Abstract
Conditionally activated, caged morpholino antisense agents (cMOs) are tools that enable the temporal and spatial investigation of gene expression, regulation, and function during embryonic development. Cyclic MOs are conformationally gated oligonucleotide analogs that do not block gene expression until they are linearized through the application of an external trigger, such as light or enzyme activity. Here, we describe the first examples of small molecule-responsive cMOs, which undergo rapid and efficient decaging via a Staudinger reduction. This is enabled by a highly flexible linker design that offers opportunities for the installation of chemically activated, self-immolative motifs. We synthesized cyclic cMOs against two distinct, developmentally relevant genes and demonstrated phosphine-triggered knockdown of gene expression in zebrafish embryos. This represents the first report of a small molecule-triggered antisense agent for gene knockdown, adding another bioorthogonal entry to the growing arsenal of gene knockdown tools.
Graphical Abstract
INTRODUCTION
Oligonucleotide-based tools have found extensive application in the elucidation of gene function and genetic networks that dictate biological processes such embryogenesis and disease progression.1–3 Morpholino oligonucleotides (MOs) are well-established reagents that are used to sequence-specifically control gene expression during embryonic development by blocking translational start sites, sequestering microRNAs, and inhibiting mRNA splice sites.4–7
MOs induce gene (or microRNA) silencing almost immediately after injection into fertilized oocytes and persist in the developing embryo for up to 5 days due to their nuclease resistance.8 This lack of external control over their basal activity has been addressed through the development of caged MOs (cMOs), which render the MO inactive until it is stimulated with an external trigger. We and others have devised various MO caging strategies, including photolabile inhibitory duplexes,9–12 incorporation of photocaged nucleobases,13,14 and macrocyclization with photocleavable15–17 and enzyme-responsive linkers.18 The curvature induced by the end-to-end macrocyclization of MOs diminishes target mRNA binding, as the MO/mRNA heteroduplex has limited flexibility. Furthermore, cyclic cMOs can be generated in a few synthetic steps with virtually any MO and conditionally controlled linkers equipped with the proper reactive handles, thereby rendering this approach highly modular. Additionally, cyclic cMOs can be activated following a single triggering event, obviating the need for modified nucleobases or auxiliary oligonucleotides.
Herein, we report the first chemically activated MO. Small molecules have enormous potential as conditional triggers, as they are minimally invasive and can be applied with high temporal resolution. Furthermore, they do not require irradiation equipment and they can be easily applied to large numbers of embryos in parallel. Various bioorthogonal small molecule pairs have been proven to be useful probes, enabling studies into biological processes without perturbing the endogenous system.19–21 The Staudinger reduction and its variants are among the most bioorthogonal reactions and have consistently shown success in various demanding biological environments.21–25 Based on their biorthogonality and history of use in vivo,21 we selected the azide/phosphine pair to generate a cMO linker that is cleaved through a Staudinger reduction-induced self-immolation.
RESULTS AND DISCUSSION
The cyclic p-azidobenzyl cMO is reduced following exposure to a phosphine trigger, forming a p-aminobenzyl intermediate. This intermediate then collapses, undergoing a 1,6-elimination and subsequent decarboxylation to cleave the carbamate linkage. The resulting iminoquinone methide intermediate is then quenched with water.26–28 Upon cMO cleavage the active, linear MO is generated, allowing for hybridization to its target mRNA and silencing of gene expression (Figure 1). The mutual reactivity and enhanced orthogonality of the azide/triarylphosphine pair can be credited to its generally “soft” chemical nature, whereas most competing biological nucleophiles and electrophiles are primarily “hard” in character.21 Thus, this chemoselective pair is particularly attractive for achieving small molecule-mediated control of antisense function.
Figure 1.
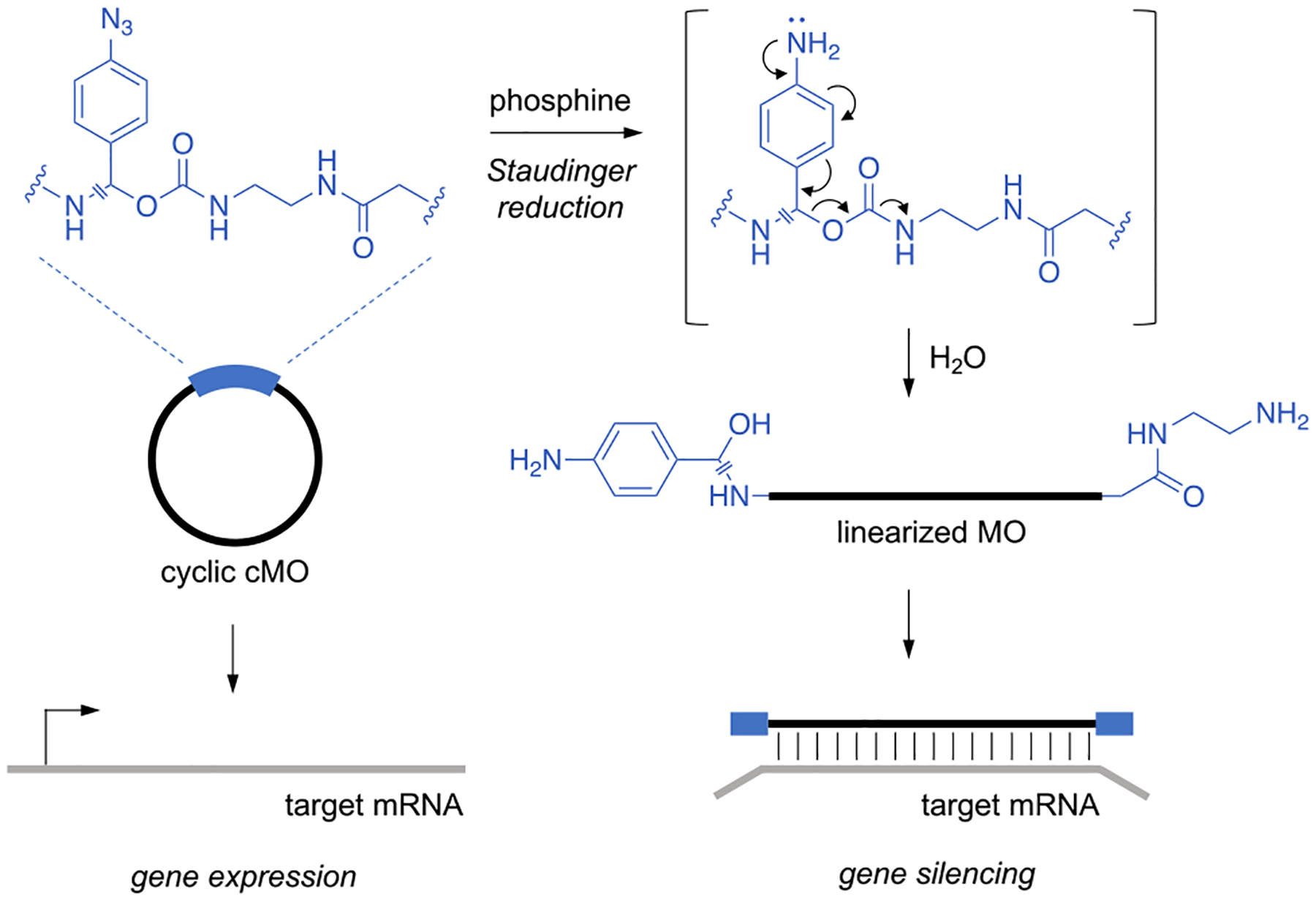
Following exposure to a phosphine trigger, the p-azidobenzyl linker undergoes a Staudinger reduction, forming a p-aminobenzyl intermediate. The linker then rapidly self-cleaves via a 1,6-elimination and subsequent decarboxylation to generate the active, linearized MO, which can then hybridize to target mRNAs and silence gene expression.
The linker is based on a p-azidobenzyl motif which, after exposure to a phosphine trigger, will undergo a self-immolative cleavage via a Staudinger reduction. We placed the azido group in the 6-position rather than the 4-position, as 1,6-eliminations occur nearly twice as quickly as 1,4-eliminations,28,29 and we expected reduced steric hindrance for the phosphine. In designing the linker, we utilized a carbamate leaving group, as it is stable to physiological conditions and well-established for self-immolation.28,30 Furthermore, the benzylic position of the linker was substituted, as this has been shown to accelerate self-immolation by stabilizing the partial positive charge that develops during elimination.28 These design considerations are supported by recent comparisons of azidobenzyl carbamates as phosphine-removable protecting groups in biological settings.25,31 Aside from the substituted p-azidobenzyl carbamate core, we functionalized the linker with N-hydroxysuccinimide (NHS) ester and chloroacetamide handles for cyclization of MOs modified at the termini with the proper amino and thiol handles.
The linker was synthesized in 10 steps from commercially available 4-aminoacetophenone (1) (Figure 2A). Diazotization and azide substitution of 1 yielded the aryl azide 2.25 Bromination of 2 provided the α-bromo ketone 3,32 which was then transformed into the corresponding primary amine 4 utilizing hexamine in a Delépine reaction. The primary amine was then reacted with methyl adipoyl chloride to form the amide 5.33 Treatment with sodium borohydride reduced the ketone, affording alcohol 6, which was conjugated to ethylenediamine after activation with 1,1-carbonyldiimidazole (CDI) and was subsequently capped with 2-chloroacetyl chloride to form the chloroacetamide 7.15 Saponification of the methyl ester 7 with aqueous lithium hydroxide afforded the corresponding carboxylic acid 8, which was activated to the NHS-ester using O-(N-succinimidyl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TSTU) to provide the final linker 9.
Figure 2.
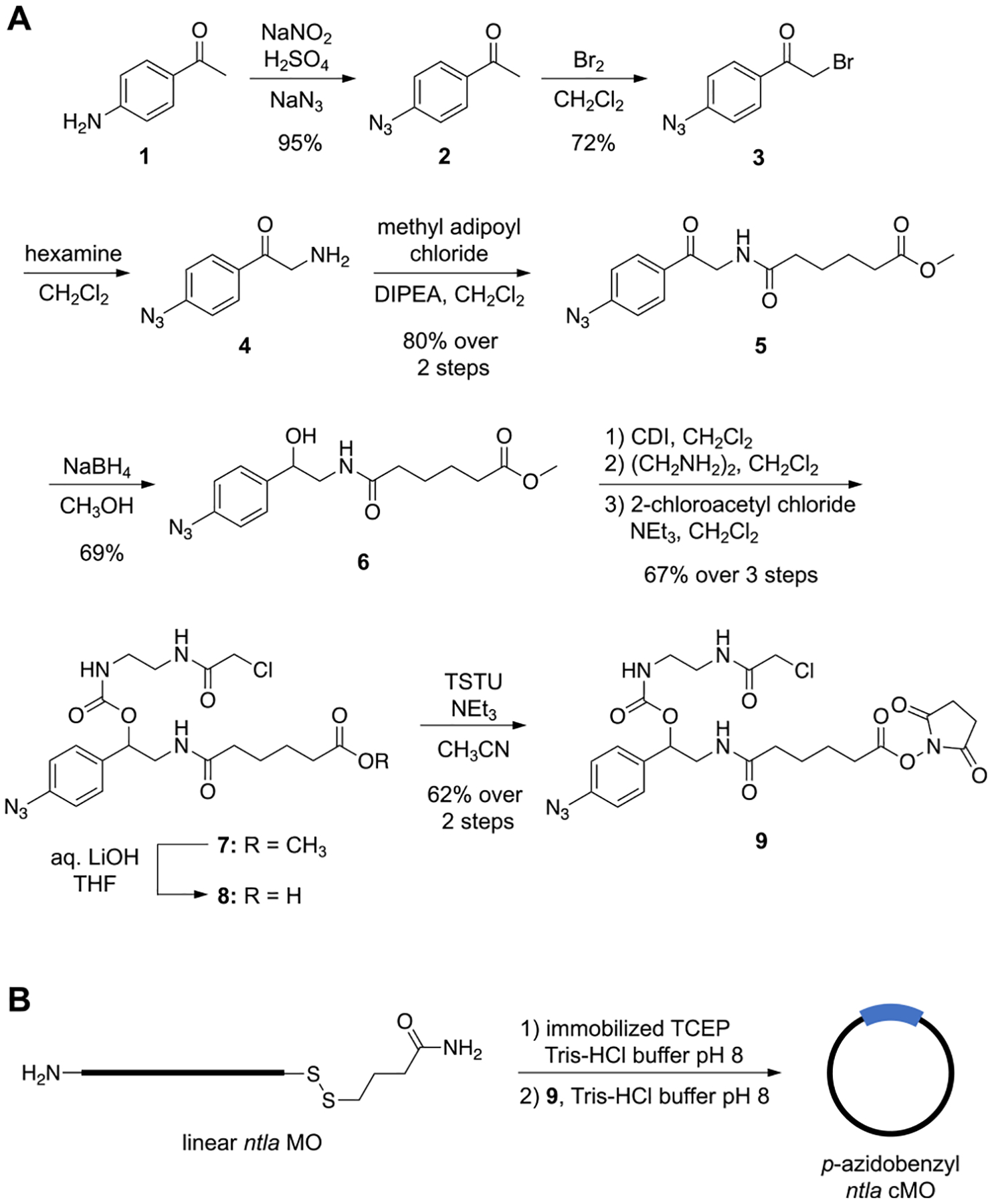
(A) Synthesis of p-azidobenzyl MO linker 9. (B) Application of the linker in the synthesis of the cMO targeting ntla.
The linker 9 was then used to prepare a cyclic p-azidobenzyl cMO targeting the T-box transcription factor Ta (tbxta), also commonly referred to as no tail a (ntla, 5′-GACTTGAGGCAGACATATTTCCGAT-3′, anti-start codon underlined) (Figure 2B). The ntla MO was purchased (GeneTools, LLC) with 5′-amine and 3′-disulfide modifications to allow for bioconjugation to the small molecule linker. In previously reported macrocyclization protocols, the amine was first reacted with the NHS ester prior to thiol reduction and subsequent cyclization via an intramolecular thioether formation.15,16 However, to avoid premature reduction of the azide linker, the reduction step must be performed first. As such, the MO was first incubated with resin-immobilized tris(2-carboxyethyl)phosphine (TCEP) to reduce the 3′-disulfide, generating the free thiol. The reduced MO was then reacted with excess linker 9. Analysis of the reaction progression by MALDI-TOF MS revealed that the NHS-ester moiety of 9 first reacted with the primary amine on the MO, forming an amide linkage to the 5′-termini (Figure S1 in the Supporting Information). The conjugated product then underwent a spontaneous, intramolecular cyclization through the reaction of the free 3′-thiol on the MO with the chloroacetamide moiety on the linker to generate the desired cMO macrocycle. Undesired, linear MO reaction species were removed from the reaction mixture using iodoacetyl- and NHS-functionalized resins and purification by high-performance liquid chromatography (HPLC).
We first sought to evaluate phosphine-triggered decaging using a model fluorescence-based system. We designed and synthesized the dextran-conjugated caged Q-rhodamine sensor 10. We chose Q-rhodamine as the fluorescent core due to its excellent photostability, high extinction coefficient (>80000 M−1 cm−1),34 quantum yield of almost 1,35 and the ability to utilize the near total loss of fluorescence upon amino group acylation for our caging approach.36 We installed two p-azidobenzyl carbamates, which upon exposure to a phosphine undergo a Staudinger reduction-mediated self-immolation as previously described, thus generating the free fluorophore 11 (Figure 3A). With the sensor 10a in hand, a small panel of phosphines were screened for fluorescence activation of the sensor (Figure 3B,C). While inactivation of phosphines by oxidation is well-documented, the relatively low solubility of oxygen in water,37 coupled with the slow rate of oxidation for triarylphosphines, makes these reagents useful for biological studies on the time scale of regulation and perturbation of gene function during development. Based on previous reports,23,31,38 2-(diphenylphosphino)benzamide (2DPBM) was selected, as it has repeatedly demonstrated superior activation kinetics. Further, it is relatively stable to oxidation, soluble up to 100 μM in water, and nontoxic, making it practical for use in biological experiments. These properties stem from the ortho-amido group that enhances solubility and improves hydrolysis kinetics via a neighboring-group effect.38 The corresponding carboxylic acid, 2DPBA, has enhanced solubility, in addition to the same set of desirable properties, and was evaluated alongside 2DPBM. In an effort to enhance the neighboring-group effect by increasing electron density at the amide with minimal perturbation to the P(III) center, a methoxy group was installed at the 4-position (4-OMe-2DPBM). Finally, 4DPBA and 4DPBM were evaluated as controls that are unable to exhibit the neighboring-group effect. Overall, 2DPBM showed the best activation kinetics, decaging the sensor rapidly (t1/2 ≈ 11 min) in comparison to other phosphines tested at the same concentration (50 μM).
Figure 3.
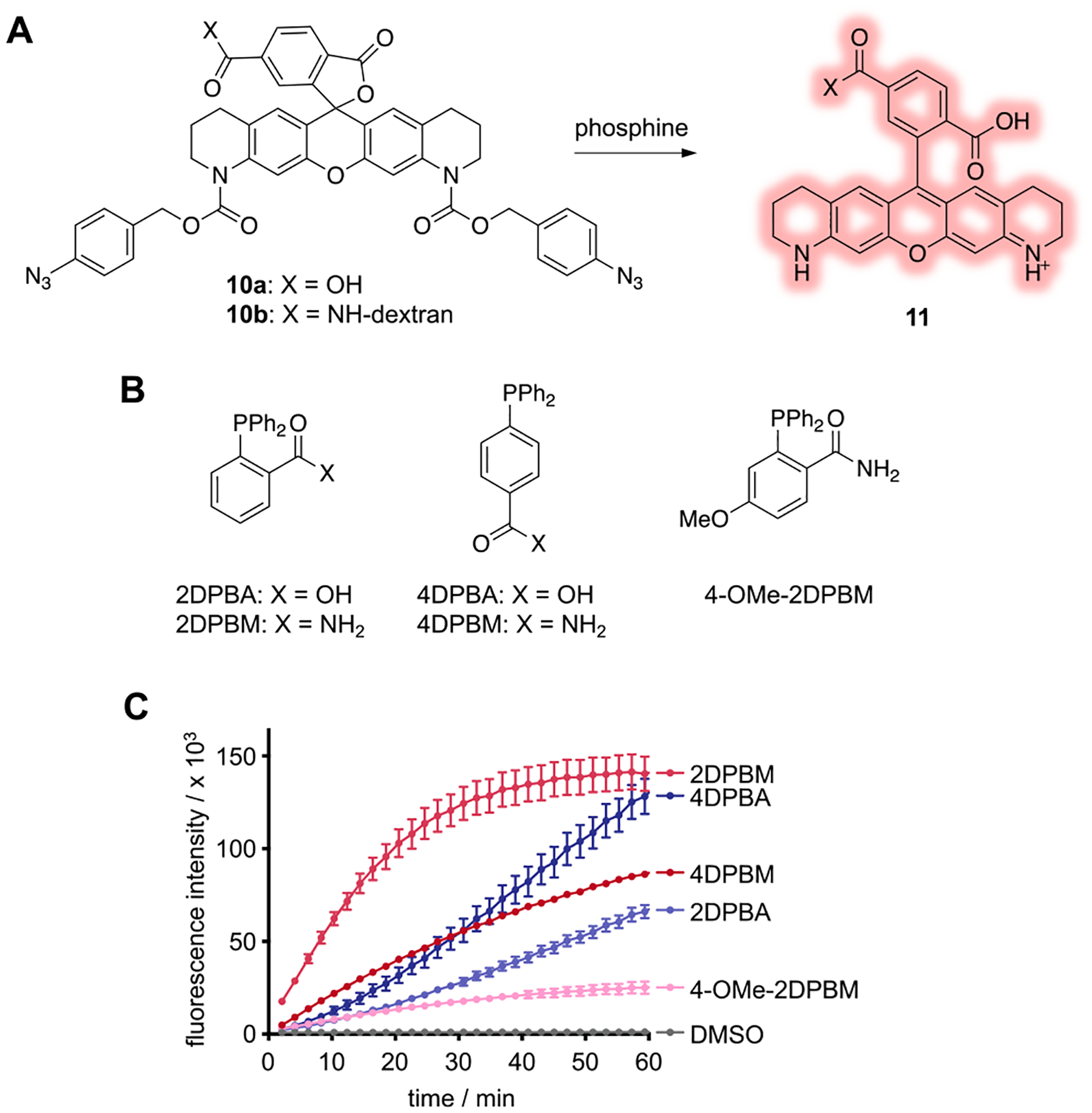
(A) The Q-rhodamine fluorophore is caged with p-azidobenzyl carbamates. Following treatment with phosphine, fluorescence is activated via Staudinger reduction-induced decaging. (B) Structures of phosphines. (C) The fluorescence activation of the sensor (5 μM) was monitored over time following incubation with various phosphines (50 μM) at 29 °C. Error bars represent standard deviations from the average of three independent experiments.
For testing of our chemical decaging strategy in zebrafish embryos, the rhodamine sensor was conjugated to an aminemodified dextran polymer via NHS-activation of the carboxylate functionality on the pendant aryl ring, generating 10b. Dextran-conjugated fluorophores have a long history of use in zebrafish embryos,39–43 as they are nontoxic, hydrophilic,44 and prevent diffusion out of the blastomeres.41,45
2DPBM was chosen as the phosphine trigger, as it demonstrated the fastest kinetics in our initial screening and has been previously been used for Staudinger reductions in mammalian cells.23,25 As discussed above, phosphines are susceptible to oxidation, and while the half-life of 2DPBM under aquaculture conditions is ~200 min (Figure S2 in the Supporting Information), we anticipate the rate of phosphine oxidation to be accelerated within a metabolically active zebrafish embryo. To ensure the integrity and availability of the phosphine trigger, treated embryos were soaked in E3 water supplemented with 2DPBM and the medium was refreshed once after the initial treatment. Previous reports demonstrate rapid diffusion of small molecules into zebrafish embryos, suggesting this method to be a quick and efficient means of compound delivery.46 Furthermore, pulsed phosphine dosing has been demonstrated as a more effective regimen than single dosing in the release of azido-modified prodrugs in mammalian cell culture.27 Accordingly, no toxicity was observed in zebrafish embryos treated with 2DPBM (100 μM), for varying durations of phosphine exposure (Figure S3 in the Supporting Information).
Zebrafish embryos at the 1-cell stage were microinjected in the yolk with the sensor 10b, and fluorescence activation was monitored over the first 8 h of embryonic development (Figure 4A). Reagents microinjected into the yolk prior to the 8-cell stage are globally distributed to the developing embryo via cytoplasmic bridging between the yolk and the dividing cells.41 At 2 h post-fertilization (hpf), the injected zebrafish embryos were transferred to E3 water supplemented with 2DPBM (100 μM) and were incubated at 29 °C for the indicated amount of time (30, 60, or 90 min). The fish water was refreshed with 2DPBM-supplemented water, and the embryos were soaked for an amount of time equivalent to that of the first exposure before being transferred into phosphine-free E3 water. Fluorescence activation of the sensor was observed within 30 min of 2DPBM exposure, suggesting efficient delivery through embryo soaking (Figure 4B and Figure S4 in the Supporting Information). Furthermore, no fluorescence increase was detected in injected embryos in the absence of 2DPBM. These results indicate that the p-azidobenzyl carbamate is stable in the developing zebrafish embryo environment, despite previous reports speculating that aromatic azides are susceptible to metabolic reduction in vivo.47 Together, these results demonstrate that 2DPBM permeates through the chorion into the embryo and validates the quick Staudinger reduction-mediated decaging in vivo.
Figure 4.
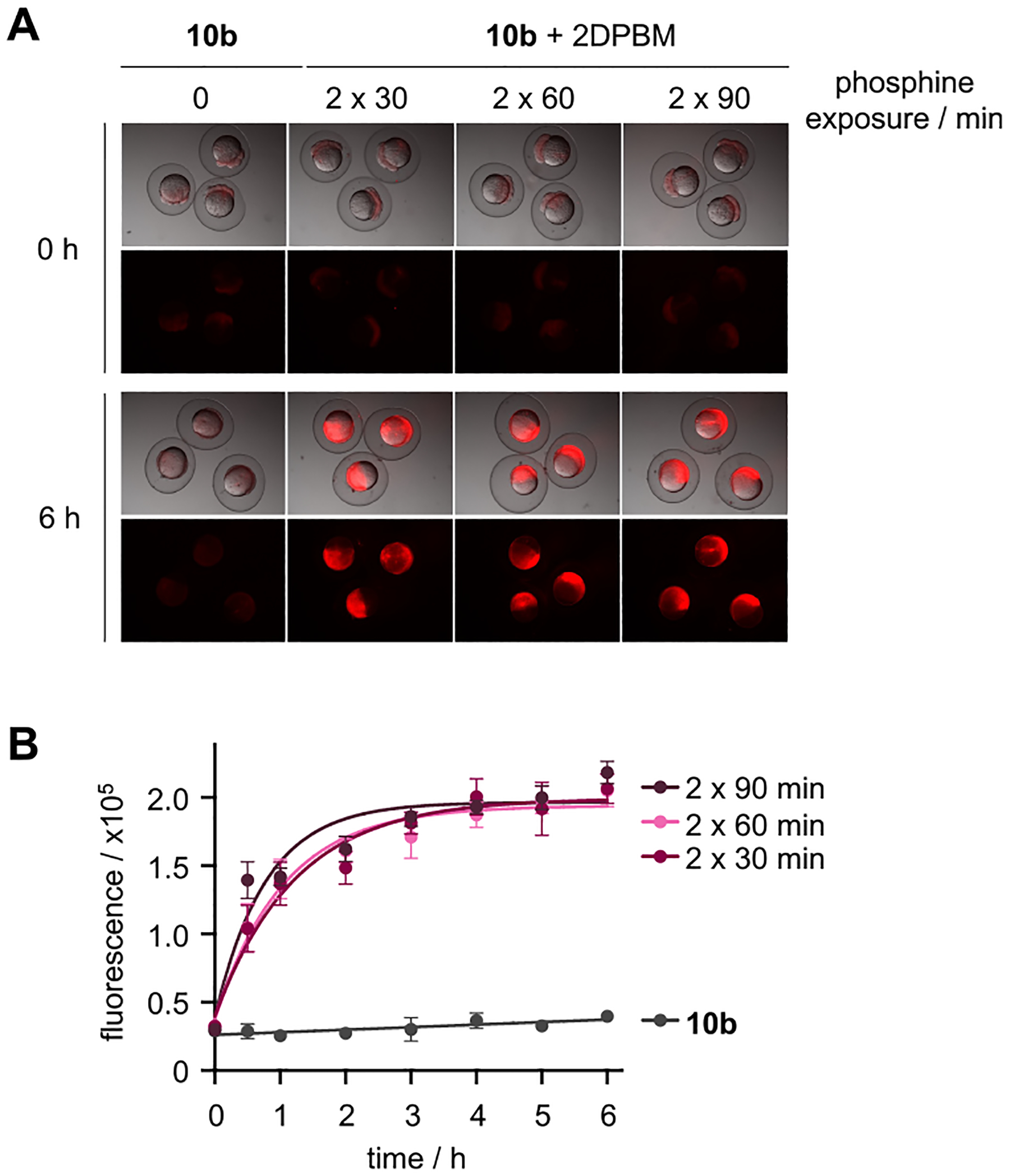
(A) Representative images of zebrafish embryos injected with 10b and soaked in E3 water supplemented with 2DPBM (100 μM) for various durations. (B) Quantification of rhodamine fluorescence (n = 5 embryos/condition) demonstrates efficient activation of 10b following phosphine treatment and supports the micrograph findings. Error bars represent standard deviations from the average of 5 embryos.
Having validated the chemical activation of the p-azidobenzyl-caged fluorescent sensor, we next sought to evaluate small molecule-triggered activation of MO function. ntla encodes the zebrafish ortholog of Brachyury, a T-box transcription factor required for axial mesoderm development.48–50 We first analyzed phosphine-triggered linearization of our p-azidobenzyl ntla cMO in vitro (Figure S5 in the Supporting Information). The cMO was diluted to a final concentration of 6 μM (so as to mimic the conditions utilized for zebrafish studies)51 and incubated in the presence of DMSO or 2DPBM at 29 °C. Cleavage of the cMO was assessed by MALDI-TOF mass spectrometry. Gratifyingly, in the presence of 2DPBM full linearization of the ntla cMO was observed, as indicated by the detection of the masses consistent with the cleaved iminoquinone methide intermediate and the quenched product. The detection of these linearized products suggests that the self-immolative mechanism of linker cleavage proceeds via the anticipated Staudinger reduction-induced 1,6-elimination discussed previously (Figure 1). Furthermore, no linearized MO products are detected when the cMO is treated with vehicle (DMSO) alone, suggesting that the cMO is stable and linearization is chemoselectively induced through a phosphine-mediated azide reduction.
We next sought to determine if phosphine-triggered linearization would also rescue the MO silencing function. The ability of photocaged cMOs to bind to their target sequences following irradiation has previously been exclusively evaluated with biophysical assays, including melt temperature measurements, fluorescence activation of molecular beacons, and gel shift assays.13,15,17 However, these assays solely measure target hybridization and do not directly reflect how a cyclic, and subsequently linearized, MO will function within the context of mRNA translation. Thus, we adopted a lysate-based translation system to screen phosphine-triggered MO function in vitro.52,53 We generated a luciferase reporter construct in which the ntla MO binding sequence (ntlaBS) was cloned directly upstream of and in frame with the firefly luciferase coding sequence (ntlaBS-Fluc). In the presence of a linear ntla MO, the expression of the corresponding ntlaBS-Fluc mRNA is repressed due to sequence-specific hybridization of the MO to the target mRNA, blocking the translational machinery from accessing the start codon (Figure 5A). We observed a dose-dependent inhibition of luciferase expression with increasing concentrations of linear ntla MO and nearly full silencing at 2 μM (Figure 5B). When it was treated at the same concentration, the cyclic p-azidobenzyl ntla cMO exhibited no significant reduction in luciferase expression, indicating that the target binding and silencing activity is impeded by the cMO curvature. Further, the reticulocyte lysate is supplemented with thiol-based reducing agents, dithiothreitol and cysteine, further supporting the stability of the p-azidobenzyl carbamate to the endogenous reducing environment. It is only after activation with 2DPBM that nearly full silencing of luciferase expression, identical with that observed with the linear ntla MO, is observed (Figure 5C). These results indicate that the developed cMO provides an excellent small molecule ON to OFF switch of gene function, based on a Staudinger reduction.
Figure 5.
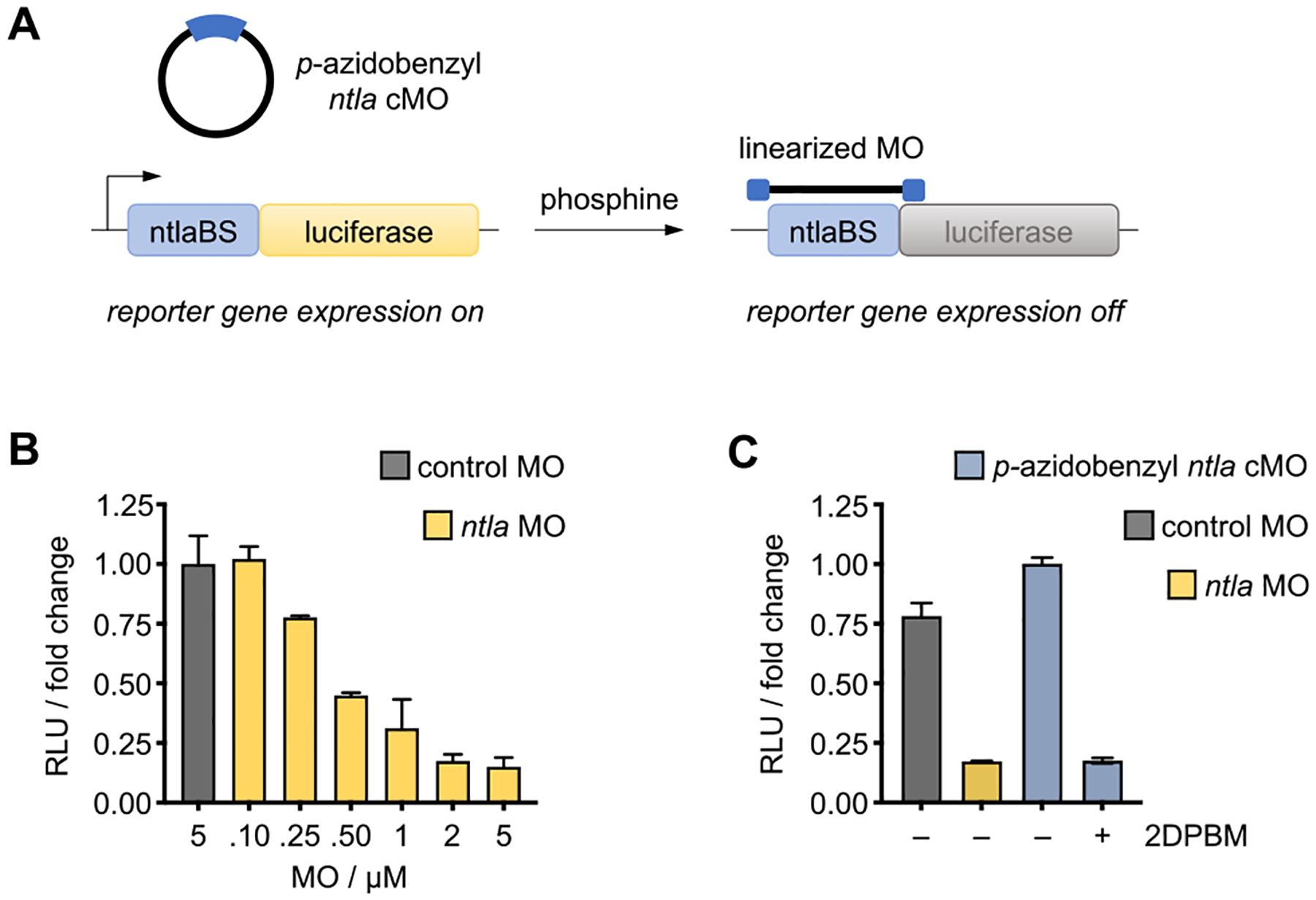
(A) In the presence of the cyclic p-azidobenzyl ntla cMO, luciferase is expressed, as cyclization impedes target binding and masks antisense activity. Treatment with phosphine induces cMO cleavage, generating the active, linearized MO which represses luciferase expression. (B) Titration of linear ntla MO induces dose-dependent silencing of Fluc expression. (C) Nearly complete silencing of Fluc expression is observed following 2DPBM-mediated activation of the cyclic p-azidobenzyl ntla cMO, while no effect on reporter expression is observed with cMO alone. Data represent the means ± standard deviation from at least three independent experiments.
Having established efficient, conditional control of ntlaBS-driven reporter gene expression in vitro, we next investigated the ability of our cyclic p-azidobenzyl ntla cMO to regulate endogenous ntla expression in zebrafish embryos using small molecule control. Silencing of ntla expression during the early stages of embryo development induces distinct morphological defects, including a loss of notochord cells, posterior truncation, and U-shaped somites, which are evident at 24 hpf (Figure 6A).48,49 Zebrafish embryos were microinjected with 100 pg of a negative control MO, a linear ntla MO, or our cyclic ntla cMO. At ~2–3 hpf, embryos were transferred to E3 water supplemented with either DMSO (0.2%) or 2DPBM (100 μM) and soaked following the 2 × 90 min treatment regimen that we identified as highly efficient in Figure 4A. Following treatment, the embryos were transferred to E3 water and incubated until 24 hpf, at which point they were analyzed for the ntla loss-of-function phenotype (Figure 6A). Since the hallmark indicator of the ntla morphant phenotype includes loss of posterior tissue, the phenotypic strength was scored based on the quantification of shortened embryo body length (Figure S6 in the Supporting Information). Embryos injected with the cyclic p-azidobenzyl ntla cMO and exposed to DMSO exhibited minimal phenotypic changes. These results are consistent with our luciferase reporter assay (Figure 5C), indicating that the curvature induced through macrocyclization with our p-azidobenzyl linker is sufficient to block MO activity in vivo. The cMO remains functionally inert until treatment with 2DPBM, as evidenced by 82% of cMO-injected embryos exhibiting a strong ntla morphant phenotype after treatment with the small molecule phosphine trigger. Furthermore, wild-type zebrafish embryos treated with 2DPBM at the same concentration and duration exhibit no obvious toxicity or developmental defects (Figure S3 in the Supporting Information), supporting that the phenotypic result can be specifically attributed to MO-induced gene silencing and establishing the Staudinger reduction as a potentially broadly applicable conditional control mechanism for biological processes in the zebrafish model.
Figure 6.
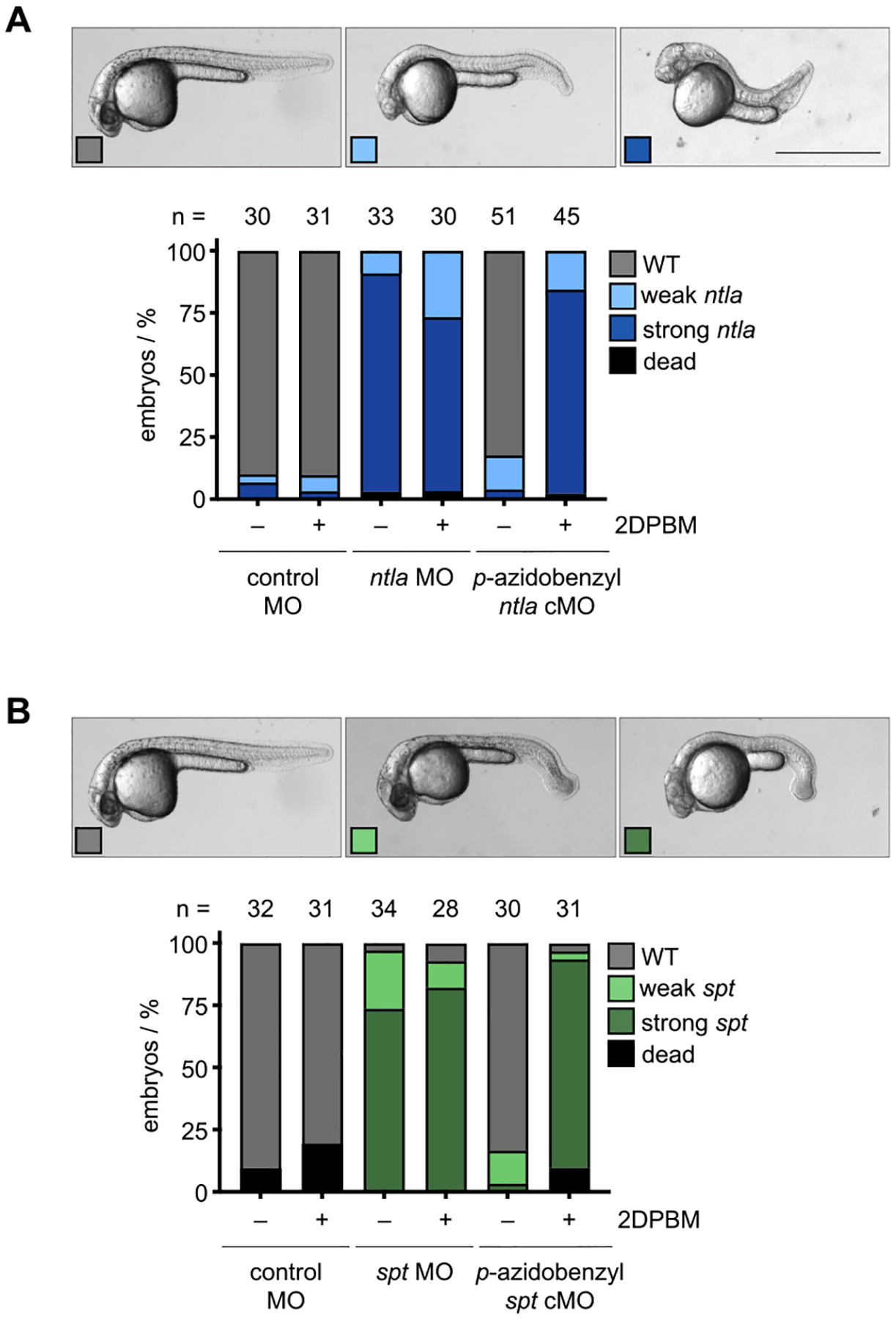
(A) Representative images of ntla morphant phenotypes in zebrafish embryos at 24 hpf and phenotypic scoring of embryos injected with the indicated morpholino reagent and soaked in E3 water supplemented with DMSO (0.2%) or 2DPBM (100 μM). (B) Representative images of spt morphant phenotypes in zebrafish embryos at 24 hpf and phenotypic scoring of zebrafish embryos injected with the indicated morpholino reagents and soaked in E3 water supplemented with DMSO (0.2%) or 2DPBM (100 μM). The scale bar equals 1 mm.
To demonstrate the robustness of our approach, we synthesized a second cyclic p-azidobenzyl cMO targeting tbx16. tbx16, also commonly known as spadetail (spt), is a T-box transcription factor in zebrafish that works in tandem with ntla to regulate the fates of paraxial mesoderm progenitor cells during trunk development.50,54–56 Embryos lacking spt function fail to form trunk somites, and the progenitor cells that would normally contribute to those mesodermal derivatives are mislocalized to the tail. The resulting tail morphology serves as the predominant indicator of the spt phenotype (Figure 6B).57,58
Zebrafish embryos were injected with the cyclic spt cMO and soaked in E3 water supplemented with DMSO (negative control) or 2DPBM (100 μM), following the same treatment regimen established for conditional ntla silencing. At 24 hpf, embryos were scored for a loss-of-function spt phenotype (Figure 6B). Zebrafish embryos injected with the cMO and soaked with vehicle (0.2% DMSO) demonstrate minimal phenotypic response (16%), consistent with that observed following injection of the cyclic ntla cMO. Following soaking with the phosphine trigger, MO-mediated spt silencing is efficiently rescued, as evidenced by the majority of embryos exhibiting a strong spt morphant phenotype (84%), comparable to that observed in embryos injected with the linear spt MO. Taken together, these results validate that bioorthogonal small molecules, such as azides and phosphines, can be effectively engineered into molecular switches to regulate gene expression in live animals.
CONCLUSION
In summary, we have successfully demonstrated the first example of a chemically controlled oligonucleotide-based tool to regulate gene function in vitro and in vivo. By exploiting the biorthogonality of the Staudinger reduction, we designed and synthesized a p-azidobenzyl linker that can undergo self-cleavage following exposure to a small molecule phosphine trigger. The use of a Staudinger reduction as an effective and rapid chemical decaging strategy was established through the successful, phosphine-triggered activation of a rhodamine-based fluorescent sensor. Furthermore, the caged fluorophore demonstrated the stability of aryl azide-based caging groups in an aquatic embryo. Based on these results, we then demonstrated the efficient rescue of MO-mediated silencing function following small molecule exposure, as evidenced by the silencing of reporter gene expression in a lysate translation system. Finally, the functionality and robustness of our modular approach for controlling endogenous gene expression with p-azidobenzyl cMOs were validated through small molecule-triggered silencing of two separate, developmentally relevant genes in zebrafish embryos.
The strategy disclosed herein complements the current suite of optically and enzymatically triggered antisense agents and introduces, for the first time, small molecule control of nucleic acid function. The phosphine trigger induces rapid cMO activation with precise temporal resolution and enables user-controlled gene silencing in deep and opaque tissues, overcoming some of the limitations of optochemical and optogenetic approaches. Additionally, the modularity of our azidobenzyl linker allows applications to other antisense reagents and small molecule control of oligonucleotide function in general. The approach presented herein should be amenable for regulating gene expression in cultured cells and non-aquatic animal models.59 We anticipate that small molecule-activated cMOs will nicely complement current cyclic cMO strategies and can be used in combination with other methods to independently control the expression of a single gene within synchronous multi-gene networks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Institutes of Health (GM112728) and the University of Pittsburgh. The authors wish to thank Chasity Hankinson for assistance in phosphine synthesis.
Footnotes
Supporting Information
Supporting Information is available free of charge at [link to DOI]. The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c08723.
Supporting figures as described in the text and an experimental section, including chemical synthesis procedures, full characterization of all new compounds, and biological protocols (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.1c08723
The authors declare no competing financial interest.
Contributor Information
Kristie Darrah, Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States.
Joshua Wesalo, Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States.
Bradley Lukasak, Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States.
Michael Tsang, Department of Developmental Biology, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States.
James K. Chen, Department of Chemical and Systems Biology, Stanford University School of Medicine, Stanford, California 94305, United States.
Alexander Deiters, Department of Chemistry, University of Pittsburgh, Pittsburgh, Pennsylvania 15260, United States.
REFERENCES
- (1).Chakraborty S; Mehtab S; Krishnan Y The predictive power of synthetic nucleic acid technologies in RNA biology. Acc. Chem. Res 2014, 47 (6), 1710–9. [DOI] [PubMed] [Google Scholar]
- (2).Khvorova A; Watts JK The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol 2017, 35 (3), 238–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Setten RL; Rossi JJ; Han SP The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discovery 2019, 18 (6), 421–446. [DOI] [PubMed] [Google Scholar]
- (4).Summerton JE Morpholino, siRNA, and S-DNA compared: impact of structure and mechanism of action on off-target effects and sequence specificity. Curr. Top. Med. Chem 2007, 7 (7), 651–60. [DOI] [PubMed] [Google Scholar]
- (5).Timme-Laragy AR; Karchner SI; Hahn ME Gene knockdown by morpholino-modified oligonucleotides in the zebrafish (Danio rerio) model: applications for developmental toxicology. Methods Mol. Biol 2012, 889, 51–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Corey DR; Abrams JM Morpholino antisense oligonucleotides: tools for investigating vertebrate development. Genome Biol. 2001, 2 (5), reviews1015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Shestopalov IA; Chen JK Oligonucleotide-based tools for studying zebrafish development. Zebrafish 2010, 7 (1), 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Shestopalov IA; Chen JK Chemical technologies for probing embryonic development. Chem. Soc. Rev 2008, 37 (7), 1294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Tomasini AJ; Schuler AD; Zebala JA; Mayer AN PhotoMorphs: a novel light-activated reagent for controlling gene expression in zebrafish. Genesis 2009, 47 (11), 736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Tallafuss A; Gibson D; Morcos P; Li Y; Seredick S; Eisen J; Washbourne P Turning gene function ON and OFF using sense and antisense photo-morpholinos in zebrafish. Development 2012, 139 (9), 1691–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Shestopalov IA; Sinha S; Chen JK Light-controlled gene silencing in zebrafish embryos. Nat. Chem. Biol 2007, 3 (10), 650–1. [DOI] [PubMed] [Google Scholar]
- (12).Ouyang X; Shestopalov IA; Sinha S; Zheng G; Pitt CL; Li WH; Olson AJ; Chen JK Versatile synthesis and rational design of caged morpholinos. J. Am. Chem. Soc 2009, 131 (37), 13255–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Bardhan A; Deiters A; Ettensohn CA Conditional gene knockdowns in sea urchins using caged morpholinos. Dev. Biol 2021, 475, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Deiters A; Garner RA; Lusic H; Govan JM; Dush M; Nascone-Yoder NM; Yoder JA Photocaged morpholino oligomers for the light-regulation of gene function in zebrafish and Xenopus embryos. J. Am. Chem. Soc 2010, 132 (44), 15644–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Yamazoe S; Shestopalov IA; Provost E; Leach SD; Chen JK Cyclic caged morpholinos: conformationally gated probes of embryonic gene function. Angew. Chem., Int. Ed 2012, 51 (28), 6908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yamazoe S; Liu Q; McQuade LE; Deiters A; Chen JK Sequential gene silencing using wavelength-selective caged morpholino oligonucleotides. Angew. Chem., Int. Ed 2014, 53 (38), 10114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Wang Y; Wu L; Wang P; Lv C; Yang Z; Tang X Manipulation of gene expression in zebrafish using caged circular morpholino oligomers. Nucleic Acids Res. 2012, 40 (21), 11155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Yamazoe S; McQuade LE; Chen JK Nitroreductase-activatable morpholino oligonucleotides for in vivo gene silencing. ACS Chem. Biol 2014, 9 (9), 1985–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Nguyen SS; Prescher JA Developing bioorthogonal probes to span a spectrum of reactivities. Nature Reviews Chemistry 2020, 4 (9), 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Agard NJ; Baskin JM; Prescher JA; Lo A; Bertozzi CR A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol 2006, 1 (10), 644–8. [DOI] [PubMed] [Google Scholar]
- (21).Sletten EM; Bertozzi CR From mechanism to mouse: a tale of two bioorthogonal reactions. Acc. Chem. Res 2011, 44 (9), 666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Prescher JA; Dube DH; Bertozzi CR Chemical remodelling of cell surfaces in living animals. Nature 2004, 430 (7002), 873–7. [DOI] [PubMed] [Google Scholar]
- (23).Luo J; Liu Q; Morihiro K; Deiters A Small-molecule control of protein function through Staudinger reduction. Nat. Chem 2016, 8 (11), 1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Shah L; Laughlin ST; Carrico IS Light-Activated Staudinger-Bertozzi Ligation within Living Animals. J. Am. Chem. Soc 2016, 138 (16), 5186–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Wesalo JS; Luo J; Morihiro K; Liu J; Deiters A Phosphine-Activated Lysine Analogues for Fast Chemical Control of Protein Subcellular Localization and Protein SUMOylation. ChemBioChem 2020, 21 (1–2), 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Azoulay M; Tuffin G; Sallem W; Florent JC A new drug-release method using the Staudinger ligation. Bioorg. Med. Chem. Lett 2006, 16 (12), 3147–9. [DOI] [PubMed] [Google Scholar]
- (27).van Brakel R; Vulders RC; Bokdam RJ; Grull H; Robillard MS A doxorubicin prodrug activated by the staudinger reaction. Bioconjugate Chem. 2008, 19 (3), 714–8. [DOI] [PubMed] [Google Scholar]
- (28).Alouane A; Labruere R; Le Saux T; Schmidt F; Jullien L Self-immolative spacers: kinetic aspects, structure-property relationships, and applications. Angew. Chem., Int. Ed 2015, 54 (26), 7492–509. [DOI] [PubMed] [Google Scholar]
- (29).Erez R; Shabat D The azaquinone-methide elimination: comparison study of 1,6- and 1,4-eliminations under physiological conditions. Org. Biomol. Chem 2008, 6 (15), 2669–2672. [DOI] [PubMed] [Google Scholar]
- (30).Mosey RA; Floreancig PE Versatile approach to α-alkoxy carbamate synthesis and stimulus-responsive alcohol release. Org. Biomol. Chem 2012, 10 (39), 7980–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lukasak B; Morihiro K; Deiters A Aryl Azides as Phosphine-Activated Switches for Small Molecule Function. Sci. Rep 2019, 9 (1), 1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Habeeb AG; Praveen Rao PN; Knaus EE Design and Synthesis of Celecoxib and Rofecoxib Analogues as Selective Cyclooxygenase-2 (COX-2) Inhibitors: Replacement of Sulfonamide and Methylsulfonyl Pharmacophores by an Azido Bioisostere. J. Med. Chem 2001, 44 (18), 3039–3042. [DOI] [PubMed] [Google Scholar]
- (33).Ouyang X; Shestopalov IA; Sinha S; Zheng G; Pitt CLW; Li W; Olson AJ; Chen JK Versatile Synthesis and Rational Design of Caged Morpholinos. J. Am. Chem. Soc 2009, 131, 13255–13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Mitchison TJ; Sawin KE; Theriot JA; Gee K; Mallavarapu A Caged fluorescent probes. Methods Enzymol. 1998, 291, 63–78. [DOI] [PubMed] [Google Scholar]
- (35).Karstens T; Kobs K Rhodamine B and rhodamine 101 as reference substances for fluorescence quantum yield measurements. J. Phys. Chem 1980, 84 (14), 1871–1872. [Google Scholar]
- (36).Wysocki LM; Grimm JB; Tkachuk AN; Brown TA; Betzig E; Lavis LD Facile and general synthesis of photoactivatable xanthene dyes. Angew. Chem., Int. Ed 2011, 50 (47), 11206–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Sato T; Hamada Y; Sumikawa M; Araki S; Yamamoto H Solubility of Oxygen in Organic Solvents and Calculation of the Hansen Solubility Parameters of Oxygen. Ind. Eng. Chem. Res 2014, 53 (49), 19331–19337. [Google Scholar]
- (38).Saneyoshi H; Ochikubo T; Mashimo T; Hatano K; Ito Y; Abe H Triphenylphosphinecarboxamide: An Effective Reagent for the Reduction of Azides and Its Application to Nucleic Acid Detection. Org. Lett 2014, 16 (1), 30–33. [DOI] [PubMed] [Google Scholar]
- (39).Gee KR; Weinberg ES; Kozlowski DJ Caged Q-rhodamine dextran: a new photoactivated fluorescent tracer. Bioorg. Med. Chem. Lett 2001, 11 (16), 2181–2183. [DOI] [PubMed] [Google Scholar]
- (40).Kozlowski DJ; Weinberg ES, Photoactivatable (Caged) Fluorescein as a Cell Tracer for Fate Mapping in the Zebrafish Embryo. In Developmental Biology Protocols; Walker JM, Tuan RS, Lo CW, Eds. Humana Press: 2000; Vol. I, pp 349–355. [DOI] [PubMed] [Google Scholar]
- (41).Kimmel CB; Law RD Cell lineage of zebrafish blastomeres: I. Cleavage pattern and cytoplasmic bridges between cells. Dev. Biol 1985, 108 (1), 78–85. [DOI] [PubMed] [Google Scholar]
- (42).Woo K; Fraser SE Order and coherence in the fate map of the zebrafish nervous system. Development 1995, 121 (8), 2595–2609. [DOI] [PubMed] [Google Scholar]
- (43).Clanton JA; Shestopalov I; Chen J; Gamse JT Lineage Labeling of Zebrafish Cells with Laser Uncagable Fluorescein Dextran. J. Visualized Exp 2011, 50, No. e2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wetts R; Fraser SE Microinjection of fluorescent tracers to study neural cell lineages. Development 1991, 113, 1–8. [PubMed] [Google Scholar]
- (45).Strehlow D; Gilbert W A fate map for the first cleavages of the zebrafish. Nature 1993, 361 (6411), 451–453. [Google Scholar]
- (46).Burns CG; Milan DJ; Grande EJ; Rottbauer W; MacRae CA; Fishman MC High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol 2005, 1 (5), 263–4. [DOI] [PubMed] [Google Scholar]
- (47).Sasmal PK; Carregal-Romero S; Han AA; Streu CN; Lin Z; Namikawa K; Elliott SL; Koster RW; Parak WJ; Meggers E Catalytic azide reduction in biological environments. ChemBioChem 2012, 13 (8), 1116–20. [DOI] [PubMed] [Google Scholar]
- (48).Schulte-Merker S; van Eeden FJ; Halpern ME; Kimmel CB; Nusslein-Volhard C no tail (ntl) is the zebrafish homologue of the mouse T (Brachyury) gene. Development 1994, 120 (4), 1009–15. [DOI] [PubMed] [Google Scholar]
- (49).Halpern ME; Ho RK; Walker C; Kimmel CB Induction of muscle pioneers and floor plate is distinguished by the zebrafish no tail mutation. Cell 1993, 75 (1), 99–111. [PubMed] [Google Scholar]
- (50).Showell C; Binder O; Conlon FL T-box genes in early embryogenesis. Dev. Dyn 2004, 229 (1), 201–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Nasevicius A; Ekker SC Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet 2000, 26 (2), 216–20. [DOI] [PubMed] [Google Scholar]
- (52).Taylor MF; Paulauskis JD; Weller DD; Kobzik L In vitro efficacy of morpholino-modified antisense oligomers directed against tumor necrosis factor-alpha mRNA. J. Biol. Chem 1996, 271 (29), 17445–52. [DOI] [PubMed] [Google Scholar]
- (53).Kamachi Y; Okuda Y; Kondoh H Quantitative assessment of the knockdown efficiency of morpholino antisense oligonucleotides in zebrafish embryos using a luciferase assay. Genesis 2008, 46 (1), 1–7. [DOI] [PubMed] [Google Scholar]
- (54).Ruvinsky I; Silver LM; Ho RK Characterization of the zebrafish tbx16 gene and evolution of the vertebrate T-box family. Dev. Genes Evol 1998, 208 (2), 94–9. [DOI] [PubMed] [Google Scholar]
- (55).Warga RM; Mueller RL; Ho RK; Kane DA Zebrafish Tbx16 regulates intermediate mesoderm cell fate by attenuating Fgf activity. Dev. Biol 2013, 383 (1), 75–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Amacher SL; Draper BW; Summers BR; Kimmel CB The zebrafish T-box genes no tail and spadetail are required for development of trunk and tail mesoderm and medial floor plate. Development 2002, 129 (14), 3311–23. [DOI] [PubMed] [Google Scholar]
- (57).Griffin KJ; Amacher SL; Kimmel CB; Kimelman D Molecular identification of spadetail: regulation of zebrafish trunk and tail mesoderm formation by T-box genes. Development 1998, 125 (17), 3379–88. [DOI] [PubMed] [Google Scholar]
- (58).Ho RK; Kane DA Cell-autonomous action of zebrafish spt-1 mutation in specific mesodermal precursors. Nature 1990, 348 (6303), 728–30. [DOI] [PubMed] [Google Scholar]
- (59).Morcos PA; Li Y; Jiang S Vivo-Morpholinos: a non-peptide transporter delivers Morpholinos into a wide array of mouse tissues. BioTechniques 2008, 45 (6), 613–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


