Abstract
In people living with HIV (PLWH) on antiretroviral therapy (ART), virus persists in a latent form where there is minimal transcription or protein expression. Latently infected cells are a major barrier to curing HIV. Increasing HIV transcription and viral production in latently infected cells could facilitate immune recognition and reduce the pool of infected cells that persist on ART. Given that programmed cell death protein 1 (PD-1) expressing CD4+ T cells are preferentially infected with HIV in PLWH on ART, we aimed to determine whether administration of antibodies targeting PD-1 would reverse HIV latency in vivo. We therefore evaluated the impact of intravenous administration of pembrolizumab every 3 weeks on HIV latency in 32 PLWH and cancer on ART. After the first infusion of anti–PD-1, we observed a median 1.32-fold increase in unspliced HIV RNA and 1.61-fold increase in unspliced RNA:DNA ratio in sorted blood CD4+ T cells compared to baseline. We also observed a 1.65-fold increase in plasma HIV RNA. The frequency of CD4+ T cells with inducible virus evaluated using the tat/rev limiting dilution assay was higher after 6 cycles compared to baseline. Phylogenetic analyses of HIV env sequences in a participant who developed low concentrations of HIV viremia after 6 cycles of pembrolizumab did not demonstrate clonal expansion of HIV-infected cells. These data are consistent with anti–PD-1 being able to reverse HIV latency in vivo and support the rationale for combining anti–PD-1 with other interventions to reduce the HIV reservoir.
INTRODUCTION
Despite the success of antiretroviral therapy (ART) in reducing morbidity and mortality for people living with HIV (PLWH), there is still a need for a cure (1). ART inhibits HIV replication, but latent HIV persists as an integrated genome in long-lived CD4+ T cells capable of proliferation with minimal or no antigen expression (2). Latency reversal is a proposed approach to induce expression of HIV antigens or virions to allow for immune recognition and elimination of infected cells (reviewed by Zerbato et al.) (3). Together with enhanced immune clearance, this approach could potentially eliminate cells that contain replication-competent HIV (4). Latency-reversing agents evaluated to date in PLWH on ART, including histone deacetylase inhibitors and toll-like receptor agonists, have not consistently demonstrated latency reversal, and no intervention other than allogeneic stem cell transplant has yet shown a sustained decrease or elimination of cells that contain replication-competent virus (5).
Programmed cell death protein 1 (PD-1) is an inhibitory checkpoint molecule expressed on T cells that inhibits immune responses against cancers and viral infections. Monoclonal antibodies targeting PD-1 or its ligand, PD-L1, are approved to treat a growing number of cancers, including several HIV-associated malignancies (6). PD-1 is up-regulated on CD4+ and CD8+ T cells in PLWH on and off ART (7) and, along with other immune checkpoints, is preferentially expressed on latently infected cells in blood and tissue (8–12). Ex vivo, engagement of PD-1 by its ligand PD-L1 inhibits T cell receptor–mediated activation, allowing for the establishment of HIV latency (12). In vitro, anti–PD-1 antibodies enhanced viral production from infected cells when used in combination with a submaximal activating stimulus (10, 13).
On the basis of preclinical data, we hypothesized that anti–PD-1 would lead to latency reversal in vivo. Several small studies of PLWH on ART who have received anti–PD-1 or other immune checkpoint blockers, such as anti–PD-L1 or anti–cytotoxic T lymphocyte–associated protein 4 (CTLA-4) have previously explored this issue. Most, but not all, suggest that immune checkpoint blockade may perturb the HIV reservoir (12, 14–16). Cancer Immunotherapy Trials Network 12 (CITN-12; NCT02595866) is a prospective clinical trial in PLWH on ART and cancer across a range of CD4+ T cell counts evaluating the safety and anticancer activity of pembrolizumab, a humanized immunoglobulin G4 (IgG4) monoclonal antibody. Participants from CITN-12 were enrolled in a prospective virology substudy to conduct a detailed characterization of the longitudinal effects of anti–PD-1 on HIV transcription.
RESULTS
Participants had a range of cancers and well-controlled HIV at baseline
Thirty-two participants were included in the analysis (Fig. 1). There were 9 participants in cohort 1 (100 to 199 CD4+ T cells/µl), 12 in cohort 2 (200 to 350 CD4+ T cells/µl), and 11 in cohort 3 (>350 CD4+ T cells/µl) (Table 1). One (13%) participant in cohort 1 had an AIDS-defining cancer, compared to four (33%) in cohort 2 and five (46%) in cohort 3. No other substantial differences were noted between the cohorts. At baseline, plasma HIV RNA concentrations were below the limit of quantification (20 copies/ml) in 29 participants and detectable in the remaining 3 participants (at 21, 21, and 168 copies/ml). Individual participant HIV, cancer, and treatment characteristics are noted in table S1.
Fig. 1. Study schema of specimen collection in relation to pembrolizumab administration.
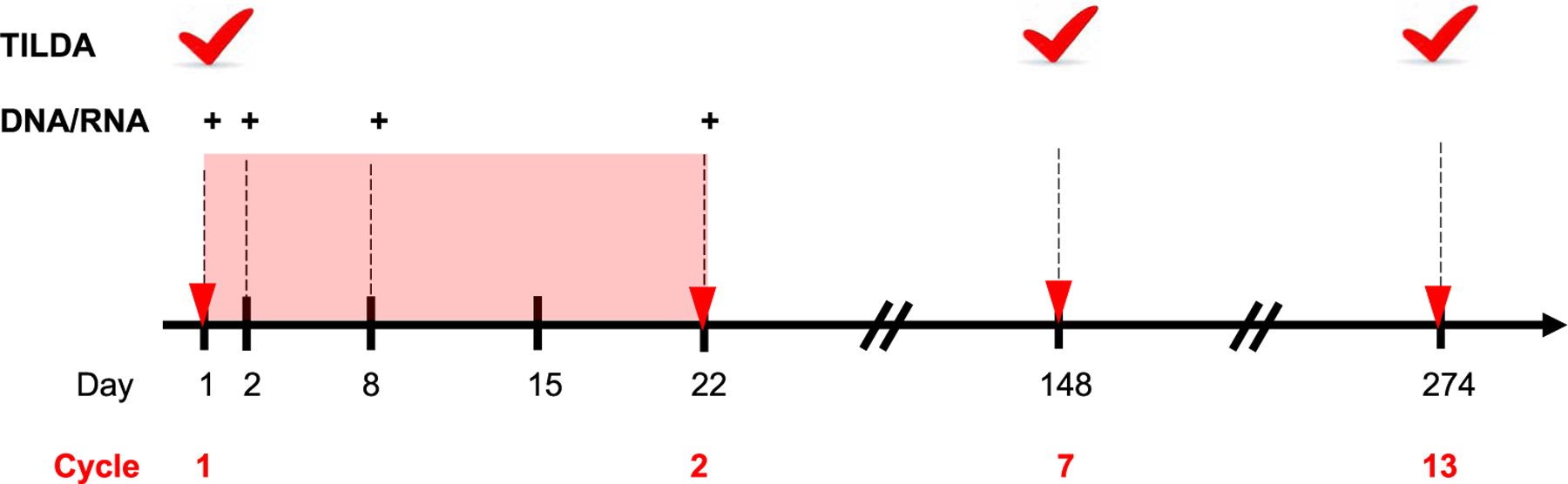
Cycles of pembrolizumab (red triangles) were administered every 3 weeks on day 1 of a cycle for up to 35 cycles for participants benefiting from therapy. Participants discontinued treatment in the setting of unacceptable adverse events, progressive cancer, or participant preference. Blood was collected at regular time points (shown as dashed vertical lines), including before pembrolizumab administration on day 1, and plasma and peripheral blood mononuclear cells (PBMCs) were processed and cryopreserved. More frequent collection of specimens occurred after the first dose (red shaded box). DNA and RNA were extracted from CD4+ T cells isolated from PBMCs, and HIV DNA and unspliced and multiply spliced HIV RNA were quantified by PCR (shown as +). Plasma HIV RNA was quantified at the same time points. PBMCs collected at baseline, before cycle 7, and before cycle 13 were used for the inducible HIV using the tat/rev induced limiting dilution assay (TILDA; red tick). For participants receiving less than 7 cycles of therapy, TILDA was evaluated at the end of therapy.
Table 1.
Participant baseline characteristics.
| Characteristic | Number or median | Percent or IQR |
|---|---|---|
| Cohort, n (% of cohort) | ||
| 100 to 199 CD4+/µl | 9 | 28% |
| 200 to 350 CD4+/µl | 12 | 38% |
| >350 CD4+/µl | 11 | 34% |
| Age, median (IQR) | 55 | (48, 61) |
| Sex, n (% of cohort) | ||
| Men | 29 | 91% |
| Women | 3 | 9% |
| Race, n (% of cohort) | ||
| White | 19 | 59% |
| Black/African-American | 9 | 28% |
| Other/unknown/more than 1 | 4 | 13% |
| Ethnicity | ||
| Hispanic | 3 | 9% |
| Non-Hispanic | 28 | 88% |
| Not reported | 1 | 3% |
| Cancer, n (% of cohort) | ||
| AIDS-defining | 10 | 31% |
| Non–AIDS-defining | 22 | 69% |
| CD4:CD8 ratio, n (% of cohort) | ||
| <0.5 | 16 | 50% |
| ≥0.5 | 16 | 50% |
| Plasma HIV RNA, n (% of cohort) | ||
| Below limit of detection | 29 | 91% |
| Detected | 3 | 9% |
Anti–PD-1 induces transient declines in HIV DNA and increases in unspliced HIV RNA and the HIV RNA:DNA ratio
Baseline intracellular unspliced HIV RNA, HIV DNA, and multiply spliced HIV RNA did not differ by CD4 cohort. Baseline median [interquartile range (IQR)] plasma HIV RNA by the HIV Molecular and Monitoring Core gag (HHMCgag) assay was 0.31 (0.08, 0.54), 2.60 (0.18, 9.90), and 1.25 (0.49, 2.10) copies/ml in cohorts 1 to 3, respectively (P = 0.03; Fig. 2). Baseline plasma HIV RNA by HHMCgag assay was positively correlated with unspliced HIV RNA (Spearman ρ = 0.42, P = 0.049) but not with multiply spliced HIV RNA (ρ = 0.04, P = 0.9) or HIV DNA (ρ = 0.28, P = 0.15). Plasma and unspliced HIV RNA remained correlated at day 8 (ρ = 0.52, P = 0.01). Unspliced HIV RNA, multiply spliced HIV RNA, and HIV DNA were not correlated on a pairwise basis at baseline or day 8 after pembrolizumab administration (table S2).
Fig. 2. Baseline measures demonstrated persistent forms of HIV on ART.
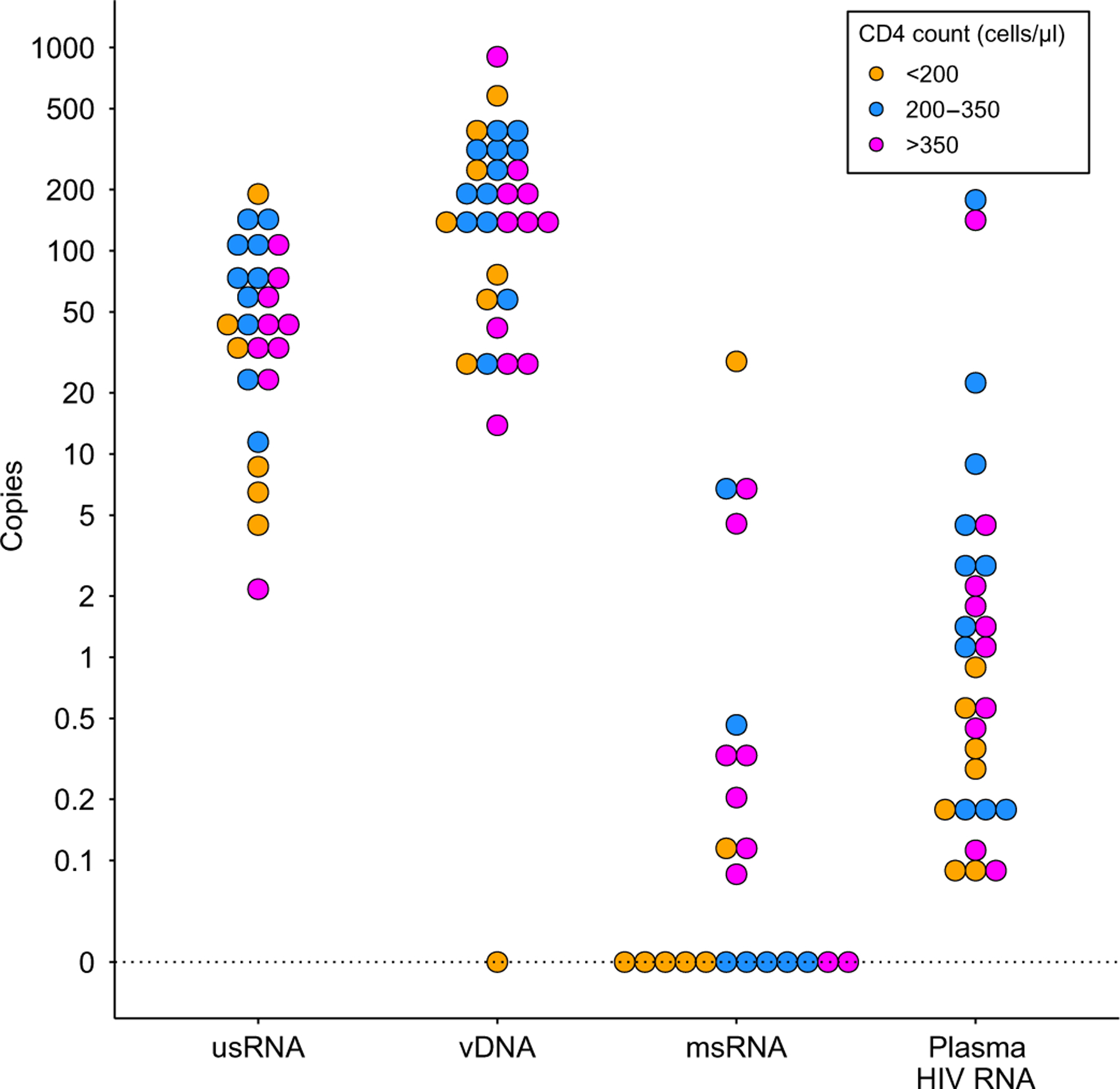
Baseline (day 1) plasma HIV RNA (copies/ml), unspliced HIV RNA (usRNA; copies per 106 cells), HIV DNA (vDNA; copies per 106 cells), and multiply spliced HIV RNA (msRNA; copies per 106 cells) are shown for participants stratified by CD4+ T cell count. Values greater than zero were transformed by log10(y) and grouped in 10 bins per log10 unit, with exact zeros set apart for visualization.
On day 8 after anti–PD-1, evaluation of intraparticipant changes demonstrated that unspliced HIV RNA was 1.32-fold higher than baseline values (P = 0.03; Table 2 and Fig. 3A). By day 22, unspliced HIV RNA remained elevated, although it was no longer different from baseline. In contrast, cell-associated HIV DNA decreased 1 day after pembrolizumab infusion to 0.78-fold that of baseline (P = 0.005; Fig. 3B) and trended back toward the baseline value by day 22. As a result, the HIV unspliced RNA:DNA ratio increased on day 8 after first administration of pembrolizumab to become 1.61-fold of baseline (P = 0.001; Fig. 3C). In a subset that included 23 participants, no changes in multiply spliced HIV RNA were noted compared to baseline at any time point (Table 2). Whereas plasma HIV was moderately correlated with unspliced HIV RNA, plasma HIV did not increase during the first cycle of therapy (Table 2 and Fig. 3D). Individual-level changes in measures of HIV are noted in waterfall plots (Fig. 4), with increased unspliced RNA:DNA ratio noted in 75% of study participants at day 8. We also evaluated trends in plasma HIV at later time points. Compared to baseline, plasma HIV RNA was not significantly (P = 0.29) higher at end of treatment (EOT) among the 12 participants for whom EOT was less than 6 months after baseline. However, it was 5.8-fold (P = 0.04) higher among four participants for whom EOT was greater than 6 months from baseline.
Table 2. Effects of pembrolizumab on measures of HIV.
C1, cycle 1; EOT, end of treatment; <C7, before cycle 7; ≥C7, on or after cycle 7; C13, cycle 13; 2H, 2 hours after treatment.
| Time point |
HIV measure |
|||
|---|---|---|---|---|
| Plasma HIV RNA | ||||
| C1, day | N * | Fold change† | (95% CI) | P |
| 1 | 28 | Ref. | ||
| 1 (2H) | 28 | 0.87 | (0.71, 1.07) | 0.20 |
| 2 | 29 | 0.91 | (0.70, 1.19) | 0.50 |
| 8 | 26 | 1.65 | (0.97, 2.81) | 0.07 |
| 22 | 27 | 1.68 | (0.74, 3.84) | 0.22 |
| 22 (2H) | 25 | 1.42 | (0.62, 3.23) | 0.40 |
| Cell-associated unspliced HIV RNA | ||||
| C1, day | N * | Fold change† | (95% CI) | P |
| 1 | 25 | Ref. | ||
| 2 | 26 | 1.13 | (0.79, 1.61) | 0.50 |
| 8 | 27 | 1.32 | (1.02, 1.70) | 0.03 |
| 22 | 23 | 1.46 | (0.98, 2.17) | 0.06 |
| Cell-associated HIV DNA | ||||
| C1, day | N * | Fold change† | (95% CI) | P |
| 1 | 31 | Ref. | ||
| 2 | 31 | 0.78 | (0.66, 0.93) | 0.005 |
| 8 | 31 | 0.85 | (0.72, 1.01) | 0.06 |
| 22 | 26 | 0.91 | (0.77, 1.08) | 0.28 |
| Ratio of cell-associated unspliced HIV RNA/HIV DNA | ||||
| C1, day | N * | Fold change† | (95% CI) | P |
| 1 | 31 | Ref. | ||
| 2 | 31 | 1.68 | (0.99, 2.87) | 0.06 |
| 8 | 31 | 1.61 | (1.21, 2.14) | 0.001 |
| 22 | 26 | 1.34 | (0.84, 2.14) | 0.22 |
| Cell-associated multiply spliced HIV RNA | ||||
| C1, day | N * | Fold change† | (95% CI) | P |
| 1 | 23 | Ref. | ||
| 2 | 21 | 0.59 | (0.20, 1.69) | 0.33 |
| 8 | 21 | 0.39 | (0.14, 1.11) | 0.08 |
| 22 | 20 | 2.08 | (0.63, 6.88) | 0.23 |
| Tat/rev induced limiting dilution assay (TILDA) | ||||
| Visit | N * | Fold change† | (95% CI) | P |
| C1, day 1 | 23 | Ref. | ||
| EOT < C7 | 13 | 0.77 | (0.61, 0.97) | 0.02 |
| C7 | 10 | 1.40 | (1.09, 1.80) | 0.008 |
| EOT ≥ C7 | 6 | 1.22 | (0.76, 1.95) | 0.41 |
| C13 | 5 | 0.75 | (0.55, 1.03) | 0.07 |
Number of participants contributing to the results at each time.
Estimate of within-participant fold change compared to baseline (C1, day 1), 95% confidence interval (CI), and P value from Wald test of fold change equal to 1, from a multilevel mixed-effects regression model.
Fig. 3. Anti–PD-1 affects measures of HIV in CD4-sorted T cells.
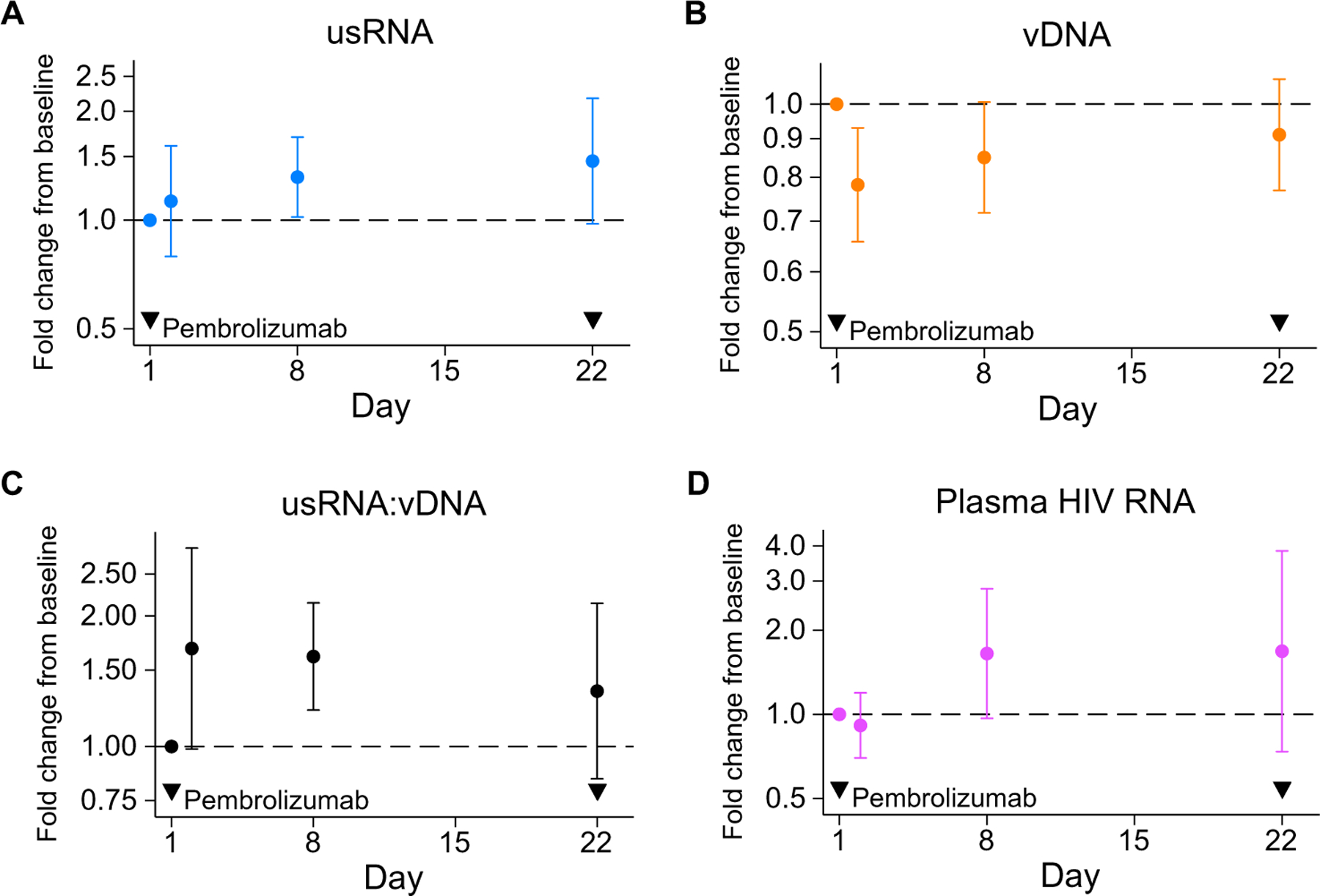
Estimated within-participant changes from cycle 1, day 1 administration of pembrolizumab of (A) unspliced HIV RNA, (B) HIV DNA, (C) the ratio of unspliced HIV RNA:HIV DNA, and (D) plasma HIV RNA are shown. The y axis scale is log-transformed. Dots represent point estimates from regression analyses, and error bars indicate 95% confidence intervals; dashed line indicates no change from baseline; and exclusion of dashed line from confidence interval indicates P < 0.05 by Wald test of regression coefficient.
Fig. 4. After a single infusion of pembrolizumab, there is an increase in unspliced HIV RNA and the ratio of unspliced HIV RNA:HIV DNA.
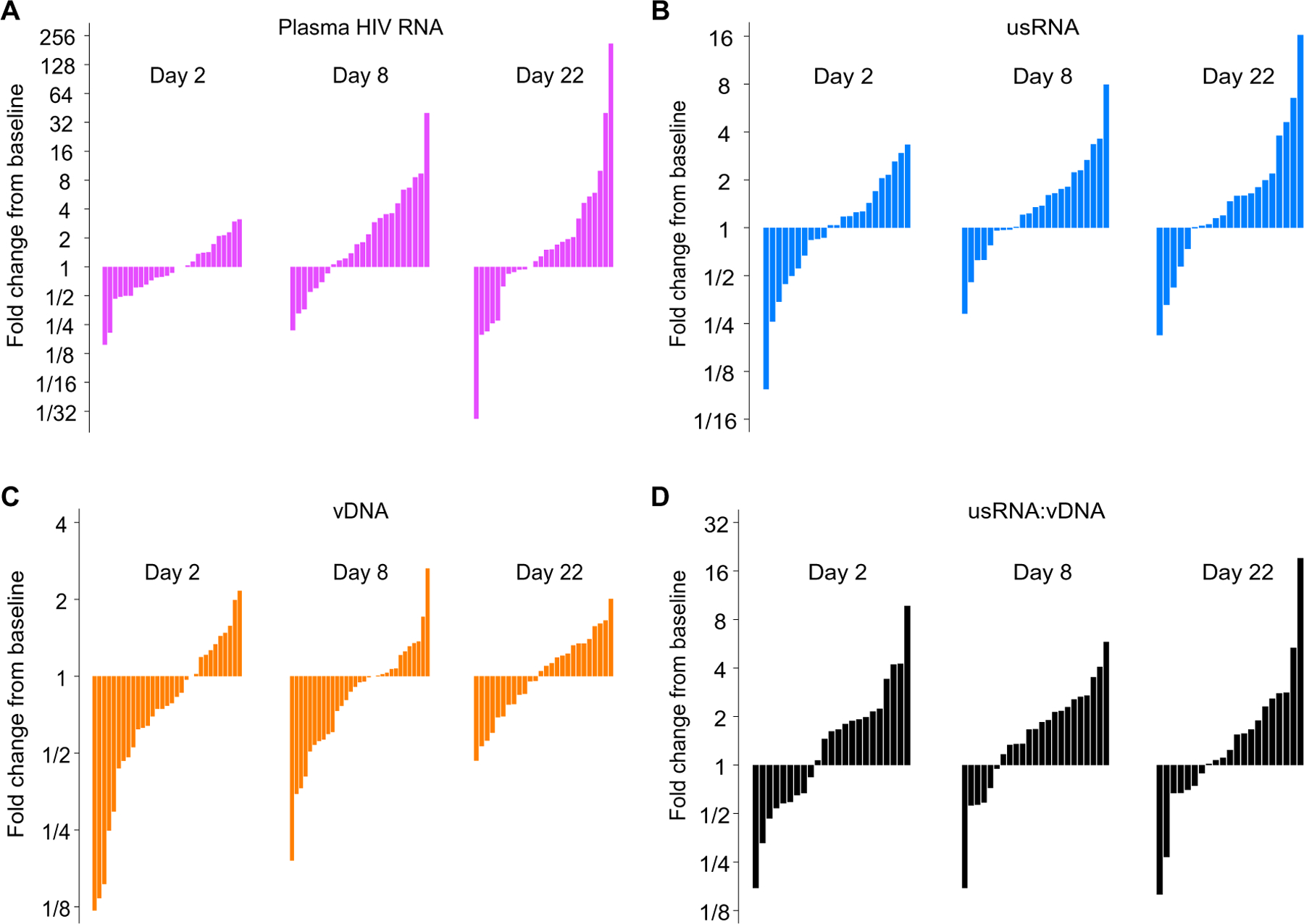
Waterfall plots of changes in (A) plasma HIV RNA, (B) unspliced HIV RNA, (C) HIV DNA, and (D) ratio of unspliced HIV RNA:HIV DNA from baseline administration of pembrolizumab on day 1 are shown for individual patients. Participants missing baseline or visit measures were excluded from the corresponding waterfall plots.
Post hoc analysis suggested that perturbation of the reservoir was related to the CD4:CD8 ratio. The increase in unspliced HIV RNA, unspliced HIV RNA:DNA ratio, and HIV plasma RNA by HMMCgag assay between baseline and day 8 was larger among participants with CD4:CD8 ratios above 0.5 (P = 0.004, 0.041, and 0.053, respectively; table S3), whereas the decrease in HIV DNA from baseline to day 2 was larger in participants with CD4:CD8 ratios below 0.5 (P = 0.017; table S3). Change in HIV DNA from baseline to day 8 was similar among all CD4:CD8 ratio strata (P = 0.8; table S3). Changes in unspliced HIV RNA, unspliced HIV RNA:DNA ratio, and HIV plasma RNA by HMMCgag assay between baseline and day 8 did not differ between participants with AIDS-defining or non–AIDS-defining cancers or by cohort.
Administration of anti–PD-1 over 6 cycles led to increased measures of an inducible HIV reservoir by the tat/rev induced limiting dilution assay
TILDA (tat/rev induced limiting dilution assay) was performed in 23 participants with longitudinal samples, including 13 participants who completed less than 7 cycles of anti–PD-1 therapy and 10 participants analyzed per protocol who completed at least 7 cycles of therapy (Fig. 5). Of the 10 participants completing at least 7 cycles of therapy (pink circles), 5 were further evaluated after 13 cycles of therapy (red circles) and 6 were evaluated at a subsequent EOT time point (orange triangles). By the seventh cycle of anti–PD-1 therapy, the frequency of cells with inducible virus was 1.44-fold higher than the baseline frequency (P = 0.008). Follow-up of these same participants demon-strated a subsequent decrease in the frequency of cells with inducible HIV before the 13th cycle to a ratio of 0.75 (P = 0.07) compared to baseline. In participants who discontinued pembrolizumab before completing 6 cycles of therapy, there was a decrease in the frequency of cells with inducible virus at the end of anti–PD-1 therapy (Table 2 and Fig. 5, blue triangles). For participants who completed 6 cycles of therapy, those with a CD4:CD8 ratio greater than 0.5 exhibited increased inducible HIV transcription (P = 0.025). Stratification by baseline CD4 count, AIDS-defining cancer, baseline HIV DNA, or evidence of HIV latency reversal at day 8 did not modify results (table S3).
Fig. 5. Repeated administration of anti–PD-1 over 6 cycles increased the inducible HIV reservoir in CD4+ T cells.
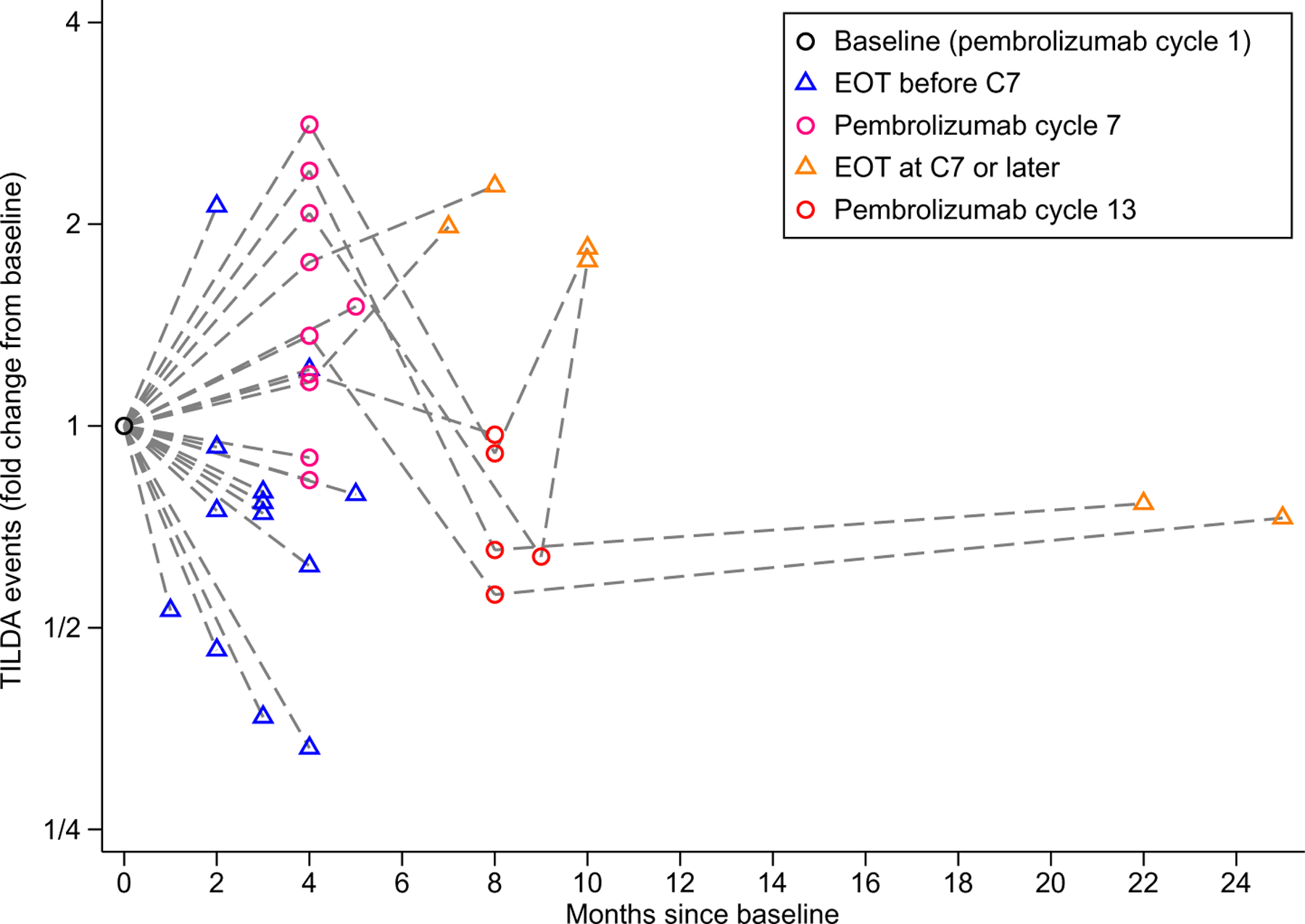
Changes in the frequency of cells with inducible HIV as measured by TILDA are shown stratified by initial comparisons performed at the prespecified cycle 7 (C7) time point versus an earlier time point due to dis-continuation of therapy. Orange triangles represent the final study time point. Cessation of the study participation occurred because of toxicity or progression of malignancy. EOT indicates end of therapy.
HIV single-genome sequencing from a participant with low concentrations of plasma HIV RNA after anti–PD-1 despite adherence to ART revealed no evidence of clonal expansion
Given that anti–PD-1 can cause substantial T cell proliferation of both CD4+ and CD8+ T cells [reviewed in (17)] and that we and others have shown that HIV latency is enriched in PD-1+ CD4+ T cells in PLWH on ART (8, 18), we asked whether anti–PD-1 could drive clonal expansion of infected CD4+ T cells. We chose to investigate this in the one participant who had a low degree of persistent viremia (<400 copies/ml) after anti–PD-1 treatment. Therefore, HIV sequence analyses were performed in a participant who was aviremic pre-pembrolizumab and who had persistent low concentrations of plasma HIV RNA (20 to 400 copies/ml), first noted after 6 cycles despite adherence to ART (19). We performed HIV env single-genome sequencing from sorted CD4+ T cell subsets at baseline, cycle 4, cycle 7, and EOT after 24 months of therapy. As with others in the cohort, this participant had decreased HIV DNA on day 2, increased unspliced HIV RNA on day 8, increased inducible HIV after 6 cycles, and decreased inducible HIV after 12 cycles. Despite adherence to ART, we detected HIV RNA (20 to 400 copies/ml) in plasma from this participant between cycles 6 and 31 of therapy, measured by a commercial assay performed during routine safety monitoring. We hypothesized that anti–PD-1 therapy may have induced the proliferation of a specific proviral clone, contributing to the persistent viremia, as has been previously observed in PLWH on ART (20). We observed several infected clones that persisted across multiple time points during treatment (Fig. 6A) including several clones present in cycles 4 and 7 that were not detected in the baseline sample (cycle 1). A large clone that was infected with the predominant sequence detected at baseline (clone 12; 34.4% of all detected variants) declined in prevalence over the treatment period (Fig. 6B). We also did not observe an increase in env diversity over the time (Figs. 6C and 7). There was insufficient plasma remaining from this participant to analyze the diversity and potential evolution of the circulating viruses.
Fig. 6. Comparison of HIV env sequences and clonal composition over time revealed no evidence of clonal expansion.
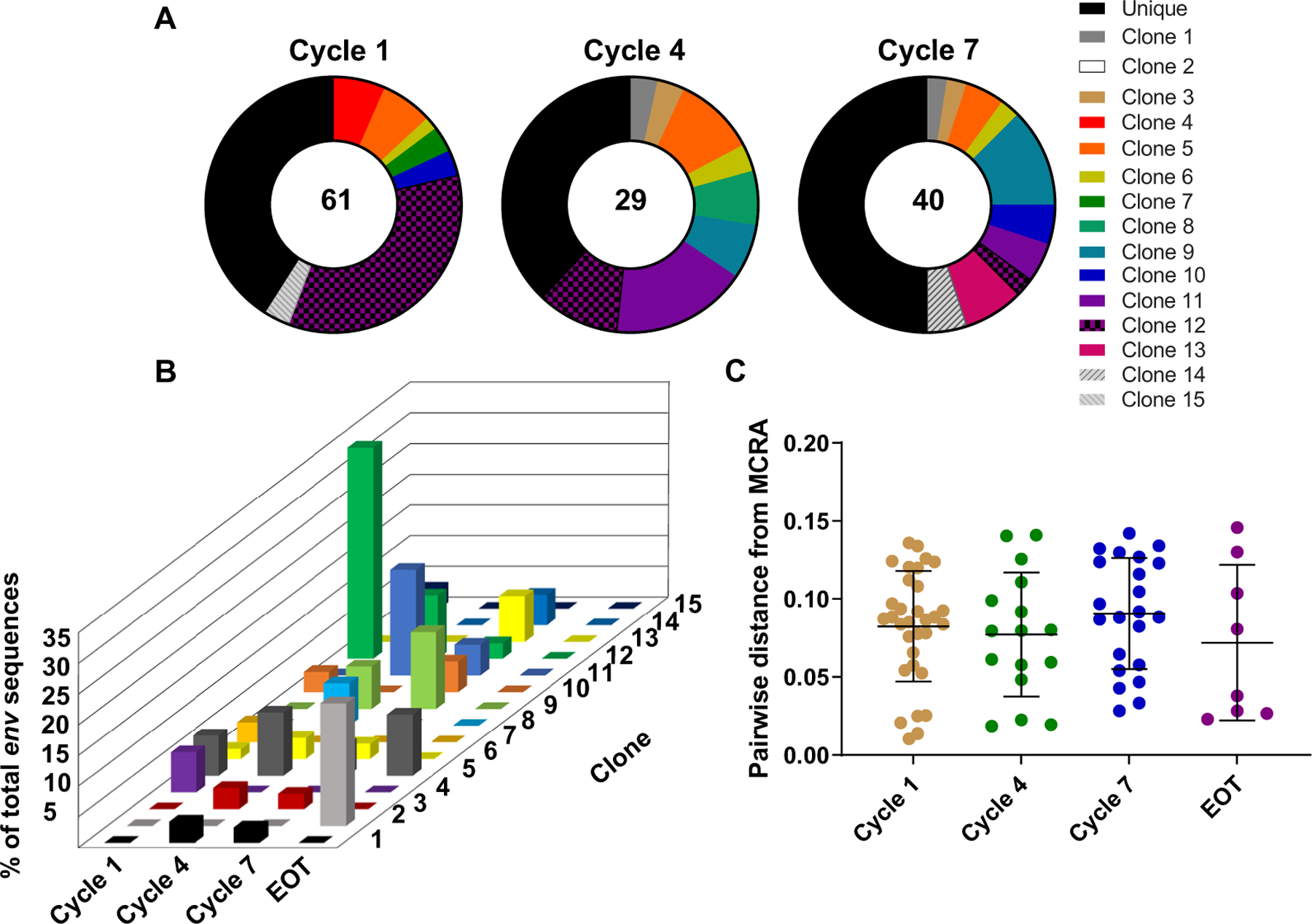
(A) The proportional distribution of 13 clones over four time points (cycle 1, cycle 4, cycle 7, and EOT) is shown. Clones are represented by color. (B) The percent of total env sequence at each time point are shown. Clone 12, which represented 34.4% of all clones at baseline, declined proportionally over time. (C) Pairwise distance analyses of distance from most recent common ancestor (MRCA) reveal no significant change over time. Data are presented as means and SD.
Fig. 7. Phylogenetic analysis of HIV env sequences isolated from CD4+ T cell subsets over 2 years in a participant receiving pembrolizumab reveals no evidence of clonal expansion.
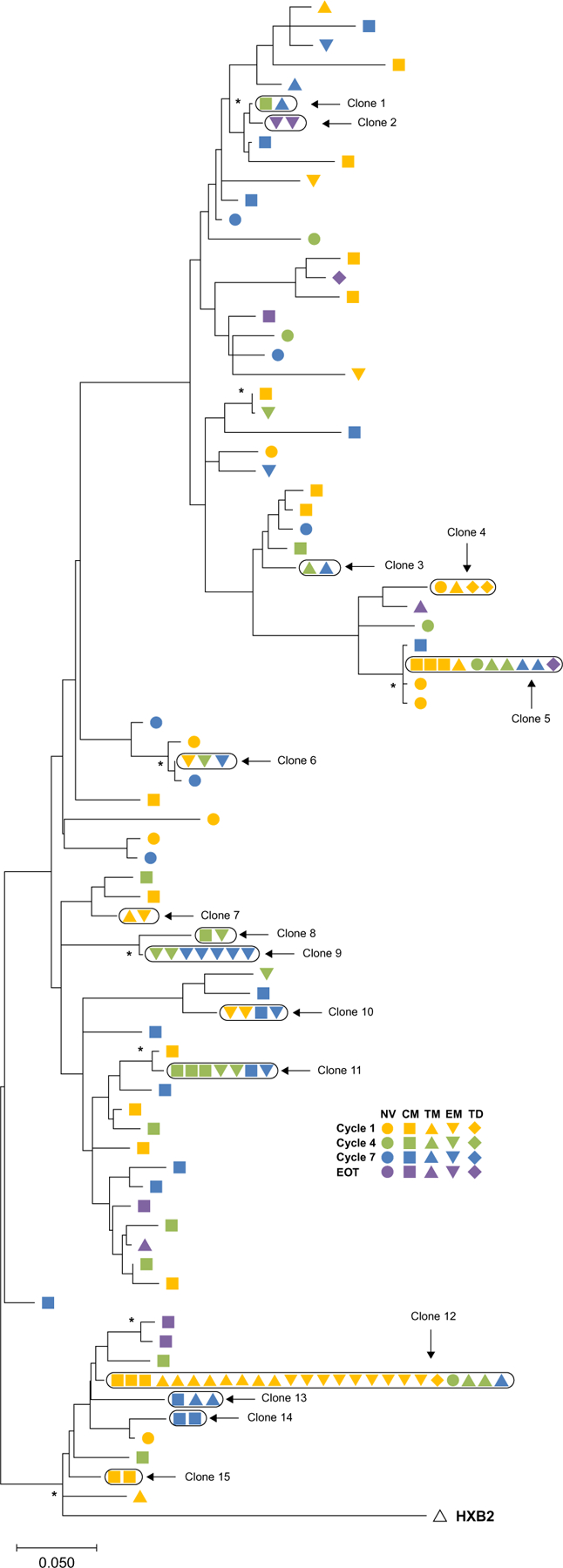
Sequences were categorized by time point (color) and T cell subset (shape). The outgroup is the prototypical HIV type 1 subtype B (isolate HXB2) (empty triangle). The scale indicates the number of nucleotide substitutions per site. Asterisk (*) indicates branches supported by >74% bootstrap replicates. NV, naïve; CM, central memory; TM, transitional memory; EM, effector memory; TD, terminally differentiated.
DISCUSSION
During long-term ART, HIV is enriched in CD4+ T cells that express PD-1. Leveraging a clinical trial of pembrolizumab in PLWH on ART with advanced cancer, we prospectively evaluated the effects of anti–PD-1 on HIV transcription and persistence. We found evidence of modest latency reversal defined by increased unspliced HIV RNA 1 week after administration of anti–PD-1 treatment, consistent with the induction of HIV transcription (10). By contrast, multiply spliced HIV RNA did not change after pembrolizumab administration, and values tended to be lower 8 days after the anti–PD-1 infusion. After multiple infusions of anti–PD-1, there was an increase in the frequency of cells with inducible virus as measured by TILDA in several participants, consistent with enhanced capacity for HIV transcription.
Our findings differ from a recent report showing no evidence of latency reversal after administration of another anti–PD-1 monoclonal antibody, nivolumab, in PLWH and cancer (15). The different findings in relation to HIV latency reversal may be due to several factors related to study design. First, participants in CITN-12 had lower baseline unspliced HIV RNA and HIV DNA, suggesting better control of HIV. In addition, our post hoc analyses show that induction of viral transcription was greatest in participants with higher CD4:CD8 ratios, suggesting that unmeasured differences in chronic inflammation between the cohorts may be responsible for differences in findings. Last, we adjusted our analyses for HIV DNA that decreased after PD-1 in both studies, which strengthened the inferred case for latency reversal. We cannot exclude that differences between studies may be due to differences between pembrolizumab and nivolumab. Both are IgG4 monoclonal antibodies with variable regions that bind different epitopes on PD-1 [reviewed in (21)]. Their relative effect on CD4+ T cell activation, which could affect HIV latency reversal, has, to our knowledge, not been directly compared. As IgG4 monoclonal antibodies, both are inefficient in eliciting complement-dependent cytotoxicity and antibody-induced cytotoxicity, and therefore, unmeasured differences in the Fc function are unlikely to be contributing to observed differences.
We used TILDA, an assay that measures the frequency of cells expressing tat/rev after exposure to T cell activation with phorbol 12-myristate 13-acetate (PMA) and ionomycin, which represents an inducible provirus population. We observed an increase in the frequency of cells with inducible virus after 6 cycles and recognize at least two potential interpretations. One possibility is the expansion of the number of infected cells potentially through proliferation, resulting in more infected cells with inducible virus. We think that this is unlikely because this would require infected cells to proliferate more than uninfected cells after anti–PD-1 treatment. Alternatively, the finding could be explained by the number of infected cells being unchanged, but that HIV transcription was easier to induce after stimulation with PMA and ionomycin ex vivo. We are unable to distinguish between these two possible explanations currently; however, we favor the latter interpretation given our prior studies where we examined CD4+ T cells from PLWH on ART ex vivo and showed that PD-1 engagement directly inhibited the positive transcriptional elongation factor b (P-TEFb) signaling pathway, which is usually activated after T cell stimulation (10) and enhances HIV transcription (22). Therefore, in the presence of anti–PD-1, we would expect to see greater activity of P-TEFb and enhancement of HIV transcription in each latently infected cell, which means an increase in the frequency of cells with inducible virus, as we observed with the TILDA assay. Ideally, we would measure the number of intact viruses using the intact proviral DNA assay (IPDA), but sufficient material was not available to complete this assay. It is intriguing that, in participants that did not complete 6 cycles because of adverse events or disease progression, there was a decline in the frequency of cells with inducible virus. This could indicate a PD-1–independent mechanism of T cell suppression leading to lack of response to anti–PD-1 that is common to both cancer responses and HIV latency reversal.
Most CITN-12 participants had undetectable HIV at all time points using a commercial plasma HIV RNA assay, and we did not observe any consistent changes in plasma HIV RNA. We did, however, observe a sustained increase in plasma HIV RNA in one participant. To evaluate the possibility of clonal expansion on long-term anti–PD-1 administration, we conducted HIV sequencing in this individual, who clinically exhibited a complete clinical oncologic response to pembrolizumab and who reported adherence to ART throughout his treatment. Sequencing of HIV env within peripheral blood mononuclear cells (PBMCs) collected at four time points did not demonstrate emergence of a predominant variant as might be expected if anti–PD-1 therapy was inducing the proliferation of a proviral clone. In the absence of sequencing of the plasma RNA, we are unable to conclude whether any of the minor emergent variants were the source of the persistent viremia observed. Our results did not show evidence of ongoing viral replication on the basis of lack of sequence evolution, suggesting that low concentrations of plasma HIV may be due to latency reversal in this participant. We acknowledge that the analysis of env sequences in only one participant does not exclude the possibility of clonal expansion of HIV-infected CD4+ T cells or ongoing virus replication after anti–PD-1, but in this one participant, new onset viremia could not be explained by an adverse effect of anti–PD-1 on the HIV reservoir. Collectively, these data support the hypothesis that PD-1 signaling in cells helps maintain HIV in a latent state and that inhibition of this signaling can contribute to latency reversal.
Our study also confirmed a decrease in HIV DNA in CD4+ T cells observed 24 hours after anti–PD-1 therapy (15). Our results may be due to generalized decreases in lymphocytes 24 hours after anti–PD-1 administration as has been observed in other studies and has been attributed to cell trafficking to tissues (23). However, anti–PD-1 therapy may also lead to proliferation and activation of HIV-specific CD8+ T cells (24). Ongoing studies in our laboratories are evaluating the effects of pembrolizumab on immune responses and HIV-specific cytotoxicity.
Our study has important strengths and limitations. It was prospectively designed, with clinical data and biologic specimens collected specifically to address the role of PD-1 blockade on the HIV reservoir. We recruited PLWH who were adherent to ART. The main limitation of the study was that we studied individuals with cancer and hence were limited in the types of biologic specimens obtained. In addition, because of the cancer and its treatment, the immune system in our cohort was not normal and the results may not be generalizable. The reservoir assays used also have their limitations. A quantitative viral outgrowth assay, the gold standard for quantitating the HIV reservoir, was not feasible within a multicenter cancer study because of leukapheresis requirements. Future studies could use the IPDA, which measures intact proviruses and is not expected to be affected by inducibility of HIV by anti–PD-1 (25). Last, we were unable to obtain sufficient plasma to assess the env sequence of cell-free virus, which might be another informative way to characterize latency reversal.
Development of immunotherapy remains a high scientific priority for HIV cure research (26). One of the main challenges for developing anti–PD-1 as a strategy for an HIV cure is the associated immune-related adverse events well described with all immune checkpoint blockers (27). Although we have shown that the frequency and spectrum of immune-related adverse events in PLWH compared to people living without HIV is similar (19), there is a very low tolerance for any grade 3 or 4 adverse events in PLWH, given that the quality of life is high on ART and life expectancy is similar to people without HIV (28). Therefore, although we have shown a clear effect of a single dose of anti–PD-1 on reversing latency in this study, further dose-finding studies will be needed in PLWH to determine whether toxicity could be reduced through reduction in dose without reducing activity, as recently demonstrated in a clinical trial of low-dose nivolumab in chronic hepatitis B virus (29). Whether a low dose of anti–PD-1 alone will overcome all immune-related adverse events is unclear, as recently demonstrated in a dose-escalating study of cemiplimab in PLWH on ART without cancer (30). Alternate strategies include subcutaneous administration of anti–PD-1 monoclonal antibodies (clinicaltrials.gov, NCT03316274) or combination of low-dose anti–PD-1 with other interventions that can reverse latency or enhance HIV-specific T cell function.
In summary, monoclonal antibodies targeting PD-1 led to reversal of HIV latency in CD4+ T cells after the first infusion. With repeated dosing in the setting of cancer therapy, we observed an increase in the frequency of cells with inducible virus. Because limited exposure of anti–PD-1 therapy in PLWH without cancer is desirable given possible immune-related toxicities, it is promising that latency reversal was observed even after a single infusion.
MATERIALS AND METHODS
Study design
Within a multicenter, open-label, nonrandomized phase 1 clinical trial of pembrolizumab for PLWH on ART and advanced cancer, CITN-12, we conducted a prospective correlative study to test the hypothesis that anti–PD-1 induced latency reversal and to evaluate the effects of anti–PD-1 on measures of HIV persistence. Participant characteristics and primary clinical results have been previously described (19). Briefly, the study enrolled PLWH on ART with advanced cancer, on ART for at least 4 weeks with plasma HIV RNA of <200 copies/ml, and CD4 of >100 cells/µl. The sample size determination for this study was based on the evaluation of safety in three cohorts of up to 12 participants with HIV and advanced cancer defined by baseline CD4+ T cell count (100 to 199, 200 to 350, or >350 cells/µl). CITN-12 was approved by institutional review boards at the Fred Hutchinson Cancer Research Center and each participating center. All participants provided written informed consent. Pembrolizumab (200 mg) was administered intravenously every 3 weeks for up to 35 doses with continuous ART. Treatment was discontinued for progressive disease as defined by disease-specific criteria, unacceptable adverse events as graded by the Common Terminology Criteria for Adverse Events version 4.0, other serious illness, investigator decision, consent withdrawal, or completion of 2 years of therapy (19).
Biospecimen collection was prespecified (Fig 1). Peripheral blood was collected in acid citrate dextrose tubes and shipped overnight for central processing of PBMCs. PBMCs were isolated from whole blood using Ficoll-Hypaque and cryopreserved as described (31). Frozen PBMC aliquots were stored in temperature-monitored liquid N2 vapor-phase freezers. Plasma was processed locally within 4 hours of collection for plasma HIV RNA quantification using an ultracentrifugation procedure as previously described (32). Blood was collected before each pembrolizumab administration, 1 day after administration (day 2), and 7 days after administration (day 8). For the first cycle, blood was also collected 2 hours after administration. Additional blood samples were collected at the EOT and at a follow-up visit 30 days after EOT (denoted FU).
Samples with at least 10 million banked PBMCs (CD4+ T cell–associated HIV RNA and HIV DNA assays) or at least 15 million banked PBMCs (TILDA) or 8 ml of plasma were processed for analyses. For measures of CD4+ T cell–associated HIV RNA (unspliced or multiply spliced) and CD4+ T-cell associated HIV DNA, results from experimental wells with fewer than 5000 or 1000 input CD4+ T cells, respectively, were excluded from analysis. Laboratory experiments included technical replicates. Analyses for unspliced and multiply spliced HIV RNA were performed in quadruplicate and for HIV DNA in triplicate, as previously described (33, 34). Samples from different time points from each patient were run together on the same plate to reduce interassay variability. For TILDA analyses, flow cytometry measures confirmed efficient activation of CD4+ T cells (CD69+ CD4+ T cells average frequency = 94%) and cell viability (87% LIVE/DEAD staining), and no samples were excluded from the analysis. For each sample evaluated by TILDA, activated CD4+ T cells were distributed in limiting dilutions (four dilutions, up to 24 replicates per dilution, average number of replicate wells per dilution = 21.3). All measurements of HIV were made blinded to the clinical correlation including CD4+ T cell count and time point in relation to administration to pembrolizumab.
Measurement of HIV RNA and DNA
Cell-associated unspliced and multiply spliced HIV RNA and HIV DNA were quantified in CD4+ T cells isolated from PBMCs collected at baseline and during the first 2 cycles of therapy as previously described (33, 34). The lower limit of detection of the total HIV DNA and cell-associated HIV RNA assays was one copy per well. If there was a signal detected but unable to be quantified because it was less than 1.0, then it was included as 0.5. If there was no signal, then this was included as zero.
Plasma HIV RNA was quantified using a previously described quantitative real-time polymerase chain reaction (PCR) single-copy assay (HMMCgag) targeting a noncoding 5′ region of HIV gag (32). In addition, plasma HIV RNA was monitored by clinical assays before each of the first 4 cycles of pembrolizumab, and then every 3 cycles, using commercial assays with lower limits of detection of either 20 or 40 copies/ml, depending on the treating site.
tat/rev induced limiting dilution assay
Measurement of the frequency of CD4+ T cells harboring inducible viruses was measured by TILDA as previously described at baseline, after 6 and 12 cycles of therapy, and at EOT (35). CD4+ T cells were isolated from PBMCs by negative magnetic selection (STEMCELL Technologies) per the manufacturer’s instructions and then stimulated for 12 hours with PMA (100 ng/ml) and ionomycin (1 µg/ml) (Sigma-Aldrich). Cells were plated in limiting dilutions, and HIV tat/rev transcripts were measured by real-time PCR. The number of positive wells at each dilution was counted, and Poisson law was used to calculate for frequencies of HIV RNA+ cells per million CD4+ T cells using extreme limiting dilution analysis (ELDA) software (http://bioinf.wehi.edu.au/software/elda/).
Cell sorting, nucleic acid isolation, single-genome sequencing, and sequence analysis
HIV sequence analysis was performed on CD4+ T cell subsets isolated from blood collected at baseline, cycle 4, cycle 7, and EOT. Cryopreserved PBMCs were thawed and incubated with a CD4+ T cell subset identification cocktail [anti-CD3 phycoerythrin (PE), anti-CD4 PE–Texas Red, anti-C-C motif chemokine receptor 7 (CCR7) Alexa Fluor 488, anti-CD27 Brilliant Violet 711, anti-CD45RA PE-Cy7, and LIVE/DEAD fixable dead cell aqua; table S4]. CD4+ T cell subsets were defined as naïve (CD4+CD45RA+CD27+CCR7+), central memory (CD4+CD45RA−CD27+CCR7+), transitional memory (CD4+ CD45RA−CD27+CCR7−), effector memory (CD4+CD45RA−CD27−CCR7−), and terminally differentiated (CD4+CD45RA+CD27−CCR7−). (36) Sorting was performed on a FACSAria sorter (BD Biosciences). Sorted cells were resuspended in lysis buffer [10 mM tris-HCl, 0.5% NP-40, 0.5% Tween 20, and proteinase K (300 µg/ml)]. Samples were incubated at 55°C for 1 hour followed by an inactivation phase for 15 min at 85°C. Lysates were used as templates for single-genome sequencing of the gp120 V1-V3 region as previously described, and PCR amplimers were sequenced with Sanger sequencing (36).
Hypermutants were identified using the Hypermut2.0 tool (hiv.lanl.gov), and nonintact env sequences were removed from analyses. Alignments were performed using Clustal W in CLC Main Workbench (QIAGEN), and maximum likelihood trees were constructed on Molecular Evolutionary Genetics Analysis (MEGA) X (http://megasoftware.net) using the general time reversible plus gamma model of nucleotide substitution and supported by 100 bootstrap replicates. Identical sequences representing expansion of infected cells were identified using the UniSeq program (https://indra.mullins.microbiol.washington.edu/cgi-bin/UniSeq/uniseq.cgi) and confirmed with individual sequence alignments. Divergence of unique sequences related to the most common recent ancestor was estimated using DIVEIN (37). Images were created with MEGA X (phylogenetic tree) or PRISM (graphs) and further edited using Adobe Illustrator.
Statistical analysis
Raw, individual-level data can be found in data file S1. For each study visit, measurements of plasma HIV RNA, as well as unspliced HIV RNA, multiply spliced HIV RNA and HIV DNA, and the ratio of unspliced HIV RNA to DNA were summarized by median and IQR. Comparison of baseline measurements between cohorts was evaluated by the Kruskal-Wallis test. Fold change compared to the baseline of individual measures of plasma HIV RNA, unspliced HIV RNA, and HIV DNA per 106 CD4+ T cells and the RNA:DNA ratio were computed for each participant at each visit. Pairwise Spearman rank correlation coefficients of these measures were calculated for baseline and day 8.
Within-participant longitudinal changes in HIV RNA and DNA were assessed by multilevel mixed-effects negative binomial regression models in which each virological measure was the dependent variable (33, 38). Log of the cellular input (18S ribosomal RNA copies for unspliced HIV RNA and CCR5 DNA copies for HIV DNA) was included in the fixed effects model as an offset. The clustered sandwich estimator was used for SEs from regression models. For analysis of plasma HIV RNA, input was assumed fixed. Time was parameterized as a categorical variable by visit. Two-tailed P < 0.05 was interpreted as statistically significant. CD4 cohort, CD4:CD8 strata (<0.5 versus ≥0.5), and tumor type (AIDS-defining malignancy versus non–AIDS-defining malignancy) were evaluated for effect modification of HIV outcomes by adding a multiplicative interaction term to regression models. All analyses were conducted with Stata v15 and v16. Data for Figs. 2 to 5 were visualized with the ggplot2 package.
For TILDA analyses, because not all participants completed 6 cycles of therapy, EOT samples were categorized as before or after 6 cycles. Within-participant fold change of TILDA from baseline was estimated by nesting TILDA within a multilevel mixed-effects model with complementary log-log link function (39). For one participant with no positive replicates at one time point, we imputed a single positive replicate to allow the model to converge. In addition, we evaluated whether baseline HIV DNA elevated unspliced HIV RNA:DNA ratio at day 8, tumor type, baseline CD4+ T cell stratum, and baseline CD4:CD8 stratum for effect modification of TILDA by adding a multiplicative interaction term to the regression model.
Supplementary Material
Acknowledgments:
We dedicate this manuscript to the memory of the late M. A. “Mac” Cheever, Principal Investigator of the CITN, who was instrumental in developing CITN-12. We thank the study participants; staff from the CITN: J. Kaiser, L. Lundgren, A. Claeys, A. Wright, B. Bertoldi, A. LaCroix, and D. Schullery; as well as CITN sites that enrolled participants in CITN-12 [National Cancer Institute (NCI) HIV and AIDS Malignancy Branch at the Center for Cancer Research, University of California San Francisco, Yale, Roswell Park, and Louisiana State University Health Science Center]. We also thank S. Townson, E. Chartash, and D. Kaufman from Merck as well as E. Sharon, W. Merritt, and R. Little from the NCI’s Cancer Treatment Evaluation Program. We thank A. Pagliuzza from Université de Montréal for technical assistance.
Funding:
This study was funded in part by the Intramural Research Program of the National Institutes of Health (NIH), NCI ZIABC011700 (to T.S.U.), and ZIABC010888 (to R.Y.). Additional U.S. federal support came from the NCI, UM1CA154967 (to M.A.C.), NIH contracts HHSN261200800001E and 75N91019D00024 (to J.D.L.), the National Institutes of Health Intramural Program, and the National Institutes for Allergy and Infectious Diseases UM1AI126611 [Delaney AIDS Research Enterprise (DARE) Collaboratory to S.G.D., S.R.L., R.S., and N.C.]. Additional study funding was obtained from the National Health and Medical Research Council (NHMRC) of Australia including an NHMRC program grant (to S.R.L.) and practitioner fellowship (to S.R.L.). Merck & Co. Inc., Kenilworth, NJ, USA provided funding to the NCI Cancer Therapy Evaluation Program and CITN in support of clinical trial NCT02595866.
Footnotes
Competing interests: T.S.U., M.A.C., S.P.F., and R.Y. receive research support from Merck; T.S.U., R.R., K.L., and R.Y. receive research support from Celgene/Bristol-Myers Squibb; and R.R., K.L., and R.Y. receive research support from EMD Serano and CTI Biopharma, all through cooperative research and development agreements (CRADA) with the NCI. T.S.U. received research support from Roche through a clinical trial agreement with the Fred Hutchinson Cancer Research Center and consulted for AbbVie and Seattle Genetics. T.S.U. and P.H.G. are current employees of Regeneron and have stock options. R.Y. received drug for research from Janssen Pharmaceuticals under a material transfer agreement (MTA). T.S.U. and R.Y. are co-inventors on U.S. patent 10 001,483 titled “Methods for the treatment of Kaposi’s sarcoma or KSHV-induced lymphoma using immunomodulatory compounds and uses of biomarkers.” R.Y. is also a co-inventor on patents on a peptide vaccine for HIV (U.S. patent 9474793) and the treatment of Kaposi sarcoma with IL-12 (U.S. patents 6509321 and 6423308), and an immediate family member of R.Y. is a co-inventor on patents related to internalization of target receptors (U.S. patent 8420620), KSHV vIL-6 (U.S. patents 7374756, 7235365, 7108981, and 6939547), and the use of calreticulin and calreticulin fragments to inhibit angiogenesis (U.S. patent 7488711). All rights, title, and interest to these patents have been or should, by law, be assigned to the U.S. Department of Health and Human Services; the government conveys a portion of the royalties it receives to its employee inventors under the Federal Technology Transfer Act of 1986 (P.L. 99–502). S.R.L. has received investigator-initiated research funding from Merck, Gilead Sciences, Viiv, and Leidos and is a member of the scientific advisory boards of Merck, Viiv, Gilead, Immunocore, and Aelix. J.D.L. has received research funding from and served as a scientific advisor to Gilead Sciences. S.G.D. receives research support from Gilead and Merck; is a member of the scientific advisory boards for BryoLogyx, Enochian Biosciences, and Tendel; and has consulted for AbbVie, Eli Lilly, GSK/ViiV, and Immunocore. N.C. has received investigator-initiated research funding from Merck and EMD Serono. C.-c.J.W. has received investigator-initiated research support from Bristol-Myers Squibb. All other authors declare that they have no competing interests.
Data and materials availability:
All data associated with this study are present in the paper or the Supplementary Materials. Sequences generated in this study have been deposited in GenBank under accession number MZ344692-MZ344769. Deidentified data and available biospecimens may be made available to the scientific community after scientific review. Inquiries can be made by contacting CITN@fredhutch.org and mta@fredhutch.org. Completion of an MTA is required for all shared materials.
REFERENCES AND NOTES
- 1.Dybul M, Attoye T, Baptiste S, Cherutich P, Dabis F, Deeks SG, Dieffenbach C, Doehle B, Goodenow MM, Jiang A, Kemps D, Lewin SR, Lumpkin MM, Mathae L, McCune JM, Ndung’u T, Nsubuga M, Peay HL, Pottage J, Warren M, Sikazwe I; Sunnylands 2019 Working Group, The case for an HIV cure and how to get there. Lancet HIV 8, e51–e58 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simonetti FR, Zhang H, Soroosh GP, Duan J, Rhodehouse K, Hill AL, Beg SA, McCormick K, Raymond HE, Nobles CL, Everett JK, Kwon KJ, White JA, Lai J, Margolick JB, Hoh R, Deeks SG, Bushman FD, Siliciano JD, Siliciano RF, Antigen-driven clonal selection shapes the persistence of HIV-1-infected CD4+ T cells in vivo. J. Clin. Invest 131, e145254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zerbato JM, Purves HV, Lewin SR, Rasmussen TA, Between a shock and a hard place: Challenges and developments in HIV latency reversal. Curr. Opin. Virol 38, 1–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Margolis DM, Garcia JV, Hazuda DJ, Haynes BF, Latency reversal and viral clearance to cure HIV-1. Science 353, aaf6517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta RK, Abdul-Jawad S, McCoy LE, Mok HP, Peppa D, Salgado M, Martinez-Picado J, Nijhuis M, Wensing AMJ, Lee H, Grant P, Nastouli E, Lambert J, Pace M, Salasc F, Monit C, Innes AJ, Muir L, Waters L, Frater J, Lever AML, Edwards SG, Gabriel IH, Olavarria E, HIV-1 remission following CCR5∆32/∆32 haematopoietic stem-cell transplantation. Nature 568, 244–248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lurain K, Ramaswami R, Yarchoan R, Uldrick TS, Anti-PD-1 and anti-PD-L1 monoclonal antibodies in people living with HIV and cancer. Curr. HIV/AIDS Rep 17, 547–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerham LR, Jain V, Sinclair E, Glidden DV, Hartogenesis W, Hatano H, Hunt PW, Martin JN, Pilcher CD, Sekaly R, McCune JM, Hecht FM, Deeks SG, Programmed death-1 expression on CD4(+) and CD8(+) T cells in treated and untreated HIV disease. AIDS 28, 1749–1758 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sekaly RP, Chomont N, CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLOS Pathog 12, e1005761 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banga R, Procopio FA, Noto A, Pollakis G, Cavassini M, Ohmiti K, Corpataux JM, de Leval L, Pantaleo G, Perreau M, PD-1(+) and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat. Med 22, 754–761 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, Hoh R, Deeks SG, Hazuda DJ, Lewin SR, Routy JP, Sekaly RP, Chomont N, PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat. Commun 10, 814 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardons M, Baxter AE, Massanella M, Pagliuzza A, Fromentin R, Dufour C, Leyre L, Routy JP, Kaufmann DE, Chomont N, Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLOS Pathog 15, e1007619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, Palmer S, Fromentin R, Chomont N, Sekaly RP, Cameron PU, Lewin SR, Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 32, 1491–1497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van der Sluis RM, Kumar NA, Pascoe RD, Zerbato JM, Evans VA, Dantanarayana I, Anderson JL, Sekaly RP, Fromentin R, Chomont N, Cameron PU, Lewin SR, Combination immune checkpoint blockade to reverse HIV latency. J. Immunol 204, 1242–1254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guihot A, Marcelin AG, Massiani MA, Samri A, Soulie C, Autran B, Spano JP, Drastic decrease of the HIV reservoir in a patient treated with nivolumab for lung cancer. Ann. Oncol 29, 517–518 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen TA, Rajdev L, Rhodes A, Dantanarayana A, Tennakoon S, Chea S, Spelman T, Lensing S, Rutishauser R, Bakkour S, Busch M, Siliciano JD, Siliciano RF, Einstein MH, Dittmer DP, Chiao E, Deeks S, Durand C, Lewin SR, Impact of anti-PD-1 and anti-CTLA-4 on the human immunodeficiency virus (HIV) reservoir in people living with HIV with cancer on antiretroviral therapy: The AIDS Malignancy Consortium 095 study. Clin. Infect. Dis 73, e1973–e1981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scully EP, Rutishauser RL, Simoneau CR, Delagreverie H, Euler Z, Thanh C, Li JZ, Hartig H, Bakkour S, Busch M, Alter G, Marty FM, Wang CC, Deeks SG, Lorch J, Henrich TJ, Inconsistent HIV reservoir dynamics and immune responses following anti-PD-1 therapy in cancer patients with HIV infection. Ann. Oncol 29, 2141–2142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Waldman AD, Fritz JM, Lenardo MJ, A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol 20, 651–668 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel M-R, Ghattas G, Brenchley JM, HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med 15, 893–900 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uldrick TS, Goncalves PH, Abdul-Hay M, Claeys AJ, Emu B, Ernstoff MS, Fling SP, Fong L, Kaiser JC, Lacroix AM, Lee SY, Lundgren LM, Lurain K, Parsons CH, Peeramsetti S, Ramaswami R, Sharon E, Sznol M, Wang CJ, Yarchoan R, Cheever MA; Cancer Immunotherapy Trials Network (CITN)-12 Study Team, Assessment of the safety of pembrolizumab in patients With HIV and advanced cancer-a phase 1 study. JAMA Oncol 5, 1332–1339 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halvas EK, Joseph KW, Brandt LD, Guo S, Sobolewski MD, Jacobs JL, Tumiotto C, Bui JK, Cyktor JC, Keele BF, Morse GD, Bale MJ, Shao W, Kearney MF, Coffin JM, Rausch JW, Wu X, Hughes SH, Mellors JW, HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J. Clin. Invest 130, 5847–5857 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fessas P, Lee H, Ikemizu S, Janowitz T, A molecular and preclinical comparison of the PD-1-targeted T-cell checkpoint inhibitors nivolumab and pembrolizumab. Semin. Oncol 44, 136–140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mbonye UR, Gokulrangan G, Datt M, Dobrowolski C, Cooper M, Chance MR, Karn J, Phosphorylation of CDK9 at Ser175 enhances HIV transcription and is a marker of activated P-TEFb in CD4(+) T lymphocytes. PLOS Pathog 9, e1003338 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardiner D, Lalezari J, Lawitz E, DiMicco M, Ghalib R, Reddy KR, Chang KM, Sulkowski M, Marro SO, Anderson J, He B, Kansra V, McPhee F, Wind-Rotolo M, Grasela D, Selby M, Korman AJ, Lowy I, A randomized, double-blind, placebo-controlled assessment of BMS-936558, a fully human monoclonal antibody to programmed death-1 (PD-1), in patients with chronic hepatitis C virus infection. PLOS ONE 8, e63818 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rust BJ, Kean LS, Colonna L, Brandenstein KE, Poole NH, Obenza W, Enstrom MR, Maldini CR, Ellis GI, Fennessey CM, Huang ML, Keele BF, Jerome KR, Riley JL, Kiem HP, Peterson CW, Robust expansion of HIV CAR T cells following antigen boosting in ART-suppressed nonhuman primates. Blood 136, 1722–1734 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Mohsen M, Richman D, Siliciano RF, Nussenzweig MC, Howell BJ, Martinez-Picado J, Chomont N, Bar KJ, Yu XG, Lichterfeld M, Alcami J, Hazuda D, Bushman F, Siliciano JD, Betts MR, Spivak AM, Planelles V, Hahn BH, Smith DM, Ho YC, Buzon MJ, Gaebler C, Paiardini M, Li Q, Estes JD, Hope TJ, Kostman J, Mounzer K, Caskey M, Fox L, Frank I, Riley JL, Tebas P, Montaner LJ; BEAT-HIV Delaney Collaboratory to Cure HIV-1 infection, Recommendations for measuring HIV reservoir size in cure-directed clinical trials. Nat. Med 26, 1339–1350 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deeks SG, Archin N, Cannon P, Collins S, Jones RB, de Jong M, Lambotte O, Lamplough R, Ndung’u T, Sugarman J, Tiemessen CT, Vandekerckhove L, Lewin SR; International AIDS Society (IAS) Global Scientific Strategy working group, Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat. Med 27, 2085–2098 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Zhou S, Yang F, Qi X, Wang X, Guan X, Shen C, Duma N, Vera Aguilera J, Chintakuntlawar, Price KA, Molina JR, Pagliaro LC, Halfdanarson TR, Grothey A, Markovic SN, Nowakowski GS, Ansell SM, Wang ML, Treatment-related adverse events of PD-1 and PD-L1 inhibitors in clinical trials: A systematic review and meta-analysis. JAMA Oncol 5, 1008–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewin SR, Attoye T, Bansbach C, Doehle B, Dubé K, Dybul M, SenGupta D, Jiang A, Johnston R, Lamplough R, McCune JM, Nabel GJ, Ndung’u T, Pottage J, Ripin D, Rooney JF, Sikazwe I, Nsubuga M, Warren M, Deeks SG, Multi-stakeholder consensus on a target product profile for an HIV cure. Lancet HIV 8, e42–e50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gane E, Verdon DJ, Brooks AE, Gaggar A, Nguyen AH, Subramanian GM, Schwabe C, Dunbar PR, Anti-PD-1 blockade with nivolumab with and without therapeutic vaccination for virally suppressed chronic hepatitis B: A pilot study. J. Hepatol 71, 900–907 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Gay CL, Bosch RJ, McKhann A, Moseley KF, Wimbish CL, Hendrickx SM, Messer M, Furlong M, Campbell DM, Jennings C, Benson C, Turner Overton E, Macatangay BJC, Kuritzkes DR, Miller E, Tressler R, Eron JJ, David Hardy W, Suspected immune-related adverse events with an anti-PD-1 inhibitor in otherwise healthy people with HIV. J. Acquir. Immune Defic. Syndr 87, e234–e236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhardwaj N, Friedlander PA, Pavlick AC, Ernstoff MS, Gastman BR, Hanks BA, Curti BD, Albertini MR, Luke JJ, Blazquez AB, Balan S, Bedognetti D, Beechem JM, Crocker S, D’Amico L, Danaher P, Davis TA, Hawthorne T, Hess BW, Keler T, Lundgren LM, Morishima C, Ramchurren N, Rinchai DS, Salazar AM, Salim BA, Sharon E, Vitale LA, Wang E, Warren S, Yellin MJ, Doisis ML, Cheever MA, Fling SP, Flt3 ligand augments immune responses to anti-DEC-205-NY-ESO-1 vaccine through expansion of dendritic cell subsetts. Nat. Cancer 1, 1204–1217 (2020). [DOI] [PubMed] [Google Scholar]
- 32.Somsouk M, Dunham RM, Cohen M, Albright R, Abdel-Mohsen M, Liegler T, Lifson J, Piatak M, Gorelick R, Huang Y, Wu Y, Hsue PY, Martin JN, Deeks SG, McCune JM, Hunt PW, The immunologic effects of mesalamine in treated HIV-infected individuals with incomplete CD4+ T cell recovery: A randomized crossover trial. PLOS ONE 9, e116306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR, Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLOS Pathog 10, e1004473 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerbato JM, Khoury G, Zhao W, Gartner MJ, Pascoe RD, Rhodes A, Dantanarayana, Gooey M, Anderson J, Bacchetti P, Deeks SG, McMahon J, Roche M, Rasmussen TA, Purcell DF, Lewin SR, Multiply spliced HIV RNA is a predictive measure of virus production ex vivo and in vivo following reversal of HIV latency. EBioMed 65, 103241 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O’Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJ, Miller MD, Hazuda DJ, Deeks SG, Sekaly RP, Chomont N, A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMed 2, 874–883 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roche M, Tumpach C, Symons J, Gartner M, Anderson JL, Khoury G, Cashin K, Cameron PU, Churchill MJ, Deeks SG, Gorry PR, Lewin SR, CXCR4-using HIV strains predominate in naive and central memory CD4(+) T cells in people living with HIV on antiretroviral therapy: Implications for how latency is established and maintained. J. Virol 94, e01736–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI, DIVEIN: A web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 48, 405–408 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bacchetti P, Bosch RJ, Scully EP, Deng X, Busch MP, Deeks SG, Lewin SR, Statistical analysis of single-copy assays when some observations are zero. J. Virus Erad 5, 167–173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu Y, Smyth GK, ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in the paper or the Supplementary Materials. Sequences generated in this study have been deposited in GenBank under accession number MZ344692-MZ344769. Deidentified data and available biospecimens may be made available to the scientific community after scientific review. Inquiries can be made by contacting CITN@fredhutch.org and mta@fredhutch.org. Completion of an MTA is required for all shared materials.


