Abstract
Short interfering RNAs (siRNAs) show promise as gene-silencing therapeutics, but their cellular uptake remains a challenge. We have recently shown the synthesis of siRNAs bearing a single neutral phenylethyl phosphotriester linkage within the sense strand. Here, we report the synthesis of siRNAs bearing three different hydrophobic phosphate triester linkages at key positions within the sense strand and assess their gene silencing in the absence of a transfection carrier. The best siRNAs bearing hydrophobic phosphate triester tails were not aromatic and exhibited effective gene silencing (IC50 ≈ 56–141 nM), whereas the aromatic derivative with three hydrophobic tails did not exhibit carrier-free gene silencing.
Keywords: Short interfering RNA, Backbone modification, RNAi, Carrier-free delivery, Phosphate triester
Short interfering RNAs (siRNAs) work within the native RNA interference pathway to specifically suppress the expression of target genes by cleaving a complementary messenger RNA (mRNA) sequence and inhibiting translation.1−3 Because of this gene-silencing capability, siRNAs have been investigated as therapeutics in the treatment of rare genetic disorders, with much promise.4,5 Since 2018, three siRNA-based drugs have been approved by the U.S. Food and Drug Administration (FDA), and many other candidates are in late and early clinical trials.6−8 Despite this success, there are still numerous challenges associated with the inherent properties of siRNAs, including susceptibility to nuclease cleavage and poor cellular uptake due to the presence of the anionic phosphodiester backbone.9,10 Chemical modifications can be incorporated within the siRNA backbone and can mitigate some of these limitations.11,12 One of the most common types of backbone modifications is the phosphorothioate modification, which has been used in FDA-approved siRNA and antisense drugs.13,14
This modification improves the siRNA’s resistance against enzymatic degradation by nucleases, as the native phosphate backbone is the substrate for these enzymes.15 Despite the increase in stability, phosphorothioate linkages retain a negative charge. One way to overcome this negative charge within siRNAs involves replacing the phosphate backbone with neutral backbone moieties such as amides, triazoles, and other functional groups within siRNAs.16−20 Another way involves using neutral triester phosphate backbones.21 These modifications more closely resemble the natural phosphodiester backbone by retaining the overall structure of the phosphate group. Despite these properties, these groups have largely not been studied within siRNAs until relatively recently. In 2020, our group developed a chemical synthesis for siRNAs bearing a neutral phenylethyl phosphotriester linkage within the sense strand.22 The resulting siRNAs showed excellent gene silencing and improved siRNA strand selection. However, these siRNAs required a transfection agent to localize them into the cell to elicit their biological effect. Thus, improvements in the structure of neutral triester phosphate backbones are necessary to potentially improve gene silencing without a transfection agent carrier.
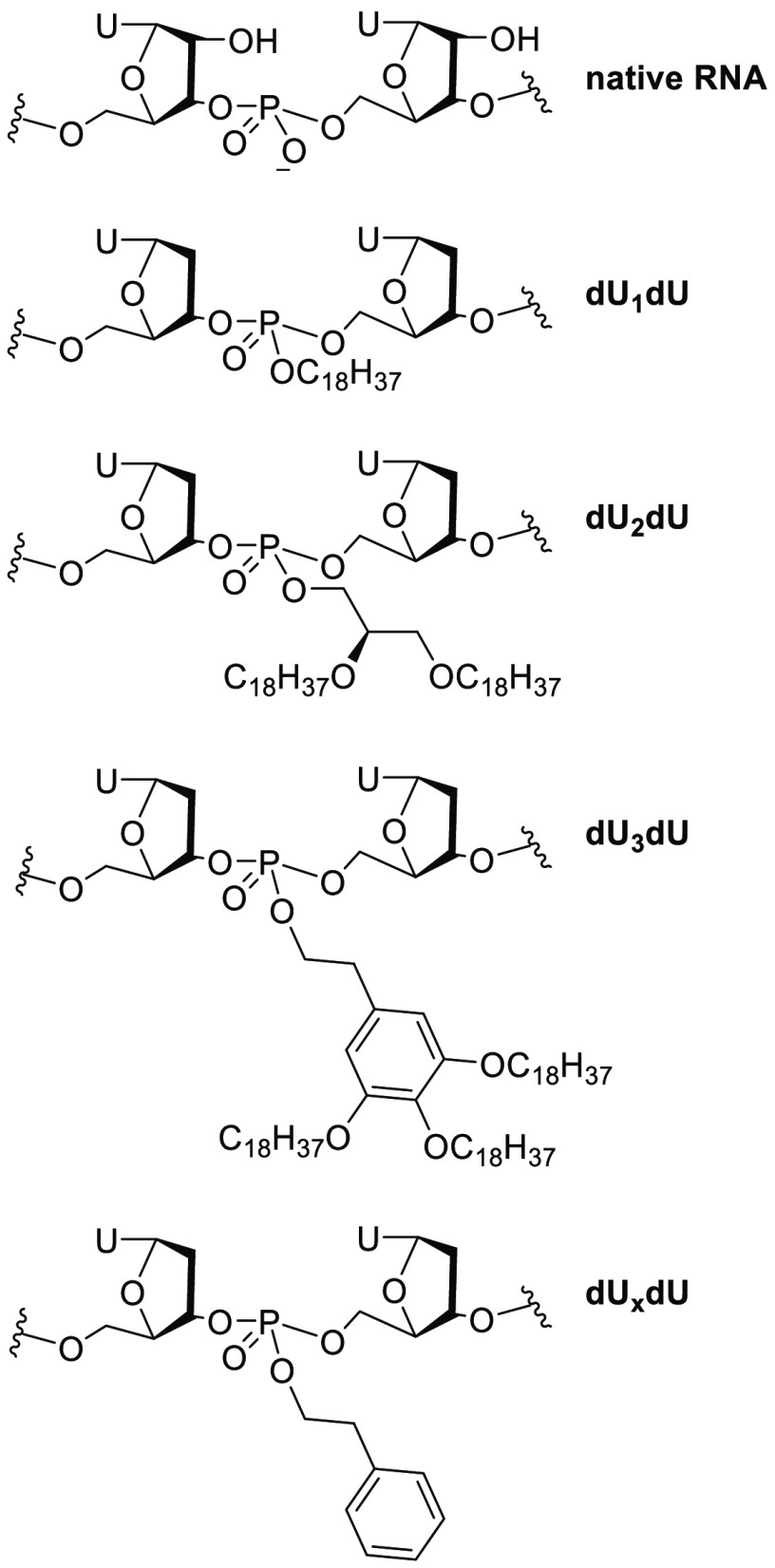
To expand and compare from this previous study, this paper reports the synthesis and biological evaluation of a small library of siRNAs bearing three novel hydrophobic neutral phosphate triester linkages at key positions within the sense strand (Figure 1). The first derivative, dU1dU, includes a stearyl phosphate ester group. The second derivative, dU2dU, contains a 2,3-distearyloxypropyl phosphate ester group. Finally, the third derivative, dU3dU, contains a 3,4,5-tris(stearyloxy)phenethyl phosphate ester group. The dUxdU modification contains a phenylethyl modification that was based on our prior study.21 Given our success with triester phosphate backbone siRNAs with the phenylethyl modification, the goal of this study is to expand the carbon repertoire of phosphate triesters within siRNAs to examine siRNA strand selection and to improve gene silencing without a transfection carrier. Our data suggests that many of these siRNAs displayed excellent gene-silencing activity and improved siRNA strand selection when compared to wild-type (wt) siRNA. Furthermore, several of them displayed effective gene silencing without the use of a transfection carrier reagent.
Figure 1.
Structure of chemical modifications incorporated within siRNAs in this study (dU1dU, dU2dU, and dU3dU). The dUxdU modification is from our prior study.22
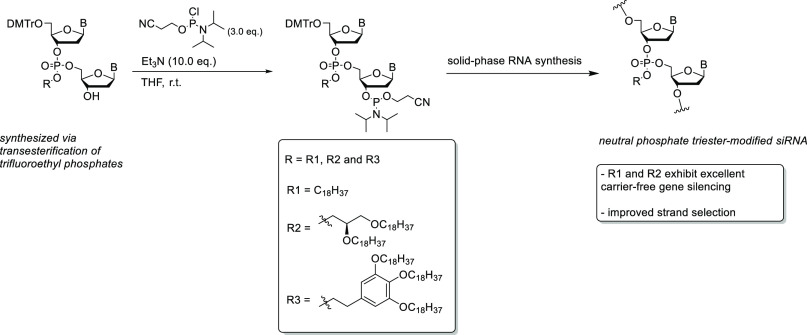
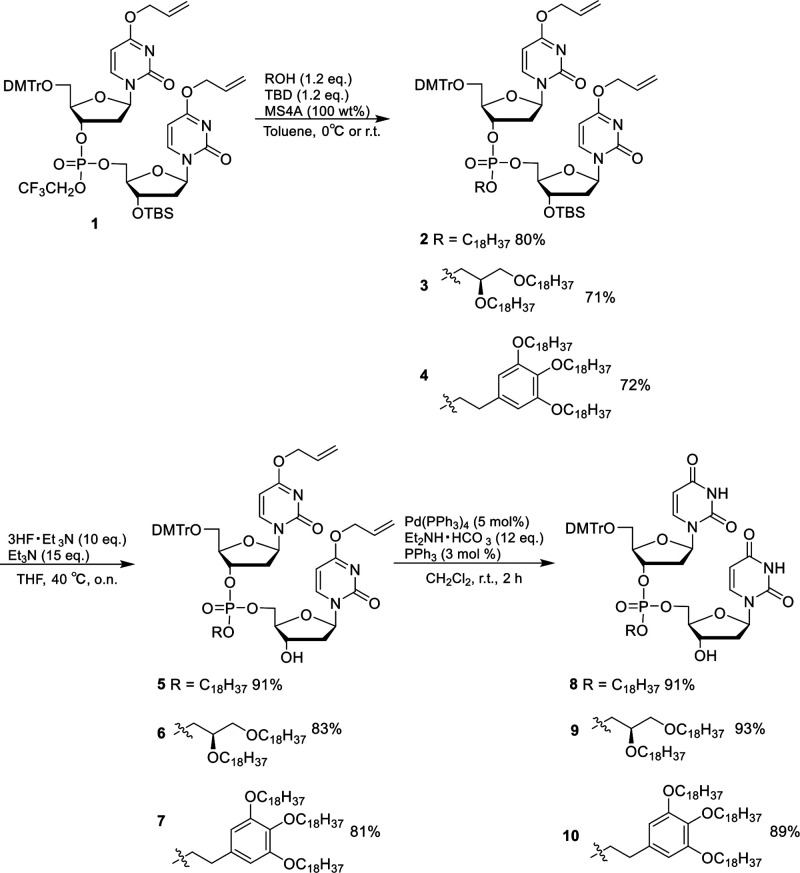
In order to incorporate the hydrophobic moieties within the siRNA backbone, three novel monoalcohols (compounds 5–7) were first synthesized (Scheme 1), starting with our recently published methodology to generate unsymmetrical dinucleotide 1.23 In order to generate the compounds 2–4 from 1, a solution of toluene with the alcohol, 1,5,7-triazabicyclo[4.4.0]dec-5-ene (TBD), and MS4A were added and allowed to mix for 30 min, followed by the addition of the dinucleotide 1 at 0 °C. To afford compound 2, stearyl alcohol was added to the solution, and the reaction was then quenched after 3 h with an acetic acid/toluene solution to generate the compound in an excellent 80% yield. In order to make compound 3, the stearyl alcohol was substituted with (S)-2,3-bis(octadecyloxy)propan-1-ol, which has a branched chain hydrophobic tail. After the reaction was quenched with acetic acid, compound 3 was obtained in a yield of 71%. The final monoalcohol 4 was produced using 2-(3,4,5-tris(octadecyloxy)phenyl)ethan-1-ol to give an aromatic ring substituted with three hydrophobic tails in a yield of 72%. Each compound was then treated with hydrofluoric acid (HF) to remove the TBS protecting group to afford the monoalcohols 5–7 in excellent yields (81–91%). Each compound 5–7 was then further deprotected by treatment with tetrakis(triphenylphosphine)palladium, triphenylphosphine, and diethylammonium hydrogen carbonate to restore the uracil nitrogenous base back to its original form to generate compounds 8–10 in high yields (89–93%).
Scheme 1. Synthesis of DMT-Protected Hydrophobic Phosphotriester Dinucleotide Monoalcohols.
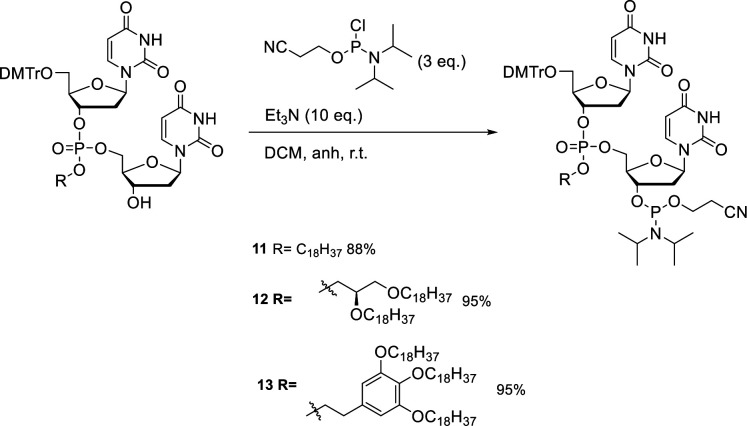
Once obtained, each monoalcohol 8, 9, and 10 was treated separately with a phosphitylating reagent under inert conditions to produce our DMT-phosphoramidite building blocks in good yields (Scheme 2). Each reaction had an overall yield of 88%, 95%, and 95% for phosphoramidite compounds 11, 12, and 13, respectively. Each phosphoramidite was immediately used for solid-phase RNA synthesis.
Scheme 2. Synthesis of DMT-Protected Hydrophobic Phosphotriester Dinucleotide Phosphoramidites.
The resulting oligonucleotides were cleaved from the solid support and deprotected following standard procedures. Oligonucleotide sequences are reported in Table 1. We then purified each single strand using reverse-phase HPLC and characterized it by mass spectrometry (Supporting Information Table S-1) prior to annealing it with its complementary antisense sequence. Circular dichroism studies were then performed to confirm that each duplex adopted the typical A-form helical conformation of siRNAs (Supporting Information Figure S-1). The A-form major groove of the helix is recognized by the RNA-induced silencing complex (RISC) during the RNA interference pathway, making this conformation desirable for proper siRNA activity.24,25 We also assessed the thermal stability of each duplex by measuring its melting temperature (Tm). The incorporation of the dU1dU modification at the 3′-end of the sequence (siRNA 1) did not cause a significant destabilizing effect (ΔTm = −4 °C). Additionally, this modification had only a moderate effect on the duplex’s thermal stability when placed internally (siRNA 2) or at the 5′-end (siRNA 3) (ΔTm = −8 and −6 °C, respectively). On the other hand, the incorporation of the dU2dU modification led to significant thermal destabilization at all positions (siRNAs 4–6) (ΔTm between −18 and −14 °C). In contrast, the siRNA bearing the aromatic dU3dU modification (siRNA 7) showed a slight increase in thermal stability (ΔTm = +1 °C), whereas the aromatic dUxdU modification (siRNAs 8 and 9) imparted only a slight destabilizing effect (ΔTm between −4 and −2 °C). In general, the long alkyl modifications on siRNAs 1–3 had moderate destabilization at all positions. The modifications on siRNAs 4–6 had high levels of thermal destabilization. And finally, the aromatic modifications from siRNAs 7–9 had minimal effects on destabilization, perhaps due to favorable aromatic stacking interactions with neighboring bases. Only one siRNA from the dU3dU modification was generated (siRNA 7: 3′-end of the sense strand) due to low stability issues with the phosphoramidite 13.
Table 1. siRNA Sequences, IC50, and Tm Valuesa.
dU1dU (in red) corresponds to the deoxyuridine-based modification with a single long alkyl chain (−C18H27); dU2dU (in blue) corresponds to the deoxythymidine-based modification with two long alkyl chains (−OC18H27); dU3dU (in purple) corresponds to the deoxythymidine-based modification with three long alkyl chains (−OC18H27) attached to an aromatic group; dUxdU (in brown) corresponds to the phenylethyl modification from our previous study.22 The top strand is the sense strand; the bottom strand is the antisense strand. The antisense strand is 5′-phosphorylated. IC50 values were calculated from luciferase data in the absence of a transfection carrier. NA = not applicable.
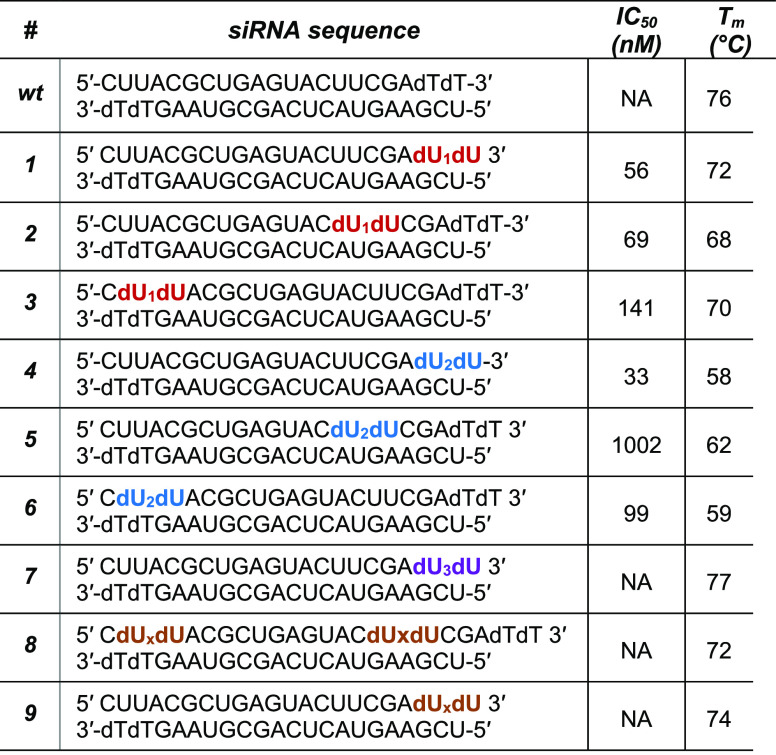
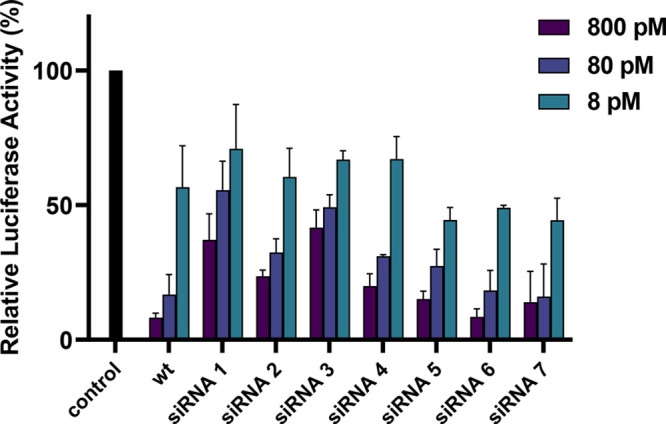
Since all siRNAs target firefly luciferase, we assessed their gene-silencing activity using a standard Dual-Luciferase Reporter Assay (Promega). We first confirmed the biological activity of all siRNAs in HeLa cells using Lipofectamine 2000 as a transfection carrier. All siRNAs showed dose-dependent knockdown comparable to that observed with wt siRNA, as illustrated in Figure 2. Given our interest in the design of these neutral phosphate triester derivatives, we assessed their ability to silence gene expression in the absence of a transfection carrier. As can be seen in Figure 3, siRNAs containing the single long alkyl chain triester phosphate (the dU1dU modification) exhibited effective gene silencing, thus suggesting that they were easily taken up by HeLa cells without the need of a transfection reagent. Regardless of the position of the dU1dU modification on the sense strand, this led to potent gene-silencing activity (IC50 = 56, 69, and 141 nM for all three siRNAs 1, 2, and 3, respectively). With respect to the 2,3-distearyloxypropyl phosphate ester group, the incorporation of the dU2dU modification at the 3′-end (siRNA 4) or at the 5′-end (siRNA 6) led to potent activity (IC50 = 33 and 99 nM, respectively). However, placing this modification within the central region impacted the siRNA’s gene-silencing activity compared to the other positions (IC50 = 1002 nM). It is not entirely clear why this is the case with the centrally modified siRNA, but perhaps the large, sterically demanding long alkyl chain at the central region is interfering with RISC loading. Despite the reduction in potency compared to that of siRNAs 4 and 6, siRNA 5 still exhibits a remarkable potency and a low IC50. In contrast to the above siRNAs, siRNAs incorporating both the aromatic modifications dU3dU (siRNA 7) and dUxdU (siRNAs 8 and 9) did not exhibit any noticeable gene silencing, thus suggesting that they were not taken up by HeLa cells in the absence of a transfection reagent.
Figure 2.
Relative expression of firefly luciferase after treatment with modified wild-type and modified siRNAs 1–7 using Lipofectamine 2000. Each value is the average of at least three biological replicates, and error bars indicate standard deviation.
Figure 3.
Relative expression of firefly luciferase after treatment with modified wild-type and modified siRNAs in the absence of a transfection carrier. Each value is the average of at least three biological replicates, and error bars indicate standard deviation.
We were not surprised that siRNAs 8 and 9 did not exhibit any gene-silencing activity, because the phenylethyl group is relatively small. However, we were surprised that the 3,4,5-tris(stearyloxy)phenethyl phosphate ester modification (dU3dU) did not exhibit activity. Given that siRNA 7 does work well in the gene-silencing assay with the transfection agent Lipofectamine 2000 (Figure 2), it is likely that siRNA 7 does not pass the cell membrane well, perhaps due to the three hydrophobic tails adopting a conformation that prevents its uptake. Finally, as a control and as expected, the wt siRNA, which lacks a hydrophobic tail to aid in cellular uptake, did not show any activity.
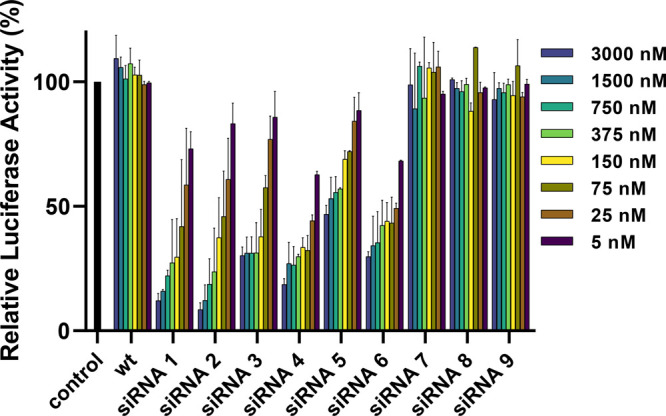
We then examined the effect of the dU1dU and dU2dU modifications on RISC strand selection using an assay we developed that quantifies the relative amounts of antisense- and sense-mediated gene silencing using quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR).26 The activity of a strand depends on efficient siRNA recognition, preferential selection during RISC maturation, and target recognition of the intended mRNA target.27 Chemical modifications to an siRNA can either enhance or interfere with any of these stages. Several modifications have been designed specifically for this controlling strand activity; however, the impact that novel modifications have on strand activity is often hard to predict.28 Therefore, siRNAs which showed carrier-free uptake were investigated for their impact on strand activity, and the RNA was measured by qRT-PCR (Figure 4). To omit any siRNA delivery variables, the strand activity assay was performed with Lipofectamine 2000.
Figure 4.
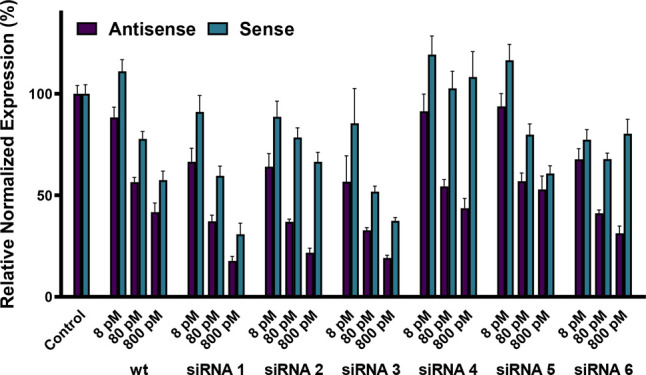
Relative abundance of antisense and sense strand mRNA target treatments with modified wild-type and modified siRNAs 1–6.
The goal for an applied siRNA is for the desired antisense strand to be incorporated in higher abundance into the RISC over the sense strand to minimize off-target effects. As shown in Figure 4, the unmodified wt siRNA exhibits a large preference for antisense strand uptake over sense strand uptake at 8 pM. However, at concentrations of 80 pM and above, the selectivity between antisense and sense strand uptake is negligible, meaning that both strands are contributing to their own gene silencing in roughly equal amounts.
SiRNAs bearing the single alkyl chain (dU1dU) modification exhibited increased selectivity between the desired antisense strand and the non-desired sense strand at all concentrations and positions. The largest and best difference was with siRNA 2, where excellent gene silencing occurred at 80 pM and above for the desired antisense strand but minimal gene silencing with the sense strand. This is a large improvement in selectivity compared to wt siRNA. SiRNAs bearing the larger double alkyl modifications (dU2dU) exhibited patterns similar to those of siRNAs 1–3; however, the largest difference in gene silencing occurs with siRNA 4 from 80 to 800 pM. The modification of this double alkyl chain of siRNA 4 is located at the 3′-end of the siRNA.22 In general, improvements in strand selection occur with these modifications when they are placed at the 3′-end and 5′-end positions of the sense strand. Finally, we investigated the nuclease stability of the 3′-modified siRNAs 1, 4, and 7. However, there was no significant improvement in nuclease resistance compared to the wt siRNA (Supporting Information Figure S-2).
To conclude, we report the synthesis of three novel dinucleotide phosphoramidite building blocks, with hydrophobic phosphate ester linkages, which were then used to synthesize a small library of siRNAs. Although all siRNAs showed good to excellent gene-silencing activity after transfection with Lipofectamine 2000, only those bearing one or two hydrophobic tail modifications (dU1dU and dU2dU) displayed potent activity in our carrier-free studies. This is an important observation because the delivery of large siRNAs across the cellular membrane remains a current challenge in the field, despite some recent breakthroughs with conjugates such as GalNAc29 and fatty acids.30 The addition of a third hydrophobic tail appears to interfere with cellular uptake during the carrier-free assays. It is not entirely clear why this is the case, but one possible explanation for this is that the increase in hydrophobic tails (to n = 3) interferes with the formation of a vesicle for endocytosis-mediated uptake or prevents binding to the receptor in the case of receptor-mediated uptake. We report that siRNAs 1–3, bearing a single hydrophobic chain, displayed the lowest IC50 values. Adding a second carbon chain led to a small loss of activity, but the resulting siRNAs were still quite effective in the absence of a transfection carrier. In contrast, siRNA 7, which possessed three long carbon chains attached to an aromatic phosphate triester linker, showed no activity without a transfection carrier. Previous data from siRNAs 8 and 9, bearing no long carbon chains at the phosphate triester moiety, also failed to penetrate the cell membrane.22 In addition, improved selectivity for antisense to sense strand-mediated gene silencing was observed for most siRNAs studied, thus highlighting improvements in strand selection and minimizing off-target effects. Future work will include trying to further explore the mechanism of carrier-free uptake through the use of imaging and novel chemical modifications to the triester phosphate moieties.
Acknowledgments
We acknowledge the Natural Sciences and Engineering Research Council (NSERC) and the Institute of Rheological Functions of Food in Japan for funding.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.2c00027.
Experimental procedures and 1H, 13C, and 31P NMR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hannon G. J. RNA interference. Nature 2002, 418, 244–51. 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Ameres S. L.; Martinez J.; Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell 2007, 130, 101–12. 10.1016/j.cell.2007.04.037. [DOI] [PubMed] [Google Scholar]
- Fire A.; Xu S. Q.; Montgomery M. K.; Kostas S. A.; Driver S. E.; Mello C. C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Bumcrot D.; Manoharan M.; Koteliansky V.; Sah D. W. Y. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. 2006, 2, 711–719. 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setten R. L.; Rossi J. J.; Han S.-P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discovery 2019, 18, 421–446. 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- Adams D.; Gonzalez-Duarte A.; O’Riordan W. D.; Yang C. C.; Ueda M.; Kristen A. V.; Tournev I.; Schmidt H. H.; Coelho T.; Berk J. L.; Lin K. P.; Vita G.; Attarian S.; Planté-Bordeneuve V.; Mezei M. M.; Campistol J. M.; Buades J.; Brannagan T. H. 3rd; Kim B. J.; Oh J.; Parman Y.; Sekijima Y.; Hawkins P. N.; Solomon S. D.; Polydefkis M.; Dyck P. J.; Gandhi P. J.; Goyal S.; Chen J.; Strahs A. L.; Nochur S. V.; Sweetser M. T.; Garg P. P.; Vaishnaw A. K.; Gollob J. A.; Suhr O. B. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21. 10.1056/NEJMoa1716153. [DOI] [PubMed] [Google Scholar]
- Scott L. J. Givosiran: First Approval. Drugs 2020, 80, 335–339. 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- Garrelfs S. F.; Frishberg Y.; Hulton S. A.; Koren M. J.; O’Riordan W. D.; Cochat P.; Deschênes G.; Shasha-Lavsky H.; Saland J. M.; Van’t Hoff W. G.; Fuster D. G.; Magen D.; Moochhala S. H.; Schalk G.; Simkova E.; Groothoff J. W.; Sas D. J.; Meliambro K. A.; Lu J.; Sweetser M. T.; Garg P. P.; Vaishnaw A. K.; Gansner J. M.; McGregor T. L.; Lieske J. C. Lumasiran, an RNAi Therapeutic for Primary Hyperoxaluria Type 1. N. Engl. J. Med. 2021, 384, 1216–1226. 10.1056/NEJMoa2021712. [DOI] [PubMed] [Google Scholar]
- Gavrilov K.; Saltzman W. M. Therapeutic siRNA: principles, challenges, and strategies. Yale J. Biol. Med. 2012, 85, 187–200. [PMC free article] [PubMed] [Google Scholar]
- Chernikov I. V.; Vlassov V. V.; Chernolovskaya E. L. Current Development of siRNA Bioconjugates: From Research to the Clinic. Front Pharmacol. 2019, 10, 444. 10.3389/fphar.2019.00444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S.; Sumaria C. S.; Pradeepkumar P. I. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. ChemMedChem. 2010, 5, 328–49. 10.1002/cmdc.200900444. [DOI] [PubMed] [Google Scholar]
- Braasch D. A.; Jensen S.; Liu Y.; Kaur K.; Arar K.; White M. A.; Corey D. R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 2003, 42, 7967–75. 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Detzer A.; Overhoff M.; Mescalchin A.; Rompf M.; Sczakiel G. Phosphorothioate-stimulated cellular uptake of siRNA: a cell culture model for mechanistic studies. Curr. Pharm. Des. 2008, 14, 3666–73. 10.2174/138161208786898770. [DOI] [PubMed] [Google Scholar]
- Eckstein F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic. Acid. Ther. 2014, 24, 374–87. 10.1089/nat.2014.0506. [DOI] [PubMed] [Google Scholar]
- Behlke M. A. Chemical modification of siRNAs for in vivo use. Oligonucleotides 2008, 18, 305–19. 10.1089/oli.2008.0164. [DOI] [PubMed] [Google Scholar]
- Mutisya D.; Selvam C.; Lunstad B. D.; Pallan P. S.; Haas A.; Leake D.; Egli M.; Rozners E. Amides are excellent mimics of phosphate internucleoside linkages and are well tolerated in short interfering RNAs. Nucleic Acids Res. 2014, 42, 6542–6551. 10.1093/nar/gku235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotikam V.; Rozners E. Amide-Modified RNA: Using Protein Backbone to Modulate Function of Short Interfering RNAs. Acc. Chem. Res. 2020, 53, 1782–1790. 10.1021/acs.accounts.0c00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiou T. C.; Peel B.; Huynh V.; Desaulniers J. P. Evaluation of siRNAs that contain internal variable-length spacer linkages. Bioorg. Med. Chem. Lett. 2012, 22, 5590–4. 10.1016/j.bmcl.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Efthymiou T. C.; Huynh V.; Oentoro J.; Peel B.; Desaulniers J. P. Efficient synthesis and cell-based silencing activity of siRNAS that contain triazole backbone linkages. Bioorg. Med. Chem. Lett. 2012, 22, 1722–1726. 10.1016/j.bmcl.2011.12.104. [DOI] [PubMed] [Google Scholar]
- Peel B. J.; Hagen G.; Krishnamurthy K.; Desaulniers J. P. Conjugation and Evaluation of Small Hydrophobic Molecules to Triazole-Linked siRNAs. ACS Med. Chem. Lett. 2015, 6, 117–122. 10.1021/ml500260j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade B. R.; Gogoi K.; Hamil A. S.; Palm-Apergi C.; van den Berg A.; Hagopian J. C.; Springer A. D.; Eguchi A.; Kacsinta A. D.; Dowdy C. F.; Presente A.; Lönn P.; Kaulich M.; Yoshioka N.; Gros E.; Cui X.-S.; Dowdy S. F. Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat. Biotechnol. 2014, 32, 1256–1261. 10.1038/nbt.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki K.; Hammill M. L.; Varley A. J.; Kitamura M.; Okauchi T.; Desaulniers J.-P. Synthesis and Evaluation of Neutral Phosphate Triester Backbone-Modified siRNAs. ACS Med. Chem. Lett. 2020, 11, 1457–1462. 10.1021/acsmedchemlett.0c00232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubaki K.; Shimooka H.; Kitamura M.; Okauchi T. Selective Transesterification of 2,2,2-Trifluoroethyl Phosphates: Synthesis of Mixed Unsymmetrical Phosphates. Org. Lett. 2019, 21, 9779–9783. 10.1021/acs.orglett.9b04003. [DOI] [PubMed] [Google Scholar]
- Hernández A. R.; Peterson L. W.; Kool E. T. Steric restrictions of RISC in RNA interference identified with size-expanded RNA nucleobases. ACS Chem. Biol. 2012, 7, 1454–1461. 10.1021/cb300174c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard L.; Rossi J. J. RNAi therapeutics: Principles, prospects and challenges. Adv. Drug Delivery Rev. 2007, 59, 75–86. 10.1016/j.addr.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley A. J.; Hammill M. L.; Salim L.; Desaulniers J.-P. Effects of Chemical Modifications on siRNA Strand Selection in Mammalian Cells. Nucleic Acid Ther. 2020, 30, 229–236. 10.1089/nat.2020.0848. [DOI] [PubMed] [Google Scholar]
- Noland C. L.; Doudna J. A. Multiple sensors ensure guide strand selection in human RNAi pathways. RNA 2013, 19, 639–48. 10.1261/rna.037424.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley A. J.; Desaulniers J.-P. Chemical strategies for strand selection in short-interfering RNAs. RSC Adv. 2021, 11, 2415–2426. 10.1039/D0RA07747J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair J. K.; Willoughby J. L.; Chan A.; Charisse K.; Alam M. R.; Wang Q.; Hoekstra M.; Kandasamy P.; Kel’in A. V.; Milstein S.; Taneja N.; O’Shea J.; Shaikh S.; Zhang L.; van der Sluis R. J.; Jung M. E.; Akinc A.; Hutabarat R.; Kuchimanchi S.; Fitzgerald K.; Zimmermann T.; van Berkel T. J.; Maier M. A.; Rajeev K. G.; Manoharan M. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014, 136, 16958–61. 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- Biscans A.; Coles A.; Echeverria D.; Khvorova A. The valency of fatty acid conjugates impacts siRNA pharmacokinetics, distribution, and efficacy in vivo. J. Controlled Release 2019, 302, 116–125. 10.1016/j.jconrel.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.