Abstract
Genomes have complex three-dimensional structures. High-resolution population-based biochemical studies over the last decade have painted a mostly static picture of the genome characterized by universal organizational features, such as chromatin domains and compartments. Yet, when analyzed at the single cell level, these architectural elements are highly variable. The heterogeneity in genome organization is in line with the inherent stochasticity of transcription that shows high variation between individual cells. We highlight recent findings on single-cell variability in genome organization and describe a framework for how the stochastic nature of chromatin organization may relate to transcription dynamics.
Keywords: Genome organization, transcription, gene expression, heterogeneity, stochastic bursting
Introduction
Much progress has been made in mapping genomes in space and time (**Misteli 2020). In particular, biochemical ensemble studies using chemical crosslinking methods and single-cell imaging have over the past decade elucidated the organizational structure of genomes at unprecedented resolution and have uncovered several ubiquitous architectural features associated with gene regulation (Kempfer and Pombo 2020; Misteli 2020; Bohrer and Larson 2021).
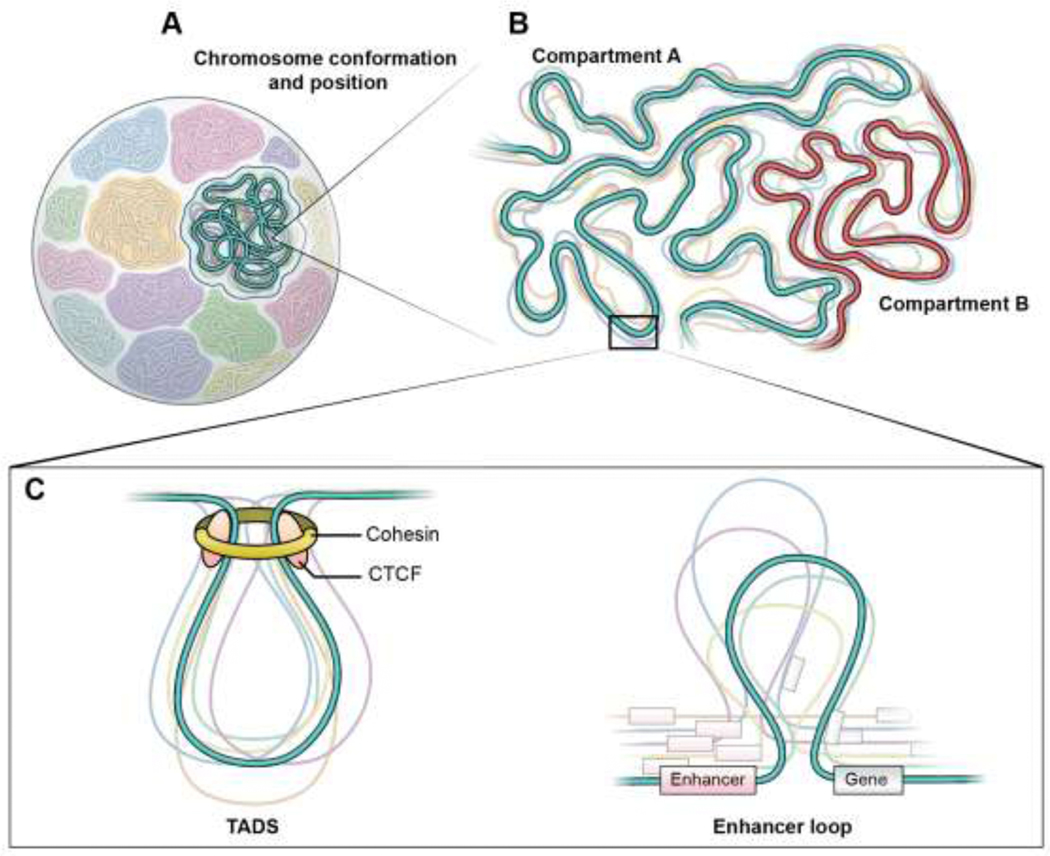
At the lowest level of organization, the negatively supercoiled DNA is wrapped around a histone octamer to form nucleosomes, which have recently been shown to organize into tightly associated regularly phased arrays of 3–10 ‘clutches’ arranged in a zig-zag pattern that maximizes local interactions (**Hsieh et al. 2020; **Krietenstein et al. 2020). These nucleosomal arrays loop to bring distal elements in close vicinity and often occur within self-interacting regions of the genome, referred as topologically associating domains (TADs) (Nora et al. 2012; Sexton et al. 2012; Rao et al. 2014; **Hsieh et al. 2020; **Krietenstein et al. 2020) (Fig. 1). Various methods to detect chromatin-chromatin interactions, either using chemical ligation of proximal DNA regions, such as Hi-C and micro-C, or ligation-independent techniques, have defined TADs as largely tissue invariant structures that promote by ∼ 2-fold the enrichment of chromatin contacts within 100Kb-2Mb regions defined by insulating borders relative to adjacent domains (Akdemir et al. 2020; Kempfer and Pombo 2020; McArthur and Capra 2021) (Fig. 1). TADs are functionally and evolutionarily conserved units of genome organization that often coincide with epigenetically marked regions and are thought to promote interactions between distal regulatory elements with target genes within their boundaries (**Hoencamp et al. 2021; McArthur and Capra 2021). When mapped at higher resolution, TADs are subdivided into locally interacting sub-TADs that are variable across cell types and are related to transcriptional states (**Hsieh et al. 2020; **Krietenstein et al. 2020). On a larger scale, regions of active and inactive chromatin, even located on distinct chromosomes, self-associate into mutually exclusive A and B compartments, respectively (Nora et al. 2017; Brown et al. 2018; **Su et al. 2020) (Fig. 1). However, compartments form mostly independent of TADs as indicated by the fact that compartmental changes do not corelate with TAD alterations during differentiation and intra-compartmental interactions increase upon loss of TADs (Nora et al. 2017; Rao et al. 2017; Chathoth and Zabet 2019). Ultimately, at a global level, genomes are organized in chromosomes territories which are non-randomly positioned with respect to nuclear landmarks such as the nuclear periphery (**Misteli 2020; Takei et al. 2021) (Fig. 1).
Figure 1: Extensive heterogeneity across all levels of genome organization:
(A) Within the eukaryotic nucleus the chromosomes occupy non-random territories but show variation their overall folding and their location between single cells. (B) Chromosomes are organized into mutually exclusive active and inactive compartments, referred as compartments A and B, respectively. Compartments across both the same and different chromosomes self-associate but vary in conformation and both the levels of compaction and segregation. (C) Compartments are further organized into loop domains that are driven by loop extrusion using the DNA motor protein, cohesin, and usually flanked by the insulator protein, CTCF, at their boundaries. These loops, referred as topological associated domains (TADs), show variation in loop size and boundary position between single cells. (D) TADs encompass smaller regulatory sub-loops that promote interactions of a gene with its distal enhancers. Such loops show high variability in size and anchor positions, resulting in variations in three-dimensional distances between enhancers and promoters amongst single cells.
The organization of genomes is evolutionarily conserved in eukaryotes (**Hoencamp et al. 2021; McArthur and Capra 2021) and has functional implications as suggested by the observation that structural reorganizations correlate with in vivo transcriptional changes during development in mouse (Williamson et al. 2019; Winick-Ng et al. 2020), in Drosophila (Mateo et al. 2019) and during differentiation processes (Oudelaar et al. 2020). Furthermore, disruption of genome architecture can lead to disease-specific repositioning of genes, congenital defects associated with deletion of regulatory boundary and ectopic activation of proto-oncogene upon loss of insulated neighborhoods (Krumm and Duan 2019). Finally, in silico models that incorporate both compartmentalization and the formation of domains are sufficient to recapitulate the in vivo genome organization and structural changes associated with mutations of architectural proteins (Nuebler et al. 2018). Thus, it seems that organizational features of the genome are linked to genome function.
The stochasticity of genome organization
The series of well-defined structural features of the genome might give the impression that transcription regulation occurs in the context of a rigid and static genome organization. However, recent findings have made it clear that all aspects of genome organization and function are dynamic and stochastic. Cryo-EM (Trzaskoma et al. 2020), single cell biochemical mapping (Tan et al. 2018), high-throughput imaging (**Finn et al. 2019) and high-resolution light microscopy (Bintu et al. 2018; Szabo et al. 2018; Mateo et al. 2019; **Luppino et al. 2020; **Takei et al. 2021) have revealed extensive heterogeneity of nuclear organization between single cells (Fig. 1). These methods find that chromosomes occur in territories but show variation in structure, position and levels of trans-chromosomal contacts, even between the two homologs, amongst individual cells (**Su et al. 2020; **Takei et al. 2021) (Fig. 1A). Similarly, the segregation between ensembledefined A and B compartments also varies amongst single cells (**Johnsone et al. 2020; **Su et al. 2020) (Fig. 1B). Notably, extensive single-cell heterogeneity exists for the occurrence of loops and the precise location of TAD boundaries (Bintu et al. 2018; **Finn et al. 2019; Mateo et al. 2019; **Luppino et al. 2020; Szabo et al. 2020), subTADs (Rao et al. 2014; Tan et al. 2018) and interactions between enhancers and promoters (**Rodriguez et al. 2019) (Fig. 1C, D). Thus, variation in architectural features of the genome seems to be the rule rather than the exception and, as a result, the study of the stochastic nature of genome organization, its origin and implications for genome function have recently come into focus.
Several studies across different cell types in human (Bintu et al. 2018; **Finn et al. 2019; **Luppino et al. 2020; **Su et al. 2020), mouse (Szabo et al. 2020) and Drosophila (Mateo et al. 2019) show that TADs reflect a probabilistic structure rather than a stable domain. While, single cells have sharp TAD boundaries, widespread heterogeneity in the boundary position concomitant with intermingling across population defined boundaries, is seen between individual cells (Bintu et al. 2018; **Finn et al. 2019; Mateo et al. 2019; **Luppino et al. 2020; Su et al. 2020; Szabo et al. 2020) (Fig. 1C). Indeed, high capture frequency in ensemble HiC may not accurately reflect the actual variation in 3D distances between locus pairs in single cells. For example, the top 20% of HiC interactors show perfect overlap in <10% of alleles in single cell FISH assays capable of resolving loci separated by 250kb at 200nm resolution. Furthermore, genomic loci with similar HiC capture frequencies may show several fold differences in the frequency of physical overlap (**Finn et al. 2019). In addition, TADs vary in their physical size and end-to-end distance across single cells and show heterogeneity in contact frequency depending on gene density (**Cardozo Gizzi et al. 2019; **Finn et al. 2019; **Su et al. 2020). Variations in TAD structures are even also apparent in some population-based biochemical crosslinking assays that map interactions at high resolution, such as Micro-C, to uncover low frequency structures around the ensemble average (**Hsieh et al. 2020; **Krietenstein et al. 2020). Reassuringly, averaging the single cell-derived structures recapitulates both the ensemble TAD structure and the typically observed 2-fold enrichment of contacts within TADs compared to the surrounding chromatin regions (Bintu et al. 2018; **Finn et al. 2019; **Su et al. 2020). Taken together, these observations document extensive variability in the most prominent features of genome organization and raise the question how these variations are generated.
Heterogeneity in TADs seems to be driven by the opposing actions of the two major architectural proteins, CTCF and cohesin, which are involved in the formation of TADs (Mirny et al. 2019). Cohesin, which is a ring-shaped complex that holds sister chromatids together, drives the extrusion of chromatin loops to form a TAD, and CTCF, which binds to boundary elements and genetic insulators, hinders looping across its binding sites (Nora et al. 2012; Rao et al. 2014; **Luppino et al. 2020; Szabo et al. 2020). In line with the well-established dynamic nature of many chromatin proteins (**Misteli 2020), CTCF and cohesin, form a dynamic complex with residence times of 1–2min and ∼22min, respectively, and widely different reloading kinetics (Hansen et al. 2017). The transient fluctuations of these binding events likely generate heterogeneity in boundary positions between single cells. Indeed, ensemble TADs are lost upon depletion of CTCF as chromatin interactions become more promiscuous whereas upon depletion of cohesion, with or without CTCF, interactions decreased overall (**Luppino et al. 2020; Szabo et al. 2020). However, TAD-like structures are still observed in single cells depleted of the cohesin complex but their boundary positions show more variations and lack a preference for CTCF sites (Bintu et al. 2018). Thus, the inherently stochastic nature of the coupled CTCF- and cohesion-binding kinetics are a likely source of variability in TAD structure.
Single cells also show widespread variation in both the arrangement and the segregation of A and B compartments (**Su et al. 2020). Super-resolution imaging and ultra-deep Hi-C shows that the A compartment is often split, diffusely spread within the B compartment (**Gu et al., 2021; **Su et al. 2020), consistent with higher strength of B-B interactions than A-A interactions (Falk et al. 2019) (Fig. 1B). Furthermore, the distinction between A and B compartments is not complete, with full segregation in only ∼80% of single cells (**Su et al. 2020). This heterogeneity is partly reflected in bulk measurements that detect an intermediate compartment between the A and B compartments, particularly in cancer cells (**Johnsone et al. 2020). Since compartments are, at least in part, driven by the transcriptional states of the genome regions they contain (Cavalli and Misteli 2013; Luo et al. 2020; **Misteli 2020), the heterogeneity in compartmentalization is likely a reflection of single-cell variability in gene expression programs (see below).
Taken together, single cell imaging and biochemical studies have highlighted pervasive variation across all levels of genome organization around the ensemble structure seen in population-based studies (Fig. 1). The observed variability in genome structure is intriguing in the light of the stochastic nature of gene expression and transcription (Rodriguez and Larson 2020).
Transcription bursting and stochastic enhancer interactions
Heterogeneity in transcription has been appreciated and studied for decades (Rodriguez and Larson 2020). A major source of intrinsic heterogeneity arises from stochastic pulses of transcription called transcriptional bursts (Elowitz et al. 2002). During transcriptional bursts, RNA polymerases engage to generate RNA for a while (On-time), but transcriptional activity then ceases for a period of time (Off-time). Stochasticity in bursting is a fundamental aspect of transcription that can push gene expression in single cells away from the population average. The transcriptional heterogeneity arising from bursting can impact critical functions such as cell fate and development (Abranches et al. 2014), responses to environmental stimuli (Fritzsch et al. 2018) and diseases (Shaffer et al. 2020). Bursting heterogeneity can be regulated at the level of the duration of the On-time, the frequency of the burst, which is reflected in the Off-time, or the size of the burst (Rodriguez and Larson 2020). Based on live cell analysis of 770 genes, modulation of the burst frequency appears to be the major source of heterogeneity (**Wan et al. 2021). While average off-times for genes can vary from 20 min to 100 min for slow and fast bursting genes, respectively, (**Wan et al. 2021) in extreme cases individual allele can stay in the “Off” state for days (**Rodriguez et al. 2019) and the combination of slow bursting and small burst size can lead to apparent monoallelic expression (Symmons et al. 2019). Alleles in the same nucleus often show coupled bursting (**Rodriguez et al. 2019; Stavreva et al. 2019) further increasing the heterogeneity between cells. Thus, the stochastic nature of gene bursting makes gene expression inherently heterogeneous.
Mechanisms that regulate bursting operate via two modes: One, proximal regulation via dynamic binding of transcription factors and coactivators to the promoters and two, distal regulation involving enhancer elements at large genomic distances (**Donovan et al. 2019; Stavreva et al. 2019; Rodriguez and Larson 2020). Transcription factor binding at promoters strongly correlates with bursting in both yeast and humans in real time (**Donovan et al. 2019; Stavreva et al. 2019). Burst duration and burst sizes are regulated by transcription factor dwell times, which in turn are determined by the binding affinities and nucleosome occupancy (**Donovan et al. 2019; Stavreva et al. 2019). Burst frequency on the other hand is regulated by the binding rates of transcription factors as demonstrated in both single-gene live cell-studies (**Donovan et al. 2019; Stavreva et al. 2019) and sequencing-based genome-wide measurements (Larsson et al. 2019). Distal regulation by enhancers however primarily modulates burst frequency, which has been shown in both single-cell studies where enhancer deletions increased the Off-time and in genome-wide studies where single nucleotide polymorphisms in enhancers from different mice strains were associated with changes in burst frequencies (Fukaya et al. 2016; Larsson et al. 2019; **Rodriguez et al. 2019). Thus, transcriptional bursting is a result of stochasticity in factor binding at the promoter and gene interactions with distal regulatory elements.
How enhancers modulate target expression over large genomic distance in the context of stochastic fluctuations in architecture is still unclear. The traditional view suggests that an enhancer must physically contact a target promoter. Indeed, looping to juxtapose enhancers and promoters has been shown to be necessary for gene activation, observed to be concordant with global expression changes, and in some cases, to be sufficient for activation of expression (Weintraub et al. 2017; Williamson et al. 2019; **Johnsone et al. 2020). However, several recent studies have challenged the need of a direct chromatin-chromatin interaction for transcriptional activation (Alexander et al. 2019; Benabdallah et al. 2019; Oudelaar et al. 2020; ****Espinola et al. 2021). Both in situ hybridization and live cell imaging show widespread heterogeneity in the enhancer-promoter distances amongst individual cells (Alexander et al. 2019; **Rodriguez et al. 2019), highlighting the stochastic nature of these interactions. Even the classic looping model, based on the alpha-globin locus, shows multiple interactions within its self-interacting domain (Davies et al. 2016). It now appears that enhancers interact with a broad range of target sites rather than a single locus and often explore a subset of conformational space over short time scales (Symmons et al. 2016; Alexander et al. 2019; Symmons et al. 2019). This search process may be aided by the formation of phase separated aggregates of transcriptional activators, such as Mediator or BRD4, around enhancers, which may serve to concentrate relevant transcription factors near the target promoter (Hnisz et al. 2017; Feric and Misteli 2021).
The stochastic nature of enhancer-promoter interactions is consistent with gene bursting. Active bursting occurs for a short fraction of the total time (<5%) (Alexander et al. 2019) and it is possible that the stochastically occurring proximity events trigger the individual bursts. Alignment of bursts with enhancer-promoter proximity has shown both strong and weak correlations depending on the experimental system used (Chen et al. 2018; Alexander et al. 2019). For example, the even-skipped enhancer in Drosophila showed sustained proximity to the target promoter around bursting events (Chen et al. 2018), whereas bursting of Sox2 in mouse ES cells did not necessarily require physical proximity of the Sox2 promoter to its control region (Alexander et al. 2019). A recent hypothesis potentially reconciles these discrepancies in a model where each enhancer interaction event leaves a ‘tag’ that contributes towards activation of a gene and repeat interactions over time lead to the accumulation of enough ‘tags’ to reach a threshold for gene activation (Xiao et al. 2020). It has been postulated that these tags could be chromatin modifications or buildup of coactivators that promote transcription (Xiao et al. 2020). In line with this interpretation, induction of the TFF1 gene in MCF7 cells increased the burst frequency by two-fold, however, the enhancer interaction did not scale proportionally (**Rodriguez et al. 2019). Instead, the interactions became more focused around the target gene, which resulted in decreased local entropy for chromatin organization that was proportional to the strength of the induction (**Rodriguez et al. 2019). These observations suggest that transient enhancer-promoter proximity and interactions contribute to stochastic transcriptional bursts but are not the sole determinants of bursting.
Stochasticity in gene expression in the context of heterogeneous TADs
TADs have been suggested to contribute to control of gene expression programs (van Steensel and Furlong 2019). Given the structural heterogeneity of chromatin domains and the stochastic nature of transcription, it is important to understand whether, and how, the structure of a TAD relates to the activity of the genes it contains.
The primary proposed mode of action for TADs in regulating gene expression has been via promoting chromatin-chromatin interactions, such as enhancers and target promoters, within their boundaries but disfavoring promoter enhancer interactions across TAD boundaries (Yokoshi et al. 2020) (Fig. 2). Indeed, enhancer-promoter interactions are less sensitive to alterations in linear distances within TADs as opposed to inversions across boundaries (Symmons et al. 2016). TADs also deter interactions across boundaries as indicted by the fact that deletion or inversion of CTCF sites at TAD boundaries can promote crosstalk between neighboring TADs and rewire enhancer-promoter contacts in Drosophila and humans (Despang et al. 2019; Arzate-Mejia et al. 2020) (Fig. 2). Deletion of TAD boundaries can also result in ectopic gene regulation that occasionally leads to diseases in humans (Krumm and Duan 2019). Furthermore, concordant changes between TADs and gene expression are seen during early development in flies (Mateo et al. 2019) and during brain development in mouse (Winick-Ng et al. 2020).
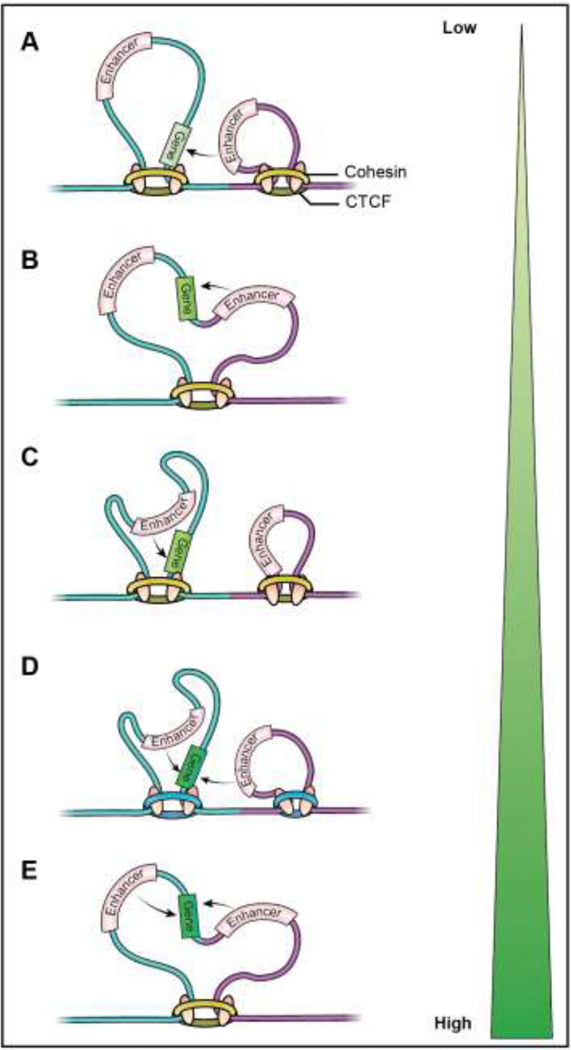
Figure 2. Distal regulation of gene activity in the context of heterogeneity in TAD structure:
A gene may experience various types of interactions with enhancers leading to a gradient of gene activation. (A) Enhancers from a neighboring TAD infrequently interact with a gene resulting in weak transcriptional activation. (B) Stochastic variations in the TAD boundary or a deletion of boundary element can transiently extrude enhancers from adjoining TADs into a single TAD to transiently increase the activation from such enhancers. (C) When TAD boundaries are stable, an enhancer within the same TAD interacts frequently with the gene and strongly upregulates its expression. (D) Simultaneous interaction with enhancers on both same and adjacent TADs can additively contribute to gene activation that is modulated by variation in TAD structure (D and E).
However, accumulating observations challenge this role for TADs. Gene expression patterns do not appear to strictly dependent on TAD structure since loss of TADs upon depletion of CTCF and cohesin leads to minimal changes in genome wide expression with very few genes, enriched for cell type specific transcription factors, showing more than two-fold change (Nora et al. 2017; Rao et al. 2017, Hsieh et al. 2021). In addition, TAD structures are both largely invariant across different germ layers despite differing in chromatin modifications and expression profiles and are formed after the enhancer-promoter loops are established during Drosophila development (**Espinola et al. 2021; **Ing-Simmons et al. 2021). Strikingly, synthetic or pathological alterations of TADs do not lead to widespread changes in gene expression (**Ghavi-Helm et al. 2019; Akdemir et al. 2020). In flies, nested inversions, deletions, and recessive lethal mutations showed very limited expression changes during embryonic development (**Ghavi-Helm et al. 2019). Along the same lines, structural variations that affect TAD boundaries across 2658 cancers, showed that only 14% of boundary rearrangements were associated with changes in gene expression (Akdemir et al. 2020). Furthermore, it has been observed in both gene specific and genome wide studies that some enhancers can activate gene expression across TAD boundaries, in some cases up to 20% of all enhancer-promoter interactions (Beccari et al. 2021, Hsieh et al. 2021). Orthogonally supporting these results is the observation of largely invariant TAD structures despite global changes in gene expression between normal and tumor cells (**Johnsone et al. 2020). Finally, early gene activation can precede TAD formation upon mitotic exit (Zhang et al. 2019) and TAD reorganization does not alter transcription for hours to days after differentiation or reprogramming of somatic cells to iPCS (Brown et al. 2018). These results suggest that tissue-specific chromatin conformation is not necessary for tissue-specific gene expression but rather the domain structure merely provides a structural framework to promote gene expression when enhancers become active (Misteli and Finn 2021). This interpretation is in line with the notion that the primary effect of chromatin structure is to facilitate and amplify functional states by increasing the probability of transcriptional events to occur, but not to acts as a binary regulator (Misteli 2020).
Transcription by itself, on the other hand, does not generally have a major impact on TAD structure or alter TAD boundaries (van Steensel and Furlong 2019). Indeed, ectopic expression of repressed genes is not sufficient to create new TAD boundaries (Bonev et al. 2017) and TADs form mostly independent of zygotic gene activation during development in Drosophila, mouse and zebrafish (Vallot and Tachibana 2020). Furthermore, while early zygotic genes serve as nucleation sites for TAD boundaries in Drosophila, the appearance of TAD boundaries is independent of transcription (Hug et al. 2017). However, more locally, at the sub-TAD level, transcription can lead to changes in intra-TAD subdomains to stabilize enhancer–promoter interactions in human and mouse embryonic cell lines (**Hsieh et al. 2020; **Krietenstein et al. 2020). Subdomain boundaries are enriched for transcriptional activity, invariably depleted of nucleosomes and centered around promoters, enhancers and architectural proteins like CTCF and YY1 (**Hsieh et al. 2020; **Krietenstein et al. 2020). Thus, it appears that fine-scale genome organization and transcription are able to mutually modulate and reinforce each other (**Misteli, 2020).
Given the relatively small effect of TAD structure on enhancer-promoter interactions and the probabilistic nature of TADs, how does the variation in TAD structure relate to bursting in single cells? One scenario is that stochastic enhancer-promoter interactions are superimposed over the heterogeneity in TAD organization (Fig. 2). In this scenario, enhancer-promoter pairs within TADs still have a higher likelihood of interacting relative to adjacent TADs, but variations in TAD boundaries may expose promoters to regulatory elements from neighboring TADs (Fig. 2B, E)). Intriguingly, the few genes with altered bursting properties upon depletion of cohesin, which leads to loss of TAD boundaries, are enriched at TAD boundaries (**Luppino et al. 2020), suggesting that stochastic intermingling might have a regulatory role for this subset of genes. Unlike TADs, stochastic fluctuations in the local chromatin environment may play a more dominant role in transcriptional bursting. In support, the ratio of A/B compartment around the transcription start site in single cells, which serves as a proxy for local chromatin environment, predicts bursting with >80% accuracy (**Su et al. 2020). The relatively stronger contribution of the chromatin environment over genome architecture in regulating transcription is further supported by the transcriptional and architectural dynamics upon mitotic exit. Genes marked for early expression during cytokinesis are bookmarked with acetylation of H3K27 and form short enhancer promoter loops (Pelham-Webb et al. 2021), while TADs are formed much later (Kang et al. 2020). Thus, heterogeneity in TADs likely affects the interaction of the regulatory regions with target genes, but it is the fluctuations in the chromatin environment in single cells that guides heterogeneity in compartmentalization and plays a dominant role in stochastic bursting.
Summary
The foundational principles of genome organization and gene expression are now well established (**Misteli, 2020). How genome structure relates to function, however, remains one of the central open questions in the field. The recent realization that both are stochastic adds a new dimension to relating genome organization to function. Many of the insights into the stochastic nature of the genome have come from single cell analysis and although these methods are sufficient to probe individual processes, such as organization or gene expression, exploring their interplay will require the simultaneous monitoring of structural and functional fluctuations in living cells all while controlling for the chromatin states and trans-factors associated with genes to track events leading to transcriptional bursts. While challenging, these approaches should make it clearer how accurately the current view of gene function, which is strongly based on ensemble analysis, reflects true genome behavior.
Acknowledgements
Research in the Misteli lab is supported by funding from the Intramural Research Program of the National Institutes of Health (NIH), National Cancer Institute, and Center for Cancer Research (1ZIA-BC010309).
Footnotes
Conflict of interest
All the authors approve the submitted version of the manuscript and have no conflict of interests and or competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abranches E, Guedes AM, Moravec M, Maamar H, Svoboda P, Raj A, Henrique D. 2014. Stochastic NANOG fluctuations allow mouse embryonic stem cells to explore pluripotency. Development 141: 2770–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akdemir KC, Le VT, Chandran S, Li Y, Verhaak RG, Beroukhim R, Campbell PJ, Chin L, Dixon JR, Futreal PA et al. 2020. Disruption of chromatin folding domains by somatic genomic rearrangements in human cancer. Nat Genet 52: 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander JM, Guan J, Li B, Maliskova L, Song M, Shen Y, Huang B, Lomvardas S, Weiner OD. 2019. Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzate-Mejia RG, Josue Cerecedo-Castillo A, Guerrero G, Furlan-Magaril M, Recillas-Targa F. 2020. In situ dissection of domain boundaries affect genome topology and gene transcription in Drosophila. Nat Commun 11: 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccari L, Jaquier G, Lopez-Delisle L, Rodriguez-Carballo E, Mascrez B, Gitto S, Woltering J, Duboule D. 2021. Dbx2 regulation in limbs suggests interTAD sharing of enhancers. Dev Dyn 250: 1280–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabdallah NS, Williamson I, Illingworth RS, Kane L, Boyle S, Sengupta D, Grimes GR, Therizols P, Bickmore WA. 2019. Decreased Enhancer-Promoter Proximity Accompanying Enhancer Activation. Mol Cell 76: 473–484 e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintu B, Mateo LJ, Su JH, Sinnott-Armstrong NA, Parker M, Kinrot S, Yamaya K, Boettiger AN, Zhuang X. 2018. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohrer CH, Larson DR. 2021. The Stochastic Genome and Its Role in Gene Expression. Cold Spring Harb Perspect Biol. [DOI] [PMC free article] [PubMed]
- Bonev B, Mendelson Cohen N, Szabo Q, Fritsch L, Papadopoulos GL, Lubling Y, Xu X, Lv X, Hugnot JP, Tanay A et al. 2017. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell 171: 557–572 e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Roberts NA, Graham B, Waithe D, Lagerholm C, Telenius JM, De Ornellas S, Oudelaar AM, Scott C, Szczerbal I et al. 2018. A tissue-specific self-interacting chromatin domain forms independently of enhancer-promoter interactions. Nat Commun 9: 3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo Gizzi AM, Cattoni DI, Fiche JB, Espinola SM, Gurgo J, Messina O, Houbron C, Ogiyama Y, Papadopoulos GL, Cavalli G et al. 2019. Microscopy-Based Chromosome Conformation Capture Enables Simultaneous Visualization of Genome Organization and Transcription in Intact Organisms. Mol Cell 74: 212–222 e215. **The authors describe a sequential imaging method, Hi-M, for single cell assaying of genome architecture and transcription in Drosophila that shows high corelation with Hi-C.
- Cavalli G, Misteli T. 2013. Functional implications of genome topology. Nat Struct Mol Biol 20: 290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chathoth KT, Zabet NR. 2019. Chromatin architecture reorganization during neuronal cell differentiation in Drosophila genome. Genome Res 29: 613–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Levo M, Barinov L, Fujioka M, Jaynes JB, Gregor T. 2018. Dynamic interplay between enhancer-promoter topology and gene activity. Nat Genet 50: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JO, Telenius JM, McGowan SJ, Roberts NA, Taylor S, Higgs DR, Hughes JR. 2016. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods 13: 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despang A, Schopflin R, Franke M, Ali S, Jerkovic I, Paliou C, Chan WL, Timmermann B, Wittler L, Vingron M et al. 2019. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat Genet 51: 1263–1271. [DOI] [PubMed] [Google Scholar]
- Donovan BT, Huynh A, Ball DA, Patel HP, Poirier MG, Larson DR, Ferguson ML, Lenstra TL. 2019. Live-cell imaging reveals the interplay between transcription factors, nucleosomes, and bursting. EMBO J 38. **The authors identify how binding kinetics of the transcription factor Gal4 affects the bursting of target gene GAL1. They show that TF binding frequncy/fraction bound affect the frequency while the the binding duration affects the burst size.
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186. [DOI] [PubMed] [Google Scholar]
- Espinola SM, Gotz M, Bellec M, Messina O, Fiche JB, Houbron C, Dejean M, Reim I, Cardozo Gizzi AM, Lagha M et al. 2021. Cis-regulatory chromatin loops arise before TADs and gene activation, and are independent of cell fate during early Drosophila development. Nat Genet 53: 477–486. **The authors use a single-cell spatial genomics approach using sequential imaging, Hi-M, to detect intra-TAD looping interactions between enhancers and promoters during early Drosophila development. Comparison of these loops across cells types showed looping does not correlate with expression and occurs before TADs or expression patterns are established.
- Falk M, Feodorova Y, Naumova N, Imakaev M, Lajoie BR, Leonhardt H, Joffe B, Dekker J, Fudenberg G, Solovei I et al. 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Misteli T. 2021. Phase Separation in Genome Organization across Evolution. Trends Cell Biol. [DOI] [PMC free article] [PubMed]
- Finn EH, Pegoraro G, Brandao HB, Valton AL, Oomen ME, Dekker J, Mirny L, Misteli T. 2019. Extensive Heterogeneity and Intrinsic Variation in Spatial Genome Organization. Cell 176: 1502–1515 e1510. **Using high-throughout microscopy the authors assayed interactions of over a hundred pairs of loci, within and across TADs, in single cells. While the interaction frequencies correlated well with Hi-C frequency, they were relatively low in the population and occurred in an allele-independent fashion.
- Fritzsch C, Baumgartner S, Kuban M, Steinshorn D, Reid G, Legewie S. 2018. Estrogendependent control and cell-to-cell variability of transcriptional bursting. Mol Syst Biol 14: e7678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya T, Lim B, Levine M. 2016. Enhancer Control of Transcriptional Bursting. Cell 166: 358–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. 2019. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet 51: 1272–1282. **Using Drosophila embryos with highly rearranged balancer chromosomes the authors assayed allele-specific topology and expression and found little correlation between structural alterations of TADs and gene expression.
- Gu H, Harris H, Olshansky M, Eliaz Y, Krishna A, Kalluchi A, Jacobs M, Cauer G, Pham M, Rao SPS et al. 2021. Fine-mapping of nuclear compartments using ultra-deep Hi-C shows that active promoter and enhancer elements localize in the active A compartment even when adjacent sequences do not. bioRxiv: 2021.10.03.462599 **This study demonstrates that A compartment regions are far smaller than previously thought implying that the A compartment is highly fragmented in 3D space.
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X. 2017. CTCF and cohesin regulate chromatin loop stability with distinct dynamics. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. 2017. A Phase Separation Model for Transcriptional Control. Cell 169: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoencamp C, Dudchenko O, Elbatsh AMO, Brahmachari S, Raaijmakers JA, van Schaik T, Sedeno Cacciatore A, Contessoto VG, van Heesbeen R, van den Broek B et al. 2021. 3D genomics across the tree of life reveals condensin II as a determinant of architecture type. Science 372: 984989. **This large scale study defines several distinct types of global genome organization patterns which appeared during evoluation and are related to the presence of condensin II.
- Hsieh TS, Cattoglio C, Slobodyanyuk E, Hansen AS, Rando OJ, Tjian R, Darzacq X. 2020. Resolving the 3D Landscape of Transcription-Linked Mammalian Chromatin Folding. Mol Cell 78: 539–553 e538. **Using Micro-C, which uncovers fine-scale TAD features in mESCs, the authors show a link between chromatin structure and transcription. Combinatorial binding of transcription factors and chromatin modifiers organize finer-scale interaction within TADs between enhancer-promoter and promoter-promoter. Acute transcriptional inhibition disrupts these gene-related foldings without altering higher order structures.
- Hsieh T- HS, Cattoglio C, Slobodyanyuk E, Hansen AS, Darzacq X, Tjian R. 2021. Enhancer-promoter interactions and transcription are maintained upon acute loss of CTCF, cohesin, WAPL, and YY1. bioRxiv: 2021.2007.2014.452365. [DOI] [PMC free article] [PubMed]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin Architecture Emerges during Zygotic Genome Activation Independent of Transcription. Cell 169: 216–228 e219. [DOI] [PubMed] [Google Scholar]
- Ing-Simmons E, Vaid R, Bing XY, Levine M, Mannervik M, Vaquerizas JM. 2021. Independence of chromatin conformation and gene regulation during Drosophila dorsoventral patterning. Nat Genet 53: 487–499. **Using Drosophila embryo mutants for dorsoventral patterning as a model the authors show that tissue-specific chromatin states do not correspond to a tissue-specifc chromatin conformation and conclude that chromatin conformation is not essential but rather acts as a scaffold for facilitating tissue-specific expression when enhancers get activated.
- Johnstone SE, Reyes A, Qi Y, Adriaens C, Hegazi E, Pelka K, Chen JH, Zou LS, Drier Y, Hecht V et al. 2020. Large-Scale Topological Changes Restrain Malignant Progression in Colorectal Cancer. Cell 182: 1474–1489 e1423. **The authors compared architectural, epigenetic and transcription changes between normal and tumor intestinal cells and tissue. They find mostly unchanged TADs, but spatial partitioning of open and closed compartments was profoundly compromised in tumors, especially at the interface between A and B compartments.
- Kang H, Shokhirev MN, Xu Z, Chandran S, Dixon JR, Hetzer MW. 2020. Dynamic regulation of histone modifications and long-range chromosomal interactions during postmitotic transcriptional reactivation. Genes Dev 34: 913–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempfer R, Pombo A. 2020. Methods for mapping 3D chromosome architecture. Nat Rev Genet 21: 207–226. [DOI] [PubMed] [Google Scholar]
- Krietenstein N, Abraham S, Venev SV, Abdennur N, Gibcus J, Hsieh TS, Parsi KM, Yang L, Maehr R, Mirny LA et al. 2020. Ultrastructural Details of Mammalian Chromosome Architecture. Mol Cell 78: 554–565 e557. **The authors use Mirco-C, a high resolution Hi-C technique, and find thousands of new loops with boundaries at nucleosome precision. Many of these loops are localized along extrusion stripes consistent with their anchors impeding cohesin-dependent loop extrusion.
- Krumm A, Duan Z. 2019. Understanding the 3D genome: Emerging impacts on human disease. Semin Cell Dev Biol 90: 62–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson AJM, Johnsson P, Hagemann-Jensen M, Hartmanis L, Faridani OR, Reinius B, Segerstolpe A, Rivera CM, Ren B, Sandberg R. 2019. Genomic encoding of transcriptional burst kinetics. Nature 565: 251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Wang X, Jiang H, Wang R, Chen J, Chen Y, Xu Q, Cao J, Gong X, Wu J et al. 2020. Reorganized 3D Genome Structures Support Transcriptional Regulation in Mouse Spermatogenesis. iScience 23: 101034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luppino JM, Park DS, Nguyen SC, Lan Y, Xu Z, Yunker R, Joyce EF. 2020. Cohesin promotes stochastic domain intermingling to ensure proper regulation of boundary-proximal genes. Nat Genet 52: 840–848. **The authors use Oligopaint with high resolution imaging to show heterogeneity in TAD boundaries and inter-TAD interactions amongst single cells. Using depletion assays they show that cohesin promotes both inter- and intra-TAD interactions and CTCF prevents inter-TAD interactions. Loss of TAD boundaries was shown to have an impact on bursting of genes close to the boundary.
- Mateo LJ, Murphy SE, Hafner A, Cinquini IS, Walker CA, Boettiger AN. 2019. Visualizing DNA folding and RNA in embryos at single-cell resolution. Nature 568: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArthur E, Capra JA. 2021. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am J Hum Genet 108: 269–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirny LA, Imakaev M, Abdennur N. 2019. Two major mechanisms of chromosome organization. Curr Opin Cell Biol 58: 142–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T 2020. The Self-Organizing Genome: Principles of Genome Architecture and Function. Cell 183: 28–45. **This comprehensive review on the interplay between genome function and structure highlights how genome function is a driver of structure and structure in turn is a modulator, rather than a binary determinant, of function.
- Misteli T, Finn EH. 2021. Chromatin architecture is a flexible foundation for gene expression. Nat Genet 53: 426–427. [DOI] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell 169: 930–944 e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuebler J, Fudenberg G, Imakaev M, Abdennur N, Mirny LA. 2018. Chromatin organization by an interplay of loop extrusion and compartmental segregation. Proc Natl Acad Sci U S A 115: E6697–E6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudelaar AM, Beagrie RA, Gosden M, de Ornellas S, Georgiades E, Kerry J, Hidalgo D, Carrelha J, Shivalingam A, El-Sagheer AH et al. 2020. Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat Commun 11: 2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham-Webb B, Polyzos A, Wojenski L, Kloetgen A, Li J, Di Giammartino DC, Sakellaropoulos T, Tsirigos A, Core L, Apostolou E. 2021. H3K27ac bookmarking promotes rapid post-mitotic activation of the pluripotent stem cell program without impacting 3D chromatin reorganization. Mol Cell 81: 1732–1748 e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID et al. 2017. Cohesin Loss Eliminates All Loop Domains. Cell 171: 305–320 e324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Larson DR. 2020. Transcription in Living Cells: Molecular Mechanisms of Bursting. Annu Rev Biochem 89: 189–212. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Ren G, Day CR, Zhao K, Chow CC, Larson DR. 2019. Intrinsic Dynamics of a Human Gene Reveal the Basis of Expression Heterogeneity. Cell 176: 213–226 e218. **Using estrogen-induced expression of the TFF1 gene as model for transcriptional bursting the authors show by live-cell imaging that Off-times can vary from minutes to days. The long Off- times, which represents a deep repressive state, lead to heterogeneity in expression. They derive a mathematical model that relates transcription, chromosome structure and sensitivity to estrogen.
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- Shaffer SM, Emert BL, Reyes Hueros RA, Cote C, Harmange G, Schaff DL, Sizemore AE, Gupte R, Torre E, Singh A et al. 2020. Memory Sequencing Reveals Heritable Single-Cell Gene Expression Programs Associated with Distinct Cellular Behaviors. Cell 182: 947–959 e917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavreva DA, Garcia DA, Fettweis G, Gudla PR, Zaki GF, Soni V, McGowan A, Williams G, Huynh A, Palangat M et al. 2019. Transcriptional Bursting and Co-bursting Regulation by Steroid Hormone Release Pattern and Transcription Factor Mobility. Mol Cell 75: 1161–1177 e1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su JH, Zheng P, Kinrot SS, Bintu B, Zhuang X. 2020. Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 182: 1641–1659 e1626. **The authors coupled sequential hybridization with high resoultion imaging to trace multiple chromosomes and uncover pervasive heterogeneity in structure between cells. They employed MERFISH for simultaneous genome scale chromatin tracing and nascent RNA FISH and propose a framework describing trans-chromatin environment regulation of gene bursting.
- Symmons O, Chang M, Mellis IA, Kalish JM, Park J, Susztak K, Bartolomei MS, Raj A. 2019. Allele-specific RNA imaging shows that allelic imbalances can arise in tissues through transcriptional bursting. PLoS Genet 15: e1007874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symmons O, Pan L, Remeseiro S, Aktas T, Klein F, Huber W, Spitz F. 2016. The Shh Topological Domain Facilitates the Action of Remote Enhancers by Reducing the Effects of Genomic Distances. Dev Cell 39: 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Donjon A, Jerkovic I, Papadopoulos GL, Cheutin T, Bonev B, Nora EP, Bruneau BG, Bantignies F, Cavalli G. 2020. Regulation of single-cell genome organization into TADs and chromatin nanodomains. Nat Genet 52: 1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo Q, Jost D, Chang JM, Cattoni DI, Papadopoulos GL, Bonev B, Sexton T, Gurgo J, Jacquier C, Nollmann M et al. 2018. TADs are 3D structural units of higher-order chromosome organization in Drosophila. Sci Adv 4: eaar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Yun J, Zheng S, Ollikainen N, Pierson N, White J, Shah S, Thomassie J, Suo S, Eng CL et al. 2021. Integrated spatial genomics reveals global architecture of single nuclei. Nature 590: 344–350. **This study reports the comprehensive mapping of genome organization features along with multiple chromatin marks, subnuclear structures and expression profiling in mouse ES cells. They find multiple distinct subpopulations of cells and show that highly expressed genes are pre-positioned to active nuclear zones, independent of bursting dynamics in single cells.
- Tan L, Xing D, Chang CH, Li H, Xie XS. 2018. Three-dimensional genome structures of single diploid human cells. Science 361: 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzaskoma P, Ruszczycki B, Lee B, Pels KK, Krawczyk K, Bokota G, Szczepankiewicz AA, Aaron J, Walczak A, Sliwinska MA et al. 2020. Ultrastructural visualization of 3D chromatin folding using volume electron microscopy and DNA in situ hybridization. Nat Commun 11: 2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallot A, Tachibana K. 2020. The emergence of genome architecture and zygotic genome activation. Curr Opin Cell Biol 64: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Furlong EEM. 2019. The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 20: 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Anastasakis DG, Rodriguez J, Palangat M, Gudla P, Zaki G, Tandon M, Pegoraro G, Chow CC, Hafner M et al. 2021. Dynamic imaging of nascent RNA reveals general principles of transcription dynamics and stochastic splice site selection. Cell 184: 2878–2895 e2820. **The authors tagged intron RNA of over 700 endogenous gene to track bursting and splicing in live cells and found that genes differ primarily in their Off- times of bursting and splice long before the polymerase reaches the 3’ splice sites. Using a combination of nascent RNAseq and live cell imaging they show hundreds of genes are spliced recursively from cryptic 3’ splice sites within the intron.
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, Abraham BJ, Cohen MA, Nabet B, Buckley DL et al. 2017. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 171: 1573–1588 e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson I, Kane L, Devenney PS, Flyamer IM, Anderson E, Kilanowski F, Hill RE, Bickmore WA, Lettice LA. 2019. Developmentally regulated Shh expression is robust to TAD perturbations. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winick-Ng W, Kukalev A, Harabula I, Redondo LZ, Meijer M, Serebreni L, Bianco S, Szabo D, Chiariello AM, Azcarate II et al. 2020. Cell-type specialization in the brain is encoded by specific long-range chromatin topologies. bioRxiv: 2020.2004.2002.020990.
- Xiao J, Hafner A, Boettiger AN. 2020. How subtle changes in 3D structure can create large changes in transcription. bioRxiv: 2020.2010.2022.351395. [DOI] [PMC free article] [PubMed]
- Yokoshi M, Segawa K, Fukaya T. 2020. Visualizing the Role of Boundary Elements in Enhancer-Promoter Communication. Mol Cell 78: 224–235 e225. [DOI] [PubMed] [Google Scholar]