (Fig. 1). This sequence was found to contain five putative open reading frames (ORFs) (ORF1 to ORF5). Three ORFs (ORF1, ORF4, and ORF5) were functionally assigned, based on significant sequence similarities to genes encoding proteins with known functions from other organisms (Fig. 1). ORF4, nucleotides 2798 to 5530, was assigned as the gyrA gene of M. hominis (see below).
Sequence analysis of the M. hominis GyrA subunit.
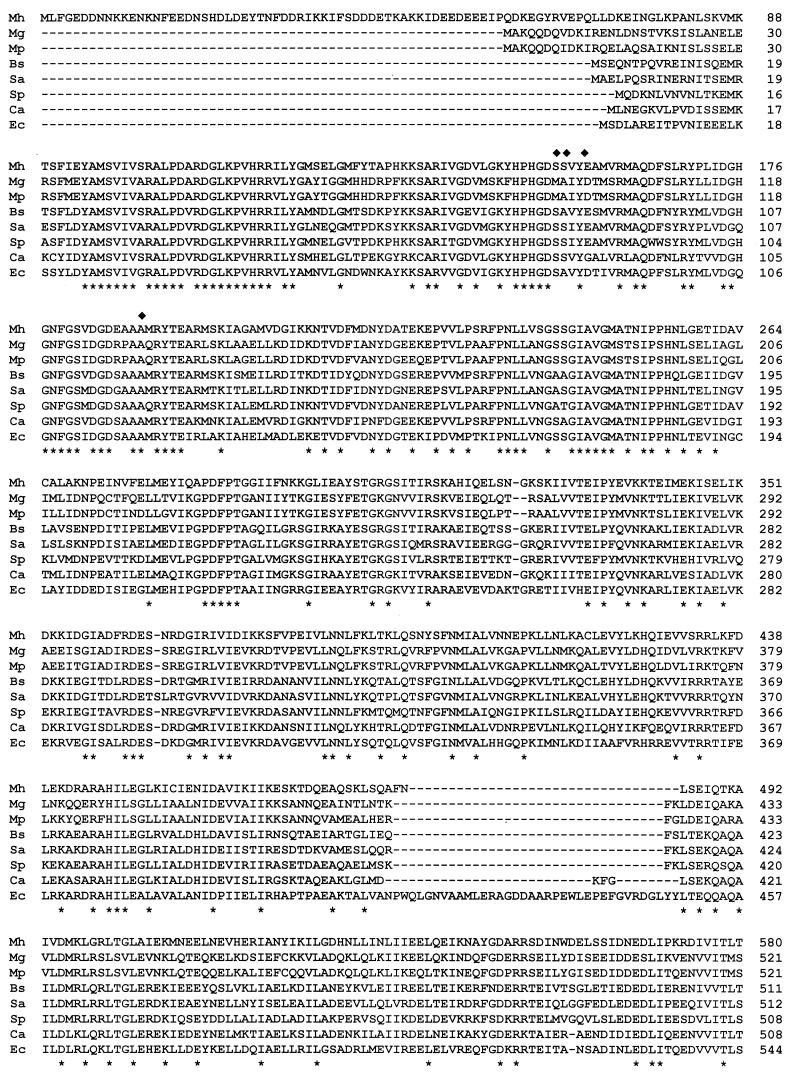
The predicted GyrA polypeptide contains 911 amino acids (aa) and has a calculated molecular mass of 102.5 kDa. It contains only one UGA-encoded tryptophan residue and has a G+C content of 31%. AT-rich sequences characteristic of putative −10 and −35 promoter sequences and a putative ribosome binding site were found upstream of the ATG initiation codon. The M. hominis GyrA subunit, with 911 residues, seems to be the largest GyrA subunit sequenced so far among organisms related to gram-positive bacteria. Only three gram-negative bacteria have a larger GyrA protein, Neisseria gonorrhoeae (916 aa) (8), Aeromonas salmonicida (922 aa) (33), and Pseudomonas aeruginosa (923 aa) (27). It is noteworthy that M. hominis ParC, 866 aa long, is also the largest topoisomerase IV ParC subunit known (4). Compared to other GyrA subunits, the M. hominis protein contains an additional stretch of 58 aa at the N-terminal end (see Fig. 3).
FIG. 3.
Alignment of the M. hominis (Mh) GyrA amino acid sequence with those of its counterparts in M. genitalium (Mg) (16), M. pneumoniae (Mp) (22), B. subtilis (Bs) (26), S. aureus (Sa) (30), S. pneumoniae (Sp) (1), C. acetobutylicum (Ca) (44), and E. coli (Ec) (42). An asterisk indicates a residue identical in all eight proteins. Residues involved in quinolone resistance in M. hominis are indicated by diamonds. Dashes indicate gaps.
Compared to the GyrA and ParC proteins of M. genitalium (16), M. pneumoniae (22), Ureaplasma urealyticum (accession no. AF222894), Bacillus subtilis (26), Staphylococcus aureus (13, 30), Streptococcus pneumoniae (1, 34), Clostridium acetobutylicum (44), and E. coli (38, 42), the M. hominis GyrA polypeptide exhibits a higher percentage of identity with the GyrA subunits than with the ParC subunits. The identity of M. hominis GyrA with the other GyrA proteins varies between 39.2% (E. coli) and 47.8% (U. urealyticum), while its identity with the ParC proteins ranges from 28.3% (M. pneumoniae) to 35.4% (B. subtilis). Like the topoisomerase IV ParC and ParE subunits (4), M. hominis GyrA shows higher homology with its counterpart in the gram-positive bacteria B. subtilis (45% identity), S. pneumoniae (44.1%), and S. aureus (42.9%) than with that in the gram-negative bacterium E. coli (39.2%).
Among the eubacteria, the best identity score, found with C. acetobutylicum GyrA, is in agreement with the phylogenetic origin of the Mollicutes, believed to have arisen from ancestors of low-G+C-content gram-positive bacteria, such as Clostridium (48). In a comparison with other human mycoplasmas, M. hominis GyrA was found to share 47.8, 45.3, and 43.9% identical amino acids with the GyrA subunits of U. urealyticum, M. genitalium, and M. pneumoniae, respectively. From these data, we assigned the ORF4-encoded polypeptide as the GyrA subunit of M. hominis. An overall identity of 33.3% was found between the GyrA and ParC peptide sequences of M. hominis. This percentage is lower than that of the GyrB-ParE comparison (44.2%) (4).
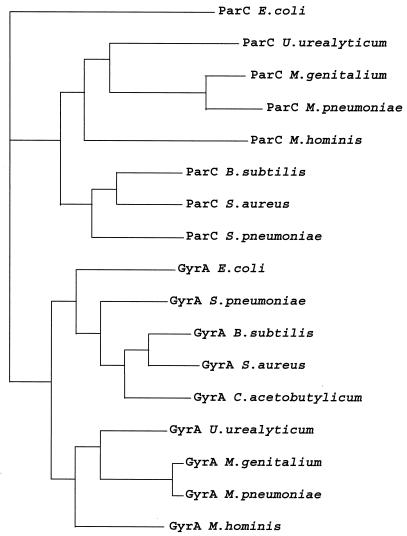
A protein tree was constructed from the GyrA and ParC sequences of the nine bacteria listed above. As shown in Fig. 2, the GyrA and ParC sequences clearly clustered in two groups. The topoisomerase II sequences of the gram-positive bacteria with low G+C contents (B. subtilis, S. aureus, S. pneumoniae, and C. acetobutylicum) and of the class Mollicutes (M. pneumoniae, M. genitalium, U. urealyticum, and M. hominis) formed differentiated clusters, as previously shown by Huang (24). For mycoplasmas, phylogenetic data obtained with topoisomerase II are in good agreement with those obtained with 16S rRNA (29). Indeed, M. pneumoniae GyrA and M. genitalium GyrA belong to the same phylogenetic group, while M. hominis GyrA and U. urealyticum GyrA form two distinct groups. As in the 16S rRNA tree, U. urealyticum formed on group and M. pneumoniae and M. genitalium formed another group arising from the same branch.
FIG. 2.
Protein tree for full-length GyrA and ParC subunits from nine bacteria: M. hominis (4; this study), M. genitalium (16), M. pneumoniae (22), U. urealyticum, B. subtilis (26), S. aureus (13, 30), S. pneumoniae (1, 34), C. acetobulylicum (44), and E. coli (38, 42). The tree was compiled by using the CLUSTAL W multiple-alignment program.
In Fig. 3, the GyrA amino acid sequence of M. hominis was compared to those of M. genitalium, M. pneumoniae, B. subtilis, S. aureus, S. pneumoniae, C. acetobutylicum, and E. coli. The highest homology among the GyrA proteins of all eight bacteria is located at the N-terminal moiety, while the C-terminal region is much less conserved. Amino acid residues Ser153, Ser154, and Glu157 of M. hominis are the equivalents of Ser83, Ser84, and Glu87 of E. coli GyrA, which have been shown to be hot spots for quinolone resistance. Indeed, we have reported substitutions of these three amino acids in fluoroquinolone-resistant mutants of M. hominis selected in vivo and in vitro (3, 5, 7).
Location of the DNA gyrase (gyrA) gene on the genomic map of M. hominis PG21.
PFGE and Southern blot hybridization with probe MH3-MH4 containing the gyrA QRDR of M. hominis confirmed that the gyrA gene was located within SmaI, BamHI, XhoI, and SalI genomic DNA fragments of 100, 84.5, 124, and 410 kbp, respectively (4, 28). Hence, the gyrA gene is located within the 74-kpb region where these restriction fragments overlap. It is noteworthy that this region is quite distant from the topoisomerase II genes gyrB and parC-parE.
In many bacteria, gyrB lies close to the origin of replication, where genes are organized in the following order: dnaA, dnaN, recF, gyrB, and gyrA. A similar gene organization has been described for M. pneumoniae (22) and M. genitalium (16). In U. urealyticum, for which the complete genomic sequence is now available (http://genome.microbio.uab.edu/uu/), gyrB and gyrA are contiguous and are located between dnaN and recA but about 100 kbp downstream from dnaA. However, in M. hominis, gyrA and gyrB were shown not to be coupled. Instead, gyrA mapped at least 35 kbp downstream of gyrB (28). These results were confirmed by our PFGE data showing a gyrA gene 47 and 31 kbp distant from gyrB and parE-parC, respectively (data not shown). Furthermore, we found the following gene organization around the gyrA gene; ftsH homolog, hypothetical MG347-ORF homolog, gyrA homolog, and dnaE homolog. In M. genitalium and M. pneumoniae, ftsH and dnaE homologs are located within 30- and 40-kbp regions, respectively, surrounding the origin of replication. In addition, the M. hominis DNA primase motif homolog found downstream from gyrA shared homology with only the 3′-end parts of other bacterial dnaE or dnaG genes. These genes encode DNA primases that synthesize small RNA primers at replication forks during DNA synthesis. In M. genitalium (16) and M. pneumoniae (22), they are located close to the origin of replication. Such a situation in M. hominis, with gyrA being found between ftsH and dnaE homologs but not downstream from the gyrB and dnaA regions, could be explained by chromosomal rearrangement. Thus, it is tempting to speculate that M. hominis gyrB and gyrA were first contiguous and located in the vicinity of the initiation site of replication, before being separated by chromosomal rearrangement during evolution. It should be noted that M. hominis oriC has not yet been identified.
Characterization of trovafloxacin-resistant mutants selected in a stepwise manner.
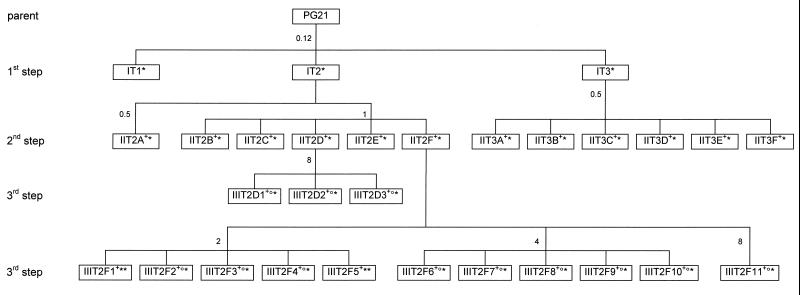
Trovafloxacin-resistant mutants were selected stepwise in vitro to further examine the role of topoisomerase IV and DNA gyrase in the development of resistance. The scheme used for the selection of trovafloxacin-resistant mutants is summarized in Fig. 4. Following this procedure, three independent sets of first- and second-step mutants and four independent sets of third-step mutants were obtained with mutation frequencies ranging from 10−7 to 10−4. Except for the 10−4 frequency obtained with mutant IT3 on a trovafloxacin concentration corresponding to the MIC, mutation frequencies were similar for all steps and all concentrations used for selection (one to four times the MIC). These frequencies were relatively high (1 × 10−7 to 2.5 × 10−6) and were equivalent to those found for the selection of ofloxacin- and sparfloxacin-resistant mutants (5, 25), confirming the high rate of mutation of M. hominis (12). The 29 trovafloxacin-resistant strains obtained were characterized for their susceptibilities to six fluoroquinolones and for the QRDR status of their gyrA, parC, gyrB, and parE genes (Table 1).
FIG. 4.
Relationships between M. hominis PG21 and fluoroquinolone-resistant mutants IT1 to IIIT2F11 selected by stepwise exposure to trovafloxacin. The numbers outside the boxes indicate the trovafloxacin concentrations (in micrograms per milliliter) used in the selection steps. The superscripts +, °, and ∗ indicate the presence of mutations in GyrA, ParC, and ParE, respectively (Table 1).
TABLE 1.
Characteristics of trovafloxacin-selected mutants of M. hominis
| Strain | Amino acid change at the indicated position in the QRDR ofa:
|
MIC (μg/ml)b of:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GyrA
|
ParC
|
ParE
|
TVA | SPX | CIP | OFX | NOR | PEF | ||||||
| 153 | 157 | 189 | 91 | 92 | 95 | 446 | 466 | |||||||
| PG21 (reference strain) | Ser | Glu | Ala | Ser | Ser | Glu | Leu | Glu | 0.06 | 0.06 | 2 | 1 | 32 | 4 |
| First-step mutants | ||||||||||||||
| IT1 | —c | — | — | — | — | — | — | Lys | 0.25 | 0.12 | 8 | 2 | 128 | 4 |
| IT2 | — | — | — | — | — | — | — | Lys | 0.25 | 0.12 | 8 | 2 | 128 | 4 |
| IT3 | — | — | — | — | — | — | — | Lys | 0.25 | 0.12 | 8 | 2 | 128 | 4 |
| Second-step mutants | ||||||||||||||
| IIT2A | — | — | Val | — | — | — | — | Lys | 0.5 | 0.12 | 8 | 1 | 128 | 4 |
| IIT2B | Leu | — | — | — | — | — | — | Lys | 2 | 1 | 32 | 2 | 128 | 4 |
| IIT2C | Leu | — | — | — | — | — | — | Lys | 2 | 1 | 32 | 2 | 128 | 4 |
| IIT2D | Leu | — | — | — | — | — | — | Lys | 2 | 1 | 32 | 2 | 128 | 4 |
| IIT2E | Leu | — | — | — | — | — | — | Lys | 2 | 1 | 32 | 2 | 128 | 4 |
| IIT2F | Leu | — | — | — | — | — | — | Lys | 2 | 1 | 32 | 2 | 128 | 4 |
| IIT3A | — | — | Glu | — | — | — | — | Lys | 1 | 0.5 | 16 | 2 | 128 | 4 |
| IIT3B | — | — | Glu | — | — | — | — | Lys | 1 | 0.5 | 16 | 2 | 128 | 4 |
| IIT3C | — | Lys | — | — | — | — | — | Lys | 1 | 1 | 16 | 2 | 128 | 4 |
| IIT3D | — | — | Glu | — | — | — | — | Lys | 1 | 0.5 | 16 | 2 | 128 | 4 |
| IIT3E | — | — | Glu | — | — | — | — | Lys | 1 | 0.5 | 16 | 2 | 128 | 4 |
| IIT3F | — | — | Glu | — | — | — | — | Lys | 1 | 0.5 | 16 | 2 | 128 | 4 |
| Third-step mutants | ||||||||||||||
| IIIT2D1 | Leu | — | — | — | Pro | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2D2 | Leu | — | — | Ile | — | — | — | Lys | 16 | 32 | 128 | 64 | >128 | 32 |
| IIIT2D3 | Leu | — | — | — | — | Gln | — | Lys | 8 | 8 | 32 | 16 | >128 | 16 |
| IIIT2F1 | Leu | — | — | — | — | — | Phe | Lys | 16 | 32 | 128 | 64 | >128 | 32 |
| IIIT2F2 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | 128 | 64 | >128 | 32 |
| IIIT2F3 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F4 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F5 | Leu | — | — | — | — | — | Phe | Lys | 16 | 64 | 128 | 64 | >128 | 32 |
| IIIT2F6 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F7 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F8 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F9 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F10 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | >128 | 64 | >128 | 32 |
| IIIT2F11 | Leu | — | — | Ile | — | — | — | Lys | 16 | 64 | 128 | 64 | >128 | 32 |
GyrA, ParC, and ParE residue positions are based on the respective gene sequences for M. hominis (4; this study). GyrA positions 153, 157, and 189 correspond to E. coli coordinates 83, 87, and 119, respectively. ParC positions 91, 92, and 95 correspond to E. coli coordinates 80, 81, and 84, respectively. ParE positions 446 and 466 correspond to E. coli coordinates 440 and 460, respectively.
TVA, trovafloxacin; SPX, sparfloxacin; CIP, ciprofloxacin; OFX, ofloxacin; NOR, norfloxacin; PEF, pefloxacin.
—, identical to that in the reference strain, PG21.
For each of the first-step mutants, IT1 to IT3, there was only a ParE Glu466→Lys change associated with a fourfold significant increase in the MICs of trovafloxacin, ciprofloxacin, and norfloxacin, and there was no significant increase (one- to twofold) in the MICs of sparfloxacin, ofloxacin, and pefloxacin. A twofold increase usually is considered not significant within experimental error for the twofold dilution method used for MIC determinations.
When first-step mutants IT2 and IT3 were used as parental strains, 12 second-step mutants were selected; all had acquired an additional GyrA substitution. Six of them, IIT2B to IIT2F, bearing a Ser153→Leu substitution, and IIT3C, bearing a Glu157→Lys substitution, showed four- to eightfold increases in the MICs of trovafloxacin, sparfloxacin, and ciprofloxacin but no changes in the MICs of ofloxacin, norfloxacin, and pefloxacin. The six remaining mutants, IIT2A, IIT3A, IIT3B, and IIT3D to IIT3F, all carried an amino acid change at Ala189 in GyrA. For second-step mutants harboring the Ala→Glu change, the MICs of trovafloxacin, sparfloxacin, and ciprofloxacin were two- to fourfold higher than those for their parental strains. Surprisingly, mutant IIT2A did not shown any significant fluoroquinolone MIC increase even though it had acquired a Ala→Val substitution at the same position, 189.
Finally, 14 third-step mutants generated from two different parental strains, IIT2D and IIT2F (ParE Glu466→Lys and GyrA Ser153→Leu), all had acquired an additional alteration in topoisomerase IV subunits, either ParC or ParE. Ten of them, IIIT2D2, IIIT2F2 to IIIT2F4, and IIIT2F6 to IIIT2F11, were found to carry the ParC Ser91→Ile substitution, while mutants IIIT2D1 and IIIT2D3 had Ser92→Pro and Glu95→Gln changes, respectively. Except for strain IIIT2D3, all of these third-step mutants were characterized by significant increases in the MICs of trovafloxacin (8-fold), sparfloxacin (32- to 64-fold), and ciprofloxacin (≥4-fold) and especially by dramatic increases in the MICs of ofloxacin (32-fold and pefloxacin (8-fold). Strain IIIT2D3, bearing the Glu95→Gln substitution, exhibited globally two- to eightfold smaller increases in the MICs of the quinolones tested, compared to the other third-step parC mutant strains. Two mutants, IIIT2F1 and IIIT2F5, harbored an additional substitution in ParE, corresponding to a Leu446→Phe change and associated with the same fluoroquinolone MIC increases as those seen with the ParC Ser91-mutated third-step mutants. It should be noted that the increased ofloxacin and pefloxacin MICs were found associated only with ParC and ParE amino acid changes at position 446 and not with the ParE substitution at position 466, contained in first-step mutants.
In summary, the development of a high level of resistance to trovafloxacin (MIC, ≥16 μg/ml) in M. hominis occurred in three steps, each associated with a mutation in the topoisomerase gene, beginning with a ParE alteration and involving alternating changes in DNA gyrase and topoisomerase IV.
Recent studies with the gram-positive bacteria S. aureus and S. pneumoniae indicated that different quinolones can have different preferential targets, depending on the bacterial species and on whether the studies are based on genetic or biochemical enzymatic data (9, 13, 15, 18, 20, 31, 32, 35–37, 43, 45). Trovafloxacin was reported to initially target topoisomerase IV by several genetic and enzymatic data for both S. aureus (14, 20) and S. pneumoniae (18, 19, 39, 45). For M. hominis, a low-G+C-content organism related to gram-positive bacteria, we and others showed by genetic studies that the primacy for fluoroquinolones of the target enzyme seemed to be drug specific (5, 25). DNA gyrase is the primary target of sparfloxacin, whereas topoisomerase IV is the primary target of pefloxacin, ofloxacin, and ciprofloxacin. In this study, we have determined the target specificity of trovafloxacin in M. hominis through analysis of mutants selected in a stepwise manner. All the first-step trovafloxacin-resistant mutants harbored a change in the ParE QRDR, while gyrA mutations were detected only in second-step mutants. These results indicate that, in M. hominis, as in S. pneumoniae or S. aureus, topoisomerase IV is the primary target of trovafloxacin.
GyrA and ParC mutations selected by trovafloxacin in M. hominis were predominantly those described previously for other fluoroquinolones. GyrA positions Ser153 (Ser83) and Glu157 (Asp87) (E. coli coordinates) and ParC positions Ser91 (Ser80) and Ser92 (Ala81) were found to be hot spots for quinolone resistance in many bacteria (23) and were already described as being mutated in M. hominis (5, 7, 25). In contrast, GyrA Ala189 (Ala119), ParC Glu95 (Glu84), and ParE Leu446 (Leu440) and Glu466 (Glu460) substitutions are novel. The GyrA Ala119→Val or Glu substitution was previously described for quinolone-resistant isolates of Salmonella enterica serovar Typhimurium (21). In M. hominis, only the Ala→Glu amino acid change was associated with significant increases in fluoroquinolone MICs. One explanation could be the charge difference induced by the amino acid change. Indeed, the Ala→Val substitution does not lead to a change in the residue charge (Ala and Val are both nonpolar), while the Ala→Glu change substitutes a nonpolar residue with a larger, negatively charged one. The new Glu84→Gln substitution in ParC has also been found to occur at the same position in the GyrA subunit of S. pneumoniae clinafloxacin-resistant mutants (36).
In contrast to the GyrA and ParC changes, to our knowledge, the ParE mutations acquired in the first- and third-step mutants have never been reported. First, the Glu466→Lys change in the first-step mutants does not occur in the EGDSA and PLRGK stretches designed as the GyrB QRDR (49). However, this position is located in a motif already associated with fluoroquinolone resistance. An Asn470→Asp mutation, 2 aa upstream from Glu466, has been described for S. aureus ParE (15). Moreover, the Glu474→Lys substitution recently described for S. pneumoniae GyrB (36) corresponds to the amino acid position just before Glu466 in M. hominis. The ParE Glu466 alteration, occurring in all M. hominis mutants, may be significant in quinolone resistance. A provocative experiment would be to point mutagenize back to wild type the ParE position 466 Lys mutants to see if the effect on the ParE subunit affects trovafloxacin resistance. The second mutation found in ParE, Leu446→Phe, lies in the second QRDR motif, PLRGK. Two ParE alterations have already been described for this motif; they concern the proline residue—Pro451→Ser or Gln in S. aureus (20, 41) and Pro454→Ser in S. pneumoniae (36). These data confirm that GyrB or ParE QRDR limits may require extension compared to the first QRDR, described for E. coli (49). It is interesting that, while the Glu466 substitution was associated with resistance to trovafloxacin, ciprofloxacin, and norfloxacin, only the substitution of Leu446 led to significant increases in the MICs of ofloxacin and pefloxacin. The functional role of the ParE QRDR is unknown, but it is tempting to speculate that some mutations could interfere with quinolone action, depending on the molecule and the mutated position.
The results described here indicate that only one mutation in parE or parC is necessary to reach a high level of resistance (MIC, ≥8 μg/ml) of M. hominis to ciprofloxacin, norfloxacin, and pefloxacin. In contrast, for sparfloxacin and trovafloxacin, at least three sequential mutational events, two in topoisomerase IV and one in DNA gyrase, are required to lead to high-level resistance in M. hominis, as previously described for trovafloxacin resistance in S. aureus (14) and coagulase-negative staphylococci (11). However, for sparfloxacin, the presence of GyrA Ser83 and ParC Ser80 or Glu84 mutations was shown to be associated with high-level resistance in sparfloxacin-selected mutants of M. hominis (5, 25). Furthermore, as previously reported for M. hominis in vitro and clinical isolates resistant to fluoroquinolones (6), our data suggest that trovafloxacin could retain activity against parE and parE-gyrA mutants (MICs, 0.12 to 2 μg/ml).
In conclusion, these data clearly confirm the enhanced activity of new fluoroquinolones, such as trovafloxacin, against mycoplasmas and indicate that susceptibility testing with ciprofloxacin or ofloxacin would not suffice to evaluate the activity of this antimicrobial class against these microorganisms. Furthermore, knowing the complete sequences of the four topoisomerase genes of M. hominis is the starting point for further enzymatic studies of DNA gyrase or topoisomerase IV preferential targeting of different fluoroquinolones.
ACKNOWLEDGMENTS
This study was supported in part by a grant from Pfizer and a grant from Pôle Aquitaine Santé.
We thank John Glass and Gail Cassell from the University of Alabama at Birmingham for kindly providing gyrase and topoisomerase IV sequences of U. urealyticum serovar 3. The complete genome sequence is available at the following website: http://genome.microbio.uab.edu/uu/. We thank Joel Renaudin for helpful comments and critical reading of the manuscript.
REFERENCES
- 1.Balas D, Fernandez-Moreira E, De La Campa A G. Molecular characterization of the gene encoding the DNA gyrase A subunit of Streptococcus pneumoniae. J Bacteriol. 1998;180:2854–2861. doi: 10.1128/jb.180.11.2854-2861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bébéar C, Robertson J. Determination of minimal inhibitory concentration. In: Tully J G, Razin S, editors. Molecular and diagnostic procedures in mycoplasmology. II. San Diego, Calif: Academic Press, Inc.; 1996. pp. 189–199. [Google Scholar]
- 3.Bébéar C M, Bové J M, Bébéar C, Renaudin J. Characterization of Mycoplasma hominis mutations involved in resistance to fluoroquinolones. Antimicrob Agents Chemother. 1997;41:269–273. doi: 10.1128/aac.41.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bébéar C M, Charron A, Bové J M, Bébéar C, Renaudin J. Cloning and nucleotide sequences of the topoisomerase IV parC and parE genes of Mycoplasma hominis. Antimicrob Agents Chemother. 1998;42:2024–2031. doi: 10.1128/aac.42.8.2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bébéar C M, Renaudin H, Charron A, Bové J M, Bébéar C, Renaudin J. Alterations in topoisomerase IV and DNA gyrase in quinolone-resistant mutants of Mycoplasma hominis obtained in vitro. Antimicrob Agents Chemother. 1998;42:2304–2311. doi: 10.1128/aac.42.9.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bébéar C M, Renaudin H, Charron A, Gruson D, Lefrançois M, Bébéar C. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob Agents Chemother. 2000;44:2557–2560. doi: 10.1128/aac.44.9.2557-2560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bébéar C M, Renaudin J, Charron A, Renaudin H, de Barbeyrac B, Schaeverbeke T, Bébéar C. Mutations in the gyrA, parC, and parE genes associated with fluoroquinolone resistance in clinical isolates of Mycoplasma hominis. Antimicrob Agents Chemother. 1999;43:954–956. doi: 10.1128/aac.43.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belland R J, Morrison S G, Ison C, Huang W M. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol Microbiol. 1994;14:371–380. doi: 10.1111/j.1365-2958.1994.tb01297.x. [DOI] [PubMed] [Google Scholar]
- 9.Blanche F, Cameron B, Bernard F X, Maton L, Manse B, Ferrero L, Ratet N, Lecoq C, Goniot A, Bisch D, Crouzet J. Differential behaviors of Staphylococcus aureus and Escherichia coli type II DNA topoisomerases. Antimicrob Agents Chemother. 1996;40:2714–2720. doi: 10.1128/aac.40.12.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drlica K, Zhao X L. DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol Mol Biol Rev. 1997;61:377–392. doi: 10.1128/mmbr.61.3.377-392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubin D T, Fitzgibbon J E, Nahvi M D, John J F. Topoisomerase sequences of coagulase-negative staphylococcal isolates resistant to ciprofloxacin or trovafloxacin. Antimicrob Agents Chemother. 1999;43:1631–1637. doi: 10.1128/aac.43.7.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dybvig K, Voelker L L. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- 13.Ferrero L, Cameron B, Manse B, Lagneaux D, Crouzet J, Famechon A, Blanche F. Cloning and primary structure of Staphylococcus aureus DNA topoisomerase IV: a primary target of fluoroquinolones. Mol Microbiol. 1994;13:641–653. doi: 10.1111/j.1365-2958.1994.tb00458.x. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgibbon J E, John J F, Delucia J L, Dubin D T. Topoisomerase mutations in trovafloxacin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:2122–2124. doi: 10.1128/aac.42.8.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier B, Hooper D C. Mutations in topoisomerase IV and DNA gyrase of Staphylococcus aureus: novel pleiotropic effects on quinolone and coumarin activity. Antimicrob Agents Chemother. 1998;42:121–128. doi: 10.1128/aac.42.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 17.Freundt E A. Culture media for classic mycoplasmas. In: Razin S, Tully J G, editors. Methods in mycoplasmology. Vol. 1. New York, N.Y: Academic Press, Inc.; 1983. pp. 127–135. [Google Scholar]
- 18.Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gootz T D, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Polzer R J. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gootz T D, Zaniewski R P, Haskell S L, Kaczmarek F S, Maurice A E. Activities of trovafloxacin compared with those of other fluoroquinolones against purified topoisomerases and gyrA and grlA mutants of Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43:1845–1855. doi: 10.1128/aac.43.8.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griggs D J, Gensberg K, Piddock L J. Mutations in gyrA gene of quinolone-resistant Salmonella serotypes isolated from humans and animals. Antimicrob Agents Chemother. 1996;40:1009–1013. doi: 10.1128/aac.40.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hooper D C. Bacterial topoisomerases, anti-topoisomerases, and anti-topoisomerase resistance. Clin Infect Dis. 1998;27:S54–S63. doi: 10.1086/514923. [DOI] [PubMed] [Google Scholar]
- 24.Huang W M. Bacterial diversity based on type II DNA topoisomerase genes. Annu Rev Genet. 1996;30:79–107. doi: 10.1146/annurev.genet.30.1.79. [DOI] [PubMed] [Google Scholar]
- 25.Kenny G E, Young P A, Cartwright F D, Sjostrom K E, Huang W M. Sparfloxacin selects gyrase mutations in first-step Mycoplasma hominis mutants, whereas ofloxacin selects topoisomerase IV mutations. Antimicrob Agents Chemother. 1999;43:2493–2496. doi: 10.1128/aac.43.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Danchin A, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 27.Kureishi A, Diver J M, Beckthold B, Schollaardt T, Bryan L E. Cloning and nucleotide sequence of Pseudomonas aeruginosa DNA gyrase gyrA gene from strain PAO1 and quinolone-resistant clinical isolates. Antimicrob Agents Chemother. 1994;38:1944–1952. doi: 10.1128/aac.38.9.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ladefoged S A, Christiansen G. Sequencing analysis reveals a unique gene organization in the gyrB region of Mycoplasma hominis. J Bacteriol. 1994;176:5835–5842. doi: 10.1128/jb.176.18.5835-5842.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniloff J. Phylogeny of mycoplasmas. In: Maniloff J, McElhaney R N, Finch L R, Baseman J, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C.: American Society for Microbiology; 1992. pp. 549–559. [Google Scholar]
- 30.Margerrison E E, Hopewell R, Fisher L M. Nucleotide sequence of the Staphylococcus aureus gyrB-gyrA locus encoding the DNA gyrase A and B proteins. J Bacteriol. 1992;174:1596–1603. doi: 10.1128/jb.174.5.1596-1603.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrissey I, George J. Activities of fluoroquinolones against Streptococcus pneumoniae type II topoisomerases purified as recombinant proteins. Antimicrob Agents Chemother. 1999;43:2579–2585. doi: 10.1128/aac.43.11.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munoz R, De La Campa A G. ParC subunit of DNA topoisomerase IV of Streptococcus pneumoniae is a primary target of fluoroquinolones and cooperates with DNA gyrase A subunit in forming resistance phenotype. Antimicrob Agents Chemother. 1996;40:2252–2257. doi: 10.1128/aac.40.10.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppegaard H, Soren H. Cloning and nucleotide sequence of the DNA gyrase gyrA gene from the fish pathogen Aeromonas salmonicida. Antimicrob Agents Chemother. 1996;40:1126–1133. doi: 10.1128/aac.40.5.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pan X S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan X S, Fisher L M. Targeting of DNA gyrase in Streptococcus pneumoniae by sparfloxacin: selective targeting of gyrase or topoisomerase IV by quinolones. Antimicrob Agents Chemother. 1997;41:471–474. doi: 10.1128/aac.41.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan X S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan X S, Fisher L M. Streptococcus pneumoniae DNA gyrase and topoisomerase IV: overexpression, purification, and differential inhibition by fluoroquinolones. Antimicrob Agents Chemother. 1999;43:1129–1136. doi: 10.1128/aac.43.5.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng H, Marians K J. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J Biol Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 39.Pestova E, Beyer R, Cianciotto N P, Noskin G A, Peterson L R. Contribution of topoisomerase IV and DNA gyrase mutations in Streptococcus pneumoniae to resistance to novel fluoroquinolones. Antimicrob Agents Chemother. 1999;43:2000–2004. doi: 10.1128/aac.43.8.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 41.Schmitz F J, Jones M E, Hofmann B, Hansen B, Scheuring S, Luckefahr M, Fluit A, Verhoef J, Hadding U, Heinz H P, Kohrer K. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob Agents Chemother. 1998;42:1249–1252. doi: 10.1128/aac.42.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanberg S L, Wang J C. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of DNA gyrase. J Mol Biol. 1987;197:729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- 43.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ullmann S, Dürre P. Nucleotide sequence and molecular characterization of the DNA gyrase genes from Clostridium acetobutylicum. Anaerobe. 1996;2:239–248. [Google Scholar]
- 45.Varon E, Janoir C, Kitzis M D, Gutmann L. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:302–306. doi: 10.1128/aac.43.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waites K B, Taylor-Robinson D. Mycoplasma and ureaplasma. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. pp. 782–794. [Google Scholar]
- 47.Williamson D L, Renaudin J, Bové J M. Nucleotide sequence of the Spiroplasma citri fibril protein gene. J Bacteriol. 1991;173:4353–4362. doi: 10.1128/jb.173.14.4353-4362.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshida H, Bogaki M, Nakamura M, Yamanaka L M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrB gene of Escherichia coli. Antimicrob Agents Chemother. 1991;35:1647–1650. doi: 10.1128/aac.35.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]