Abstract
Avian pathogenic Escherichia coli (APEC) is a pathotype of extraintestinal pathogenic E. coli and one of the most serious infectious diseases of poultry. It not only causes great economic losses to the poultry industry, but also poses a serious threat to public health worldwide. Here, we examined the role of YqeH, a transcriptional regulator located at E. coli type III secretion system 2 (ETT2), in APEC pathogenesis. To investigate the effects of YqeH on APEC phenotype and virulence, we constructed a yqeH deletion mutant (APEC40-ΔyqeH) and a complemented strain (APEC40-CΔyqeH) of APEC40. Compared with the wild type (WT), the motility and biofilm formation of APEC40-ΔyqeH were significantly reduced. The yqeH mutant was highly attenuated in a chick infection model compared with WT, and showed severe defects in its adherence to and invasion of chicken embryo fibroblast DF-1 cells. However, the mechanisms underlying these phenomena were unclear. Therefore, we analyzed the transcriptional effects of the yqeH deletion to clarify the regulatory mechanisms of YqeH, and the role of YqeH in APEC virulence. The deletion of yqeH downregulated the transcript levels of several flagellum-, biofilm-, and virulence-related genes. Our results demonstrate that YqeH is involved in APEC pathogenesis, and the reduced virulence of APEC40-ΔyqeH may be related to its reduced motility and biofilm formation.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13567-022-01049-6.
Keywords: Avian pathogenic Escherichia coli, yqeH, motility, biofilm formation, virulence, pathogenicity
Introduction
Avian pathogenic Escherichia coli (APEC), a pathotype of extraintestinal pathogenic E. coli (ExPEC), causes serious infectious diseases in poultry [1]. Different APEC serotypes cause local or systemic infections in poultry, including respiratory infections, sepsis, polyserositis, coligranuloma, cellulitis, yolk sac infection, omphalitis, and swollen head syndrome, resulting in significant economic losses to the poultry industry [2]. Understanding the underlying molecular mechanisms of APEC pathogenicity is crucial for controlling avian colibacillosis.
There are many transcriptional regulators in Gram-negative pathogenic bacteria, which play important roles in regulating bacterial metabolism and the expression of virulence genes. More than 250 transcription factors are known to regulate gene expression in E. coli, and coordinate the expression of numerous promoters in response to specific environmental cues [3–5]. Although many virulence factors are known to be associated with APEC pathogenicity, the regulation of their expression is still not fully understood.
In E. coli, two distinct type III secretion systems (T3SSs) have been identified and characterized. The locus of enterocyte effacement (LEE), which encodes T3SS in intestinal E. coli, such as enteropathogenic E. coli and enterohemorrhagic E. coli (EHEC), is essential for the formation of attaching and effacing (A/E) lesions. A second T3SS, E. coli type III secretion system 2 (ETT2), plays an important role in the invasion, intracellular survival, and virulence of ExPEC, including APEC and newborn meningitis E. coli [6, 7]. We speculated that the transcriptional regulators located in the ETT2 cluster play an important role in APEC pathogenicity. Many reviews have described the various transcriptional regulator genes located in the ETT2 cluster, including yqeK/etrB, etrA, and eivF, which encode transcription factors that regulate the expression of virulence genes in intestinal pathogenic E. coli [8]. For example, EtrA and EivF of EHEC O157 negatively affect LEE expression and bacterium adherence to epithelial cells [9]. In contrast, the regulator YqeK/EtrB activates LEE expression and promotes A/E lesion formation by directly interacting with the Ler regulatory region [10]. Identifying new transcriptional regulators and their functions will clarify ETT2 pathogenesis.
In this study, we investigated the role of the transcriptional regulator YqeH located at ETT2 locus in the pathogenesis of APEC infections. The deletion of yqeH significantly reduced the virulence of APEC in vivo and in vitro. This was associated with reductions in the expression of many virulence-associated genes regulated by YqeH, which reduced bacterium motility, biofilm formation, adhesion, and invasion, suggesting that YqeH is involved in the pathogenicity of APEC.
Materials and methods
Bacterial strains, plasmids, and growth conditions
The strains and plasmids used in this study are listed in Table 1. The nucleotide sequences of the primers (GenBank: HG738867.1) are listed in Additional file 1. APEC strain APEC40 [11] (serotype O18:K1) of the B2 and ST95 phylogenetic groups, was isolated from a chicken with clinical septicemic symptoms of colibacillosis in Anhui, China. All E. coli strains were grown in Luria–Bertani (LB) medium at 37 °C with aeration. LB medium was supplemented with ampicillin (Amp, 100 mg/mL) and chloramphenicol (Cm, 30 mg/mL), unless specified otherwise.
Table 1.
Strains and plasmids used in this study
| Strains or plasmids | Description | Reference |
|---|---|---|
| Strains | ||
| APEC40 | wild-type strain, isolated from sick chicken | This study |
| APEC40-△yqeH | yqeH deletion mutant in APEC40 | This study |
| APEC40-C△yqeH | APEC40-△yqeH with plasmid pSTV28-yqeH | This study |
| DH5a | F-,△(lacZYA-argF)U169,recA1,endA1,hsdR17 (rk-,mk +), phoA,supE44, γ- | TIANGEN |
| Plasmids | ||
| pKD46 | Amp; expresses γ red recombinase | This study |
| pKD3 | Cm gene, template plasmid | This study |
| pCP20 | Cm, Amp, yeast Flp recombinase gene, FLP | This study |
| pSTV28 | Cm, lacZ | Takara |
| pSTV28-yqeH | pSTV28 derivative harboring yqeH gene | This study |
Ethics statement
The animal experiments were approved by the Institutional Animal Care and Use Committee at the Institute of Animal Husbandry and Veterinary Science, Anhui Academy of Agricultural Sciences (Permit No: AAAS-IAHVS-Po-2020-0034). The chickens were employed in experiments that complied with the ARRIVE guidelines of laboratory animal welfare and ethics [12].
Mutant construction and complementation plasmids
A yqeH deletion mutant strain was generated with the lambda Red recombinase system, as described previously [13]. Linear PCR products were transformed into APEC40 carrying the pKD46 plasmid and the yqeH gene was replaced with a chloramphenicol resistance cassette. The chloramphenicol-resistance cassette was then eliminated by the helper plasmid pCP20 and a chloramphenicol-sensitive mutant strain was selected. The mutant strain was confirmed with PCR and DNA sequencing, and designated APEC40-ΔyqeH. To generate a complemented strain, the yqeH gene (including its putative promoter) was amplified and subcloned into plasmid pSTV28 with the primer pair yqeH Co-F and yqeH Co-R (Additional file 1). The mutant strain APEC40-ΔyqeH was then transformed with recombinant plasmid pSTV28-yqeH to generate the complemented strain APEC40-CΔyqeH.
Determination of bacterial growth, motility, and biofilm formation
Bacterial growth was measured as described previously [14]. Briefly, strains APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH were cultured in LB medium until the optical density at 600 nm (OD600) reached 1.0. An equal amount of each bacterial culture was then transferred into 100 mL of LB medium in a volumetric ratio of 1:100 (v/v) and incubated at 37 °C with shaking. OD600 was monitored at 2 h intervals with a spectrophotometer (Bio-Rad, USA). The experiment was repeated three times and all samples were measured in triplicate.
The swarming motility of strains APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH was assessed on soft-agar plates (0.25% agar), as previously described [15]. Briefly, bacteria were grown to an OD600 of 1.0 in LB medium and pelleted by centrifugation. The resulting pellet was washed and suspended in phosphate-buffered saline (PBS). The bacterial suspension (2 μL) was spread by point inoculation in the center of the soft-agar plates and incubated at 37 °C overnight. Swarming motility was assessed by measuring the diameter of migration. The experiment was repeated three times and all samples were measured in triplicate. In addition, the strains APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH were negatively stained using 2% phosphotungstic acid (Sigma). Finally, the stained bacteria were deposited on a carbon-coated grid, followed by observation under a FEI T12 transmission electron microscope (FEI, Ltd, Hillsboro, OR, USA).
An assay involving crystal violet staining was used to quantify biofilm formation by APEC, as previously described [11]. Briefly, the strains were grown overnight in 5 mL of LB medium at 37 °C with rotation at 200 rpm. The cultures were suspended in M9 minimal medium supplemented with 0.2% fructose, and diluted to an OD600 of 0.1. An aliquot (200 μL) of each cell suspension was transferred to a 96-well plate (Corning, Corning, NY, USA) and incubated at 37 °C for 36 h. The wells were washed gently three times with PBS and stained with 0.1% crystal violet for 30 min at room temperature. The wells were then rinsed three times with PBS, air-dried, and 100 μL of 95% ethanol was added to dissolve the crystal violet. OD595 was measured with a Synergy 2 micro-plate reader (BioTek, USA).
Bacterial adhesion and invasion assays
Bacterial adhesion and invasion assays were performed as described previously [16]. Chicken embryo fibroblast DF-1 cell monolayers were washed with Dulbecco’s modified Eagle’s medium (DMEM) without fetal bovine serum, and infected with bacteria at a multiplicity of infection (MOI) of 100 for 2 h at 37 °C under 5% CO2. After the cells were washed with PBS, they were lysed with 0.5% Triton X-100, and the bacteria were counted by plating them on LB agar plates. For the invasion assay, cell cultures were inoculated with bacteria as described for the bacterial adhesion assay. After incubation for 1 h, the cells were washed and treated with DMEM containing gentamicin (100 µg/mL) for 1 h to kill any extracellular bacteria. The monolayers were then washed and lysed with 0.5% Triton X-100. Serial dilutions of the cell suspensions were spread onto LB agar plates, and after overnight growth at 37 °C, the colony-forming units (CFUs) were counted. The input dilution of the bacteria was also plated to determine the CFU count for each inoculum.
Animal infection experiments
The virulence of strains APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH was determined in chicks. The APEC strains were grown to the exponential phase and collected, washed three times in PBS, and then adjusted to the appropriate concentration. For each group, eight 7-day-old Roman chicks were inoculated intratracheally with 108 CFUs of bacteria (APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH, respectively), or with PBS as negative control. Mortality was monitored daily up to 7 days post infection.
Bacterial colonization was determined during systemic infections, as described previously [17]. At 24 h post-infection, the chicks were bled, euthanized, and dissected. The bacterial loads in the blood were determined by plating the bacteria onto LB agar plates. The chick livers, spleens, and lungs were collected, weighed, and homogenized. The homogenates were diluted and plated onto LB agar to determine the bacterial numbers.
RNA-Seq transcriptomic assay
The transcriptional levels of APEC40 and APEC40-ΔyqeH cells were determined, as described previously [18], with some modification. To evaluate the effects of YqeH on the transcriptional levels in APEC40 and APEC40-ΔyqeH, the cells were collected by centrifugation at 12 000 rpm for 5 min from cultures in LB medium when OD600 attained 0.80. The collected cells were washed with PBS (pH 7.4) and centrifuged. Total RNA was extracted from the cells with TRIzol Reagent (Invitrogen, Thermo Fisher Scientific, USA), according to the manufacturer’s instructions, and the genomic DNA was removed with DNase I (TaKaRa, Japan). cDNA was synthesized with the SuperScript™ Double-Stranded cDNA Synthesis Kit (Invitrogen, Thermo Fisher Scientific) and sequenced by Shanghai Major BioBiopharm Technology Co. Ltd. (Shanghai, China) with the Illumina HiSeq2500 System (Illumina, CA, USA). The differential expression analysis was performed with the EdgeR software. Differences in expression levels between groups were considered significant after adjustment was made for multiple testing, based on a q-value < 0.05. The genes were first filtered, based on q < 0.05 and an absolute difference > twofold, when |log2fold change|> 1.0. The enrichment of the differentially expressed genes was analyzed with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed with KOBAS.
RNA isolation and reverse transcription–quantitative PCR (RT–qPCR)
RNA isolation and RT–qPCR (primers listed in Additional file 1) were performed as described previously [19]. The bacterial strains were grown to post-exponential phase (OD600 = 0.80) in the absence of antibiotics. Total RNA was isolated with the EasyPure® RNA Kit (TransGen Biotech Co., Ltd, China), according to the manufacturer’s instructions. The quality of the RNA was determined, cDNA was synthesized, and a microarray analysis performed. SYBR Green detection was used for the RT–qPCR. Each reaction was conducted in triplicate in two-step multiplex qPCR assays with SYBR® Premix Ex Taq™ (TransGen Biotech Co., Ltd), and bacterial dnaE was used as the internal reference gene. The relative expression levels were measured with the 2−ΔΔCT method [19].
Detection of transcription level of type 1 fimbriae genes by RT–qPCR
The transcription level of type 1 fimbriae genes (fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH, fimI) was detected using RT–qPCR. The total volume of the reaction system was 20 μL with one μg total RNA, and the reverse transcription was conducted using SYBR green PCR master mix. The standard cycling parameters were performed on the ABI StepOne Plus instrument, and each target gene was examined three times. The primers of the target genes were listed in Additional file 1.
Statistical analyses
Statistical analyses were performed with the GraphPad software package (GraphPad Software, LaJolla, CA, USA). One-way analysis of variance (ANOVA) was used to analyze the adhesion and invasion assay data, and two-way ANOVA was used to analyze the survival assay, and RT–qPCR results. The animal infection data were analyzed with the nonparametric Mann–Whitney U-test. Each experiment was performed three times and statistical significance indicated as follows: *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Identification of strains APEC40-ΔyqeH and APEC40-CΔyqeH
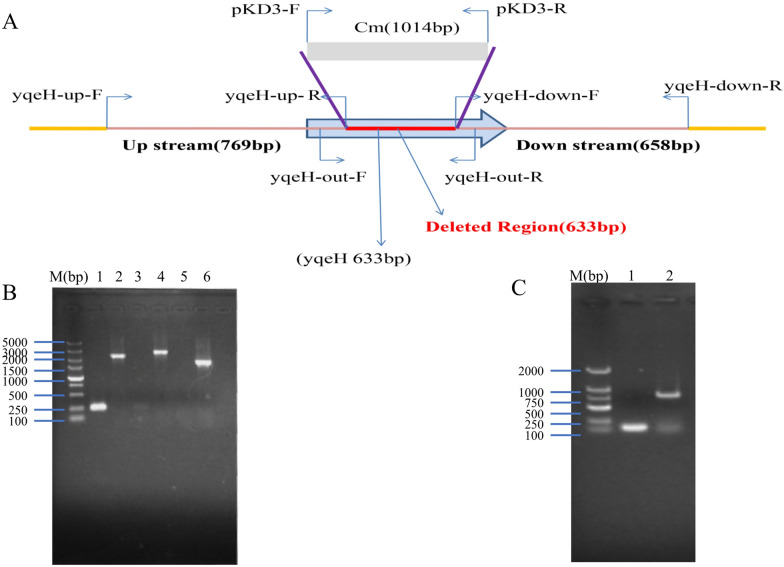
The ΔyqeH and CΔyqeH mutant strains were constructed from parental strain APEC40 with the lambda Red recombination method (Figure 1A), and were confirmed with PCR. Using primers yqeH-out-F/yqeH-out-R, a 2256-bp product was amplified from strain APEC40, whereas a 1623-bp product was amplified from ΔyqeH (Figure 1B). The nucleotide sequence of yqeH in recombinant plasmid pSTV28-yqeH was sequenced, and showed 100% nucleotide sequence identity to the yqeH gene of APEC40. The complementary strain APEC40-CΔyqeH was also confirmed by PCR (Figure 1C). A 826 bp PCR product was amplified from APEC40-CΔyqeH with PCR using primers M13F/M13R.
Figure 1.
PCR analysis of mutant strain APEC40-ΔyqeH and complemented strain APEC40-CΔyqeH. A Schematic diagram of the strategy used to construct the APEC40 yqeH deletion mutant. The yqeH gene was deleted by replacing the partial gene sequence with a chloramphenicol-resistance cassette. The primers used for confirmation of the yqeH deletion are indicated. B Confirmation of the yqeH mutant strain APEC40-ΔyqeH. Lane identities in (B)—M: 2000-bp DNA marker; 1–2: identification of wild-type APEC40-yqeH strain (lane 1, 252 bp; lane 2, 2256 bp); 3–4: identification of APEC40-ΔyqeH-Cm (lane 3, 0 bp; lane 4, 2637 bp); 5–6: identification of APEC40-ΔyqeH (lane 5, 0 bp; lane 6, 1623 bp). C Confirmation of the complemented strain APEC40-CΔyqeH. Lane identities in (C)—M: 2000-bp DNA marker; 1: Recombinant plasmid pSTV28 blank control identification, 193 bp; 2: APEC40-CΔyqeH identification, 826 bp.
Characterization of APEC40-ΔyqeH mutant strain
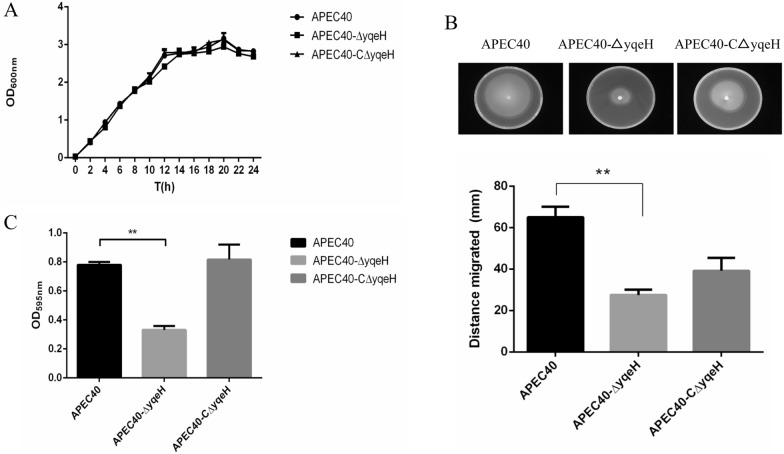
To determine whether the deletion of yqeH influenced the growth of APEC40, bacterial growth was evaluated by measuring OD600 from 0 to 24 h. After the deletion of the yqeH gene, APEC40-ΔyqeH showed a growth rate like that of WT APEC40 (Figure 2A). These data suggest that the deletion of the yqeH gene had no significant effect on the growth of APEC40.
Figure 2.
Characteristics of strains APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH. A Growth curves of APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH in LB broth. Growth rates of APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH were monitored by measuring their OD600 values in LB medium at 2 h intervals for 24 h. During culture for 24 h, there was no significant difference in the growth rates of APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH grown in LB broth (P > 0.05). Each value is the average of three independent experiments. B Bacterial motility. Swimming motility of strain APEC40-ΔyqeH was significantly lower than that of the wild-type APEC40 strain, whereas the swimming motility of complemented strain APEC40-CΔyqeH was restored. The experiment was repeated three times and all samples were measured in triplicate. C Biofilm formation by APEC strains as determined with crystal violet (CV) staining. APEC40-ΔyqeH showed significantly lower biofilm formation than wild-type strain APEC40 after crystal violet staining (**P < 0.01 for the WT vs. mutant).
The bacterial motility assay showed that APEC40-ΔyqeH mutant strain motility was significantly lower than that of the WT strain APEC40 (P < 0.05) (Figure 2B). The influence of yqeH on the bacterial morphology was examined by transmission electron microscopy. The results revealed remarkable differences in the flagella between the APEC40 and APEC40-ΔyqeH strains. Many long flagella were distributed at APEC40 periphery, but APEC40-ΔyqeH flagella were few and impaired. A few broken flagella appeared on the surface of APEC40-CΔyqeH (Additional file 2). Moreover, APEC40-ΔyqeH formed significantly less biofilm than WT strain APEC40 (Figure 2C).
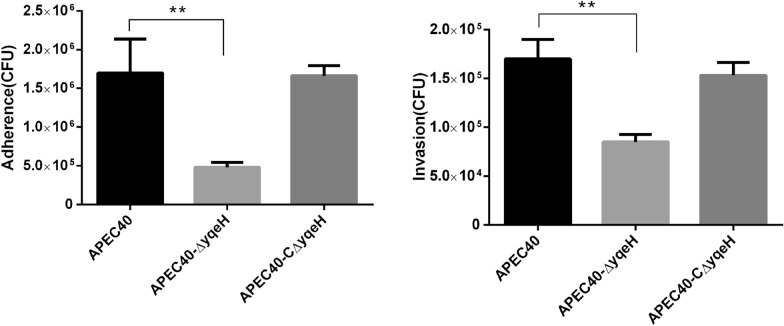
Deletion of yqeH reduces bacterial adherence and invasion
As shown in Figure 3, the APEC40-ΔyqeH mutant showed reduced adherence to and invasion of DF-1 cells compared with those of WT strain APEC40.
Figure 3.
Deletion of yqeH reduced the adherence and invasion abilities of APEC. Adherence to and invasion of DF-1 cells was significantly lower in mutant strain APEC40-ΔyqeH than in WT strain APEC40. (**P < 0.01 for WT vs. mutant).
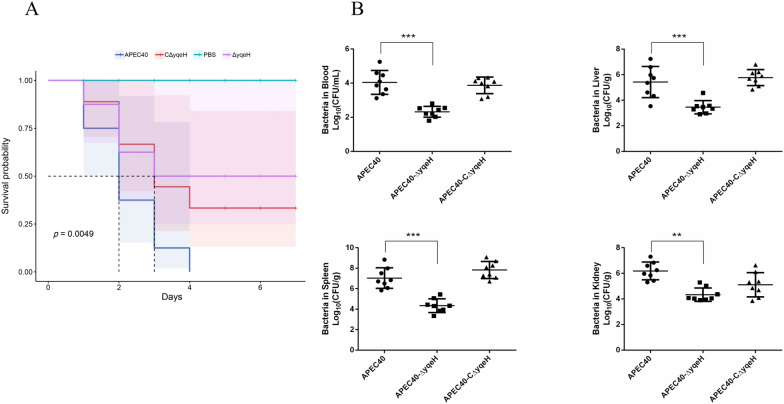
Virulence of mutant strain APEC40-ΔyqeH
To investigate the effects of YqeH on bacterial pathogenicity, the virulence of the APEC40, APEC40-ΔyqeH, and APEC40-CΔyqeH strains was compared in a chick model. As shown in Figure 4A, the mortality rate was significantly lower after infection with strain APEC40-ΔyqeH (50%, 4/8) than after infection with strain APEC40 (100%, 8/8). The mortality rate after infection with strain APEC40-CΔyqeH was restored to 75% (6/8) of the WT level. These results indicate that YqeH contributes to APEC virulence.
Figure 4.
Animal systemic infection in vivo. A Determination of bacterial virulence. Seven-day-old chicks were inoculated intratracheally with 108 colony-forming units (CFUs) of APEC40, APEC40-ΔyqeH, or APEC40-CΔyqeH. Chicks that were administered PBS were used as negative control. Survival was monitored for 7 days post-infection. B Bacterial colonization and proliferation in chicks. Groups of eight 7-day-old chicks were intratracheally infected with bacteria (108 CFUs). Bacteria were recovered from the lung, blood, liver, and spleen at 24 h post-infection. The nonparametric Mann–Whitney U-test was used to determine statistical significance (**P < 0.01, ***P < 0.001 for WT vs. mutant).
To measure the influence of YqeH on APEC bacterial numbers in blood and tissues or organs in vivo, a systemic infection experiment was performed to assess bacterial proliferation in chicks. The bacterial loads in the blood, livers, spleens, and kidneys of infected chicks were determined at 24 h post-infection. With the removal of yqeH, the bacterial loads of APEC40-ΔyqeH were significantly lower than those of WT strain APEC40 in all samples. Moreover, the virulence of APEC40-CΔyqeH was restored (Figure 4B).
Effects of YqeH on the transcriptional profile of virulence-related genes
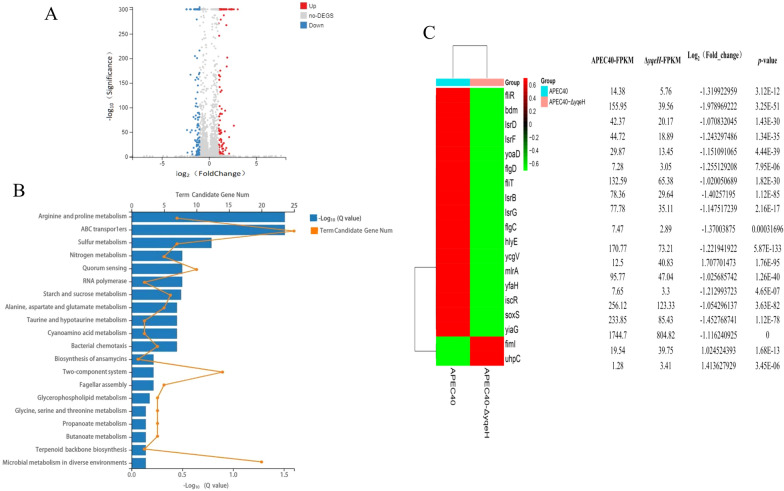
A transcriptional analysis showed that 112 genes were upregulated and 101 genes were downregulated in APEC40-ΔyqeH relative to their expression in WT strain APEC40 (Figure 5A). The up-and downregulated genes (Additional file 3) were analyzed for their enrichment in KEGG pathways. The differentially expressed genes were enriched in pathways functions related to ABC transport, quorum sensing, bacterial chemotaxis, two-component system and flagellar assembly (Figure 5B). We selected several differentially expressed genes related to biofilm formation, bacterial mobility, and virulence for further study, as shown in Figure 5C.
Figure 5.
Analysis of differentially expressed genes by RNA-seq. A Volcano plots were used to visualize the differentially expressed genes in APEC40 and APEC40-ΔyqeH. Genes with significantly upregulated expression in APEC40-ΔyqeH relative to their expression in APEC40 are indicated with red dots; those downregulated are indicated with blue dots; and grey dots represent genes with no significant difference in expression between the strains. The results showed that 112 genes were upregulated and 101 genes were downregulated in APEC40-ΔyqeH relative to their expression in wild-type strain APEC40 (differentially expressed genes were selected at fold change > 2 and q < 0.005). B KEGG enrichment analysis of differentially expressed genes. Top 20 pathways enriched in differentially expressed genes are shown in the figure. C Quantitative differences in the expression levels of 19 differentially expressed virulence-related genes between wild-type strain APEC40 and mutant strain APEC40-ΔyqeH are shown with a heatmap.
Verification of differentially expressed genes with RT–qPCR
The RNA-seq results were validated with RT–qPCR. Eight genes representing a wide range of gene expression ratios in the yqeH mutant were selected for a comparative RT–qPCR analysis. The gene expression patterns determined with RT–qPCR were highly concordant with the RNA-seq results (Figure 6).
Figure 6.
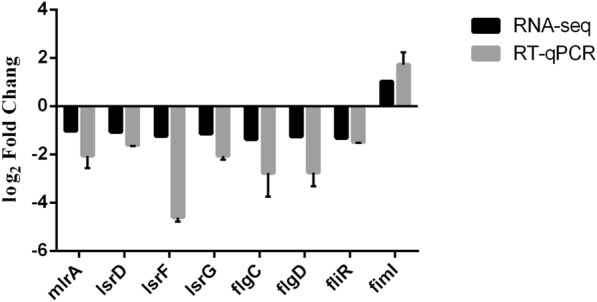
Verification of RNA-seq results with reverse transcription–quantitative real-time PCR (RT–qPCR). RT–qPCR was used to determine the expression profiles of several genes differentially expressed in the yqeH mutant strain. The x-axis shows the annotations of the selected genes. The dnaE gene was used for standardization.
Expression variations of type 1 fimbriae genes
The expression of type 1 fimbriae genes fimA, fimB, fimC, fimD, fimE, fimF, fimG, fimH and fimI showed no significant difference between the wild-type strain APEC40 and the yqeH mutant strain (Additional file 4).
Discussion
The ETT2 locus is present in most E. coli strains. However, the transcriptional regulators located in the ETT2 locus are considered key factors in the pathogenicity of E. coli [20]. Genomic studies have identified several transcriptional regulators, including YqeK/EtrB, EtrA, and EivF, located at the ETT2 locus, which regulate the expression of virulence genes in pathogenic E. coli [8]. In this report, we propose for the first time that YqeH is a transcriptional regulator located at the ETT2 locus of APEC, and describe it. We constructed a YqeH deletion mutant to identify the role of YqeH in APEC pathogenesis. However, the rescue assays for expression of YqeH in YqeH-mutant strain resulted in partial recovery. It is possibly due to deficient expression of YqeH by using a low-copy plasmid, pSTV28, as the complementation plasmid.
Biofilm formation is an important process by which bacteria establish infections, leading to host diseases [21]. In this study, APEC biofilm formation was significantly reduced after yqeH deletion compared with that of WT strain APEC40. Biofilm formation is a complex process, during which the expression of specific genes is required to produce specific substances that promote and regulate biofilm formation [22]. The effects of YqeH on the expression of biofilm-related genes in APEC were investigated by using a transcriptional analysis. In total, seven downregulated genes related to biofilm formation were enriched in differentially expressed genes of YqeH-deletion mutant compared with WT strain. Among them, mlrA, yoaD and bdM were reported for their role in c-di-GMP signaling pathway. Moreover, it was reported that lsrB, lsrD, lsrF and lsrG mediated the biosynthesis of AI-2 protein in the quorum sensing signaling pathway. As a signaling molecule, AI-2 promotes the transformation of bacteria from suspension to biofilm modes, and promotes the transcription of bacterial genes that facilitate biofilm formation [23]. The second messenger c-di-GMP regulates biofilm formation by changing the concentrations of bacteria within host cells [24]. Our results show that after the deletion of yqeH, the expression levels of genes related to the synthesis of AI-2 and c-di-GMP were lower than in WT strain APEC40, which ultimately affected the formation of the APEC biofilm.
The flagellum is a motor organelle and a protein export apparatus that controls bacterial motility and behavior [25]. In this study, bacterial motility was significantly reduced after yqeH deletion compared with that in WT strain APEC40. Transcriptomic profiling showed that the transcription of flgC, flgD, fliT, and fliR was similarly reduced. FlgC is the flagellar basal-body protein; FlgD is essential for flagellar hook formation; FliT is the flagellar synthesis and assembly chaperone protein; and FliR is the flagellar export pore protein [26, 27]. Therefore, we speculate that the reduced expression of these genes impaired flagellum synthesis in the bacteria, resulting in reduced bacterial motility.
The bacterial colonization and invasion of host cells are crucial steps in APEC infection process [28, 29]. Type 1 fimbriae contribute to bacterial adhesion and invasion of host cells, which create conditions for a series of processes, such as pathogen colonization and infection [30, 31]. However, our results (Additional file 4) showed that the transcript expression of type 1 fimbriae genes had no significant differential changes after the deletion of yqeH. The flagellum has been shown to mediate bacterial adhesion and invasion, and is implicated in the virulence of pathogenic bacteria. The motility of the flagellum is an important virulence feature of many bacterial pathogens and is necessary for the establishment of infection [32]. For example, the FlgC protein plays an important role in the binding of Salmonella enteritidis to host epithelial cells [33], and the deletion of the flagellin fliR gene affects the adhesion and pathogenicity of bacteria to host cells [34]. Biofilm formation is also strongly related to the ability of bacteria to adhere to and invade cells. Several studies have shown that the biofilm-synthesis-related gene pgaC, encoding N-glycosyltransferase, not only mediates biofilm formation, but also plays an important role in bacterial adhesion to its host cell [35]. The quorum-sensing transcriptional activator SdiA regulates the expression of the rck gene, which mediates E. coli adhesion to and invasion of epithelial cells [36]. Our results show that the loss of yqeH reduced bacterial motility and biofilm formation, which may have accounted for the diminished adhesion to and invasion of DF-1 cells by mutant APEC40-ΔyqeH. The adhesion and colonization abilities of APEC in the host blood and tissues are key factors in its pathogenesis [37]. The numbers of viable cells of deleted strain APEC40-ΔyqeH in the host blood and various tissues were significantly lower than those in the WT strain. Our experimental results demonstrate that YqeH is involved in APEC virulence.
In summary, we have demonstrated for the first time that the transcriptional regulator YqeH is involved in the motility, biofilm formation, and virulence of APEC strains. This study provides a basis for further functional research into the pathogenic role of YqeH in APEC.
Supplementary Information
Additional file 1: Primers used in this study.
Additional file 2: Bacterial micromorphology of APEC40, APEC40-ΔyqeH and APEC40-CΔyqeH observed by transmission electron microscopy.
Additional file 3: Differentially expressed genes data.
Additional file 4: Expressions of type 1 fimbriae genes of the wild-type strain APEC40, mutant strain APEC40-ΔyqeH and complementary strain APEC40-CΔyqeH were tested by RT-qPCR.
Acknowledgements
This work was supported by grants from the Natural Science Foundation of China (grant no. 31772707; 31972644); Natural Science Foundation of Anhui Province (grant no. 2108085QC137).
Author contributions
Conceptualization, KQ and LY; methodology, BC; software, XS; validation, LY, XP and KQ; formal analysis, YS; investigation, XS; resources, JT; data curation, BC; writing—original draft preparation, LY; writing—review and editing, LY; visualization, KQ; supervision, YS; project administration, XP; funding acquisition, KQ, JT and LY. All authors read and approved the final manuscript.
Availability of data and materials
The sequence was deposited in the GenBank database (Accession number PRJNA805014).
Declarations
Ethics approval and consent to participate
The animal experiments were approved by the Institutional Animal Care and Use Committee at the Institute of Animal Husbandry and Veterinary Science, Anhui Academy of Agricultural Sciences (Permit No: AAAS-IAHVS-Po-2020-0034).
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xiaocheng Pan, Email: pxcpyq@sina.com.
Kezong Qi, Email: qkz@ahau.edu.cn.
References
- 1.Apostolakos I, Laconi A, Mughini-Gras L, Yapicier Ö, Piccirillo A. Occurrence of colibacillosis in broilers and its relationship with avian pathogenic Escherichia coli (APEC) population structure and molecular characteristics. Front Vet Sci. 2021;8:737720. doi: 10.3389/fvets.2021.737720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paudel S, Fink D, Abdelhamid MK, Zöggeler A, Liebhart D, Hess M, Hess C. Aerosol is the optimal route of respiratory tract infection to induce pathological lesions of colibacillosis by a lux-tagged avian pathogenic Escherichia coli in chickens. Avian Pathol. 2021;50:417–426. doi: 10.1080/03079457.2021.1978392. [DOI] [PubMed] [Google Scholar]
- 3.Pérez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–1847. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marbaniang CN, Gowrishankar J. Transcriptional cross-regulation between Gram-negative and gram-positive bacteria, demonstrated using ArgP-argO of Escherichia coli and LysG-lysE of Corynebacterium glutamicum. J Bacteriol. 2012;194:5657–5666. doi: 10.1128/jb.00947-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slater SL, Sågfors AM, Pollard DJ, Ruano-Gallego D, Frankel G. The type III secretion system of pathogenic Escherichia coli. Curr Top Microbiol Immunol. 2018;416:51–72. doi: 10.1007/82_2018_116. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M, Guo Z, Duan Q, Hardwidge PR, Zhu G. Escherichia coli type III secretion system 2: a new kind of T3SS? Vet Res. 2014;45:32. doi: 10.1186/1297-9716-45-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Xu X, Liu X, Wang D, Liang H, Wu X, Tian M, Ding C, Wang G, Yu S. Escherichia coli type III secretion system 2 regulator EtrA promotes virulence of avian pathogenic Escherichia coli. Microbiology. 2017;163:1515–1524. doi: 10.1099/mic.0.000525. [DOI] [PubMed] [Google Scholar]
- 9.Zhang L, Chaudhuri RR, Constantinidou C, Hobman JL, Patel MD, Jones AC, Sarti D, Roe AJ, Vlisidou I, Shaw RK, Falciani F, Stevens MP, Gally DL, Knutton S, Frankel G, Penn CW, Pallen MJ. Regulators encoded in the Escherichia coli type III secretion system 2 gene cluster influence expression of genes within the locus for enterocyte effacement in enterohemorrhagic E. coli O157:H7. Infect Immun. 2004;72:7282–7293. doi: 10.1128/iai.72.12.7282-7293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzader DH, Willsey GG, Wargo MJ, Kendall MM. The type three secretion system 2-encoded regulator EtrB modulates enterohemorrhagic Escherichia coli virulence gene expression. Infect Immun. 2016;84:2555–2565. doi: 10.1128/iai.00407-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yin L, Li Q, Wang Z, Tu J, Shao Y, Song X, Pan X, Qi K. The role of Escherichia coli type III secretion system 2 chaperone protein ygeG in pathogenesis of avian pathogenic Escherichia coli. Res Vet Sci. 2021;140:203–211. doi: 10.1016/j.rvsc.2021.09.011. [DOI] [PubMed] [Google Scholar]
- 12.Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. Animal research: reporting in vivo experiments: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang S, Liu X, Xu X, Yang D, Wang D, Han X, Shi Y, Tian M, Ding C, Peng D, Yu S. Escherichia coli type III secretion system 2 ATPase EivC is involved in the motility and virulence of avian pathogenic Escherichia coli. Front Microbiol. 2016;7:1387. doi: 10.3389/fmicb.2016.01387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin L, Shen X, Zhang D, Zhao R, Dai Y, Hu X, Zhou X, Hou H, Pan X, Qi K. Flagellar rotor protein FliG is involved in the virulence of avian pathogenic Escherichia coli. Microb Pathog. 2021;160:105198. doi: 10.1016/j.micpath.2021.105198. [DOI] [PubMed] [Google Scholar]
- 16.Zuo J, Tu C, Wang Y, Qi K, Hu J, Wang Z, Mi R, Yan H, Chen Z, Han X. The role of the wzy gene in lipopolysaccharide biosynthesis and pathogenesis of avian pathogenic Escherichia coli. Microb Pathog. 2019;127:296–303. doi: 10.1016/j.micpath.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 17.Zuo J, Yin H, Hu J, Miao J, Chen Z, Qi K, Wang Z, Gong J, Phouthapane V, Jiang W, Mi R, Huang Y, Wang C, Han X. Lsr operon is associated with AI-2 transfer and pathogenicity in avian pathogenic Escherichia coli. Vet Res. 2019;50:109. doi: 10.1186/s13567-019-0725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Shulman A, Yair Y, Biran D, Sura T, Otto A, Gophna U, Becher D, Hecker M, Ron EZ. The Escherichia coli type III secretion system 2 has a global effect on cell surface. mBio. 2018 doi: 10.1128/mBio.01070-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin L, Li Q, Xue M, Wang Z, Tu J, Song X, Shao Y, Han X, Xue T, Liu H, Qi K. The role of the phoP transcriptional regulator on biofilm formation of avian pathogenic Escherichia coli. Avian Pathol. 2019;48:362–370. doi: 10.1080/03079457.2019.1605147. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Zhang W, Wu Z, Zhu X, Lu C. Functional analysis of luxS in Streptococcus suis reveals a key role in biofilm formation and virulence. Vet Microbiol. 2011;152:151–160. doi: 10.1016/j.vetmic.2011.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Zhang B, Ku X, Zhang X, Zhang Y, Chen G, Chen F, Zeng W, Li J, Zhu L, He Q. The AI-2/luxS quorum sensing system affects the growth characteristics, biofilm formation, and virulence of Haemophilus parasuis. Front Cell Infect Microbiol. 2019;9:62. doi: 10.3389/fcimb.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreira RN, Dressaire C, Barahona S, Galego L, Kaever V, Jenal U, Arraiano CM. BolA is required for the accurate regulation of c-di-GMP, a central player in biofilm formation. mBio. 2017 doi: 10.1128/mBio.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. Semin Cell Dev Biol. 2015;46:91–103. doi: 10.1016/j.semcdb.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Deme JC, Johnson S, Vickery O, Aron A, Monkhouse H, Griffiths T, James RH, Berks BC, Coulton JW, Stansfeld PJ, Lea SM. Structures of the stator complex that drives rotation of the bacterial flagellum. Nat Microbiol. 2020;5:1553–1564. doi: 10.1038/s41564-020-0788-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saijo-Hamano Y, Uchida N, Namba K, Oosawa K. In vitro characterization of FlgB, FlgC, FlgF, FlgG, and FliE, flagellar basal body proteins of Salmonella. J Mol Biol. 2004;339:423–435. doi: 10.1016/j.jmb.2004.03.070. [DOI] [PubMed] [Google Scholar]
- 28.Newman DM, Barbieri NL, de Oliveira AL, Willis D, Nolan LK, Logue CM. Characterizing avian pathogenic Escherichia coli (APEC) from colibacillosis cases, 2018. PeerJ. 2021;9:e11025. doi: 10.7717/peerj.11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellata M, Dho-Moulin M, Dozois CM, Curtiss R, 3rd, Lehoux B, Fairbrother JM. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect Immun. 2003;71:494–503. doi: 10.1128/iai.71.1.494-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lillington J, Geibel S, Waksman G. Biogenesis and adhesion of type 1 and P pili. Biochim Biophys Acta. 2014;1840:2783–2793. doi: 10.1016/j.bbagen.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 31.Spauld CN, Schreiber HL, Zheng W, Dodson KW, Hazen JE, Conover MS, Wang F, SvenmarkerLuna-RicoFrancetic PAO, Andersson M, Hultgren S, Egelman EH. Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions. Elife. 2018;7:e31662. doi: 10.7554/eLife.31662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sevrin G, Massier S, Chassaing B, Agus A, Delmas J, Denizot J, Billard E, Barnich N. Adaptation of adherent-invasive E. coli to gut environment: impact on flagellum expression and bacterial colonization ability. Gut Microbes. 2020;11:364–380. doi: 10.1080/19490976.2017.1421886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shippy DC, Eakley NM, Mikheil DM, Fadl AA. Role of the flagellar basal-body protein, FlgC, in the binding of Salmonella enterica serovar Enteritidis to host cells. Curr Microbiol. 2014;68:621–628. doi: 10.1007/s00284-014-0521-z. [DOI] [PubMed] [Google Scholar]
- 34.Ruan P, Wang XY, Sun AH, Li SJ, Yan J. Association of fliR gene in Leptospira interrogans with adhesion and pathogenicity to host cells. Zhejiang Da Xue Xue Bao Yi Xue Ban. 2008;37:572–578. doi: 10.3785/j.issn.1008-9292.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Chen KM, Chiang MK, Wang M, Ho HC, Lu MC, Lai YC. The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb Pathog. 2014;77:89–99. doi: 10.1016/j.micpath.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Smith JL, Fratamico PM, Yan X. Eavesdropping by bacteria: the role of SdiA in Escherichia coli and Salmonella enterica serovar Typhimurium quorum sensing. Foodborne Pathog Dis. 2011;8:169–178. doi: 10.1089/fpd.2010.0651. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y, Wang Y, Zhu H, Yi Z, Afayibo DJA, Tao C, Li T, Tian M, Qi J, Ding C, Yu S, Wang S. DctR contributes to the virulence of avian pathogenic Escherichia coli through regulation of type III secretion system 2 expression. Vet Res. 2021;52:101. doi: 10.1186/s13567-021-00970-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Primers used in this study.
Additional file 2: Bacterial micromorphology of APEC40, APEC40-ΔyqeH and APEC40-CΔyqeH observed by transmission electron microscopy.
Additional file 3: Differentially expressed genes data.
Additional file 4: Expressions of type 1 fimbriae genes of the wild-type strain APEC40, mutant strain APEC40-ΔyqeH and complementary strain APEC40-CΔyqeH were tested by RT-qPCR.
Data Availability Statement
The sequence was deposited in the GenBank database (Accession number PRJNA805014).