Abstract
Ability of daily sequential subcultures in subinhibitory concentrations of clinafloxacin, ciprofloxacin, and trovafloxacin to select resistant mutants was studied in 10 pneumococci (ciprofloxacin MICs, 1 to 4 μg/ml, and clinafloxacin and trovafloxacin MICs, 0.06 to 0.125 μg/ml [n = 9]; ciprofloxacin, clinafloxacin, and trovafloxacin MICs, 32, 0.5, and 2 μg/ml, respectively [n = 1]). Subculturing was done 50 times, or until MICs increased fourfold or more. Mutants for which MICs were fourfold (or more) higher than those for parent strains were selected in five strains by clinafloxacin, in six strains by trovafloxacin, and nine strains by ciprofloxacin. Sequence analysis of type II topoisomerase showed that most mutants had mutations in ParC at Ser79 or Asp83 and in GyrA at Ser81, while a few mutants had mutations in ParE or GyrB. In the presence of reserpine, the MICs of ciprofloxacin and clinafloxacin for most mutants were lower (four to eight times lower), but for none of the mutants were trovafloxacin MICs lower, suggesting an efflux mechanism affecting the first two agents but not trovafloxacin. Single-step mutation rates were also determined for eight strains for which the MICs were as follows: 0.06 μg/ml (clinafloxacin), 0.06 to 0.125 μg/ml (trovafloxacin), and 1 μg/ml (ciprofloxacin). Single-step mutation rates with drugs at the MIC were 2.0×10−9 to <1.1×10−11, 5.0×10−4 to 3.6×10−9, and 4.8×10−4 to 6.7×10−9, respectively. For two strains with clinafloxacin MICs of 0.125 to 0.5 μg/ml trovafloxacin MICs of 0.125 to 2 μg/ml, ciprofloxacin MICs of 4 to 32 μg/ml mutation rates with drugs at the MIC were 1.1×10−8−9.6×10−8, 3.3×10−6−6.7×10−8, and 2.3×10−5−2.4×10−7, respectively. Clinafloxacin was bactericidal at four times the MIC after 24 h against three parent and nine mutant strains by time-kill study. This study showed that single and multistep clinafloxacin exposure selected for resistant mutants less frequently than similar exposures to other drugs studied.
Clinafloxacin is a novel fluoroquinolone with broad-spectrum in vitro activity against gram-positive, gram-negative, and anaerobic pathogens (8). This drug is more active in vitro against Streptococcus pneumoniae than ciprofloxacin, trovafloxacin, and most other fluoroquinolones (8). Newer compounds of this class have improved activity and pharmacokinetics relative to older quinolones (3).
Fluoroquinolones present potential for empirical treatment of adult respiratory infections, in part due to the increase in the incidence of penicillin-resistant pneumococci and also because of their activity against Haemophilus influenzae, Moraxella catarrhalis, Chlamydia pneumoniae, Mycoplasma pneumoniae, and Legionella pneumophila (1, 25). However, the overuse of this class of antimicrobial agent could lead to the emergence of resistant mutants. There were no pneumococci resistant to ciprofloxacin in a 1997 U.S. surveillance study (13), and worldwide incidence of these strains is less than 1% (13). However resistance to levofloxacin (5.5%) and trovafloxacin (2.2%) was recently reported from Hong Kong (12). In Canada, the prevalence of pneumococci with reduced susceptibilities to fluoroquinolones in adults increased from 0% in 1993 to 1.7% in 1997 and 1998 (5). The prevalence of ciprofloxacin-resistant pneumococci also increased from 0.9% in 1991–1992 to 3.0% in 1997–1998 in Spain (15). It is a concern that these resistant strains may spread to other parts of the world.
The primary targets of fluoroquinolones are topoisomerase IV and DNA gyrase. Topoisomerase IV is composed of ParC and ParE subunits, which are encoded by parC and parE genes, respectively. DNA gyrase is composed of GyrA and GyrB, which are encoded by gyrA and gyrB genes, respectively. Point mutations in the quinolone resistance-determining regions (QRDRs) of topoisomerase IV and DNA gyrase genes are associated with quinolone resistance. Topoisomerase IV is the primary target for ciprofloxacin and trovafloxacin (9, 11, 19, 25), while DNA gyrase and topoisomerase IV are dual targets of clinafloxacin in S. pneumoniae (20). Mutations in the QRDRs of parE and gyrB are also believed to play a role in fluoroquinolone resistance (14, 19, 25).
In this study, we determined and compared the effects of clinafloxacin, trovafloxacin, and ciprofloxacin to select resistant pneumococcal mutants by (i) multistep resistance selection by exposure to subinhibitory concentrations of these agents and (ii) single-step resistance selection. Clinafloxacin and trovafloxacin were chosen as examples of quinolones with increased activity against S. pneumoniae. The mutations in ParC, GyrA, ParE, and GyrB of the mutants were determined, and mutant strains were also tested for the presence of an efflux mechanism by determining MICs in the presence and absence of reserpine. Furthermore, time-kill experiments of some of parent and mutant strains were performed to compare the in vitro activities of three quinolones.
MATERIALS AND METHODS
Bacteria and antimicrobial agents.
Nine strains of S. pneumoniae clinically isolated within the past 5 years were randomly selected from our collection. One strain for which the MIC of ciprofloxacin was 32 μg/ml was also used; this strain was obtained from Spain. Organisms were identified by optochin susceptibility and classified by serotyping. Antimicrobials were obtained as follows: clinafloxacin from Parke-Davis Pharmaceuticals, Ann Arbor, Mich.; trovafloxacin from Pfizer, Inc., New York, N.Y.; and ciprofloxacin from Bayer, Inc., West Haven, Conn.
Susceptibility testing.
MICs were determined by reference microdilution methodology in Muller-Hinton broth (Difco Laboratories, Detroit, Mich.) supplemented with 5% lysed horse blood (17). For the purpose of this study, strains were considered susceptible to ciprofloxacin and trovafloxacin when MICs of these drugs were ≤2 μg/ml and susceptible to clinafloxacin when the MIC was ≤1 μg/ml, based on baseline MICs of ciprofloxacin and pharmacokinetics of the other agents (3; M. A. Cohen, personal communication).
Multistep resistance selection.
Multistep resistance selection for each of the quinolones was performed as described previously (7, 23). Glass tubes containing 1 ml of caution-adjusted Muller-Hinton broth (Difco) supplemented with 5% lysed horse blood with doubling antibiotic dilutions were inoculated with approximately 5 × 105 CFU/ml at antibiotic concentrations from 3 doubling dilutions above to 3 doubling dilutions below the MIC of each agent for each strain. The initial inoculum was prepared by suspending growth from overnight Trypticase soy blood agar plate (Difco) in Muller-Hinton broth. Tubes were incubated at 35°C for 24 h. Daily passages were performed for 50 days by taking 10 μl of inoculum from the tube with the drug concentration nearest the MIC (usually the tube 1 doubling dilution below the MIC). Daily subculturing was done 50 times, or until MICs for mutants increased fourfold or more. Resistant mutants then were subcultured 10 times on drug-free medium, and the MICs were determined.
Single-step resistance selection.
The frequency of spontaneous single-step mutation was determined by spreading cultures (approximately 1010 CFU/ml) in a 100-μl volume of phosphate-buffered saline on 10% lysed horse blood. Brain heart infusion agar (Difco) plates (100-mm diameter) contained each compound at 1, 2, 4, 8, and 16 times the MIC (20). Plates were incubated aerobically at 35°C for 48 to 72 h. The resistance frequency was calculated as the number of resistant colonies per inoculum (20). MICs for parent and resistant mutant strains were determined by an agar dilution method (21, 22). Gene sequencing and efflux mechanism were determined for some single-step mutant strains (see below).
Serotyping.
Serotyping of parent and mutant strains was performed by the standard Quellung method with sera from the Statens Seruminstitut (Copenhagen, Denmark).
PFGE.
To determine whether resistant isolates obtained at the end of serial passages were derived from those used at the beginning of the study, the parent strains and the mutants for which the MICs were increased that were obtained after the last passage were tested by pulsed-field gel electrophoresis (PFGE) with a CHEF DR III apparatus (Bio-Rad, Hercules, Calif.) as previously described (7, 16).
PCR of quinolone resistance determinants and DNA sequence analysis.
To determine whether mutants that developed resistance to quinolones had alterations in topoisomerase IV or DNA gyrase compared to the parent strains, parC, parE, gyrA, and gyrB were amplified by the PCR method and sequenced as described previously (7). Mutants with mutations widely described in the literature (e.g., Ser79→Tyr or Phe in ParC and Ser83→Tyr or Phe in GyrA) were sequenced once in the forward direction. Mutants with no mutations in a particular gene or with a previously undescribed mutation were sequenced twice forward and once in the reverse direction on products of independent PCR experiments (7).
Determination of efflux mechanism.
MICs for parent and mutant strains were determined in the presence and absence of reserpine (10 μg/ml; Sigma) as described previously (4, 7). Thirty quinolone-resistant mutants for which the MICs were at least fourfold greater than those for their parent strains were tested (4). An efflux mechanism was believed to be present when the MIC of an agent in the presence of reserpine was at least fourfold less (2 doubling dilutions) than the MIC in the absence of reserpine (tests were repeated three times) (6, 7).
Time-kill methodology.
Time-kill studies were carried out for three parent strains (strains 1, 6, and 7) and three multistep mutant strains selected by each quinolone using Mueller-Hinton broth with 5% lysed horse blood as described previously (21, 22).
RESULTS
Multistep resistance selection by subculturing in subinhibitory concentrations.
For initial quinolone MICs for parent strains, there were nine strains for which the ciprofloxacin MICs were 1 to 4 μg/ml and the trovafloxacin and clinafloxacin MICs were 0.06 to 0.125 μg/ml. There was one strain for which the ciprofloxacin MIC was 32 μg/ml, the trovafloxacin MIC was 2 μg/ml, and the clinafloxacin MIC was 0.5 μg/ml. Quinolone MICs for resistant mutants resulting from serial daily subculturing in subinhibitory concentrations of antibiotics are summarized in Table 1. Subculturing in clinafloxacin selected five resistant mutants with stable resistance. Subculturing in the presence of trovafloxacin selected six resistant mutants, for which the MICs rose from 0.06 to 2 μg/ml to 4 to 16 μg/ml after 30 to 50 subcultures. Subculturing in ciprofloxacin selected eight resistant mutants, for which the MICs rose from 1 to 4 μg/ml to 4 to 128 μg/ml after 14 to 39 subcultures. Quinolone MICs for the strain for which the original ciprofloxacin MIC was 32 μg/ml rose to 128 μg/ml.
TABLE 1.
Results of multi-step resistance selection by clinafloxacin, trovafloxacin, and ciprofloxacin
| Strain | Initial MIC (μg/ml) of:
|
Selected resistance
|
MIC (μg/ml) of resistant mutant toa:
|
Mutation(s) in QRDR of resistant mutantsb
|
Difference of MIC with reserpine withg:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TROd | CIPe | CLXf | Selecting agent | No. of passagesc | MIC (μg/ml) | TRO | CIP | CLX | ParC | GyrA | ParE | GyrB | TRO | CIP | CLX | |
| 1 | 0.13 | 1 | 0.06 | TRO | 32 | 8 | 4 | 32 | 0.5 | S79→Y | S81→Y | — | — | — | — | — |
| CIP | 14 | 32 | 0.25 | 8 | 0.25 | — | — | — | — | — | 8× | — | ||||
| CLX | 32 | 16 | 1 | 64 | 1 | D83→G | S81→Y | — | — | — | 8× | — | ||||
| 2 | 0.13 | 1 | 0.06 | TRO | 50 | 1 | 0.25 | 4 | 0.13 | H102→Y | — | — | — | — | — | — |
| CIP | 14 | 64 | 0.25 | 8 | 0.25 | — | — | — | — | — | — | — | ||||
| CLX | 50 | 2 | 0.25 | 32 | 1 | — | — | — | — | — | — | — | ||||
| 3 | 0.06 | 1 | 0.06 | TRO | 32 | 8 | 8 | 32 | 0.5 | S79→Y | S81→F | — | — | — | 4× | — |
| CIP | 14 | 64 | 4 | 32 | 2 | S79→F | S81→F | — | R445→S | — | — | — | ||||
| CLX | 32 | 16 | 2 | 128 | 2 | D83→N | S81→F | — | — | — | 8× | 4× | ||||
| 4 | 0.06 | 1 | 0.06 | TRO | 50 | 2 | 1 | 4 | 0.13 | D83→G, A115→P | — | — | — | — | — | — |
| CIP | 36 | 32 | 0.25 | 2 | 0.25 | D83→H | — | — | — | — | — | — | ||||
| CLX | 50 | 4 | 0.5 | 16 | 1 | — | S81→Y | — | — | — | — | — | ||||
| 5 | 0.06 | 1 | 0.06 | TRO | 50 | 2 | 2 | 8 | 0.5 | S79→F | — | — | — | — | — | — |
| CIP | 17 | 32 | 0.13 | 4 | 0.13 | — | — | — | — | — | 8× | 4× | ||||
| CLX | 50 | 8 | 4 | 128 | 4 | S79→F | S81→Y | — | — | — | 8× | 4× | ||||
| 6 | 0.06 | 1 | 0.06 | TRO | 50 | 2 | 1 | 4 | 0.5 | — | E85→K | — | — | — | — | — |
| CIP | 14 | 32 | 0.25 | 16 | 0.5 | — | — | — | — | — | 8× | 8× | ||||
| CLX | 50 | 4 | 0.5 | 32 | 2 | — | S81→F | P454→S | — | — | 8× | 8× | ||||
| 7 | 0.06 | 1 | 0.06 | TRO | 32 | 16 | 8 | 64 | 2 | D83→N | S81→L | E474→K | — | — | — | — |
| CIP | 39 | 32 | 0.25 | 16 | 0.25 | — | — | — | — | — | 4× | — | ||||
| CLX | 50 | 2 | 0.25 | 16 | 1 | — | — | — | — | — | — | — | ||||
| 8 | 0.13 | 1 | 0.06 | TRO | 47 | 8 | 8 | 128 | 2 | D83→Y, S107→Y | S81→F | — | — | — | 4× | 4× |
| CIP | 14 | 64 | 0.5 | 32 | 0.5 | D83→N | — | — | — | — | 4× | — | ||||
| CLX | 50 | 2 | 0.5 | 32 | 2 | — | — | — | — | — | 8× | 4× | ||||
| 9 | 0.13 | 4 | 0.13 | TRO | 30 | 16 | 16 | 64 | 1 | S79→Y | S81→F | R447→C | — | — | 4× | — |
| CIP | 15 | 32 | 0.25 | 32 | 0.13 | — | — | R447→C | — | — | 4× | 4× | ||||
| CLX | 50 | 4 | 0.5 | 64 | 2 | — | — | — | — | — | 8× | 4× | ||||
| 10 | 2 | 32 | 0.5 | TRO | 50 | 8 | 8 | 64 | 2 | — | S81→Y | — | G406→S | — | 4× | — |
| CIP | 36 | 256 | 2 | 128 | 2 | — | — | — | — | — | 8× | 4× | ||||
| CLX | 50 | 1 | 4 | 32 | 1 | — | S81→F | — | G406→S | — | — | — | ||||
MICs determined after 10 days of antibiotic-free subculture.
Parent strains 4 and 9 had a mutation in ParC (K137→N); parent strains 1, 3, 4, 6, 7, 9, and 10 had a mutation in ParE (1460→V); parent strain 10 also had mutations in ParC (K50→E, S79→F) and GyrA (S81→C). —, no mutation between parent and resistant mutant strains.
Number of passages required to select resistant mutants.
TRO, trovafloxacin.
CIP, ciprofloxacin.
CLX, clinafloxacin.
—, no difference between parent and resistant mutant strains for reserpine effects. For no strains were trovafloxacin or clinafloxacin MICs for parent strains lower in the presence of reserpine. No ciprofloxacin MICs for parent strains 1 to 7 and 10 were lower. For parent strains 8 and 9 ciprofloxacin MICs were four times lower with reserpine. 8×, eight times the MIC; 4×, four times the MIC.
Resistance was usually stable (MICs remained within 1 doubling dilution after 10 serial daily passages on antibiotic-free medium) with the following exceptions: (i) strain 1, 2, 4, and 5 mutants selected by ciprofloxacin (ciprofloxacin MICs decreased from 32 to 64 μg/ml to 2 to 8 μg/ml); (ii) strain 2 mutant exposed to trovafloxacin (trovafloxacin MIC decreased from 1 to 0.25 μg/ml); (iii) strain 1, 3, and 4 mutants selected by clinafloxacin (clinafloxacin MICs decreased from 4 to 16 μg/ml to 1 to 2 μg/ml).
Cross-resistance among mutant strains.
Among 30 multistep mutant strains, 12 were resistant to trovafloxacin (five had high-level trovafloxacin resistance [MIC ≥ 8 μg/ml]), 29 were resistant to ciprofloxacin (18 had high-level ciprofloxacin resistance [MIC ≥ 43 μg/ml]), and 10 were resistant to clinafloxacin (MIC ≥ 2 μg/ml) (Table 2).
TABLE 2.
Distribution of quinolone MICs in parent and mutant strains
| Quinolone | Strain | No. of parent (n = 10) and mutant strains (n = 30) for which the MIC (μg/ml) was:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.06 | 0.125 | 0.25 | 0.5 | 1.0 | 2.0 | 4.0 | 8.0 | 16.0 | 32.0 | 64.0 | 128.0 | ||
| Trovafloxacin | Parent | 5 | 4 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mutant | 0 | 1 | 9 | 5 | 3 | 3 | 4 | 4 | 1 | 0 | 0 | 0 | |
| Ciprofloxacin | Parent | 0 | 0 | 0 | 0 | 8 | 0 | 1 | 0 | 0 | 1 | 0 | 0 |
| Mutant | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 4 | 9 | 5 | 4 | |
| Clinafloxacin | Parent | 8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mutant | 0 | 4 | 4 | 6 | 6 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | |
All five mutants with high-level resistance selected by trovafloxacin had high-level resistance to ciprofloxacin (ciprofloxacin MICs were 32 to 128 μg/ml [strains 3, 7, 8, 9, and 10]). While clinafloxacin MICs for these five mutants (0.5 to 2 μg/ml) were higher than clinafloxacin MICs for parent strains (MICs ≤ 0.12 μg/ml), none had high-level resistance to clinafloxacin. Three of these five trovafloxacin-selected high-level-resistant mutants were cross-resistant to clinafloxacin (strains 7, 8, and 10). Eight of the high-level ciprofloxacin-resistant mutants that were selected by ciprofloxacin did not have high-level resistance to either trovafloxacin or clinafloxacin. Two of four high-level ciprofloxacin-resistant mutants (strains 3 and 10) selected by ciprofloxacin were resistant to both trovafloxacin (MIC, 4 μg/ml) and clinafloxacin (MIC, 2 μg/ml). Among five clinafloxacin-resistant mutants selected by clinafloxacin, all had high-level resistance to ciprofloxacin (MICs, 32 to 128 μg/ml), and two of them were resistant to trovafloxacin (but did not have high-level resistance).
Serotyping and PFGE.
The 10 pneumococcal strains used comprised serotypes 4, 6B, 7, 9, 14, and 23F. All mutants had serotypes identical to those of the parent strains, and all had PFGE patterns identical to those the parent strains.
Mutations in topoisomerase IV and DNA gyrase.
Mutations in the QRDR that led to amino acid changes are listed in Table 1. Parent strains 4 and 9 had mutations in both ParC and ParE, and parent strains 1, 3, 6, and 7 had a variation in ParE (Ile460→Val) compared to wild-type sequences; the roles of these variations in affecting quinolone activity are unclear (6). The ciprofloxacin-resistant parent strain 10 had mutations in both ParC (Lys50→Glu and Ser79→Phe) and GyrA (Ser81→Cys), explaining the resistance of this parent strain. In all other strains the amino acid sequences for ParC, ParE, GyrA, and GyrB from the parent strains were identical to the wild-type sequences.
Sequence analysis showed that most mutants had mutations in ParC at Ser79 or Asp83 and GyrA at Ser81. Strain 4 selected by clinafloxacin (clinafloxacin MIC, 0.06 to 1 μg/ml) had the single mutation Ser81→Tyr in GyrA. Strain 6 selected by trovafloxacin (trovafloxacin MIC, 0.06 to 1 μg/ml) had a mutation of Glu85→Lys in GyrA. Few had mutations in ParE and GyrB except as follows: (i) the strain 6 mutant selected by clinafloxacin had a mutation in ParE (Pro454→Ser), (ii) the strain 7 mutant selected by trovafloxacin had a mutation in ParE (Glu474→Lys), (iii) the strain 9 mutants selected by trovafloxacin or ciprofloxacin had the same mutation in ParE (Arg447→Cys), (iv) the strain 3 mutant selected by ciprofloxacin had a mutation in GyrB (Arg445→Ser), and (v) the strain 10 mutants selected by trovafloxacin and clinafloxacin had the same mutation in GyrB (Gly406→Ser).
Efflux mechanism.
We investigated the possibility that the efflux mechanism contributed to the raised quinolone MICs for parent and mutant strains by determining MICs in the presence and absence of reserpine (Table 1). Quinolone MICs for 8 of the 10 parent strains were unaffected by the presence of reserpine; however, the ciprofloxacin MICs for strains 8 and 9 were four times lower in the presence of reserpine. For 17 of 30 mutants the ciprofloxacin MICs were four to eight times lower in presence of reserpine. Similarly, the clinafloxacin MICs for 12 mutants were lower (four to eight times lower). However, there were no mutants for which the trovafloxacin MICs were lower in the presence of reserpine. Reserpine lowered the ciprofloxacin MICs for 7 of 10 mutants for which the ciprofloxacin MICs were raised though they had no mutations in ParC, GyrA, ParE, and GyrB (strain 1, 5, 6, 7, and 10 mutants selected by ciprofloxacin and strain 8 and 9 mutants selected by clinafloxacin). Reserpine had no effect on the clinafloxacin MICs for two of four mutant strains that had no mutations in ParC, GyrA, ParE, and GyrB. Clinafloxacin MICs for a strain 1 mutant (selected after 32 passages with clinafloxacin) and a strain 3 mutant (selected after 32 passages) decreased from 16 to 1 μg/ml in the presence of reserpine. These strains had no difference in the QRDRs of parC, gyrA, parE, and gyrB compared to the parent strain by gene sequencing.
Frequency of single-step spontaneous mutation rate.
Results for each strain are listed in Table 3. For eight strains for which the clinafloxacin MICs were 0.06 μg/ml, mutation rates with clinafloxacin at the MIC were lower (2.0×10−9 to <1.1×10−11) than with trovafloxacin (5.0×10−4 to 3.6×10−9) or ciprofloxacin (4.8×10−4 to 6.7×10−9) at the MIC. For another two strains for which the clinafloxacin MICs were 0.125 to 0.5 μg/ml, mutation rates with clinafloxacin at the MIC were lower than those with the other quinolones. For all strains, single-step spontaneous mutants could not be detected with each drug at 4, 8, and 16 times the MIC. Gene sequencing in the QRDRs of ParC and GyrA and MIC testing with reserpine were carried out for single-step mutants of strain 8. Mutant strains selected at one and two times the MICs of trovafloxacin had mutations in ParC (one times the MIC of trovafloxacin, Ser79→Tyr; two times the MIC of trovafloxacin, Ser79→Phe), but not in GyrA; mutant strains selected at one and two times the MICs of ciprofloxacin and at the MIC of clinafloxacin had no mutations in either ParC or GyrA. There were no mutations in ParE and GyrB. For none of these five single-step resistant mutants were the trovafloxacin MICs lower in the presence of reserpine. However, the ciprofloxacin MICs for all of the mutants were lower (four to eightfold), and for four of them clinafloxacin MICs were lower (fourfold) in the presence of reserpine.
TABLE 3.
Frequency of single-step mutation for 10 strains of S. pneumoniae
| Strain | Selecting drug | Initial MIC (μg/ml) | Mutation frequency at drug concn showna
|
||
|---|---|---|---|---|---|
| MIC | 2×MIC | 4×MIC | |||
| 1 | Trovafloxacin | 0.125 | 1.6 × 10−5 | <2.0 × 10−10 | <2.0 × 10−10 |
| Ciprofloxacin | 1 | 1.3 × 10−4 | <2.0 × 10−10 | <2.0 × 10−10 | |
| Clinafloxacin | 0.06 | <2.0 × 10−10 | <2.0 × 10−10 | <2.0 × 10−10 | |
| 2 | Trovafloxacin | 0.125 | 5.0 × 10−4 | <1.0 × 10−11 | <1.0 × 10−11 |
| Ciprofloxacin | 1 | 4.8 × 10−4 | 2.9 × 10−9 | <1.0 × 10−11 | |
| Clinafloxacin | 0.06 | <1.0 × 10−11 | <1.0 × 10−11 | <1.0 × 10−11 | |
| 3 | Trovafloxacin | 0.06 | 1.0 × 10−5 | 1.2 × 10−6 | <5.2 × 10−10 |
| Ciprofloxacin | 1 | 1.7 × 10−4 | 5.4 × 10−6 | <5.2 × 10−10 | |
| Clinafloxacin | 0.06 | <5.2 × 10−10 | <5.2 × 10−10 | <5.2 × 10−10 | |
| 4 | Trovafloxacin | 0.06 | 1.9 × 10−5 | 1.2 × 10−7 | <2.3 × 10−10 |
| Ciprofloxacin | 1 | 4.5 × 10−5 | 3.3 × 10−7 | <2.3 × 10−10 | |
| Clinafloxacin | 0.06 | <2.3 × 10−10 | <2.3 × 10−10 | <2.3 × 10−10 | |
| 5 | Trovafloxacin | 0.06 | 7.0 × 10−6 | 1.0 × 10−7 | <2.5 × 10−10 |
| Ciprofloxacin | 1 | 1.5 × 10−5 | 5.0 × 10−9 | <2.5 × 10−10 | |
| Clinafloxacin | 0.06 | 2.0 × 10−9 | <2.5 × 10−10 | <2.5 × 10−10 | |
| 6 | Trovafloxacin | 0.06 | 1.3 × 10−7 | <8.8 × 10−10 | <8.8 × 10−10 |
| Ciprofloxacin | 1 | 3.9 × 10−6 | <8.8 × 10−10 | <8.8 × 10−10 | |
| Clinafloxacin | 0.06 | <8.8 × 10−10 | <8.8 × 10−10 | <8.8 × 10−10 | |
| 7 | Trovafloxacin | 0.06 | 3.6 × 10−9 | 1.6 × 10−10 | <1.1 × 10−11 |
| Ciprofloxacin | 1 | 6.7 × 10−9 | <1.1 × 10−11 | <1.1 × 10−11 | |
| Clinafloxacin | 0.06 | <1.1 × 10−11 | <1.1 × 10−11 | <1.1 × 10−11 | |
| 8 | Trovafloxacin | 0.125 | 3.5 × 10−6 | 1.5 × 10−8 | <6.7 × 10−10 |
| Ciprofloxacin | 1 | 1.4 × 10−4 | 1.7 × 10−7 | <6.7 × 10−10 | |
| Clinafloxacin | 0.06 | <6.7 × 10−10 | <6.7 × 10−10 | <6.7 × 10−10 | |
| 9 | Trovafloxacin | 0.125 | 3.3 × 10−6 | 8.3 × 10−8 | <3.3 × 10−10 |
| Ciprofloxacin | 4 | 2.3 × 10−5 | 2.0 × 10−7 | <3.3 × 10−10 | |
| Clinafloxacin | 0.125 | 9.6 × 10−8 | <3.3 × 10−10 | <3.3 × 10−10 | |
| 10 | Trovafloxacin | 2 | 6.7 × 10−8 | 1.2 × 10−8 | <5.6 × 10−10 |
| Ciprofloxacin | 32 | 2.4 × 10−7 | 5.6 × 10−8 | <5.6 × 10−10 | |
| Clinafloxacin | 0.5 | 1.1 × 10−8 | <5.6 × 10−10 | <5.6 × 10−10 | |
Concentration shown as multiples of the MIC of agent against parent strain.
Time-kill studies for parent strains and multistep resistant mutants.
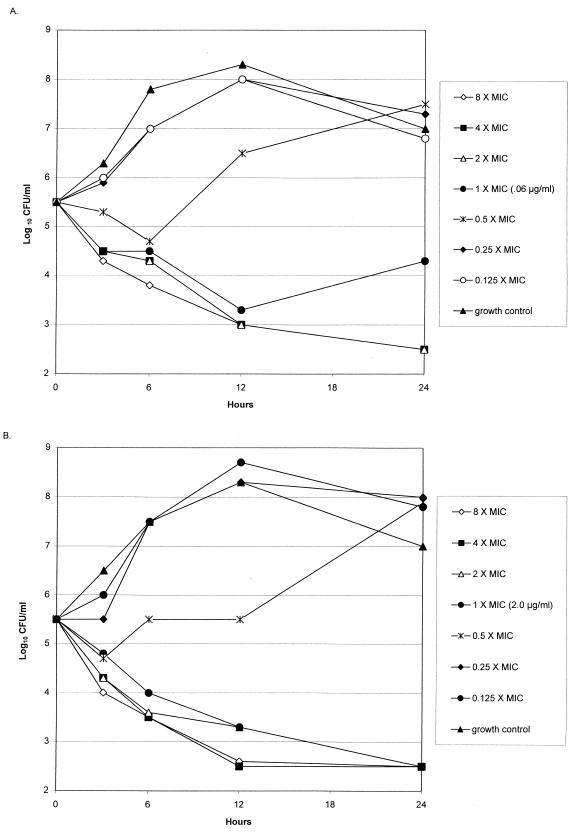
Clinafloxacin showed 99.9% killing of all three parent strains and three of the resistant mutant strains at two times the MIC after 24 h and 99% killing of all of strains after 12 h. Trovafloxacin showed 99.9% killing of all parent strains and two of three resistant mutant strains at four times the MIC after 24 h. Ciprofloxacin showed 99.9% killing of all three parent strains and three resistant mutant strains at two times the MIC after 24 h. An example of kill kinetics is presented in Fig. 1.
FIG. 1.
Time-kill study for clinafloxacin against parent 6 (clinafloxacin MIC, 0.06 μg/ml) (A) and strain 6 resistant mutant selected by clinafloxacin (clinafloxacin MIC, 2 μg/ml) (B).
DISCUSSION
This study is the first of which we are aware which simultaneously examines single and multistep selection for quinolone resistance in pneumococci. We were readily able to select quinolone-resistant S. pneumoniae mutants after serial passages in subinhibitory concentrations of trovafloxacin, ciprofloxacin, or clinafloxacin. Five mutant strains selected by ciprofloxacin became resistant (MIC ≥ 4 μg/ml) to ciprofloxacin more rapidly (14 subcultures) than mutants selected by trovafloxacin or clinafloxacin. The minimum number of subcultures required to select resistant mutants was 30 with trovafloxacin and 32 with clinafloxacin. The frequencies of spontaneous single-step mutation with clinafloxacin were lowest compared to the other two quinolones (20).
Gene sequencing of the QRDRs of parC and gyrA in this study has shown, as in previous studies, that the mutations associated with quinolone resistance were Ser79→Phe or Tyr and Asp83→Gly or His or Asn in ParC and Ser81→Phe or Tyr and Glu85→Lys in GyrA (9, 11, 18, 19, 20, 26, 27). Mutations not previously described for S. pneumoniae that were also associated with quinolone resistance were His102→Try, Ala115→Pro, or Ser107→Tyr in ParC and Ser81→Leu in GyrA. All of the five high-level trovafloxacin-resistant mutants (MICs ≥ 8 μg/ml) selected by trovafloxacin had mutations in both ParC and GyrA as described previously (7, 11, 27), except for one mutant (strain 10) selected by trovafloxacin (trovafloxacin MIC, 8 μg/ml) that had no additional mutation compared to its parent strain in ParC. Three high-level ciprofloxacin-resistant mutants selected by ciprofloxacin also had mutations in both ParC and GyrA, but one of them had mutations only in ParC; another had mutations in both ParC and ParE. A strain 9 mutant selected by clinafloxacin (clinafloxacin MIC, 2 μg/ml), for which the parent strain had a preexisting mutation in ParC, did not have any further mutations in GyrA.
Gene sequencing of the QRDRs of parE in this study has shown, as in previous studies by Pan and Fisher (20), that mutations at Pro454 are associated with quinolone resistance. A strain 6 mutant selected by clinafloxacin (clinafloxacin MIC, 2 μg/ml) had mutations in GyrA (Ser81→Tyr) and ParE (Pro454→Ser) but not ParC. This result may suggest parE mutation is also important for clinafloxacin resistance in strains with resistance with a gyrA mutation. Although ParC seems to be the target of trovafloxacin and ParC and GyrA seem to be the targets of clinafloxacin, exceptions may occur, e.g., strain 4 and 6 mutant strains, which are quinolone resistant with a single mutation in GyrA.
Mutations at Arg447 in ParE have not been described previously and were found in two quinolone-resistant mutants (strain 9 mutants selected by trovafloxacin and ciprofloxacin). The significance of this mutation is not clear, because mutants selected by trovafloxacin also had mutations in both ParC and GyrA and the mutant selected by ciprofloxacin also had a mutation in ParC, which was reported as the primary ciprofloxacin target by Pan and Fisher (19). A similar interpretation applies to strains with mutations in the QRDR of GyrB. A strain 3 mutant selected by ciprofloxacin had a mutation in GyrB (Arg445→Ser) and had mutations in both ParC and GyrA, and strain 10 mutants selected by trovafloxacin and clinafloxacin had a mutation in GyrB (Gly406→Ser) and had mutations in both ParC and GyrA. Therefore, it is unclear what role GyrB mutations have in quinolone resistance. Time-kills showed that all resistant clones were killed by concentrations at or above two times the MIC after 24 h.
For two parent strains and 17 of the 30 mutants the MICs of at least one of the quinolones was lower in the presence of reserpine, which suggests the involvement of an efflux mechanism described previously by Baranova and Neyfakh and Brenwald et al. (2, 4). However, this effect was not seen equally for the three quinolones studied. The ciprofloxacin MIC for parent strain 9, for which the ciprofloxacin MICs were higher (4 μg/ml), was four times lower in the presence of reserpine. However, this parent strain also had a mutation in ParC, which is the target for ciprofloxacin resistance (19). The role of efflux in mutant strain 8 was unclear because parent strain 8 (ciprofloxacin MIC, 1 μg/ml) also had an efflux mechanism. Trovafloxacin MICs were not affected by reserpine in this study, suggesting that trovafloxacin is a poor substrate for PmrA or other reserpine-inhibitable pumps in those strains of pneumococci in which there was a substantial reserpine effect for the other drugs (10).
For strain 2 and 8 mutants selected by clinafloxacin, the clinafloxacin MICs were 16 to 32 times greater than the clinafloxacin MICs for parent strains, but no mutations in the QRDR of ParC, GyrA, ParE, and GyrB were found in these mutants. The clinafloxacin MICs of a strain 8 mutant selected by clinafloxacin was four times lower in the presence of reserpine. This suggests that an efflux mechanism plays an important role in clinafloxacin resistance, while this mechanism may play a limited role for trovafloxacin. The trovafloxacin, ciprofloxacin, or clinafloxacin MICs for the strain 2 mutant did not change in the presence of reserpine. This result suggests that quinolone resistance mechanisms other than mutations in the QRDR and efflux mechanisms exist or that reserpine did not affect the efflux mechanism of this strain. Two strains (strain 1 and 3 mutants selected with clinafloxacin) which showed unstable increased resistance had the efflux mechanism, which was lost with subsequent subcultures.
In our study, most high-level clinafloxacin-resistant mutant strains had mutations in ParC at Ser79 or Asp83 and GyrA at Ser81 as previously reported (20). The efflux mechanism was also thought to be a resistance mechanism for clinafloxacin. Single and multistep testing showed clinafloxacin selected resistant mutants less frequently than trovafloxacin and ciprofloxacin. These in vitro results may suggest that clinafloxacin would be less likely than trovafloxacin or ciprofloxacin to result in the development of resistance.
Although the quinolone MICs for mutants selected by all three agents increased equally above those for parent strains, MICs of clinafloxacin and trovafloxacin remained below the susceptible breakpoints of these agents in about two-thirds of the mutants. Results of this study indicate that this class of compound has potential for treatment of pneumococcal infections, including those caused by strains resistant to older quinolones.
ACKNOWLEDGMENT
This study was supported by a grant from Parke-Davis Pharmaceuticals, Inc., Ann Arbor, Mich.
REFERENCES
- 1.Appelbaum P C. Antimicrobial resistance in Streptococcus pneumoniae: an overview. Clin Infect Dis. 1992;15:77–83. doi: 10.1093/clinids/15.1.77. [DOI] [PubMed] [Google Scholar]
- 2.Baranova N N, Neyfakh A A. Apparent involvement of a multidrug transporter in the fluoroquinolone resistance of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:1396–1398. doi: 10.1128/aac.41.6.1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blondeau J M. A review of the comparative in-vitro activities of 12 antimicrobial agents, with a focus on five new ‘respiratory quinolones.’. J Antimicrob Chemother. 1999;43(Suppl. B):1–11. doi: 10.1093/jac/43.suppl_2.1. [DOI] [PubMed] [Google Scholar]
- 4.Brenwald N P, Gill M J, Wise R. Prevalence of putative efflux mechanism among fluoroquinolone-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2032–2035. doi: 10.1128/aac.42.8.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen D K, McGeer A, de Azavedo J C, Low D E. Decreased susceptibility of Streptococcus pneumoniae to fluoroquinolone in Canada. N Engl J Med. 1999;22:233–239. doi: 10.1056/NEJM199907223410403. [DOI] [PubMed] [Google Scholar]
- 6.Davies T A, Kelly L M, Pankuch G A, Credito K L, Jacobs M R, Appelbaum P C. Antipneumococal activities of gemifloxacin compared to those of nine other agents. Antimicrob Agents Chemother. 2000;44:304–310. doi: 10.1128/aac.44.2.304-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies T A, Pankuch G A, Dewasse B E, Jacobs M R, Appelbaum P C. In vitro development of resistance to five quinolones and amoxicillin-clavulanate in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:1177–1182. doi: 10.1128/aac.43.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ednie L M, Jacobs M R, Appelbaum P C. Comparative activities of clinafloxacin against gram-positive and -negative bacteria. Antimicrob Agents Chemother. 1998;42:1269–1273. doi: 10.1128/aac.42.5.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda H, Hiramatsu K. Primary targets of fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:410–412. doi: 10.1128/aac.43.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill M J, Brenwald N P, Wise R. Identification of an efflux pump gene, pmrA, associated with fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:187–189. doi: 10.1128/aac.43.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gootz T D, Zaniewski R, Haskell S, Schmieder B, Tankovic J, Girard D, Courvalin P, Polzer R J. Activity of the new fluoroquinolone trovafloxacin (CP-99,219) against DNA gyrase and topoisomerase IV mutants of Streptococcus pneumoniae selected in vitro. Antimicrob Agents Chemother. 1996;40:2691–2697. doi: 10.1128/aac.40.12.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho P-L, Que T-L, Tsang D N-C, Ng T-K, Chow K-H, Seto W-H. Emergence of fluoroquinolone resistance among multiply resistant strains of Streptococcus pneumoniae in Hong Kong. Antimicrob Agents Chemother. 1999;43:1310–1313. doi: 10.1128/aac.43.5.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs M R, Bajaksouzian S, Zilles A, Lin G, Pankuch G A, Appelbaum P C. Susceptibilities of Streptococcus pneumoniae and Haemophilus influenzae to 10 oral antimicrobial agents based on pharmacodynamic parameters: 1997 U.S. surveillance study. Antimicrob Agents Chemother. 1999;43:1901–1908. doi: 10.1128/aac.43.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janoir C, Zeller V, Kitzis M, Moreau N J, Gutmann L. High-level fluoroquinolone resistance in Streptococcus pneumoniae requires mutation in parC and gyrA. Antimicrob Agents Chemother. 1996;40:2760–2764. doi: 10.1128/aac.40.12.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liñares J, Campa A G, Pallares R. Fluoroquinolone resistance in Streptococcus pneumoniae. N Engl J Med. 1999;20:1546–1548. doi: 10.1056/nejm199911113412013. [DOI] [PubMed] [Google Scholar]
- 16.Moissenet D, Valcin M, Marchand V, Garabédian E-N, Geslin P, Garbarg-Chenon A, Vu-Thien H. Molecular epidemiology of Streptococcus pneumoniae with decreased susceptibility to penicillin in a Paris children's hospital. J Clin Microbiol. 1997;35:298–301. doi: 10.1128/jcm.35.1.298-301.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing, 8th informational supplement. M100-S8. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 18.Pan X-S, Fisher L M. Cloning and characterization of the parC and parE genes of Streptococcus pneumoniae encoding DNA topoisomerase IV: role in fluoroquinolone resistance. J Bacteriol. 1996;178:4060–4069. doi: 10.1128/jb.178.14.4060-4069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X-S, Ambler J, Mehtar S, Fisher L M. Involvement of topoisomerase IV and DNA gyrase as ciprofloxacin targets in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1996;40:2321–2326. doi: 10.1128/aac.40.10.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X-S, Fisher L M. DNA gyrase and topoisomerase IV are dual targets of clinafloxacin action in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2810–2816. doi: 10.1128/aac.42.11.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pankuch G A, Lichtenberger C, Jacobs M R, Appelbaum P C. Antipneumococcal activities of RP 59500 (quinupristin-dalfopristin), penicillin G, erythromycin, and sparfloxacin determined by MIC and rapid time-kill methodologies. Antimicrob Agents Chemother. 1996;40:1653–1656. doi: 10.1128/aac.40.7.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pankuch G A, Jacobs M R, Appelbaum P C. Antipneumococcal activity of grepafloxacin compared to that of other agents by time-kill methodology. Antimicrob Agents Chemother. 1998;42:1263–1265. doi: 10.1128/aac.42.5.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pankuch G A, Jueneman S A, Davies T A, Jacobs M R, Appelbaum P C. In vitro selection of resistance to four β-lactams and azithromycin in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1998;42:2914–2918. doi: 10.1128/aac.42.11.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perichon B, Tankovic J, Courvalin P. Characterization of a mutation in the parE gene that confers fluoroquinolone resistance in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;41:1166–1167. doi: 10.1128/aac.41.5.1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piddock L J, Johnson M, Ricci V, Hill S L. Activities of new fluoroquinolones against fluoroquinolone-resistant pathogens of the lower respiratory tract. Antimicrob Agents Chemother. 1998;42:2956–2960. doi: 10.1128/aac.42.11.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tankovic J, Perichon B, Duval J, Courvalin P. Contribution of mutations in gyrA and parC genes to fluoroquinolone resistance of mutants of Streptococcus pneumoniae obtained in vivo and in vitro. Antimicrob Agents Chemother. 1996;40:2505–2510. doi: 10.1128/aac.40.11.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varon E, Janoir C, Kitzis M-D, Gutmann L. ParC and GyrA may be interchangeable initial targets of some fluoroquinolones in Streptococcus pneumoniae. Antimicrob Agents Chemother. 1999;43:302–306. doi: 10.1128/aac.43.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]