Highlights
-
•
Patients with episodic migraine showed a different methylation pattern in 2 CpG sites at the proximal promoter region of CALCA gene, that encodes the calcitonin gene-related peptide (CGRP).
-
•
Intriguingly, one of the two hypomethylated site (-1415) is located at the CREB binding site.
-
•
DNA methylation level at different CpG sites in CALCA promoter correlates with several clinical characteristics of migraine.
-
•
Our study provides evidence that the methylation of CALCA gene is reduced in patients with migraine and this hypomethylation could play a role in the disease.
Keywords: Migraine, CALCA gene, CGRP, Epigenetics, Methylation, CpG islands
Abstract
Recent studies suggested that epigenetic mechanisms, including DNA methylation, may be involved in migraine pathogenesis. The calcitonin gene-related peptide (CGRP), encoded by calcitonin gene-related peptide 1 (CALCA) gene, plays a key role in the disease. The aim of the study was to evaluate DNA methylation of CALCA gene in patients with episodic migraine. 22 patients with episodic migraine (F/M 15/7, mean age 39.7 ± 13.4 years) and 20 controls (F/M 12/8, mean age 40.5 ± 14.8 years) were recruited. Genomic DNA was extracted from peripheral blood. Cytosine-to-thymine conversion was obtained with sodium bisulfite. The methylation pattern of two CpG islands in the promoter region of CALCA gene was analyzed. No difference of methylation of the 30 CpG sites at the distal region of CALCA promoter was observed between migraineurs and controls. Interestingly, in patients with episodic migraine the methylation level was lower in 2 CpG sites at the proximal promoter region (CpG −1461, p = 0.037, and −1415, p = 0.035, respectively). Furthermore, DNA methylation level at different CpG sites correlates with several clinical characteristics of the disease, as age at onset, presence of nausea/vomiting, depression and anxiety (p < 0.05). In conclusion, we found that DNA methylation profile in two CpG sites at the proximal promoter region of CALCA is lower in migraineurs when compared to controls. Intriguingly, the −1415 hypomethylated unit is located at the CREB binding site, a nuclear transcription factor. In addition, we found a correlation between the level of CALCA methylation and several clinical features of migraine. Further studies with larger sample size are needed to confirm these results.
Introduction
Migraine is a common neurovascular disorder characterized by recurring episodes of pulsating headache associated with nausea, vomiting, photo- and phonophobia. Genetic factors play a major role in the susceptibility to the disease (Burstein et al., 2015, de Vries et al., 2009). In very rare cases, the migraine phenotype is transmitted as a monogenetic trait, but in the majority of patients, migraine is inherited as a polygenic disease, due to the contribution of variants with small effect at many genetic loci (Rainero et al., 2019a, Sutherland and Griffiths, 2017). However, at present, the number and the characteristics of genes involved in migraine are still matter of investigation.
Recently, an increased interest has been devoted to epigenetic mechanisms in migraine (Rainero et al., 2019b). Epigenetic markers are heritable changes in phenotype or gene expression in the absence of changes in DNA sequence. Epigenetic may explain how endogenous and exogenous factors, such as hormones, lights, foods and inflammation, may trigger migraine headache attacks (Sutherland et al., 2019). DNA methylation is the most common type of epigenetic modification and plays a key role in several disorders including cancer, vascular and neurodegenerative disorders (Ehrlich, 2019, Jin and Liu, 2018).
DNA methylation occurs at low abundance throughout the human genome. Biochemical process of methylation involves the addition of a methyl group to the fifth carbon of DNA cytosine residues, leading to 5-methylcytosines, and this process is mediated by a group of DNA methyltransferases (Gjaltema and Rots, 2020). The process occurs mainly in cytosines and guanines rich regions, the so-called “CpG islands”, which are enriched of CpG units. Throughout the genome there are CpG islands localized in regulatory DNA elements and are found up to 50–60% in promoter gene regions. Interestingly, the methylation of DNA within the promoter changes the functional state of regulatory regions, and methylation correlates with transcriptional silencing (Ehrlich, 2019).
A large number of studies showed that the neuropeptide calcitonin gene–related peptide (CGRP) has a key role in migraine pathogenesis (Durham, 2008, Edvinsson, 2007). CGRP and its receptors are widely expressed in trigeminal neurons (Tendl et al., 2013). In migraineurs, intravenous CGRP administration induces migraine-like symptoms and, during acute attacks, the neuropeptide is released into the cranial venous outflow, promoting a significant vasodilatation (Edvinsson, 2007, Russo, 2015). Finally, new drugs that antagonize CGRP receptor (gepants) are effective for acute migraine relief, and monoclonal antibodies against CGRP or its receptor effectively prevent migraine attacks (Edvinsson et al., 2018, Edvinsson, 2019, Russo, 2015).
CGRP is encoded by the CALCA gene, located on chromosome 11, and yields two alternatively spliced products, calcitonin and α-CGRP (Broad et al., 1989). These peptides bind to CGRP receptors, membrane heterodimer complexes composed of the calcitonin receptor like receptor protein and to an accessory protein called receptor activity modifying protein 1 (RAMP1). Surprisingly, very little is known about the methylation of the genes related to the CGRP-ergic system in patients with migraine. Interestingly, one study investigated the DNA methylation at RAMP1 promoter region, suggesting that epigenetics may play a role in migraine pathogenesis (Wan et al., 2015).
Thence, the aim of the study was to evaluate DNA methylation of CALCA gene in patients with episodic migraine and to evaluate correlation with the clinical symptoms that characterize migraine attacks as well as disease comorbidity.
Material and methods
Participants
We enrolled 22 unrelated patients with episodic migraine without aura (15 females, 7 males, mean age ± SD: 39.7 ± 13.4 years) and 20 controls (12 females, 8 males, mean age ± SD: 40.5 ± 14.8 years), attending the Headache Center of the Department of Neuroscience and Mental Health, AOU Città della Salute e della Scienza di Torino (Italy). The diagnosis of migraine was made according to ICHD-3 criteria (Headache Classification Committee of the International Headache Society (IHS), 2018). Headache characteristics were recorded using a standardized questionnaire including age at onset, headache frequency, duration and severity, pain characteristics, location, accompanying symptoms as nausea/vomiting, photophobia/phonophobia, number of medication intake, and triggers factors. In addition, neuropsychological tests as Beck’s Depression Inventory (BDI), STAI-X1 and STAI-X2 were administered. The clinical characteristics of patients and controls are summarized in Table 1. Patients and controls were of Caucasian origin and came from the same area of Northern Italy. Written informed consent was obtained from all participants, and the Internal Ethics review Board approved the study.
Table 1.
Clinical characteristics of patients with episodic migraine without aura and controls.
| Migraineurs | Controls | |
|---|---|---|
| Gender (F/M) | 15/7 | 12/8 |
| Age (yrs ± SD) | 39.7 ± 13.4 | 40.5 ± 14.8 |
| First grade positive familial history | 16 (72.7%) | – |
| Age at onset (yrs ± SD) | 18.3 ± 11.4 | – |
| Headache days per month (yrs ± SD) | 6.9 ± 3.4 | – |
| Preventive therapy (%) | 8 (36.3%) | – |
| Duration of disease (yrs ± SD) | 20.6 ± 15.6 | |
| Photophobia (%) | 19 (86.4%) | – |
| Phonophobia (%) | 17 (77.3%) | – |
| Nausea (%) | 16 (72.7%) | – |
| Vomiting (%) | 11 (50%) | – |
Genetics analysis
DNA extraction and bisulfite conversion
Genomic DNA was isolated from whole blood samples collected in an anticoagulant EDTA tube. All blood specimens were drawn via peripheral venepuncture in a pain free period (at least 12 h from the last migraine attack). DNA was extracted from 200 µL of the whole blood sample using the Gene Eluate Blood Genomic DNA Kit (SIGMA-ALDRICH, USA) according to manufacturer’s protocols.
Cytosine-to-thymine conversion by sodium bisulfite and DNA purification were performed using EZ DNA Methylation™ Kit (Zymo Research, Irvine, CA, USA). DNA recovery after conversion was quantified using Nanodrop (Thermo Scientific, Wilmington, DE, USA). After conversion, methylated C remains unchanged, while unmethylated C is read as T. Each DNA sample was sequenced following bisulfite conversion.
DNA methylation analysis
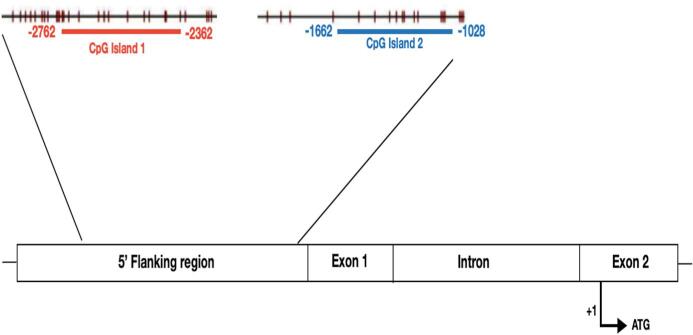
We analyzed the CpG islands in the promoter region of CALCA gene (NM_001033952.2). The promoter of CALCA includes a 2-kb long 5′ flanking region, and previous studies reported two CpG islands extending from about − 1.8 kb to exon 1 of the human CALCA gene (Broad et al, 1989). CpG islands are defined as a genomic region at least 200 bp long, with at least a GC content of 50%, and these regions have an observed-to-expected CpG ratio greater than 60% (Wan et al., 2015). In details, the promoter of CALCA contains two different CpG-rich islands: a first distal promoter region from −2762 to −2362 bp upstream (counted from ATG), and a second one encompassing the proximal promoter from −1662 to −1028 bp (Fig. 1).
Fig. 1.
CpG islands in the human CALCA promoter region. CpG island 1 (distal) is shown in red panel in the distal promoter region and CpG island 2 (proximal) in blue panel in the proximal promoter region of CALCA. Each vertical line indicates one CpG site within the indicated region (base pairs refers to the ATG site included in exon 2). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We identified and analyzed the CpG islands of the CALCA promoter using CpGplot (http:// www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/). Primers for the two target regions were designed using Primer3plus software (primers details upon request) using gDNA as template. Each canonical primer sequence (gDNA-primer) has been converted mimicking bisulfite treatment (every cytosine converted to thymine, Bs-primer), and bisulfite treatment considering possible CpG islands (cytosine converted to thymine if not located in a possible CpG island, CpG-primer). A universal M13 sequence has been added to the 5′ of each primer (M13-F in forward primers, M13-R in reverse primer). In brief, three different sequences (gDNA-, Bs- and CpG- primer) have been generated and pooled for each forward and reverse primer.
Samples were sequenced on an ABI PRISM 3130xl Genetic Analyzer, using the BigDye Terminator Cycle Sequencing Ready Reaction Kit version 3.1, following manufacturers’ instruction (Applied Biosystems). To validate the results of direct bisulfite sequencing, PCR products were amplified using the pooled primers and sequenced in both directions using universal M13 primers (M13-F and M13-R). The obtained sequences were analyzed using SeqScape version 2.5 software (Applied Biosystems).
The methylation status of each CpG was defined as unmethylated (T peak), heterozygous methylated (T/C double peaks), or fully methylated (C peak). For each subject, we obtained the total number of methylated CpGs.
Statistics
Data were analyzed using SPSS ver. 23 (Chicago, Illinois, USA). CpG sites distributions between patients and controls were analyzed using Fisher’s exact test. DNA methylation was presented as mean ± standard error, and mean differences in methylation levels were measured using Student t test. Correlations between average methylation level and clinical features of migraine were examined by Pearson test. A p < 0.05 value was considered as significant for both genetic comparisons and clinical analyses.
Results
Methylation patterns of the two different CpG islands
Distal CpG island. We analyzed the first CpG island region in the promoter of CALCA gene, in a DNA segment from −2762 to −2362 bp from ATG. After sequencing, we studied all the 30 CpGs units per this CpG island, and we found that C was methylated in a heterozygous state in 6 out of 30 CpG sites (positions −2736, −2601, −2592, −2555, −2423, −2405). The total number of methylated CpGs obtained for each sample was used to categorize samples to three methylation levels: 0 (no methylation), 1–3 CpG sites (low methylation, less than half methylated sites), and 4–6 CpG sites (high methylation, more than half methylated sites).
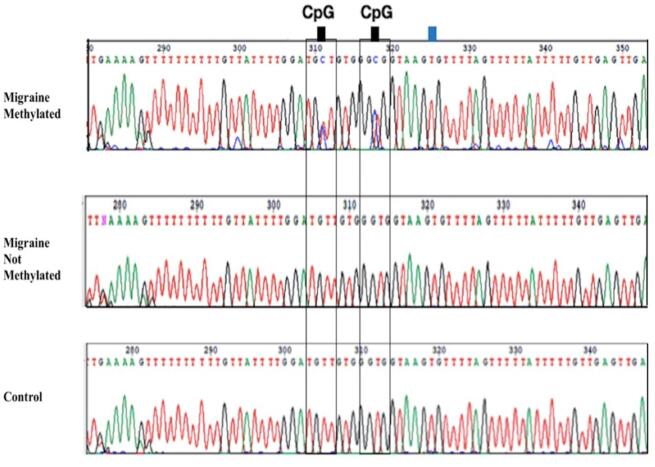
In our samples, only one male patient with episodic migraine with low frequency had high range of methylation (4 methylated sites), while the remaining patients and controls were in the low or no methylation category (0–2 methylated sites). A representative chromatogram of a sequence containing CpG sites is shown in Fig. 2.
Fig. 2.
Bisulfite sequencing. Representative chromatograms of a sequence containing two CpG-sites (indicated by black squares at the top of the sequence diagram) are shown for a patient with migraine methylated, a patient with migraine non methylated, and a control (proximal region). Non CpG cytosine (indicated by blue square) was successful converted to thymine. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
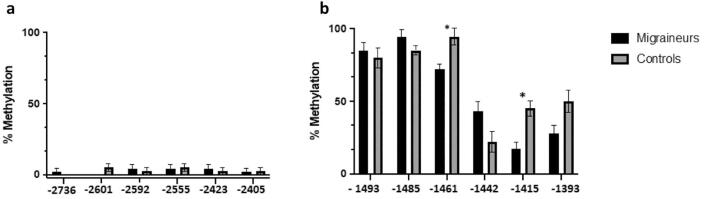
This region in the distal CpG island resulted hypomethylated in both patients and controls. In the whole study group, the mean methylation levels of all CpG sites in this region were below 10%, and our data are in accordance with the methylation profile of this region reported in the public database iMETHYL (https://imethyl.iwate-megabank.org) (Komaki et al., 2018). No significant difference in average DNA methylation was observed between migraineurs and controls (total average methylation level: 2.6% vs. 2.8%, respectively).
Furthermore, in the distal CpG island, no significant differences were found between migraineurs and controls regarding methylation levels at each of the 6 CpG sites (Fig. 3a).
Fig. 3.
Mean DNA methylation of the distal (3a) and proximal (3b) promoter region of CALCA gene in patients with episodic migraine and controls. * p < 0.05.
In both groups, methylation level in the first CpG island of CALCA promoter did not correlate with age or gender. Furthermore, no significant correlation was found between clinical characteristics of migraineurs and the level of methylation.
Proximal CpG island. We analyzed the second CpG islands region in the promoter of CALCA gene, in a DNA segment from −1662 to −1228 bp. We observed 6 CpG sites (positions −1493, −1485, −1461, −1442, −1415, −1393), in this proximal CpG island. Seven patients with episodic migraine (31%) showed a low methylation (1–3 methylated sites), whereas the remaining patients were in the high methylation category (4 or more methylated sites). In control samples, four subjects (20%) had a low methylation (1–3 methylated sites), whereas the remaining controls showed high methylation (4 or more methylated sites). No overall difference was found in the methylation level among these 6 detected CpG sites at the CALCA proximal promoter between migraineurs and controls.
Regarding the individual CpG site, a stratification analysis showed that in migraineurs the methylation level was significantly lower in respect to controls in 2 out of the 6 analyzed CpG sites (CpG −1461, p = 0.037, and −1415, p = 0.035, respectively) (Fig. 3b). No other significant differences were found between migraineurs and controls in methylation frequency of individual CpGs.
Furthermore, we estimated the association between age and DNA methylation levels, and we found that age positively correlates with DNA methylation level at −1485 CpG site (r = 0.578, p = 0.038) in the migraine group. Subsequently, we analyzed the correlation between DNA methylation and the clinical characteristics of migraine. Methylation at −1461 CpG site positively correlates with age at onset of migraine, whereas methylation at −1393 CpG site inversely correlates with the presence of nausea/vomiting during the attacks (p = 0.038, and p = 0.027, respectively) (Fig. 4). The association between the mean methylation level of −1461 and −1415 CpGs sites and episodic migraine remained still significant after adjustment for preventive therapy status (CpG −1461, p = 0.047, and −1415, p = 0.044, respectively). Finally, an inverse correlation between BDI, STAI-X1 and STAI-X2 scores and methylation levels at – 1493 unit was found (r = −0.642, p = 0.024, r = −0.707, p = 0.010, r = −0.609, p = 0.036, respectively).
Fig. 4.
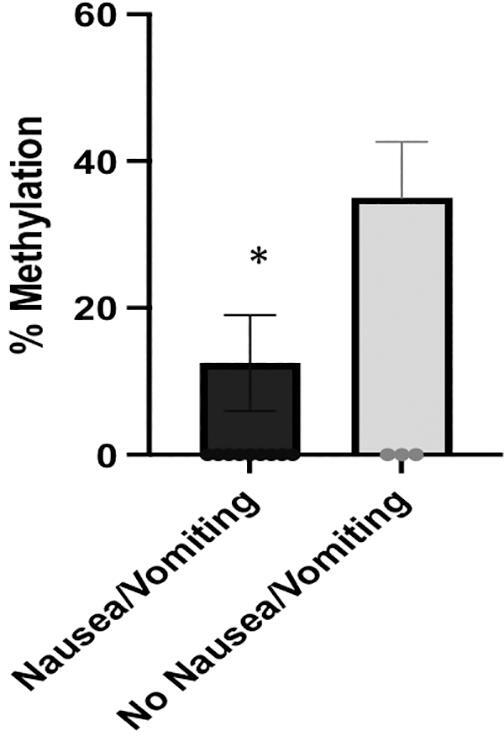
The relationship of mean methylation levels at −1393 CpG site with the presence of nausea/vomiting in patients with episodic migraine without aura. Data are presented as mean ± SE.
No other significant correlations were found between the other features of migraine and the level of methylation.
Discussion
We report the first investigation of DNA methylation at the CALCA promoter region in migraine, providing evidence that DNA hypomethylation could play a role in the disease. We found that DNA methylation profile in two CpG sites at the proximal promoter region is lower in migraineurs when compared to controls. Contrariwise, no difference of methylation in the distal region of CALCA promoter between patients with episodic migraine and healthy subjects was found. In addition, we found that the DNA methylation level at different CpG sites significantly correlates with several clinical characteristics of the disease, like age at onset, presence of nausea and vomiting, depression and anxiety scores.
To date, neurobiological mechanisms explaining DNA hypomethylation in patients with migraine remain to be clearly established. DNA methylation is crucial for maintaining tissue-specific gene expression pattern and deviation from normal DNA methylation level may affect gene expression and initiate pathologic processes (Han et al., 2008). In our sample, in accordance with multi-omics public database (https://imethyl.iwate-megabank.org), DNA methylation at the distal promotor region of CALCA gene was observed below 10% and did not show difference between migraine patients and controls. Contrariwise, we found that DNA methylation profile in two CpG sites (−1415 and −1461) at the proximal promoter region is lower in migraineurs in comparison to controls. Interestingly, the −1415 CpG site is located into a promoter region containing a cAMP responsive element (CRE), which in turn binds CREB, a nuclear transcription factor (Monla et al., 1995). In experimental models, Park et al. (2011) showed that CALCA gene expression depends upon DNA methylation at promoter region. Based on these findings, we could speculate that DNA hypomethylation observed in our patients may contribute to an increased CGRP release both during migraine attacks and in interictal phases of the disease. Several genes increase CGRP expression, enhancing intracellular cAMP signal, which in turn regulates the expression of many genes via CRE. In addition, steroid hormones, like glucocorticoids, progesterone and estrogens, may modulate DNA methylation through CREB phosphorylation (Beato et al., 1987) but their effects on the genes involved into the CGRP-ergic system still need to be investigated.
Intriguingly, in our study we found that methylation at proximal CpG units correlates with several migraine characteristics. Methylation at −1461 CpG site positively correlates with age at onset of the disease, and methylation CpG unit at −1393 inversely correlates with the presence of nausea and or vomiting during migraine attacks. Finally, in our migraine patients, anxiety and mood scores significantly correlated with the degree of methylation in promoter CALCA gene. These findings are in agreement with recent studies in experimental animals suggesting that CGRP may be involved in depression-like behaviours (Hashikawa-Hobara et al., 2015) and deserve additional studies in the complex, bidirectional comorbidity between migraine and depression.
In recent years, the role of epigenetics in migraine has become an emerging topic of interest because epigenetic marks may explain the effects of non-genetic risk factors in the disease (Cámara et al., 2017, Eising et al., 2013). Genes previously associated with migraine have direct links to epigenetic mechanisms. Several studies showed an association between single nucleotide polymorphisms (SNPs) in methylenetetrahydrofolate reductase (MTHFR), a gene required for DNA methylation, and migraine (Rainero et al., 2019b, Roecklein et al., 2013, Rubino et al., 2009). Notably, a recent paper suggested that deficiency in B6, B12 and folate, coupled with MTHFR polymorphisms, resulted in a significant DNA hypomethylation, which could have a significant role in triggering migraine attacks. More recently, genome wide association studies suggested that several SNPs linked to migraine, as MTDH, MEF2D and PRDM16, are involved in epigenetic processes (Gormley et al., 2016). A recent, pioneering epigenome-wide association study investigated methylation in women with both episodic or chronic migraine and controls. After correction for multiple testing, no significant single probe association was found. However, using a less stringent approach, the study showed that 62 independent regions, in close proximity to genes involved in solute transporter and haemostasis, were differently methylated in migraineurs and controls (Gerring et al., 2018). At present, only one study examined the DNA methylation levels at the promoter region of RAMP1, gene that encodes a key receptor subunit of CGRP. In agreement with the results of our study, a low methylation level was observed in a group of 26 migraineurs, with two GpG units significantly associated with clinical features of the disease (Wan et al., 2015).
There are some limitations of the study that suggest caution in the interpretation of the results. First, the small sample size: based on the limited samples, our findings and subsequent conclusions must be viewed cautiously, and further studies with larger sample size are needed in order to confirm our data. Second, in this pilot study the analysis was limited to patients with episodic migraine without aura. DNA methylation represents a dynamic epigenetic mechanism, and our choice was due to have a homogeneous population, and to limit the confounding factors as prophylaxis therapies or drugs abuse that could influence the DNA methylation profile. In this regard, it is well known that valproate, an approved prophylactic drug for migraine, can modulate epigenetic processes inhibiting histone deacetylation as well as DNA methylation (de Campos Vidal and Mello, 2020). Third, we analyzed CALCA methylation in peripheral blood and the correlation between peripheral blood and brain methylation pattern is still matter of debate. Finally, in our pilot study, we preferred to use sequencing instead of a next generation methylation approach. To date, a number of methods have been developed for the identification of differentially methylated regions, depending on the research goal. Notwithstanding methylation arrays provided a very high density of coverage, they do not down to each single nucleotide, and for an exploratory analysis, we prefer to use a sequence-based approach in order to determine the level of methylation in all the CpG sites in the two analyzed CpG islands of CALCA promoter.
Epigenetics in migraine represents a new unexplored field and additional studies are needed to better elucidate its role. Further investigations are needed in order to evaluate if epigenetic mechanisms are differently involved in migraine without aura and migraine with aura. In addition, whether epigenetic mechanisms could be involved in the chronification process of migraine or drug overuse needs to be adequately studied. However, the most relevant goal of these studies will be to evaluate if DNA methylation at CALCA gene may be of relevance for both acute and preventing therapies targeting CGRP.
Conclusions
In conclusion, our study provides the first evidence that the methylation of CALCA gene is reduced in patients with migraine and that this hypomethylation could play a role in the disease. Relationship between degree of methylation of the CALCA gene and pathophysiological mechanisms of migraine remains to be clearly established. Additional investigations are needed to define the epigenetic mechanisms involved in the methylation of the CALCA gene in migraine and to investigate its role in the clinical characteristic of the disease.
Data availability
The data are not publicly available because they contain information that could compromise research participant privacy/consent. Further clinical and genetic data will be available upon request from any qualified investigator.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Professor Rainero has received speaker honoraria from ElectroCore, Lusofarmaco, MSD, Novartis, and Teva.
The remaining authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was supported by Ministero dell'Istruzione, dell'Università e della Ricerca–MIUR project “Dipartimenti di Eccellenza 2018–2022” to Department of Neuroscience “Rita Levi Montalcini”, University of Torino, and AIRAlzh Onlus-ANCC-COOP (SB). The authors thank the subjects who participated in the study.
Reference
- Beato M., Arnemann J., Chalepakis G., Slater E., Willmann T. Gene regulation by steroid hormones. J. Steroid Biochem. 1987;27:9–14. doi: 10.1016/0022-4731(87)90288-3. [DOI] [PubMed] [Google Scholar]
- Broad P.M., Symes A.J., Thakker R.V., Craig R.K. Structure and methylation of the human calcitonin/alpha-CGRP gene. Nucleic Acids Res. 1989;17:6999–7011. doi: 10.1093/nar/17.17.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein R., Noseda R., Borsook D. Migraine: multiple processes, complex pathophysiology. J. Neurosci. 2015;35:6619–6629. doi: 10.1523/JNEUROSCI.0373-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cámara M.S., Martín Bujanda M., Mendioroz Iriarte M. Epigenetic changes in headache. Neurologia. 2017;S0213–4853(17):30356. doi: 10.1016/j.nrleng.2017.10.006. [DOI] [PubMed] [Google Scholar]
- de Campos Vidal, B., Mello, M.L.S., 2020. Sodium valproate (VPA) interactions with DNA and histones. Int. J. Biol. Macromol. S0141-8130(20)33716-8. 10.1016/j.ijbiomac.2020.06.265. [DOI] [PubMed]
- de Vries B., Frants R.R., Ferrari M.D., van den Maagdenberg A.M. Molecular genetics of migraine. Hum. Genet. 2009;126:115–132. doi: 10.1007/s00439-009-0684-z. [DOI] [PubMed] [Google Scholar]
- Durham P.L. Inhibition of calcitonin gene-related peptide function: a promising strategy for treating migraine. Headache. 2008;48:1269–1275. doi: 10.1111/j.1526-4610.2008.01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvinsson L. Novel migraine therapy with calcitonin gene-regulated peptide receptor antagonists. Expert Opin. Ther. Targets. 2007;11:1179–1188. doi: 10.1517/14728222.11.9.1179. [DOI] [PubMed] [Google Scholar]
- Edvinsson L., Haanes K.A., Warfvinge K., Krause D.N. CGRP as the target of new migraine therapies - successful translation from bench to clinic. Nat. Rev. Neurol. 2018;14:338–350. doi: 10.1038/s41582-018-0003-1. [DOI] [PubMed] [Google Scholar]
- Edvinsson L. Role of CGRP in Migraine. Handb. Exp. Pharmacol. 2019;255:121–130. doi: 10.1007/164_2018_201. [DOI] [PubMed] [Google Scholar]
- Ehrlich M. DNA hypermethylation in disease: mechanisms and clinical relevance. Epigenetics. 2019;14:1141–1163. doi: 10.1080/15592294.2019.1638701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eising E.A., Datson N., van den Maagdenberg A.M., Ferrari M. Epigenetic mechanisms in migraine: a promising avenue? BMC Med. 2013;11:26. doi: 10.1186/1741-7015-11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerring Z.F., McRae A.F., Montgomery G.W., Nyholt D. Genome-wide DNA methylation profiling in whole blood reveals epigenetic signatures associated with migraine. BMC Genomics. 2018;19:69. doi: 10.1186/s12864-018-4450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjaltema R.A.F., Rots M.G. Advances of epigenetic editing. Curr. Opin. Chem. Biol. 2020;57:75–81. doi: 10.1016/j.cbpa.2020.04.020. [DOI] [PubMed] [Google Scholar]
- Gormley P., Anttila V., Winsvold B.S., Palta P., Esko T., Pers T.H., Farh K.-H., Cuenca-Leon E., Muona M., Furlotte N.A., Kurth T., Ingason A., McMahon G., Ligthart L., Terwindt G.M., Kallela M., Freilinger T.M., Ran C., Gordon S.G., Stam A.H., Steinberg S., Borck G., Koiranen M., Quaye L., Adams H.H.H., Lehtimäki T., Sarin A.-P., Wedenoja J., Hinds D.A., Buring J.E., Schürks M., Ridker P.M., Hrafnsdottir M.G., Stefansson H., Ring S.M., Hottenga J.-J., Penninx B.W.J.H., Färkkilä M., Artto V., Kaunisto M., Vepsäläinen S., Malik R., Heath A.C., Madden P.A.F., Martin N.G., Montgomery G.W., Kurki M.I., Kals M., Mägi R., Pärn K., Hämäläinen E., Huang H., Byrnes A.E., Franke L., Huang J., Stergiakouli E., Lee P.H., Sandor C., Webber C., Cader Z., Muller-Myhsok B., Schreiber S., Meitinger T., Eriksson J.G., Salomaa V., Heikkilä K., Loehrer E., Uitterlinden A.G., Hofman A., van Duijn C.M., Cherkas L., Pedersen L.M., Stubhaug A., Nielsen C.S., Männikkö M., Mihailov E., Milani L., Göbel H., Esserlind A.-L., Christensen A.F., Hansen T.F., Werge T., Kaprio J., Aromaa A.J., Raitakari O., Ikram M.A., Spector T., Järvelin M.-R., Metspalu A., Kubisch C., Strachan D.P., Ferrari M.D., Belin A.C., Dichgans M., Wessman M., van den Maagdenberg A.M.J.M., Zwart J.-A., Boomsma D.I., Smith G.D., Stefansson K., Eriksson N., Daly M.J., Neale B.M., Olesen J., Chasman D.I., Nyholt D.R., Palotie A. Meta-analysis of 375,000 individuals identifies 38 susceptibility loci for migraine. Nat. Genet. 2016;48(8):856–866. doi: 10.1038/ng.3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Su B., Li W.H., Zhao Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008;9:79. doi: 10.1186/gb-2008-9-5-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashikawa-Hobara N., Ogawa T., Sakamoto Y., Matsuo Y., Ogawa M., Zamami Y., Hashikawa N. Calcitonin gene-related peptide pre-administration acts as a novel antidepressant in stressed mice. Sci. Rep. 2015;5(1) doi: 10.1038/srep12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Headache Classification Committee of the International Headache Society (IHS), 2018. The International Classification of Headache Disorders, 3rd edition. Cephalalgia. 38, 1–211. 10.1177/0333102417738202. [DOI] [PubMed]
- Jin Z., Liu Y. DNA methylation in human diseases. Genes Dis. 2018;5:1–8. doi: 10.1016/j.gendis.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaki S., Shiwa Y., Furukawa R., Hachiya T., Ohmomo H., Otomo R., Satoh M., Hitomi J., Sobue K., Sasaki M., Shimizu A. iMETHYL: an integrative database of human DNA methylation, gene expression, and genomic variation. Hum. Genome Var. 2018;5:18008. doi: 10.1038/hgv.2018.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monla Y.T., Peleg S., Gagel R.F., Monia Y.T. Cell type-specific regulation of transcription by cyclic adenosine 3’,5’-monophosphate-responsive elements within the calcitonin promoter. Mol. Endocrinol. 1995;9:784–793. doi: 10.1210/mend.9.7.7476962. [DOI] [PubMed] [Google Scholar]
- Park K.Y., Fletcher J.R., Raddant A.C., Russo A.F. Epigenetic regulation of the calcitonin gene-related peptide gene in trigeminal glia. Cephalalgia. 2011;31:614–624. doi: 10.1177/0333102410391487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainero I., Vacca A., Govone F., Gai A., Pinessi L., Rubino E. Migraine: genetic variants and clinical phenotypes. Curr. Med. Chem. 2019;26:6207–6221. doi: 10.2174/0929867325666180719120215. [DOI] [PubMed] [Google Scholar]
- Rainero I., Vacca A., Roveta F., Govone F., Gai A., Rubino E. Targeting MTHFR for the treatment of migraines. Expert Opin. Ther. Targets. 2019;23:29–37. doi: 10.1080/14728222.2019.1549544. [DOI] [PubMed] [Google Scholar]
- Roecklein K.A., Scher A.I., Smith A., Harris T., Eiriksdottir G., Garcia M., Gudnason V., Launer L.J. Haplotype analysis of the folate-related genes MTHFR, MTRR, and MTR and migraine with aura. Cephalalgia. 2013;33(7):469–482. doi: 10.1177/0333102413477738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubino E., Ferrero M., Rainero I., Binello E., Vaula G., Pinessi L. Association of the C677T polymorphism in the MTHFR gene with migraine: a meta-analysis. Cephalalgia. 2009;29:818–825. doi: 10.1111/j.1468-2982.2007.01400.x. [DOI] [PubMed] [Google Scholar]
- Russo A.F. CGRP as a neuropeptide in migraine: lessons from mice. Br. J. Clin. Pharmacol. 2015;80:403–414. doi: 10.1111/bcp.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H.G., Griffiths L.R. Genetics of Migraine: insights into the molecular basis of migraine disorders. Headache. 2017;57:537–569. doi: 10.1111/head.13053. [DOI] [PubMed] [Google Scholar]
- Sutherland H.G., Albury C.L., Griffiths L.R. Advances in genetics of migraine. J. Headache Pain. 2019;20:72. doi: 10.1186/s10194-019-1017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tendl K.A., Schulz S.MF., Mechtler T.P., Bohn A., Metz T., Greber-Platzer S., Kasper D.C., Herkner K.R., Item C.B. DNA methylation pattern of CALCA in preterm neonates with bacterial sepsis as a putative epigenetic biomarker. Epigenetics. 2013;8(12):1261–1267. doi: 10.4161/epi.26645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan D., Hou L., Zhang X., Han X., Chen M., Tang W., Liu R., Dong Z., Yu S. DNA methylation of RAMP1 gene in migraine: an exploratory analysis. J. Headache Pain. 2015;16(1) doi: 10.1186/s10194-015-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are not publicly available because they contain information that could compromise research participant privacy/consent. Further clinical and genetic data will be available upon request from any qualified investigator.