Abstract
Ligamentum flavum (LF) hypertrophy (LFH) has been recognised as one of the key contributors to lumbar spinal stenosis. Currently, no effective methods are available to ameliorate this hypertrophy. In this study, human umbilical cord mesenchymal stromal cell-derived extracellular vesicles (hUCMSC-EVs) were introduced for the first time as promising vehicles for drug delivery to treat LFH. The downregulation of miR-146a-5p and miR-221-3p expressions in human LF tissues negatively correlated with increased LF thickness. The hUCMSC-EVs enriched with these two miRNAs significantly suppressed LFH in vivo and notably ameliorated the progression of transforming growth factor β1(TGF-β1)-induced fibrosis in vitro after delivering these two miRNAs to mouse LF cells. The results further demonstrated that miR-146a-5p and miR-221-3p directly bonded to the 3′-UTR regions of SMAD4 mRNA, thereby inhibiting the TGF-β/SMAD4 signalling pathway. Therefore, this translational study determined the effectiveness of a hUCMSC-EVs-based approach for the treatment of LFH and revealed the critical target of miR-146a-5p and miR-221-3p. Our findings provide new insights into promising therapeutics using a hUCMSC-EVs-based delivery system for patients with lumbar spinal stenosis.
Keywords: Ligamentum flavum hypertrophy, Fibrosis, Umbilical cord mesenchymal stromal cells, Extracellular vesicle, miR-146a-5p, miR-221-3p
Abbreviations: LF, Ligamentum flavum; LFH, Ligamentum flavum hypertrophy; hUCMSC-EVs, human umbilical cord mesenchymal stromal cell-derived extracellular vesicles; TGF-β1, transforming growth factor-β1; LSS, Lumbar spinal stenosis; ECM, extracellular matrix; SMAD, mothers against the decapentaplegic homolog; MRI, magnetic resonance imaging
Highlights
-
•
The downregulation of miR-146a-5p and miR-221-3p expressions were negatively correlated with the development of LFH.
-
•
MiR-146a-5p and miR-221-3p enriched in hUCMSC-EVs prevent the fibrosis of LF by targeting SMAD4.
-
•
hUCMSC-EVs are effective as bioactive vehicles to ameliorate the progression of LFH.
-
•
hUCMSC-EVs-based delivery system is a promising therapy for the patients with lumbar spinal stenosis.
1. Introduction
Lumbar spinal stenosis (LSS) is one of the most common spinal diseases in the elderly worldwide. The prevalence rates of relative and absolute stenoses increase with age, reaching 47.2% and 19.4%, respectively, in the 60-to-69-year-old group [1]. The typical symptoms of LSS are intermittent claudication, back and leg pains, and limb numbness, which lead to severe disability in daily life and negative impacts on quality of life [2,3]. Intervertebral disk dehydration, annular tears, loss of disk height, and degenerative changes of the facet joint and ligamentum flavum (LF) have been determined to contribute to the progression of LSS [4]. LF hypertrophy (LFH) has been found to be one of the key contributors [5,6].
LF is an elastic connective tissue consisting of approximately 80% elastic fibres and 20% collagen fibres [6]. After the onset of hypertrophy, histopathological findings indicate that the elastic fibres are reduced and torn, with irregular arrangement, and that the collagen fibres are significantly proliferated, intercalating between the broken and reduced elastic fibres. The loss of elastic fibres and increase of collagen fibres in LF, that is, fibrotic changes, have been identified as the main pathological characteristics of LFH [6]. The repeated micro-injury caused by mechanical stress would induce chronic inflammation and the development of LF fibrosis, ultimately resulting in LFH. Increasing evidence has shown that inflammation is one of the most central events in fibrosis [7,8]. As a profibrotic cytokine, transforming growth factor β1 (TGF-β1) has been found to be associated with fibrosis in many organs and tissues [9,10]. Previous studies have found markedly upregulated TGF-β1 expression levels in the hypertrophied LF of LSS patients [11,12]. The increased synthesis of extracellular matrix (ECM) proteins after stimulation by TGF-β1 expression in LF cells [13] was observed, elucidating that the formation and accumulation of inflammatory cytokines are important pathological mechanisms of LF fibrosis.
The great properties of human umbilical cord mesenchymal stromal cells (hUCMSCs) make these cells a very effective tool in tissue repair and regeneration [[14], [15], [16]]. As a potential candidate for the treatment of fibrotic diseases, they have been successfully proven to suppress cartilage inflammation and to inhibit pulmonary, liver, and renal fibroses [[17], [18], [19]]. However, no data are available to investigate the role of hUCMSCs in preventing or ameliorating the development of LFH.
Recent studies have shown that most of the therapeutic benefits of MSCs come from the release of paracrine soluble factors [20], and extracellular vesicles (EVs) play a crucial role as natural carriers in intercellular communication [21]. As promising vehicles for drug delivery, MSC-derived EVs have been shown to possess attractive potential for the treatment of various diseases [19,22]. EVs contain proteins, mRNA, and miRNA, which can transfer functions between cells and affect protein expression in targeted cells [23]. Owing to their low tumorigenicity, immunogenicity, and ease of management, EVs could be a much safer therapeutic strategy paradigm than their parent cells in the treatment of fibrotic diseases. But whether the hUCMSC-EVs are able to prevent LFH is still unclear. Hence, the primary aim of this study was to determine the therapeutic potential of hUCMSC-EVs in the treatment of LFH.
The bioengineered EVs with the aim to improve the therapeutic effect have been increasingly explored; both the exogenous loading with the desired therapeutic cargo packaged into EVs by various manipulations such as co-incubation, electroporation and sonication, and the endogenous loading with genetically modifying the parental cell to upregulate/downregulate the target RNA or protein are efficient strategies for bioengineering EVs [24]. For instance, the engineered exosome achieved by co-incubation with 5-FU and miR-21 inhibitor by electroporation could effectively reverse drug resistance and significantly enhanced the cytotoxicity in 5-FU-resistant colon cancer cells [25]; recently, it was reported that the bone-targeted engineered exosomes (secreted by MSCs derived from human induced pluripotent stem cells) possessed the anti-osteoporosis effects after loading the siRNA of the Shn3 gene [26]. As one of the important cargoes in EVs, miRNAs are a class of endogenous small non-coding RNAs responsible for the post-transcriptional regulation of gene expressions [27]. Several microRNAs have been proven to be involved in the pathogenesis of LFH. In specimens from patients with hypertrophied LF, the miR-21 expression level increased, whereas the miR-221 expression level decreased [28,29]. Overexpression of miR-21 in LF cells could increase inflammatory cytokines level and collagen deposition [28]. Also, it was found that miR-155 could regulate the TGF-β pathway by directly targeting mothers against the decapentaplegic homolog (SMAD), eventually promoting LF fibrosis [30]. However, it is still unclear whether the specific microRNAs involved in the development of LFH could be loaded in the EVs as a new therapeutic strategy for LFH. Hence, the secondary aim was to confirm the roles of the engineered miRNAs obtained from hUCMSC-EVs in the progression of LFH and their underlying mechanisms of action.
Therefore, in this study, we firstly evaluated whether specific miRNAs were significantly changed in hypertrophied LF specimens. We found that miR-146a-5p and miR-221-3p were significantly downregulated in hypertrophied LF, but they could be enriched in the EVs harvested from hUCMSCs. After engineering EVs, the therapeutic potential in the treatment of LFH was determined using a bipedal standing mouse model and an induced fibrosis model of LF cells. Furthermore, the underlying mechanism, especially the TGF-β/SMAD4 pathway, was primarily investigated. This study would shed new light into the hUCMSC-EVs -based therapy for LFH, and unravel the therapeutic targets as well as the underlying signal pathway, offering the translational application in clinical practice.
2. Materials and methods
2.1. Ethical issues
This study was conducted according to the Declaration of Helsinki and it was approved by the institutional review board of the First Affiliated Hospital of Nanjing Medical University (2018-SR-182). All human LF specimens were harvested in the Department of Orthopedics of the First Affiliated Hospital of Nanjing Medical University. Written informed consent was obtained from all patients. The animal study was approved by the Institutional Animal Care and Use Committee of Nanjing Medical University (IACUC-1709021).
2.2. Human LF tissue collection
LF tissue samples were collected from the interlaminar region of L4/L5 of nineteen patients who were diagnosed with LSS and subsequently underwent decompressive laminectomy and lumbar interbody fusion (LFH group). Twenty-two patients who were diagnosed with lumbar disk herniation and subsequently underwent interlaminar fenestration were assigned to the control group. None of the patients from the LFH group and the control group were diagnosed with rheumatism or autoimmune diseases. The sociodemographic and clinical characteristics of the patients were described in Table 1 and Table S1. All LF specimens were collected for histological staining and RNA extraction.
Table 1.
Sociodemographic and clinical characteristics of the patients.
| Index | Control group | HLF group | P-value |
|---|---|---|---|
| Number of patients | 22 | 19 | - |
| Age (years) | 71.18 ± 9.62 | 71.84 ± 10.64 | 0.84 |
| Male/female | 10/9 | 6/13 | 0.19 |
| Level | L4/5 | L4/5 | - |
| LF thickness (mm) | 3.021 ± 0.45 | 6.565 ± 1.24 | <0. 01 |
2.3. LF thickness measurement
Before surgery, all patients received lumbar magnetic resonance imaging (MRI) scanning. The LF thickness was measured on axial T1-weighted MRI scans at the facet joint level [31]. Using the Picture Archiving and Communication System, the thickness of the LF was measured by two independent surgeons, and the average value of their measurements was considered as the final thickness.
2.4. Characterisation of hUCMSC-EVs
The hUCMSCs were purchased from Haixing Biosciences (Suzhou, Jiangsu, China). The identification of hUCMSCs is described in the supplementary materials. To prepare the hUCMSC-EVs, the cells were cultured with serum-free medium for 24 h at 37 °C and 5% CO2. The culture supernatants were collected and centrifuged at 300×g for 10 min to remove cells; the supernatant was centrifuged at 2000×g for 20 min and filtered using a 0.45 μm filter to remove cell debris. The filtered supernatant was collected and ultracentrifuged at 100,000×g at 4 °C for 90 min. The pellet was washed twice with PBS, followed by a second ultracentrifugation, and resuspended in PBS for further use.
A transmission electron microscope (JEM-1200EX, EOL Ltd., Tokyo, Japan) was used to determine the vesicle shape and size distribution. Zetasizer Nno ZS (Malvern Instruments Ltd., Malvern, UK) was used to measure the particle size distribution of the hUCMSC-EVs. NanoSight NS300 (Malvern Instruments Ltd., Malvern, UK) was used to quantify the number of hUCMSC-EVs by nanoparticle transport analysis in accordance with the operating instructions. Surface markers, including EV markers (CD63, CD73, and CD90) and the hUCMSCs marker (calreticulin), were analysed with Western blot.
2.5. hUCMSC-EVs uptake experiment
The hUCMSC-EVs were labelled with CM-DiI (red), ultracentrifuged at 100,000×g for 1 h to remove excess dye, and washed twice. Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). All reagents used were purchased from Invitrogen (Shanghai, China). A Nikon Eclipse Ti confocal laser scanning microscope was used to obtain images of EVs uptake.
2.6. Establishment of the mouse LFH model
Eight-week-old male mice (C57BL/6 background) with body weights ranging from 23 to 27 g were used in this study and housed at the Experimental Animal Center of Nanjing Medical University under a 12 h/12 h light/dark cycle. Food and water were provided to the animals ad libitum in their cages. Taking advantage of the hydrophobicity of the mice [32], the LFH model was established by inducing the mice to adopt a bipedal standing (BS) position for 6 h per day and to engage in free activity for 2 h at an interval. To verify the validity of the mouse LFH model, the LF area, the ratio of the elastic fibres to the collagen fibres, and the expression levels of Col-I, α-SMA, and Col-III were quantified after 10 weeks. Mice without any treatment were served as control. Two weeks after the bipedal standing model experiment, all the mice with LFH were randomly divided into 6 groups: (1) BS group (6 mice/group); (2) BS&EVs group (mice that received local injections of EVs [200 μg, 200 μl] after full exposure to LF as determined on microscopic examination, 6 mice/group); (3) BS&EVs-146a-inh group (mice that received local injections of EVs [200 μg, 200 μl] derived from hUCMSCs transfected with the miR-146a-5p inhibitor, 6 mice/group); (4) BS&EVs-221-inh group (mice that received local injections of EVs [200 μg, 200 μl] derived from hUCMSCs transfected with the miR-221-3p inhibitor; 6 mice/group); (5) BS&EVs-both-inh group (mice that received local injections of EVs [200 μg, 200 μl] derived from hUCMSCs transfected with both miRNA inhibitors, 6 mice/group); and (6) BS&EVs&OE SMAD4 group (mice that received local injections of hUCMSC-EVs [200 μg, 200 μl] and AAV2-SMAD4 [2.5 μl, 1.25 × 1012 vg/ml], 6 mice/group). After general anaesthesia, a 0.6-cm incision was made to expose the LF in the interlaminar region of L5/L6. EV or saline injections in the LF were performed using microliter micro-syringes (Hamilton, Switzerland) under microscopic guidance. The injection procedure was repeated every 2 weeks (injection time points: 1, 3, 5, 7, and 9 weeks after the bipedal standing experiment). After 10 weeks of modelling, all the mice were sacrificed, and samples from the intact L5/L6 vertebrae were harvested and used for the other experiments.
2.7. Isolation and culture of mouse LF cells
The 8-week-old male C57BL/6 mice, with body weights ranging from 23 to 27 g, were sacrificed under anaesthesia. Under aseptic conditions, the LF was carefully cut into 1-3 mm2 sections and digested sequentially in 0.2% type I collagenase diluted in a complete culture medium, including Dulbecco's modified Eagle's medium (Gibco, Shanghai, China) supplemented with 10% foetal bovine serum, penicillin/streptomycin, and 250 ng/mL amphotericin B (Gibco, Shanghai, China). After digestion for 17–24 h, the entire mixture was filtered through a 70-μm cell filter (Falcon, BD Biosciences, NJ, USA) and then placed in a 100-mm dish with a complete culture medium. At 80%–90% confluency, the cells were passaged using trypsin, seeded in various culture plates, and maintained in the incubator with 5% CO2 at 37 °C. All experiments were performed using LF cells at passages 2–4.
2.8. Cell viability assessment
The LF cells were cultured in a 96-well multiplate at 5 × 103 cells per well, followed by incubation with a complete culture medium in the absence or presence of various concentrations of hUCMSC-EVs or TGF-β1 for 24, 48, and 72 h after cell attachment. Cell Counting Kit-8 was applied to measure the absorbance at the 24, 48, and 72 h time points using SpectraMaxM (Molecular Devices, Shanghai, China), according to the manufacturer's instructions.
2.9. Protein extraction and western blot
Proteins were harvested from human LF tissues, mouse LF tissues, and cultured mouse LF cells using a protein extraction kit (Thermo, Massachusetts, USA). Each concentration was detected with a bicinchoninic acid protein assay kit (Beyotime, Shanghai, China). Western blot was performed in accordance with the procedure described in a previous study [33]. The same amount of total protein sample was separated using sodium dodecyl sulphate polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. After the membranes were blocked with a 5% skim milk solution and then incubated with disparate primary antibodies at 4 °C overnight, the membranes were incubated with species-matched second antibodies. Immunoreactive bands were visualised using an enhanced chemiluminescence Western blot detection kit (Beyotime, Shanghai, China) and analysed using the ImageJ software (NIH, USA). Table S2 presents a list of the primary and secondary antibodies used.
2.10. RNA isolation and analysis
Total RNAs from human LF tissues, mouse LF tissues, and cultured mouse LF cells were isolated using the Trizol reagent (Invitrogen) and reverse transcribed into complementary DNA using the HiScript III RT SuperMix for quantitative PCR (qPCR; R302-01, Vazyme, Nanjing, China), following the manufacturer's description. For miRNA, cDNA and quantitative reverse transcription PCR (qRT-PCR) syntheses were performed using the All-in-One miRNA qRT-PCR Detection Kit (GeneCopoeia, Rockville, MD, USA), following the manufacturer's protocol. Next, qPCR was conducted using ChamQ SYBR qPCR Master Mix (Low ROX Premixed; Q331-02, Vazyme) for mRNA and 2 × All-in-One qPCR Mix (GeneCopoeia) for miRNA based on the LightCycler96 PCR system (Roche, Inc., Switzerland). The mRNA quantification of qPCR was normalised to GAPDH expression, and the miRNA quantification was normalised to U48 expression. The comparative 2−△△CT method was used in the analysis. Table S3 presents a list of RT-qPCR primer sequences.
2.11. Histopathology and immunohistochemistry
The human and mouse LF samples collected were fixed in a 4% paraformaldehyde solution overnight at 4 °C; decalcified in an EDTA (ethylene diamine tetra-acetic acid)-glycerol solution for 14–21 days at 4 °C; successively dehydrated, hyalinised, and embedded in paraffin; and then sliced into 5-μm-thick sections for histological staining. The dewaxed and rehydrated sections were subjected to haematoxylin-eosin (H&E; Jiancheng, Nanjing, China) or EVG staining (Baso, Zhuhai, China), in accordance with the manufacturer's instructions. For immunohistochemical (IHC) staining, the sections were incubated overnight at 4 °C using the primary antibodies listed in Table S2. The sections were washed with PBS three times and successively incubated with biotin-conjugated secondary antibodies for an hour, Elite ABC (Vector, USA) for an hour, and diaminobenzidine (Vector) at room temperature. After counterstaining with haematoxylin, the sections were observed under a microscope.
All the images were photographed using the Leica LAS-X software, under the Leica DMi8 microscope. The IHC staining images were analysed with Image Pro Plus. In accordance with the previously described method [32], quantitative analyses of the LF areas and the ratio of the elastic fibres to the collagen fibres in mice or human LF samples were performed using the ImageJ software (NIH).
2.12. Immunofluorescence
The immunofluorescence (IF) assay was performed as previously described [33]. After washing with PBS, the LF cells were fixed with 4% paraformaldehyde and permeabilised with 0.5% Triton X-100 (T8787, Sigma-Aldrich, USA) for 30 min. After that, the cells were blocked with 5% bovine serum albumin (BSA, Sigma, USA) at room temperature for 1 h and incubated overnight at 4 °C with the specific antibodies listed in Table S2. After rinsing with PBS for three times, the appropriate Alexa Fluor 488-labelled secondary antibodies were used at room temperature, avoiding light, and the nucleus was dyed with DAPI (SouthernBiotech, Alabama, USA). Images were acquired using the Leica DMi8 fluorescent microscope with the Leica LAS-X software and analysed with the ImageJ software (NIH).
2.13. RNA sequencing and analysis
EVs were isolated using Cell Culture Media Exosome Purification kits (Norgen, CAN), and RNA was extracted using the Exosomal RNA Isolation Kit (Norgen), according to the manufacturer's instructions. The RNA quality was determined using Bioanalyzer 2100 with the RNA 6000 Nano LabChip Kit (Agilent, CA, USA). Approximately 10 ng of total RNA was used to prepare a small RNA library in accordance with the protocol for TruSeq Small RNA Sample Prep kits (Illumina, San Diego, CA, USA). Single-end sequencing (1 × 50 bp) was then performed using the Illumina Hiseq2500 at the LC-BIO (Hangzhou, China), following the vendor's recommended protocol. We utilized 2 algorithms (TargetScan 7.2 and miRanda 3.3a) to predict the genes targeted by the differentially expressed miRNAs; the VennDiagram was used to represent the common target genes predicted by both algorithms. Functional enrichment of GO (GeneOntology) terms (http://www.geneontology.org/) and KEEG (Kyoto Encyclopedia of Genes and Genomes) (http://www.genome.jp/kegg/) analyses were then performed to annotate these differentially expressed miRNA targets.
2.14. Cell transfection with miRNA mimics/inhibitor, Cy5-labelled miRNA mimics and plasmid
Hsa-miR-146a-5p and hsa-miR-221-3p mimics/mimics-negative control (NC) and their inhibitors/inhibitor-NC and Cy5-labelled miRNAs were synthesised and purified by GeneCopoeia. To prepare the engineered EVs, the hUCMSCs were plated onto 10-cm dishes; after 70–80% confluence, hUCMSCs were transfected with miR-146a-5p/miR-221-3p inhibitor or NC inhibitor (GeneCopoeia, Rockville, MD, USA) at a concentration of 50 nM using Lipofectamine 3000 in Opti-MEM (Invitrogen), and the cell culture medium was changed 6 h after transfection. All cell transfections were done according to the manufacturer's instructions. After 48 h transfection, the cell serum-free medium was used for 24 h; then the EVs were extracted according to previous description. The inhibition efficiency of miRNA in the EVs was detected by RT-qPCR.
A plasmid containing SMAD4 expression gene and control vector plasmids were produced by GeneChem (Shanghai, China). Mouse LF cells were transfected with plasmid DNA using X-treme GENE HP DNA Transfection Reagent (6366236001; Roche, Shanghai, China) and miRNA mimics/mimics NC/inhibitors/inhibitor NC using Lipofectamine 3000 in Opti-MEM (Invitrogen) according to the manufacturer's instructions.
Cy5-labelled miRNAs were transfected into hUCMSCs using Lipofectamine 3000 (Invitrogen), which was performed in agreement with as described above. hUCMSCs containing Cy5-miRNA were co-cultured with LF cells, and images were acquired with the Leica DMi8 fluorescent microscope and analysed with the ImageJ software.
2.15. Luciferase activity assay
The mouse LF cells were plated (5 × 104 cells per well) in 24-well plates overnight and then transfected with SMAD4 wild-type 3′UTR or mutant 3′UTR luciferase reporter vector and mimic-NC, miR-146a-5p mimic, miR-221-3p mimic, inhibitor-NC, miR-146a-5p inhibitor, or miR-221-3p inhibitor by using Lipofectamine 3000 (Invitrogen). Thirty-six hours after transfection, luciferase activity was detected using dual-luciferase luciferase reporter assay reagents (Promega Corp., Madison, Wisconsin, USA).
2.16. Adeno-associated virus gene transfer
The adeno-associated virus vector resulted in long-term, robust transgene expressions, with minimal toxicity, low immunogenicity, broad tropism, and ease of production in several animal models [34]. AAV2-mediated SMAD4 overexpression and vectors were produced by GeneChem (Shanghai, China). AAV2-SMAD4 was used for LF injection in the animal experiment.
2.17. Statistical analyses
All statistical analyses were performed using GraphPad Prism Version 7.0 (La Jolla, CA, USA). The results are shown as mean ± standard deviation (SD). The Student t-test or one-way analysis of variance (ANOVA) was employed for comparisons between groups. The Pearson r correlation coefficient was used to analyse the correlation between the miRNA expression levels and the LF thicknesses in the human tissue samples. The miRNA differential expression level based on normalised deep-sequencing counts was analysed by selectively using the Fisher exact test, chi-square 2 × 2 test, chi-squared N × N test, Student t-test, or ANOVA. P values < 0.05 were considered statistically significant.
3. Results
3.1. Development of LFH is strongly correlated with the decrease of miR-146a-5p and miR-221-3p
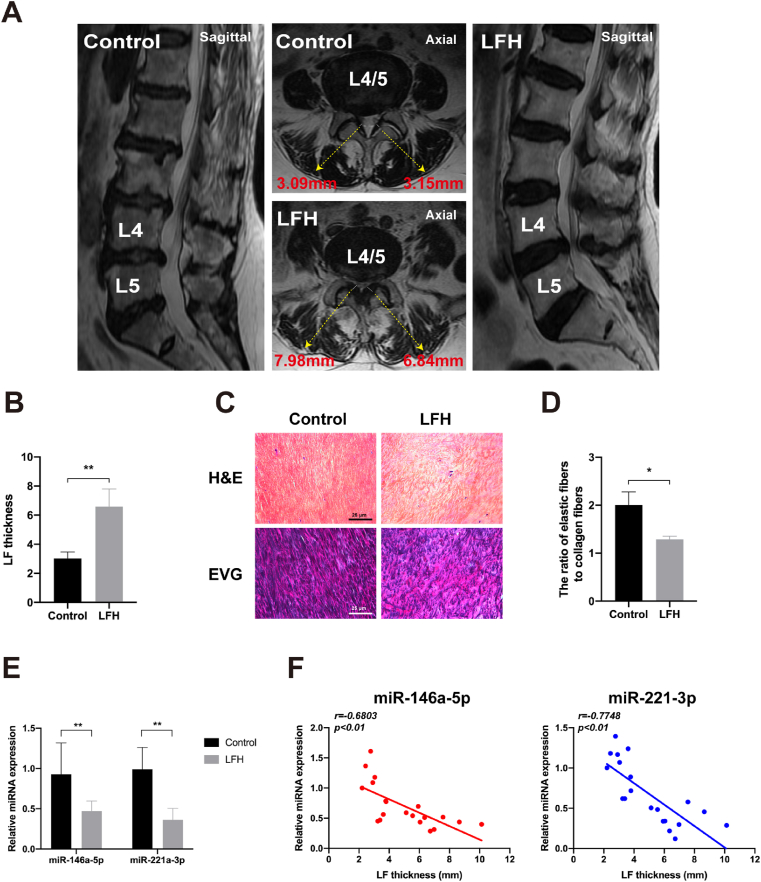
To investigate the correlation between miR-146a-5p and miR-221-3p expression levels and LF thickness, we measured the LF thickness on MRI scans and detected the expressions of miR-146a-5p and miR-221-3p using RT-qPCR. As shown in Fig. 1A, the representative MRI scans confirmed that LFH with a LF thickness of >5 mm affected most of the spinal canal and resulted in severe dural sac compression. Nevertheless, the normal LF did not compress the dural sac, and its thickness was <5 mm. In detail, as listed in Table 1 and Fig. 1B, the mean LF thickness in the LFH group was 6.565 ± 1.243 mm, much higher than that in the control group (3.021 ± 0.448 mm). The results of the H&E and EVG staining showed that in the LFH group, the elastic fibres decreased and the collagen fibres increased. The elastic fibres stained blue-black (EVG staining) were uneven, fragmented, irregularly arranged, disorganised, and partially absent. By contrast, in the control group, the elastic fibres were abundant and arrayed in a regular order, and the collagen fibres were few (Fig. 1C). The ratio of the elastic fibres to the collagen fibres in the LFH group was significantly higher than that in the control group (1.56-fold, P < 0.05; Fig. 1D). In addition, the RT-qPCR results showed that the miR-146a-5p and miR-221-3p expression levels in the LFH group were significantly lower than those in the control group (1.97-fold, P < 0.01; 2.73-fold, P < 0.01; Fig. 1E). Furthermore, the miR-146a-5p and miR-221-3p expression levels negatively correlated with LF thickness (r = −0.6803, r = −0.7748, P < 0.01; Fig. 1F). Taken together, our data indicated that miR-146a-5p and miR-221-3p strongly correlated with the development of LFH.
Fig. 1.
MiR-146a-5p and miR-221-3p negatively correlate with LF thickness in humans. (A) LF thickness measurement on representative coronal and sagittal T2-weighted MRI scans of patients diagnosed with lumbar disk herniation (LDH) and lumbar spinal stenosis (LSS). (B) Results of the quantitative analysis of LF thickness between the control and LFH groups. (C) Representative image of the H&E staining (scale bar: 200 μm) and EVG staining (scale bar: 200 μm) of the LF specimens from the control and LFH groups. (D) Results of the quantitative analysis of the ratio of the elastic fibre area to the collagen fibre area. (E) Results of the RT-qPCR analysis of the miRNA expressions of miR-146a-5p and miR-221-3p in the LF specimens of the control and LFH groups. (F) Results of the correlation analysis between miR-146a-5p and miR-221-3p expression levels and LF thickness (n = 20). Data are presented as mean ± SD, *P < 0.05, **P < 0.01, compared with the control group.
3.2. MiR-146a-5p and miR-221-3p are enriched in hUCMSC-EVs
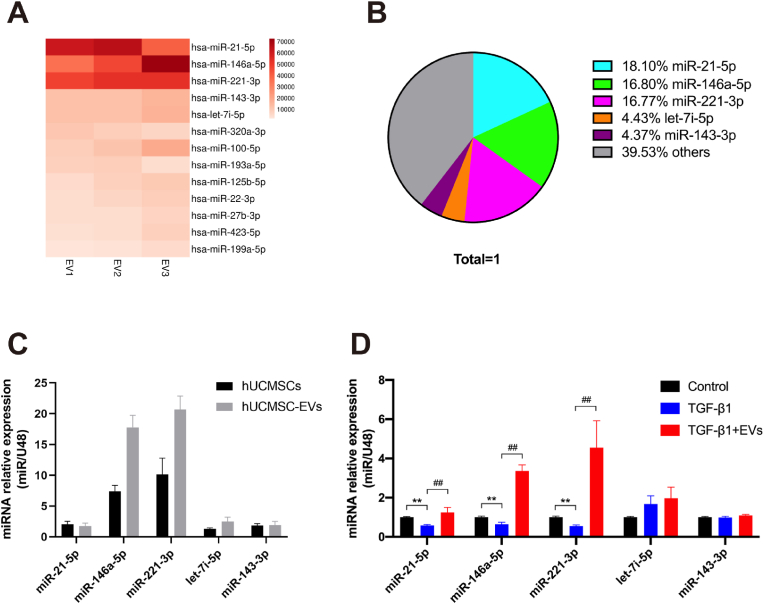
To identify and validate the miRNA expression profiles in EVs, the miRNAs in the EVs were screened using miRNA-seq. The hUCMSCs and hUCMSC-EVs were isolated and identified using optical microscopy, flow cytometry, transmission electron microscopy, nanoparticle tracking analysis, and Western blot analysis (Fig. S1). As shown in Fig. 2A and B, the top five most abundant miRNAs, namely miR-21-5p, miR-146a-5p, miR-221-3p, miR-143-3p, and let-7i-5p, accounted for approximately 60.47% of the total miRNA reads. The miR-221-3p and miR-146a-5p expression levels were highest in the hUCMSCs and hUCMSC-EVs groups, as determined by RT-qPCR (Fig. 2C).
Fig. 2.
MiR-146a-5p and miR-221-3p are enriched in hUCMSC-EVs. (A) Heat map of the top 10 most abundant miRNAs in hUCMSC-EVs by miRNA-seq. (B) Relative percentage of miRNAs in total miRNA reads. (C) Results of the RT-PCR analysis of the top five most abundant miRNAs in hUCMSCs and hUCMSC-EVs (n = 3). (D) Results of the RT-PCR analysis of the top five miRNAs in LF cells in the presence or absence of TGF-β1 and hUCMSC-EVs (n = 3). Data are presented as mean ± SD. *P < 0.05, **P < 0.01, compared with the control group. #P < 0.05, ##P < 0.01, compared with the TGF-β1 group.
To determine whether TGF-β1 could induce mouse LF cell fibrosis, we stimulated LF cells with different TGF-β1 concentrations for 3 days. As shown in Fig. S2, TGF-β1 significantly increased the viability of the LF cells in a time- and dose-dependent manner and activated ECM formation in the LF cells in a dose-dependent manner, effectively inducing the fibrosis of LF cells. Hence, we used 10 ng/ml TGF-β1 as the working concentration and 24 h as the stimulation duration in this study. We further measured the expression levels of the top five miRNAs in LF cells in the presence or absence of TGF-β1 and hUCMSC-EVs. Treatment with hUCMSC-EVs significantly increased the miR-146a-5p and miR-221-3p expression levels by 5.22- and 8.32-fold, respectively (P < 0.01; Fig. 2D). Thus, these data suggested that miR-146a-5p and miR-221-3p enriched in hUCMSC-EVs might play critical roles in the therapeutic effect of hUCMSC-EVs on TGF-β1-induced LF cell fibrosis.
3.3. hUCMSC-EVs suppress LFH in mice and ameliorate fibrosis in LF cells
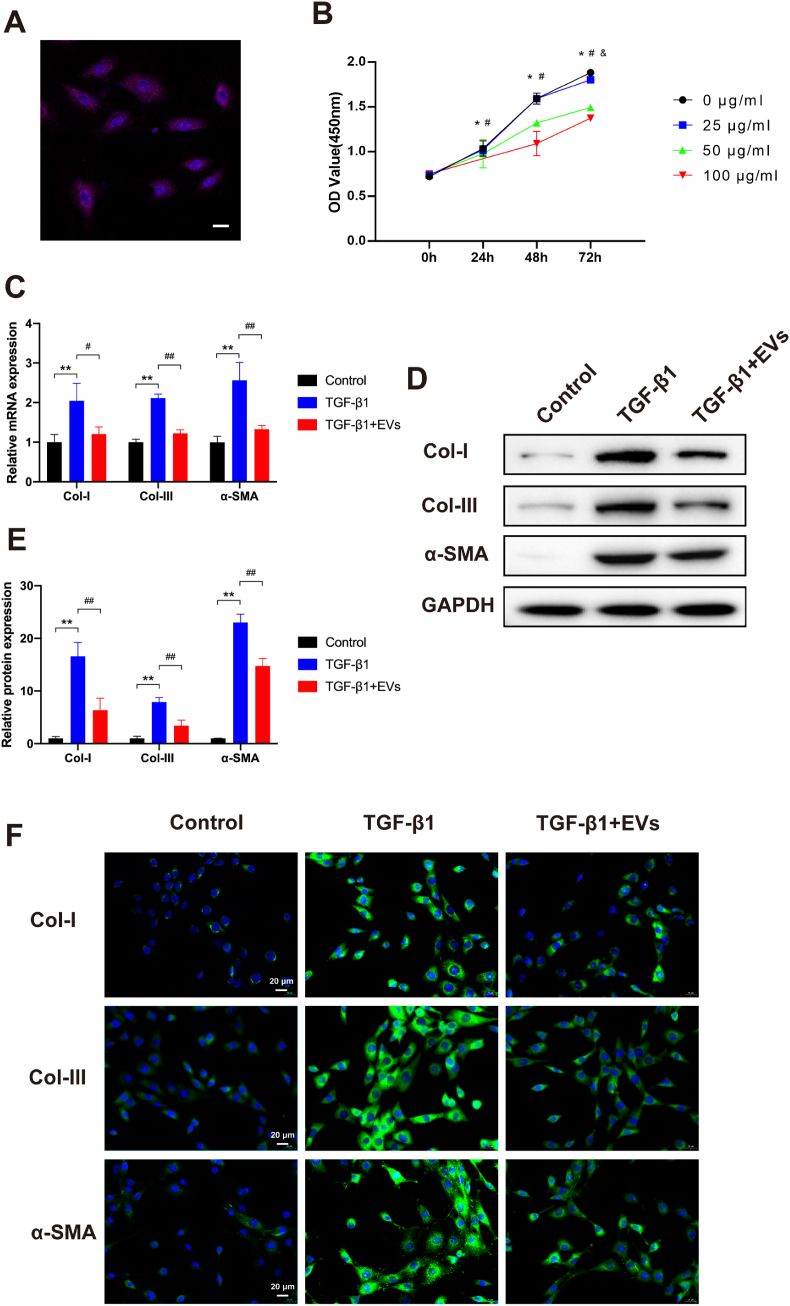
To investigate whether hUCMSC-EVs ameliorate the fibrosis of mouse LF cells in vitro, we stimulated passage 2–3 LF cells in the presence or absence of TGF-β1 and hUCMSC-EVs for 3 days. First, CM-DiI (red fluorescent lipophilic dye) was used to verify the internalisation ability of purified EVs. After the LF cells were incubated with the CM-DiI-labelled hUCMSC-EVs, red fluorescence was observed in the cytoplasm of the LF cells, which indicated that the hUCMSC-EVs could be internalised by LF cells (Fig. 3A). To investigate the effect of hUCMSC-EVs on cell proliferation, LF cells were stimulated with TGF-β1 (10 ng/ml) and different concentrations of hUCMSC-EVs (0, 25, 50, and 100 μg/ml) for 24, 48, and 72 h. No significant differences were found in any of the treatment groups before incubation with TGF-β1 and hUCMSC-EVs. As shown in Fig. 3B, suppression of cell viability was observed in the 50 and 100 μg/ml hUCMSC-EVs group at 24 h, in the 50 and 100 μg/ml hUCMSC-EVs groups at 48 h, and in all three groups at 72 h. We thus used 100 μg/ml hUCMSC-EVs as the working concentration and 24 h as the treatment duration for the following studies. The OD value of 100 μg/ml hUCMSC-EVs group at 24 h was 0.75 ± 0.03, which was much lower than that of 0 μg/ml hUCMSC-EVs group.
Fig. 3.
hUCMSC-EVs attenuate LF cell fibrosis. (A) Representative immunofluorescence images of CM-DiI (red)-labelled hUCMSC-EVs internalised by LF cells, the nuclei of which were stained with DAPI (blue). Scale bar: 20 μm. (B) Growth curves of LF cells at different hUCMSC-EVs concentrations (0, 25, 50, and 100 μg/ml) measured with the CCK8 assay at 24, 48, and 72 h (*P < 0.05, 0 μg/ml vs. 25 μg/ml; #P < 0.05, 0 μg/ml vs. 50 μg/ml; &P < 0.05, 0 μg/ml vs. 100 μg/ml). (C) Results of the RT-qPCR analysis of the mRNA expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and hUCMSC-EVs (n = 3). (D, E) Results of the Western blot analysis of the protein expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and hUCMSC-EVs (n = 3). (F) Representative immunofluorescence images of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and hUCMSC-EVs (n = 3). Data are presented as mean ± SD, *P < 0.05, **P < 0.01, compared with the control group. #P < 0.05, ##P < 0.01, compared with the TGF-β1 group. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Subsequently, RT-qPCR, western blot, and IF were performed to analyse the expression levels of fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and hUCMSC-EVs for 24 h. These results indicated that the messenger RNA (mRNA) and protein expression levels of Col-I, α-SMA, and Col-III were significantly increased in the TGF-β1 group compared with the control group but obviously decreased in the hUCMSC-EV group compared with the TGF-β1 group (Fig. 3C–F). Taken together, these data confirmed that hUCMSC-EVs ameliorated the TGF-β1-induced LF cell fibrosis.
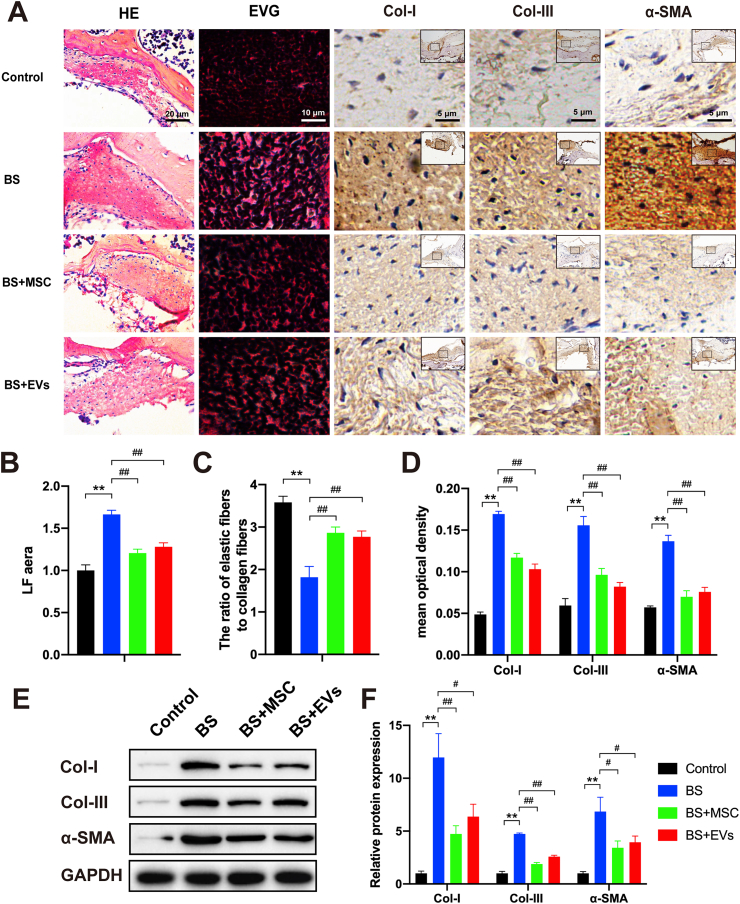
To investigate the effects of hUCMSCs and their EVs on the progression of LFH in vivo, a local injection of hUCMSCs or hUCMSC-EVs was used in the LFH group. No significant heterotopic ossification, infection or necrosis of the ligamentum flavum and surrounding muscle tissue were observed. The histological results indicated that in the 10-week bipedal standing mice, the average LF area was notably increased (1.67-fold, P < 0.01), the ratio of the elastic fibres to the collagen fibres was decreased (1.97-fold, P < 0.01), and the expression levels of Col-I, α-SMA, and Col-III were obviously increased as compared with those in the control group. These data indicated the high validity of the LFH mouse model. In the hUCMSCs and hUCMSC-EV groups (Fig. 4A–D), the average LF area and Col-I, α-SMA, and Col-III expression levels sharply decreased, and the ratio of the elastic fibres to the collagen fibres markedly increased compared with those in the LFH group. The Western blot analysis revealed that the expression levels of Col-I, α-SMA, and Col-III were significantly increased in the LFH group compared with the control group but were obviously decreased in the hUCMSCs and hUCMSC-EV groups compared with the LFH group, consistent with the IHC results (Fig. 4E and F). Taken together, these data confirmed that the hUCMSCs and hUCMSC-EVs effectively suppressed LFH formation in the bipedal standing mouse model.
Fig. 4.
hUCMSCs and their EVs suppress LFH in bipedal standing mice. (A) Representative image of the H&E staining (scale bar: 20 μm), EVG staining (scale bar: 10 μm), and immunohistochemistry staining of Col-I, α-SMA, and Col-III (scale bar: 5 μm) in LF specimens treated with hUCMSCs or their EVs in bipedal standing mice (n = 3). (B) Results of the quantitative analysis of the LF area. (C) Quantitative analysis of the ratio of the elastic fibre area to the collagen fibres area. (D) Results of the quantitative analysis of the Col-I-, α-SMA-, and Col–III–positive areas (n = 3). (E, F) Western blot analysis of the protein expressions of the fibrosis markers (Col-I, α-SMA, and Col-III; n = 3). Data are presented as mean ± SD. *P < 0.05, **P < 0.01, compared with the control group. #P < 0.05, ##P < 0.01, compared with the TGF-β1 group.
3.4. MiR-146a-5p and miR-221-3p could be delivered into LF cells via EVs and ameliorate TGF-β1-induced fibrosis
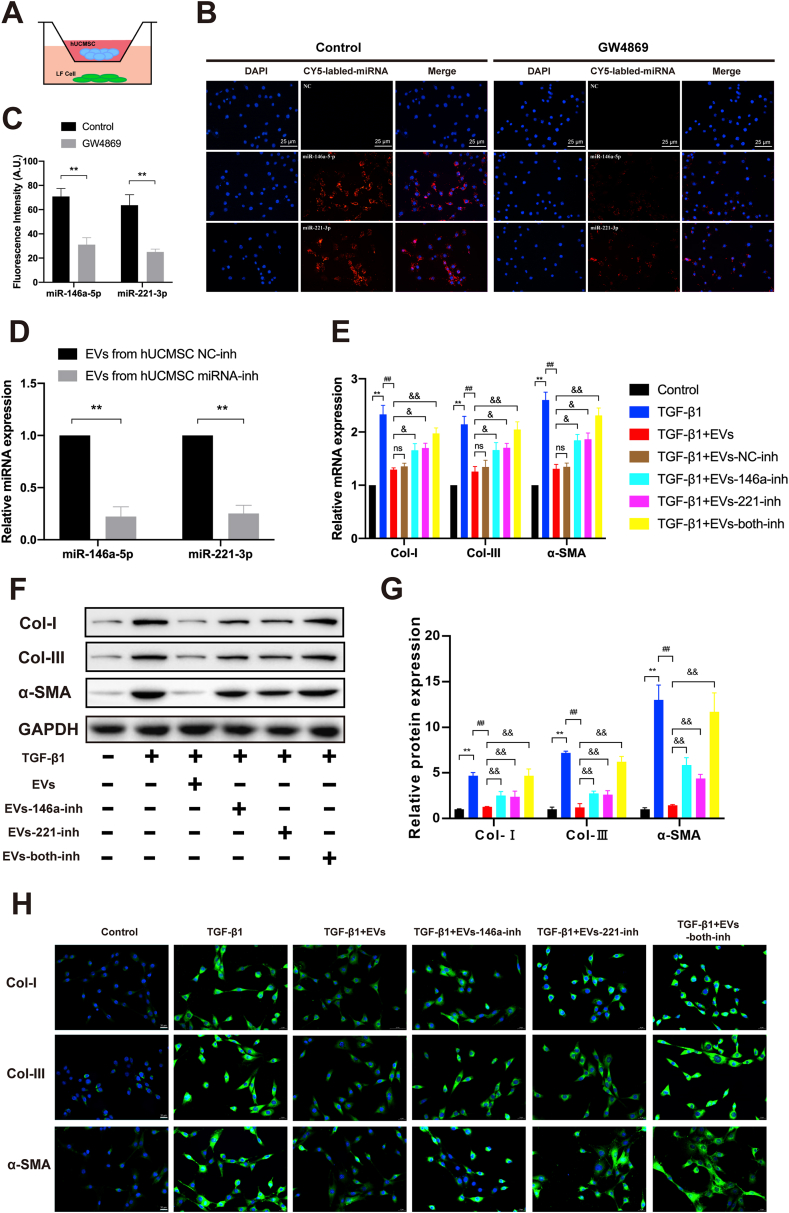
To further investigate whether miR-146a-5p and miR-221-3p could be delivered to LF cells, LF cells were co-cultured with hUCMSCs (Fig. 5A). The Cy5-labelled miR-146a-5p and miR-221-3p were first transfected into the hUCMSCs in the presence or absence of GW4869 (an inhibitor of EVs biogenesis/release) for 24 h. Subsequently, the LF cells were co-cultured with hUCMSCs using a transwell dish with a 0.4-μm pore size. After 48 h of co-culture, Cy5-positive LFs were detected using IF staining. The results showed that the fluorescence intensity in the control group was much higher than that in the GW4869 group, which confirmed that miR-146a-5p and miR-221-3p were transferred from hUCMSCs into LF cells via EVs (Fig. 5B and C). Thus, these data indicated that miR-146a-5p and miR-221-3p could be delivered into LF cells via EVs.
Fig. 5.
MiR-146a-5p and miR-221-3p could be delivered into LF cells via EVs and ameliorate TGF-β1-induced fibrosis. (A) Schematic diagram of the hUCMSC and LF cell co-culture system. (B) Representative immunofluorescence images of the Cy5-labelled miR-146 a-5p and miR-221–3p transfected into the hUCMSCs treated in the presence or absence of GW4869 (scale bar: 25 μm). (C) The results of the quantitative analysis of the average fluorescence intensity (n = 3) are also shown. (D) Results of the RT-qPCR analysis of miR-146a-5p and miR-221-3p expressions after the transfection of hUCMSCs with NC, miR-146a-5p, and miR-221-3p inhibitors. (E) Results of the RT-qPCR analysis of the mRNA expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and various modified hUCMSC-EVs (n = 3). (F, G) Western blot analysis of the protein expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and various modified hUCMSC-EVs (n = 3). (H) Representative immunofluorescence images of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and various modified hUCMSC-EVs (n = 3). Data are presented as mean ± SD. *P < 0.05, **P < 0.01, compared with the control group. #P < 0.05, ##P < 0.01, compared with the TGF-β1 group. &P < 0.05, &&P < 0.01, compared with the TGF-β1+EV group.
To further investigate the exact effects of miR-146a-5p and miR-221-3p enriched in the hUCMSC-EVs, the LF cells were stimulated with TGF-β1 (10 ng/ml) and various modified hUCMSC-EVs for 24 h. The hUCMSCs-EVs secreted from the hUCMSCs after being transfected with miR-146a-5p and miR-221-3p inhibitors were collected. RT-qPCR revealed that the miR-146a-5p and miR-221-3p expression levels were significantly decreased in the hUCMSC-EVs secreted from the hUCMSCs after being transfected with miR-146a-5p and miR-221-3p inhibitors, when compared with the expression levels in the hUCMSC-EVs secreted from the hUCMSCs without any modification (4.49-fold, P < 0.01; 3.97-fold, P < 0.01; Fig. 5D). RT-qPCR, Western blot, and IF were performed to analyse the expression levels of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1 and the modified hUCMSC-EVs. As illustrated in Fig. 5E–H, the results indicated that inhibition of miR-146a-5p and/or miR-221-3p in the EVs partially or almost completely diminished the therapeutic effect of hUCMSC-EVs on TGF-β1-induced LF cell fibrosis. Hence, these data confirmed that miR-146a-5p and miR-221-3p played critical roles in the therapeutic effect of hUCMSC-EVs on TGF-β1-induced LF cell fibrosis.
3.5. hUCMSC-EVs ameliorate LFH via inhibiting TGF-β/SMAD4 signaling
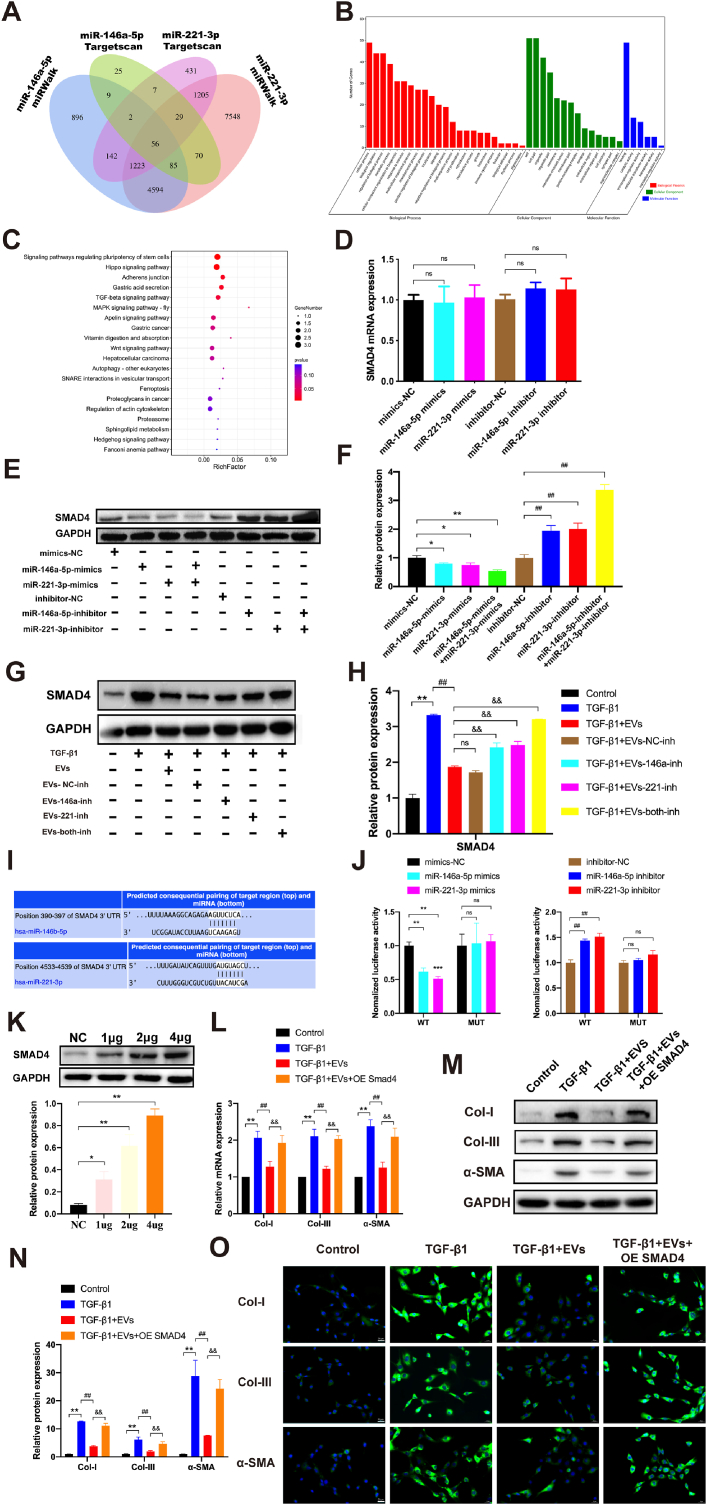
The potential targets of miR-146a-5p and miR-221-3p were predicted by in silico analysis using miRWalk (http://mirwalk.umm.uni-heidelberg.de/) and TargetScan (http://www.targetscan.org/vert_72/). As shown in Fig. 6A, the Venn diagram predicted that miR-146a-5p and miR-221-3p had 56 common targets with different algorithms. Analysis of the miRNA targets by GO revealed that 56 identified target genes were involved in diverse biological processes, including regulation of biological processes, biological regulation, regulation of cellular processes, protein binding, and metabolic processes (Fig. 6B). The pathway analysis indicated that the 56 targets were involved in six Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with significant false discovery rate-adjusted P values, including the Hippo and TGF-β signalling pathways (Fig. 6C). Among the 56 candidates, SMAD4 was selected for further analysis because of its relatively high target prediction score according to its two complementary structures with both miRNAs and GO and KEGG pathway enrichment in the bioinformatic analysis. Next, mimic-NC, miR-146a-5p mimics, miR-221-3p mimics, inhibitor-NC, miR-146a-5p inhibitor, or miR-221-3p inhibitor was transfected into the LF cells. As illustrated in Fig. 6D–F, the results showed that the SMAD4 protein expression level was markedly increased in the miR-146a-5p and miR-221-3p inhibitor-transfected LF cells but was decreased in the miR-146a-5p and miR-221-3p mimics-transfected LF cells. In addition, the co-transfection of both miRNAs mimics or inhibitors presented a stronger effect than when they were used alone. Nevertheless, the SMAD4 mRNA expression level showed no significant change, which suggested that miR-146a-5p and miR-221-3p could regulate SMAD4 expression at the post-transcriptional level in LF cells.
Fig. 6.
hUCMSC-EVs ameliorate TGF-β1-induced LF cell fibrosis by inhibiting TGF-β/SMAD4 signalling through the activities of miR-146a-5p and miR-221-3p. (A) A Venn diagram predicting all common targets of miRNAs with different algorithms. (B) Result of the Gene Ontology analysis of the putative target genes of miR-146a-5p and miR-221-3p. (C) Results of the Kyoto Encyclopedia of Genes and Genomes pathway analysis of the putative target genes of miR-146a-5p and miR-221-3p. (D) Results of the RT-qPCR analysis of the mRNA expression of SMAD4 in LF cells after transfection of miR-146a-5p and miR-221-3p mimics/mimics NC and their inhibitors/inhibitor NC (n = 3). (E, F) Results of the Western blot analysis of the protein expression of SMAD4 in LF cells after transfection of miR-146a-5p and miR-221-3p mimics/mimics NC and their inhibitors/inhibitor NC (n = 3). (G, H) Results of the Western blot analysis of the protein expression of SMAD4 in the presence or absence of TGF-β1 and various modified hUCMSC-EVs (n = 3). (I) Result of the bioinformatic analysis of the predicted binding site of miR-146a-5p and miR-221-3p targeting the 3′-UTR of SMAD4 in TargetScan (http://www.targetscan.org/vert_72/). (J) Luciferase activities detected using a dual-luciferase reporter assay system after mimic-NC, miR-146a-5p mimic, miR-221-3p mimic, inhibitor-NC, miR-146a-5p inhibitor, or miR-221-3p inhibitor and plasmid containing the wild-type or mutant 3′-UTR of SMAD4 were co-transfected into LF cells (n = 3). (K) Results of the Western blot analysis of the protein expression of SMAD4 with different amounts of transfected plasmid DNA (n = 3). (L) Results of the RT-qPCR analysis of the mRNA expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1, hUCMSC-EVs, and SMAD4 overexpression (n = 3). (M, N) Results of the Western blot analysis of the protein expressions of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1, hUCMSC-EVs, and SMAD4 overexpression (n = 3). (O) Representative immunofluorescence images of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1, hUCMSC-EVs, and SMAD4 overexpression (n = 3). Data are presented as mean ± SD. *P < 0.05, **P < 0.01, compared with the control group. #P < 0.05, ##P < 0.01, compared with the TGF-β1 group. &P < 0.05, &&P < 0.01, compared with the TGF-β1+EVs group.
After exposure of LF cells to TGF-β1, SMAD4 protein expression was markedly increased compared with that in the control group but was obviously decreased in the TGF-β1&EVs group compared with the TGF-β1 group. The protein expression levels were obviously increased in the TGF-β1&EVs-146a-inh, TGF-β1&EVs-221-inh, and TGF-β1&EVs-both-inh groups compared with the TGF-β1&EVs–NC–inh group. In addition, the EVs co-transfected with both miRNA inhibitors presented a much stronger increase than when transfected with a single inhibitor. No significant difference was observed between the TGF-β1&EVs and TGF-β1&EVs–NC–inh groups (Fig. 6G and H). The results indicated that the inhibition of miR-146a-5p and/or miR-221-3p in the EVs partially or almost completely impaired the down-regulation effect of hUCMSC-EVs on SMAD4 protein expression in LF cells.
Wild-type or miRNA binding-site mutant SMAD4 3′UTR-driven luciferase vector and both miRNA mimics/inhibitors/mimics NC/inhibitor NC were co-transfected into the LF cells to further identify whether miR-146a-5p and miR-221-3p could directly bind to the SMAD4 region. Compared with the control group, the overexpression of miR-146a-5p or miR-221-3p significantly suppressed the luciferase activity of wild-type SMAD4 3′UTR, while inhibition of miR-146a-5p or miR-221-3p enhanced luciferase activity. Nevertheless, the suppression and enhancement could be reversed by the mutant miR-146a-5p or miR-221-3p binding site of SMAD4 3′UTR (Fig. 6I and J). All these results indicated that miR-146a-5p and miR-221-3p directly targeted SMAD4 in LF cells.
To identify the role of SMAD4 in hUCMSC-EVs-mediated LF cell fibrosis, overexpressed SMAD4 and control vector plasmids (control vectors) were transfected into the LF cells. Upregulation of the SMAD4 plasmid was detected using Western blot. As shown in Fig. 6K, the SMAD4 protein expression was remarkably increased in a dose-dependent manner. Next, RT-qPCR, western blot, and IF were performed to analyse the expression levels of the fibrosis markers (Col-I, α-SMA, and Col-III) in the presence or absence of TGF-β1, hUCMSC-EVs, and SMAD4 overexpression. As illustrated in Fig. 6L–O, SMAD4 overexpression abolished the therapeutic effect of hUCMSC-EVs on TGF-β1-induced LF cell fibrosis, indicating that SMAD4 was involved in hUCMSC-EVs-mediated LF cell fibrosis. Thus, these data further confirmed that SMAD4 was the target of miR-146a-5p and miR-221-3p in hUCMSC-EVs and that SMAD4 overexpression could block the therapeutic effect of hUCMSC-EVs on the amelioration of TGF-β1-induced LF cell fibrosis.
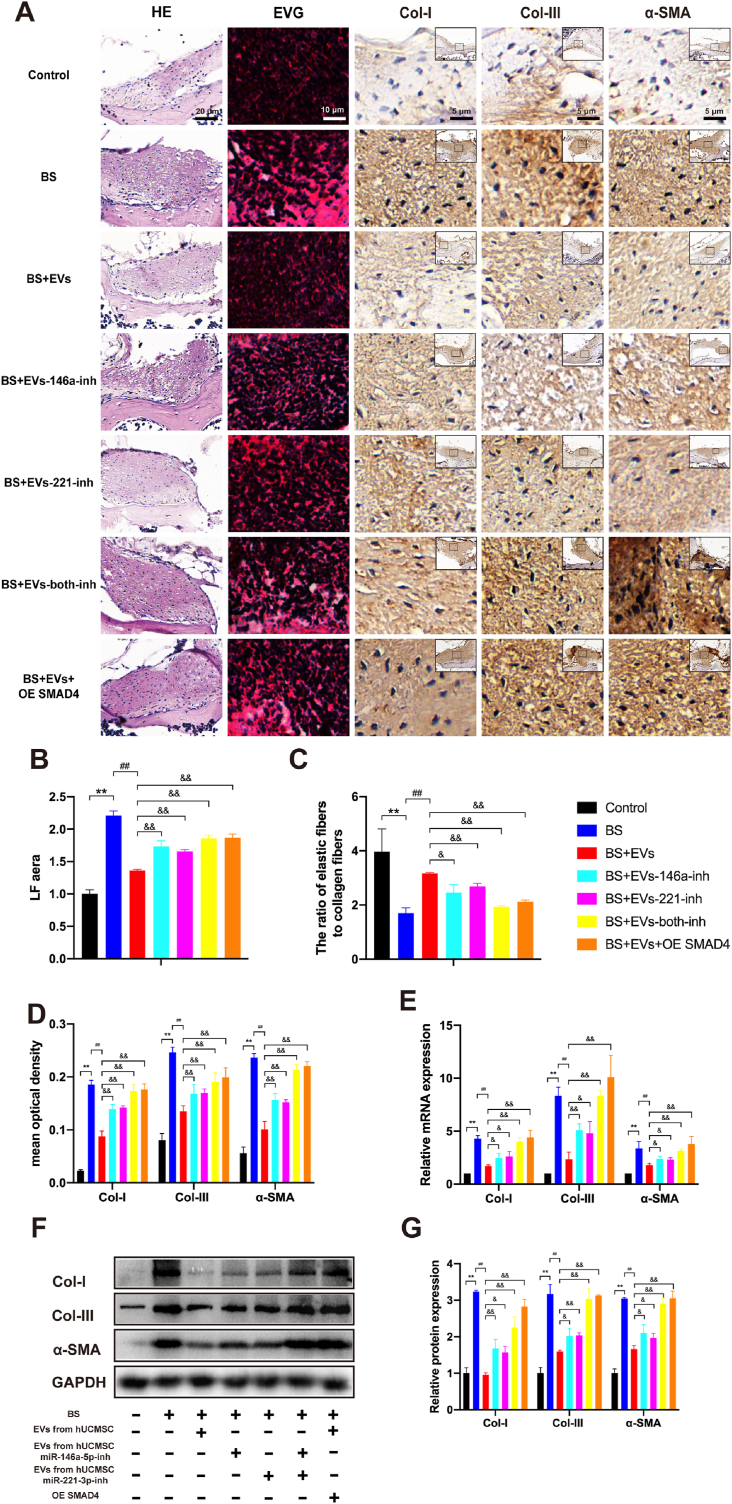
To identify the role of SMAD4 and miR-146a-5p- and miR-221-3p-enriched hUCMSC-EVs in the progression of LFH in vivo, hUCMSC-EVs with or without modifications or AAV2-SMAD4 were injected into the LF cells of bipedal standing mice. The previous data presented in Fig. 4 showed that hUCMSC-EVs could suppress LFH formation in the bipedal standing mouse model. Compared with the BS&EVs group, the BS&EVs-146a-inh, BS&EVs-221-inh, BS&EVs-both-inh, and BS&EVs&OE SMAD4 groups showed significantly increased average LF areas (1.27-, 1.22-, 1.36-, and 1.37-fold, respectively; P < 0.01). The ratios of the elastic fibres to the collagen fibres were decreased (0.78-, 0.85-, 0.61-, and 0.67-fold, respectively; P < 0.01), and the expression levels of Col-I (1.58-, 1.61-, 1.97-, and 2.00-fold, P < 0.01), α-SMA (1.24-, 1.26-, 1.41-, and 1.47-fold, respectively; P < 0.01), and Col-III (1.55-, 1.51-, 2.1-, and 2.18-fold, respectively; P < 0.01) were significantly increased. In addition, the EVs derived from the hUCMSCs co-transfected with both miRNA inhibitors presented a much stronger effect than when co-transfected with only one inhibitor (Fig. 7A–D). The RT-qPCR and Western blot analysis revealed that the mRNA and protein expression levels of Col-I, α-SMA, and Col-III exhibited the same tendency, consistent with the results of the IHC analysis (Fig. 7E, F, G). These results indicated that the inhibition of miR-146a-5p or miR-221-3p in the EVs partially diminished the therapeutic effect of hUCMSC-EVs via the suppression of LFH. If these two miRNAs expression in the EVs were inhibited simultaneously or SMAD4 was overexpressed in LF, the therapeutic effect of hUCMSC-EVs on LFH in vivo was almost abolished. Therefore, all these data demonstrated that hUCMSC-EVs suppressed LFH by inhibiting TGF-β/SMAD4 signalling through the activation of miR-146a-5p and miR-221-3p in the bipedal standing mice.
Fig. 7.
hUCMSC-EVs suppress LFH by inhibiting TGF-β/SMAD4 signalling through the activities of miR-146a-5p and miR-221-3p in bipedal standing (BS) mice. (A) Representative image of H&E staining (scale bar: 20 μm), EVG staining (scale bar: 10 μm), and immunohistochemistry staining of Col-I, α-SMA, and Col-III (scale bar: 5 μm) in LF specimens treated with hUCMSC-EVs with or without various modifications or AAV2-SMAD4 in bipedal standing mice (n = 3). Results of the quantitative analysis of the (B) LF area, (C) ratio of the elastic fibre area to the collagen fibre area, and (D) Col-I-, α-SMA-, and Col–III–positive areas. (E) Results of the RT-qPCR analysis of the mRNA expressions of the fibrosis markers (Col-I, α-SMA, and Col-III; n = 3). (F, G) Results of the Western blot analysis of the protein expressions of the fibrosis markers (Col-I, α-SMA, and Col-III; n = 3). Data are presented as mean ± SD. *P < 0.05, **P < 0.01, compared with the control group; #P < 0.05, ##P < 0.01, compared with the BS group; and &P < 0.05, &&P < 0.01, compared with the BS + EVs group.
4. Discussion
In this study, the therapeutic potential of a hUCMSC-EVs/miRNA-based strategy for the treatment of LFH and the underlying mechanism were primarily investigated. First, from the specimens of patients with LSS, we found that the miR-146a-5p and miR-221-3p expression levels were sharply downregulated and negatively correlated with LF thickness, which suggested that both miRNAs are key molecules in the development of LFH. As miR-146a-5p and miR-221-3p were the top two abundant miRNAs in hUCMSC-EVs, these findings thus encouraged us to employ the hUCMSC-EVs as a delivering system for the treatment of LFH. By using a bipedal standing mouse model, we demonstrated that hUCMSC-EVs enriched with miR-146a-5p and miR-221-3p suppressed LFH in vivo and ameliorated the progression of TGF-β1-induced fibrosis in mouse LF cells; these two findings provided important clues that miR-146a-5p and miR-221-3p were probably critical targets for the therapeutic effect of hUCMSC-EVs on the progression of LFH. Thus, the underlying mechanism was further investigated. We demonstrated that hUCMSC-EVs enriched with miR-146a-5p and miR-221-3p could be delivered to LF cells to reduce the risk of TGF-β1-induced fibrosis. Furthermore, by using the loss/gain-of-function method, we revealed that SMAD4 was the direct target of miR-146a-5p and miR-221-3p and that the downregulation of SMAD4 could prevent the proliferation and excessive deposition of connective tissue in the extracellular matrix.
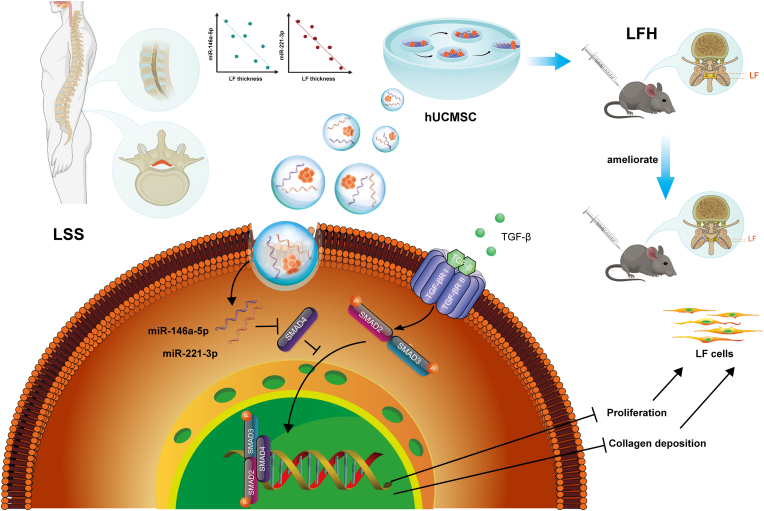
As summarised in Fig. 8, our findings indicated that hUCMSC-EVs could be used as a delivering system to ameliorate LFH by inhibiting the TGF-β/SMAD4 signalling through the activities of miR-146a-5p and miR-221-3p. To the best of our knowledge, this is the first translational study to determine the effectiveness of a hUCMSC-EVs-based approach for the treatment of LFH. Despite several studies regarding the modulation effect of miR-146a-5p on fibrosis could be found, this is the first one to determine that the miR-146a-5p and miR-221-3p delivered by engineered EVs could be administrated locally to suppress LFH in a mouse model and these two miRNAs have synergistic effect in the amelioration of LFH. This work further indicated that hUCMSC-EVs-based delivery system is a promising therapy for the patients with lumbar spinal stenosis, offering the translational application in clinical practice.
Fig. 8.
Schematic diagram illustrating the proposed mechanism of the therapeutic ability of hUCMSCs-EVs on the progression of ligamentum flavum hypertrophy. hUCMSC-EVs could suppress LFH in bipedal standing mice in vivo and attenuate the progression of LF cell fibrosis induced by TGF-β1. Moreover, miR-146a-5p and miR-221-3p enriched in hUCMSC-EVs could be delivered to LF cells to prevent proliferation and excessive deposition of connective tissue in the extracellular matrix via direct targeting of SMAD4, resulting in the attenuation of LFH.
MSCs and their derived secretomes have been extensively used as promising tools for the treatment of degenerative and inflammatory diseases [35]. Our study found no significant difference between the BS + MSC and BS + EVs groups and this result was consistent with a previous study [36], in which the adipose stem cells (ASC)-or ASC-EV-injected group exhibited similar results in the osteoporosis mouse model. Compared with MSCs, EVs have the following advantages. First, the use of EVs avoids the potential tumorigenic properties of MSCs, as unmodified MSCs may exhibit chromosomal abnormalities, leading to the formation of malignant tumours [37]. Second, EVs have lower immunogenicity than MSCs [38]. Third, EVs are highly stable and suitable for long-term storage without the addition of toxic cryopreservatives [38]. Finally, EVs possess considerable plasticity, as they can be artificially modified to alter therapeutic efficiency. In our work, after transfection of miR-221-3p and miR-146a-5p inhibitors into MSCs, the expressions of miR-221-3p and miR-146a-5p in EVs extracted from the supernatant were significantly reduced, leading to partial impairment of the fibrosis-inhibiting potential of hUCMSC-EVs. Additionally, hUCMSC-EVs acted as emerging innovative nano-sized drug delivery systems which played an important role in the progression of pulmonary, myocardial and liver fibroses [18,39,40]. Importantly, UCMSC-derived exosomes were more prominent than bone marrow MSC-derived exosomes for tissue damage repair [41]. Thus, in this study, hUCMSCs were selected as the source of EVs.
The pathological mechanism underlying LFH has not yet been fully understood. Many studies have revealed that the accumulation and formation of inflammation-related fibrosis tissue are closely related to LFH [42]. Moreover, the expression levels of TGF-β1 and collagen types I, II and V in LF cells were notably elevated in patients with LSS. Our data also showed that TGF-β1 could effectively promote the expression of Col-I, α-SMA, and Col-III and result in LF cell fibrosis in a dose-dependent manner. According to previous literatures [11,[43], [44], [45]], 10 miRNAs relative to fibrosis were selected as candidates; the RT-qPCR validation of these candidates partially matched the results reported in a previous study [46]. Since the amelioration of LFH after the EV injection was found, we hypothesised that the miRNAs enriched in the EVs would take responsibility for this amelioration. We performed RNA sequencing and bioinformatic analysis to identify the composition and function of hUCMSC-EVs. The highest expression miRNA was miR-21-5p, but this result was not reproduced by RT-qPCR analysis, despite miR-21-5p being enriched in hUCMSCs and hUCMSC-EVs. It was reported that miR-21 may positively correlate with LFH [28], however, this finding was controversial, as this correction was not found by other authors [46]. In our study, we did not observe a significant difference in miR-21-5p expression in human LF specimens between the LFH group and the control group, which was consistent with a previous study [46]. Moreover, the decreased expression of miR-21-5p in fibrotic LF cells did not correspond with the findings in human LF specimens. Thus, miR-21-5p was not the major candidate miRNA requiring further investigation in this study.
Our study showed that miR-146a-5p and miR-221-3p expressions were downregulated in the LF tissues of patients with LSS and both miRNAs negatively correlated with LF thickness. Furthermore, we measured the expression levels of the top five miRNAs in mouse LF cells in the presence or absence of TGF-β1 and hUCMSC-EVs. Compared with the control group, miR-146a-5p and miR-221-3p expression levels were significantly decreased in the TGF-β1 treatment group, which was consistent with the result in human LF tissue validation. MiR-146a-5p and miR-221-3p have been found to be involved in cell differentiation, proliferation and apoptosis, and they can modulate the inflammatory response and participate in several pathophysiological processes [[47], [48], [49], [50], [51]]. MiR-146a-5p has been demonstrated to reduce hepatic fibrosis [52], inhibit the expression of fibrosis-related markers in irradiated and TGF-β1-stimulated LX2 cells (human hepatic stellate cell line), reduce skeletal muscle fibrosis after injury, and attenuate cisplatin-induced renal fibrosis [44,45,53]. Therefore, miR-221-3p and miR-146a-5p as majorly investigated molecules might be implicated in the progression of LFH, suggesting that upregulation of the expressions of both miRNAs could be a promising strategy for the treatment of LFH.
No data are currently available to reveal the therapeutic effects of EVs on LFH. In our study, we observed that EVs as bioactive materials could attenuate the progression of fibrosis in LF cells and suppress LFH. Moreover, EVs with miR-146a-5p or miR-221-3p knockdown only partially impaired the fibrosis-inhibiting effects of hUCMSC-EVs on LF cells. However, EVs with both knockdown miRNAs further impaired the fibrosis-inhibiting effects of hUCMSC-EVs. Nevertheless, many more experiments are needed to determine whether other miRNAs, lncRNAs, proteins or lipids also suppress LF fibrosis.
Notably, EVs could be internalised by LF cells and transport miR-146a-5p and miR-221-3p; this result was consistent with the previous study where hUCMSC-EVs could be internalised by HK-2 cells (human renal tubular epithelial cell line) and transport miR-125b-5p [54]. EVs are able to transfer intraluminal cargo, including proteins, lipids and regulatory RNA between cells, which play an important role in cell–cell communication. EVs could be taken up by recipient cells in a variety of mechanisms, including direct fusion, clathrin/caveolin-dependent endocytosis, macropinocytosis, phagocytosis and lipid raft-mediated endocytosis. EV uptake is a highly specific process that only occurs when cells and EV share the right combination of ligand and receptor [55]. However, the mechanism by which EVs enter recipient LF cells needs further investigation.
EVs are natural carriers as therapeutic delivery systems and they offer many advantages due to their low immunogenicity and toxicity, biodegradability and biostability, possible intrinsic homing and ability to cross various body barriers; their unique structure, made of a hydrophobic lipid bilayer and a hydrophilic core, allows the loading of a multitude of different cargoes [56]. Our study has demonstrated that hUCMSC-EVs could ameliorate LFH, but the accumulation after local administration and clearance in vivo needs to be investigated further. Additionally, the progression of LFH commonly experiences complex and lengthy multiple phases; it is difficult to retain unconjugated or free EVs in the LF site for an extended time. Several studies have reported that a bioactive-injectable self-healing anti-inflammatory hydrogel with ultralong EV release synergistically enhances the motor functional recovery of spinal cord injury because the hydrogel can retain the EVs in the injured site for a controlled release after administration [57]. Synthetic poly (D, l-lactide)-b-poly (ethylene glycol)-b-poly (D, l-lactide) (PDLLA-PEG-PDLLA; PLEL) triblock copolymer gels carrying EVs with circRNA3503 overexpression were found to promote chondrocyte renewal and alleviate the progressive loss of chondrocytes [58]. Therefore, it is necessary to develop an innovative and biocompatible biomaterial as a sustained release carrier for EVs with the aim of efficient retention and sustained release in the LF area and to ameliorate LFH.
We acknowledge that several limitations exist in this study. First, we only detected miR-146a-5p and miR-221-3p expressions in 20 human LF specimens, a data base with a larger size is needed to further validate our major findings. Second, whether other molecules play roles in the progression of LFH remains uncertain, and this warrants further investigation. Third, in our study, we used loss/gain of function by AAV, mimics, or inhibitor transfection to evaluate the therapeutic effect of EVs. However, genetically modified animal models will provide much better information for further understanding of the underlying mechanism. Fourth, local injection of EVs was used to treat LFH, but the distribution of EVs in the mouse LF is still unclear; additionally, the easy diffusion, short duration and pain of injection site would limit its application in clinics. Future studies will be very meaningful to use the bioactive materials to load these engineering EVs and to optimize the therapeutic system for LFH. Last but not the least, an investigation to clarify whether hUCMSC-EVs can ameliorate the progression of fibrosis in human LF cells will be valuable.
5. Conclusion
Collectively, our study demonstrates that hUCMSC-EVs could be used as a bioactive material to ameliorate the progression of fibrosis in LF cells and suppressed LFH by delivering miR-146a-5p and miR-221-3p, and that both miRNAs directly bonded to the 3′‐UTR regions of SMAD4 mRNA, which led to the inhibition of the TGF-β/SMAD4 signalling pathway. Our findings not only provided new insights into the promising potential of hUCMSC-EVs/miRNA-based therapy but also demonstrated that miR-146a-5p/miR-221-3p and SMAD4 are critical therapeutic targets in the treatment of LFH. This work would significantly enhance the translational application of hUCMSC-EVs-based therapeutics for LSS.
CRediT authorship contribution statement
Cheng Ma: Conceptualization, Methodology, Investigation, Writing – original draft. Xin Qi: Investigation, Writing – original draft. Yi-Fan Wei: Methodology, Data curation, Formal analysis. Zhi Li: Data curation, Formal analysis, Validation. He-Long Zhang: Methodology, Formal analysis. He Li: Methodology. Feng-Lei Yu: Methodology. Ya-Nan Pu: Conceptualization, Supervision, Methodology, Writing – original draft, and finalizing of the study. Yong-Can Huang: Conceptualization, Methodology, Writing – original draft, and finalizing of the study. Yong-Xin Ren: Conceptualization, Supervision, Methodology, Writing – original draft, and finalizing of the study.
Declaration of competing interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (81572149 and 81600697), Guangdong Basic and Applied Basic Research Foundation (2021A1515220086), Basic Research Program of Jiangsu Province (Natural Science Foundation, K20201487), Jiangsu Province "333" Project (LGY2016001) and Sino-German Mobility programme of the Chinese-German Research Center of the National Science Foundation of China (NSFC) and the German Research Council (DFG), Grant Number M-0332.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2022.03.042.
Contributor Information
Ya-Nan Pu, Email: pyn342626@163.com.
Yong-Can Huang, Email: hycpku@hotmail.com.
Yong-Xin Ren, Email: renyongxin@njmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Kalichman L., Cole R., Kim D.H., Li L., Suri P., Guermazi A., et al. Spinal stenosis prevalence and association with symptoms: the Framingham Study. Spine J. 2009;9(7):545–550. doi: 10.1016/j.spinee.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang G., Peng Z., Li J., Song Z., Wang P. Diagnostic performance of the nerve root sedimentation sign in lumbar spinal stenosis: a systematic review and meta-analysis. Neuroradiology. 2019;61(10):1111–1121. doi: 10.1007/s00234-019-02248-3. [DOI] [PubMed] [Google Scholar]
- 3.Lai M.K.L., Cheung P.W.H., Cheung J.P.Y. A systematic review of developmental lumbar spinal stenosis. Eur. Spine J. 2020;29(9):2173–2187. doi: 10.1007/s00586-020-06524-2. [DOI] [PubMed] [Google Scholar]
- 4.Lafian A.M., Torralba K.D. Lumbar spinal stenosis in older adults. Rheum. Dis. Clin. N. Am. 2018;44(3):501–512. doi: 10.1016/j.rdc.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Sakai Y., Ito S., Hida T., Ito K., Harada A., Watanabe K. Clinical outcome of lumbar spinal stenosis based on new classification according to hypertrophied ligamentum flavum. J. Orthop. Sci. 2017;22(1):27–33. doi: 10.1016/j.jos.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Sun C., Zhang H., Wang X., Liu X. Ligamentum flavum fibrosis and hypertrophy: Molecular pathways, cellular mechanisms, and future directions. Faseb. J. 2020;34(8):9854–9868. doi: 10.1096/fj.202000635R. [DOI] [PubMed] [Google Scholar]
- 7.Weiskirchen R., Weiskirchen S., Tacke F. Organ and tissue fibrosis: Molecular signals, cellular mechanisms and translational implications. Mol. Aspect. Med. 2019;65:2–15. doi: 10.1016/j.mam.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Sziksz E., Pap D., Lippai R., Beres N.J., Fekete A., Szabo A.J., et al. Fibrosis related inflammatory Mediators: role of the IL-10 cytokine family. Mediat. Inflamm. 2015;2015 doi: 10.1155/2015/764641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landstrom M., Liu J. The 2019 FASEB science research conference on the TGF-beta superfamily: signaling in development and disease, July 28 to august 2, 2019, west Palm Beach, Florida, USA. Faseb. J. 2019;33(12):13064–13067. doi: 10.1096/fj.201902632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng X.M., Nikolic-Paterson D.J., Lan H.Y. TGF-beta: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12(6):325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 11.Hur J.W., Bae T., Ye S., Kim J.H., Lee S., Kim K., et al. Myofibroblast in the ligamentum flavum hypertrophic activity. Eur. Spine J. 2017;26(8):2021–2030. doi: 10.1007/s00586-017-4981-2. [DOI] [PubMed] [Google Scholar]
- 12.Amudong A., Muheremu A., Abudourexiti T. Hypertrophy of the ligamentum flavum and expression of transforming growth factor beta. J. Int. Med. Res. 2017;45(6):2036–2041. doi: 10.1177/0300060517711308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu L., Xu L., Chen Y., Xu G., Guo Q., Meng D., et al. Rolipram plays an anti-fibrotic effect in ligamentum flavum fibroblasts by inhibiting the activation of ERK1/2. BMC Muscoskel. Disord. 2021;22(1):818. doi: 10.1186/s12891-021-04712-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie Q., Liu R., Jiang J., Peng J., Yang C., Zhang W., et al. What is the impact of human umbilical cord mesenchymal stem cell transplantation on clinical treatment? Stem Cell Res. Ther. 2020;11(1):519. doi: 10.1186/s13287-020-02011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silini A.R., Di Pietro R., Lang-Olip I., Alviano F., Banerjee A., Basile M., et al. Perinatal derivatives: where do we stand? A roadmap of the human Placenta and consensus for tissue and cell nomenclature. Front. Bioeng. Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.610544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang F., Yu Y., Liu S., Ming L., Zhang Y., Zhou Z., et al. Advancing application of mesenchymal stem cell-based bone tissue regeneration. Bioact. Mater. 2021;6(3):666–683. doi: 10.1016/j.bioactmat.2020.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J., Peng D., You J., Zhou O., Qiu H., Hao C., et al. Type 2 alveolar epithelial cells differentiated from human umbilical cord mesenchymal stem cells alleviate mouse pulmonary fibrosis through beta-catenin-regulated cell apoptosis. Stem Cell. Dev. 2021;30(13):660–670. doi: 10.1089/scd.2020.0208. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q., Gu T., Zhang Y., Li H., Zhuansun X., Xu S., et al. Human umbilical cord mesenchymal stem cells ameliorate hepatic stellate cell activation and liver fibrosis by upregulating MicroRNA-455-3p through suppression of p21-activated Kinase-2. BioMed Res. Int. 2021;2021 doi: 10.1155/2021/6685605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Distler J.H.W., Gyorfi A.H., Ramanujam M., Whitfield M.L., Konigshoff M., Lafyatis R. Shared and distinct mechanisms of fibrosis. Nat. Rev. Rheumatol. 2019;15(12):705–730. doi: 10.1038/s41584-019-0322-7. [DOI] [PubMed] [Google Scholar]
- 20.Sun H., Pratt R.E., Hodgkinson C.P., Dzau V.J. Sequential paracrine mechanisms are necessary for the therapeutic benefits of stem cell therapy. Am. J. Physiol. Cell Physiol. 2020;319(6):C1141–C1150. doi: 10.1152/ajpcell.00516.2019. [DOI] [PubMed] [Google Scholar]
- 21.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 22.Shah R., Patel T., Freedman J.E. Circulating extracellular vesicles in human disease. N. Engl. J. Med. 2018;379(10):958–966. doi: 10.1056/NEJMra1704286. [DOI] [PubMed] [Google Scholar]
- 23.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiklander O.P.B., Brennan M.A., Lotvall J., Breakefield X.O., El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci. Transl. Med. 2019;11(492) doi: 10.1126/scitranslmed.aav8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang G., Zhu Y., Ali D.J., Tian T., Xu H., Si K., et al. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnol. 2020;18(1):10. doi: 10.1186/s12951-019-0563-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui Y., Guo Y., Kong L., Shi J., Liu P., Li R., et al. A bone-targeted engineered exosome platform delivering siRNA to treat osteoporosis. Bioact. Mater. 2022;10:207–221. doi: 10.1016/j.bioactmat.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu T., Zhang Q., Zhang J., Li C., Miao Y.R., Lei Q., et al. EVmiRNA: a database of miRNA profiling in extracellular vesicles. Nucleic Acids Res. 2019;47(D1):D89–D93. doi: 10.1093/nar/gky985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun C., Tian J., Liu X., Guan G. MiR-21 promotes fibrosis and hypertrophy of ligamentum flavum in lumbar spinal canal stenosis by activating IL-6 expression. Biochem. Biophys. Res. Commun. 2017;490(3):1106–1111. doi: 10.1016/j.bbrc.2017.06.182. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y.Q., Zhang Z.H., Zheng Y.F., Feng S.Q. MicroRNA-221 regulates hypertrophy of ligamentum flavum in lumbar spinal stenosis by targeting TIMP-2. Spine. 2016;41(4):275–282. doi: 10.1097/BRS.0000000000001226. [DOI] [PubMed] [Google Scholar]
- 30.Chen J., Liu Z., Zhong G., Qian L., Li Z., Qiao Z., et al. Hypertrophy of ligamentum flavum in lumbar spine stenosis is associated with increased miR-155 level. Dis. Markers. 2014;2014:786543. doi: 10.1155/2014/786543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.George K.M., Hernandez N.S., Breton J., Cooper B., Dowd R.S., Nail J., et al. Lumbar ligamentum flavum burden: Evaluating the role of ATTRwt amyloid deposition in ligamentum flavum thickness at all lumbar levels. Clin. Neurol. Neurosurg. 2021;206:106708. doi: 10.1016/j.clineuro.2021.106708. [DOI] [PubMed] [Google Scholar]
- 32.Zheng Z., Ao X., Li P., Lian Z., Jiang T., Zhang Z., et al. CRLF1 is a key regulator in the ligamentum flavum hypertrophy. Front. Cell Dev. Biol. 2020;8:858. doi: 10.3389/fcell.2020.00858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Che H., Li J., Li Y., Ma C., Liu H., Qin J., et al. p16 deficiency attenuates intervertebral disc degeneration by adjusting oxidative stress and nucleus pulposus cell cycle. Elife. 2020;9 doi: 10.7554/eLife.52570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21(4):255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 35.Harrell C.R., Fellabaum C., Jovicic N., Djonov V., Arsenijevic N., Volarevic V. Molecular mechanisms responsible for therapeutic potential of mesenchymal stem cell-derived secretome. Cells. 2019;8(5) doi: 10.3390/cells8050467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee K.S., Lee J., Kim H.K., Yeom S.H., Woo C.H., Jung Y.J., et al. Extracellular vesicles from adipose tissue-derived stem cells alleviate osteoporosis through osteoprotegerin and miR-21-5p. J. Extracell. Vesicles. 2021;10(12) doi: 10.1002/jev2.12152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong J.O., Han J.W., Kim J.M., Cho H.J., Park C., Lee N., et al. Malignant tumor formation after transplantation of short-term cultured bone marrow mesenchymal stem cells in experimental myocardial infarction and diabetic neuropathy. Circ. Res. 2011;108(11):1340–1347. doi: 10.1161/CIRCRESAHA.110.239848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brown C., McKee C., Bakshi S., Walker K., Hakman E., Halassy S., et al. Mesenchymal stem cells: cell therapy and regeneration potential. J Tissue Eng Regen Med. 2019;13(9):1738–1755. doi: 10.1002/term.2914. [DOI] [PubMed] [Google Scholar]
- 39.Yang D., Chen X., Wang J., Lou Q., Lou Y., Li L., et al. Dysregulated lung commensal Bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity. 2019;50(3):692–706. doi: 10.1016/j.immuni.2019.02.001. e7. [DOI] [PubMed] [Google Scholar]
- 40.Han C., Zhou J., Liang C., Liu B., Pan X., Zhang Y., et al. Human umbilical cord mesenchymal stem cell derived exosomes encapsulated in functional peptide hydrogels promote cardiac repair. Biomater Sci. 2019;7(7):2920–2933. doi: 10.1039/c9bm00101h. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z.G., He Z.Y., Liang S., Yang Q., Cheng P., Chen A.M. Comprehensive proteomic analysis of exosomes derived from human bone marrow, adipose tissue, and umbilical cord mesenchymal stem cells. Stem Cell Res. Ther. 2020;11(1):511. doi: 10.1186/s13287-020-02032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chuang H.C., Tsai K.L., Tsai K.J., Tu T.Y., Shyong Y.J., Jou I.M., et al. Oxidative stress mediates age-related hypertrophy of ligamentum flavum by inducing inflammation, fibrosis, and apoptosis through activating Akt and MAPK pathways. Aging (Albany NY) 2020;12(23):24168–24183. doi: 10.18632/aging.104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mori T., Sakai Y., Kayano M., Matsuda A., Oboki K., Matsumoto K., et al. MicroRNA transcriptome analysis on hypertrophy of ligamentum flavum in patients with lumbar spinal stenosis. Spine Surg Relat Res. 2017;1(4):211–217. doi: 10.22603/ssrr.1.2017-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu L., Rong C., Zhou Q., Zhao X., Zhuansun X.M., Wan S., et al. Bone marrow mesenchymal stem cells ameliorate cisplatin-induced renal fibrosis via miR-146a-5p/tfdp2 Axis in renal tubular epithelial cells. Front. Immunol. 2020;11:623693. doi: 10.3389/fimmu.2020.623693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Y., Li Y., Wang H., Li H., Liu S., Chen J., et al. miR-146a-5p acts as a negative regulator of TGF-beta signaling in skeletal muscle after acute contusion. Acta Biochim. Biophys. Sin. 2017;49(7):628–634. doi: 10.1093/abbs/gmx052. [DOI] [PubMed] [Google Scholar]
- 46.Xu Y-q, Zhang Z-h, Zheng Y-f, Feng S-q. MicroRNA-221 regulates hypertrophy of ligamentum flavum in lumbar spinal stenosis by targeting TIMP-2. Spine. 2016;41(4):275–282. doi: 10.1097/brs.0000000000001226. [DOI] [PubMed] [Google Scholar]
- 47.Meng Q., Liang C., Hua J., Zhang B., Liu J., Zhang Y., et al. A miR-146a-5p/TRAF6/NF-kB p65 axis regulates pancreatic cancer chemoresistance: functional validation and clinical significance. Theranostics. 2020;10(9):3967–3979. doi: 10.7150/thno.40566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li X., Liao J., Su X., Li W., Bi Z., Wang J., et al. Human urine-derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR-146a-5p which targets IRAK1. Theranostics. 2020;10(21):9561–9578. doi: 10.7150/thno.42153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu M., Li T., Wang B., Gong H., Huang T. miR-146a-5p regulated cell proliferation and apoptosis by targeting SMAD3 and SMAD4. Protein Pept. Lett. 2020;27(5):411–418. doi: 10.2174/0929866526666190911142926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu F.Y., Gan C.W., Wang M.X., Sun B.C., Li F.J., Qiu Y.H., et al. MiR-146a-5p inhibits proliferation and promotes apoptosis of oral squamous cell carcinoma cells by regulating NF-kappaB signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2020;24(7):3717–3723. doi: 10.26355/eurrev_202004_20835. [DOI] [PubMed] [Google Scholar]
- 51.Verjans R., Peters T., Beaumont F.J., van Leeuwen R., van Herwaarden T., Verhesen W., et al. MicroRNA-221/222 family counteracts myocardial fibrosis in Pressure overload-induced heart failure. Hypertension. 2018;71(2):280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 52.Zou Y., Cai Y., Lu D., Zhou Y., Yao Q., Zhang S. MicroRNA-146a-5p attenuates liver fibrosis by suppressing profibrogenic effects of TGFbeta1 and lipopolysaccharide. Cell. Signal. 2017;39:1–8. doi: 10.1016/j.cellsig.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Yuan B.Y., Chen Y.H., Wu Z.F., Zhuang Y., Chen G.W., Zhang L., et al. MicroRNA-146a-5p attenuates fibrosis-related molecules in irradiated and TGF-beta1-treated human hepatic stellate cells by regulating PTPRA-SRC signaling. Radiat. Res. 2019;192(6):621–629. doi: 10.1667/RR15401.1. [DOI] [PubMed] [Google Scholar]
- 54.Cao J.-Y., Wang B., Tang T.-T., Wen Y., Li Z.-L., Feng S.-T., et al. Exosomal miR-125b-5p deriving from mesenchymal stem cells promotes tubular repair by suppression of p53 in ischemic acute kidney injury. Theranostics. 2021;11(11):5248–5266. doi: 10.7150/thno.54550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng L., Hill A.F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 2022 doi: 10.1038/s41573-022-00410-w. [DOI] [PubMed] [Google Scholar]
- 56.Villata S., Canta M., Cauda V. EVs and bioengineering: from cellular Products to engineered nanomachines. Int. J. Mol. Sci. 2020;21(17) doi: 10.3390/ijms21176048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Wang M., Xia K., Wang J., Cheng F., Shi K., et al. A bioactive injectable self-healing anti-inflammatory hydrogel with ultralong extracellular vesicles release synergistically enhances motor functional recovery of spinal cord injury. Bioact. Mater. 2021;6(8):2523–2534. doi: 10.1016/j.bioactmat.2021.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao S.C., Huang J.Y., Gao Y., Li Z.X., Wei Z.Y., Dawes H., et al. Small extracellular vesicles in combination with sleep-related circRNA3503: a targeted therapeutic agent with injectable thermosensitive hydrogel to prevent osteoarthritis. Bioact. Mater. 2021;6(12):4455–4469. doi: 10.1016/j.bioactmat.2021.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.