Abstract
BACKGROUND:
Although pancreatic ductal adenocarcinoma (PDAC) has one of the lowest 5-year survival rates of all cancers, differences in survival exist between patients with clinically identical characteristics. The authors previously demonstrated that keratin 17 (K17) expression in PDAC, measured by RNA sequencing or immunohistochemistry (IHC), is an independent negative prognostic biomarker. Only 20% of cases are candidates for surgical resection, but most patients are diagnosed by needle aspiration biopsy (NAB). The aims of this study were to determine whether there was a correlation in K17 scores detected in matched NABs and surgical resection tissue sections and whether K17 IHC in NAB cell block specimens could be used as a negative prognostic biomarker in PDAC.
METHODS:
K17 IHC was performed for a cohort of 70 patients who had matched NAB cell block and surgical resection samples to analyze the correlation of K17 expression levels. K17 IHC was also performed in cell blocks from discovery and validation cohorts. Kaplan-Meier and Cox proportional hazards regression models were analyzed to determine survival differences in cases with different levels of K17 IHC expression.
RESULTS:
K17 IHC expression correlated in matched NABs and resection tissues. NAB samples were classified as high for K17 when ≥80% of tumor cells showed strong (2+) staining. High-K17 cases, including stage-matched cases, had shorter survival.
CONCLUSIONS:
K17 has been identified as a robust and independent prognostic biomarker that stratifies clinical outcomes for cases that are diagnosed by NAB. Testing for K17 also has the potential to inform clinical decisions for optimization of chemotherapeutic interventions.
Keywords: immunohistochemistry, keratin 17, needle aspiration biopsies, pancreatic ductal adenocarcinoma, prognostic biomarker
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC), the most common type of pancreatic cancer, has a 5-year survival rate of only 9%.1 Despite the poor overall outcome, studies have found that clinically and pathologically indistinguishable tumors have differences in disease progression, responses to chemotherapy, and survival.2,3 These observations suggest that prognostic and predictive biomarkers are urgently needed to improve the clinical management of PDAC and identify patient populations that will or will not benefit from a specific therapy in order to prevent treatment-related toxicity and improve life expectancy.
We have previously reported that in resected cases, keratin 17 (K17) expression, based on messenger RNA or immunohistochemical analyses, is as accurate as molecular subtyping, and it can serve as an independent negative prognostic biomarker that identifies subgroups of patients with advanced stage and negative surgical margin PDACs who have the shortest survival.4 In addition, we have demonstrated that K17 testing by immunohistochemistry (IHC) outperforms molecular subtyping based on an established 50-gene signature. Although these studies suggest that K17 is a prognostic biomarker with the potential to improve PDAC patient clinical management, these results are based on assays performed on surgical resection specimens. Because the vast majority of patients (80%) are not candidates for tumor resection5 and treatment decisions are made as early as the time of diagnosis, there is an urgent need to develop a method to assess the K17 status in samples taken at the time of diagnosis.
Patients with advanced-stage disease at the time of diagnosis who have locally advanced disease, major blood vessel invasion, or severe underlying medical conditions are not candidates for major surgical intervention.6 Endoscopic ultrasound–guided needle aspiration biopsy (NAB), however, can be performed even on PDACs that cannot be surgically resected and thus can provide an additional resource that can be used for multiple assessments, including molecular and immunohistochemical studies, in order to provide clinically actionable information that can help to guide treatment decisions.
The aims of our study were to compare K17 IHC expression in surgical resections and their matched NAB specimens and to demonstrate the prognostic value of K17 in NAB samples taken at the time of diagnosis. Our findings could affect chemotherapeutic interventions for all patients with PDAC, including those who are not candidates for surgical resection.
MATERIALS AND METHODS
Case Selection
Cell blocks from consecutive PDAC NABs from 2012 to 2017 were retrospectively selected from the archival collections of the Departments of Pathology at Stony Brook Medicine (n = 177) and the University of Massachusetts (n = 34).
All cytology samples obtained at the University of Massachusetts were collected in BD CytoRich Red fixative (an ethanol-based fixative; Becton, Dickinson and Company, Franklin Lakes, New Jersey). Rapid cell blocks were made with the Hologic Cellient automated cell block system (Hologic, Mississauga, Ontario, Canada). NAB rinse samples obtained at Stony Brook Medicine were fixed immediately in formalin and spun down at 1800 rpm for 10 minutes. The supernatant was discarded, and the pellet was placed in histologic tissue paper before paraffin embedding for cell block preparation.
Only cases that met the following criteria were included: a diagnosis of primary PDAC and cellularity of more than 100 viable tumor cells in the cell block section. Patients younger than 18 years at the time of diagnosis were excluded because PDAC rarely, if ever, occurs in that age group.
Out of the 211 patients, 70 had undergone surgical resection and were used to compare IHC scores between the 2 types of samples. SAS 9.4 software (SAS Institute, Cary, North Carolina) was used to randomize the cases into a discovery cohort (n = 106) or a validation cohort (n = 105) as reported in Table 1. Demographic and survival data were obtained from the respective institution’s registry (Table 1). All studies were performed in accordance with the guidelines and regulations of Stony Brook Medicine institutional review board protocol 94651. Patient written consent was waived by the institutional review board because the research was restricted to the analysis of de-identified, remnant, archived surgical pathology specimens.
Table 1.
Patient cohort demographics for the discovery and validation cohorts.
| Clinical Variables | Patient Cohorts | P-value | |
|---|---|---|---|
| Discovery | Validation | ||
| n=106 | n=105 | ||
| Age at diagnosis, mean ± SD | 68 ± 18.6 | 70 ± 11.3 | |
| Gender, number cases (%) | n=106 | n=105 | 0.0607 |
| Female | 37 (35) | 50 (48) | |
| Male | 69 (65) | 55 (52) | |
| Clinical Stage, number cases (%) | n=93 | n=88 | 0.9815 |
| I-IIA | 40 (43) | 38 (43) | |
| IIB-IV | 53 (57) | 50 (57) | |
| K17 status, number cases (%) | n=106 | n=105 | 0.5986 |
| Low K17 | 67 (63) | 70 (67) | |
| High K17 | 39 (37) | 35 (33) | |
Abbreviation: K17, keratin 17.
P-values demonstrate no differences between discovery and validation cohorts for each clinical variable.
Patients were stratified on the basis of the clinical staging, as reported by the Stony Brook Cancer Center Registry, in all cases (with or without surgical treatment) and in nonresected cases only. The clinical stage category was defined as early stage (IA-IIA) or advanced stage (IIB-IV). All analyses were performed in accordance with these criteria.
K17 Immunohistochemistry
Cell block slide sections from each case were reviewed to select those with more than 100 viable cells. An indirect immunoperoxidase method was used to identify the presence of K17 protein as previously described.6 Briefly, after incubation at 60 °C, slices were deparaffinized in xylene and rehydrated in alcohols. Antigen retrieval was performed in a citrate buffer at 120 °C for 10 minutes in a decloaking chamber. Endogenous peroxidase was blocked by 3% hydrogen peroxide, and sections were incubated overnight at 4 °C with a mouse monoclonal anti-human K17 antibody (lot A8034124; KDx, Campbell, California). After the primary antibody, biotinylated horse secondary antibodies (R.T.U. Vectastain ABC Kit; Vector Laboratories, Burlingame, California) were added. Development was performed with 3,3′-diaminobenzidine (Dako, Carpinteria, California), and counterstaining was performed with hematoxylin. Negative controls were performed on all runs with an equivalent concentration of a subclass-matched immunoglobulin.
K17 is an intermediate filament found mostly in the cytoplasm; however, we and others have reported that this protein can also be localized to the nucleus.7 The K17 staining intensity was evaluated by 2 pathologists (K.R.S and L.R.P) using our previously validated pathologic semiquantitative scoring (the PathSQ score).4,8–11 On the basis of a subjective assessment of absent (0), light (1+), or strong (2+) staining, cases were scored by consideration of the overall percentage of tumor cells with 2+ intensity staining. Results were presented as a score that reflected the percentage of strongly (2+) stained malignant cells. The PathSQ score was determined by the review of a representative immunostained cell block section from each case, with the reviewer blinded to corresponding clinical data. Discrepancies between pathologists were resolved by consensus after a joint review.
Statistical Analyses
Statistical significance between 2 categorical variables was analyzed with the χ2 test. Associations between the means of 2 groups were determined with unpaired 2-tailed Student t tests or Mann-Whitney U tests.
K17 expression was defined as a categorical variable based on the minimum Akaike information criterion with a Cox proportional hazards regression model for all possible K17 high/low cutoff points with 2+ intensity. The best and most reliable cutoff point for stratifying the cases into low and high K17 expression was identified as a PathSQ score ≥ 80. Overall survival was determined with the Kaplan-Meier method and was calculated from the date of diagnosis to the date of death. Patients still alive at the last follow-up were censored.
Univariate analyses compared survival by K17 level (low vs high), age, sex, and clinical stage (stages I-IIA vs stages IIB-IV). Adjusted for potential confounders, a multivariate analysis was performed by Cox proportional hazards regression. Statistical significance was set at a P value ≤ .05, and the analysis was performed with SAS 9.4 software and GraphPad Prism 7 (Graph Pad Software, La Jolla, California).
RESULTS
K17 Expression Detected by IHC Is Similar in NAB and Surgically Matched Resection Tissue
On the basis of our previous finding that K17 was a negative prognostic biomarker when measured at the protein level in surgical resection specimens, we set out to determine whether the level of expression was concordant in matched NABs and resected tissues. We performed K17 IHC in a cohort of 70 patient who had both NAB and surgical resection tissue samples.
To determine the level of K17 expression by IHC in cell blocks from NABs and resected PDACs, we used our previously validated semiquantitative IHC scoring method, the PathSQ score, where the percentage of strongly positively stained tumoral cells (2+) on a scale of 0% to 100% was determined within a single representative section from each case (Supporting Fig. 1).
The K17 PathSQ score in NAB significantly correlated with the K17 PathSQ score in matched resection specimens with an R value of 0.4223 (0.09884–0.665) and a P value of .0103. The mean K17 PathSQ scores in resected specimens and corresponding NABs were 65% and 57.5%, respectively (P = .962; Fig. 1). These results demonstrate that K17 testing of NAB is as accurate as performing the assay on surgical resection specimens.
Figure 1. Keratin 17 expression by IHC correlates in matched surgical resection and cell-block tissues.
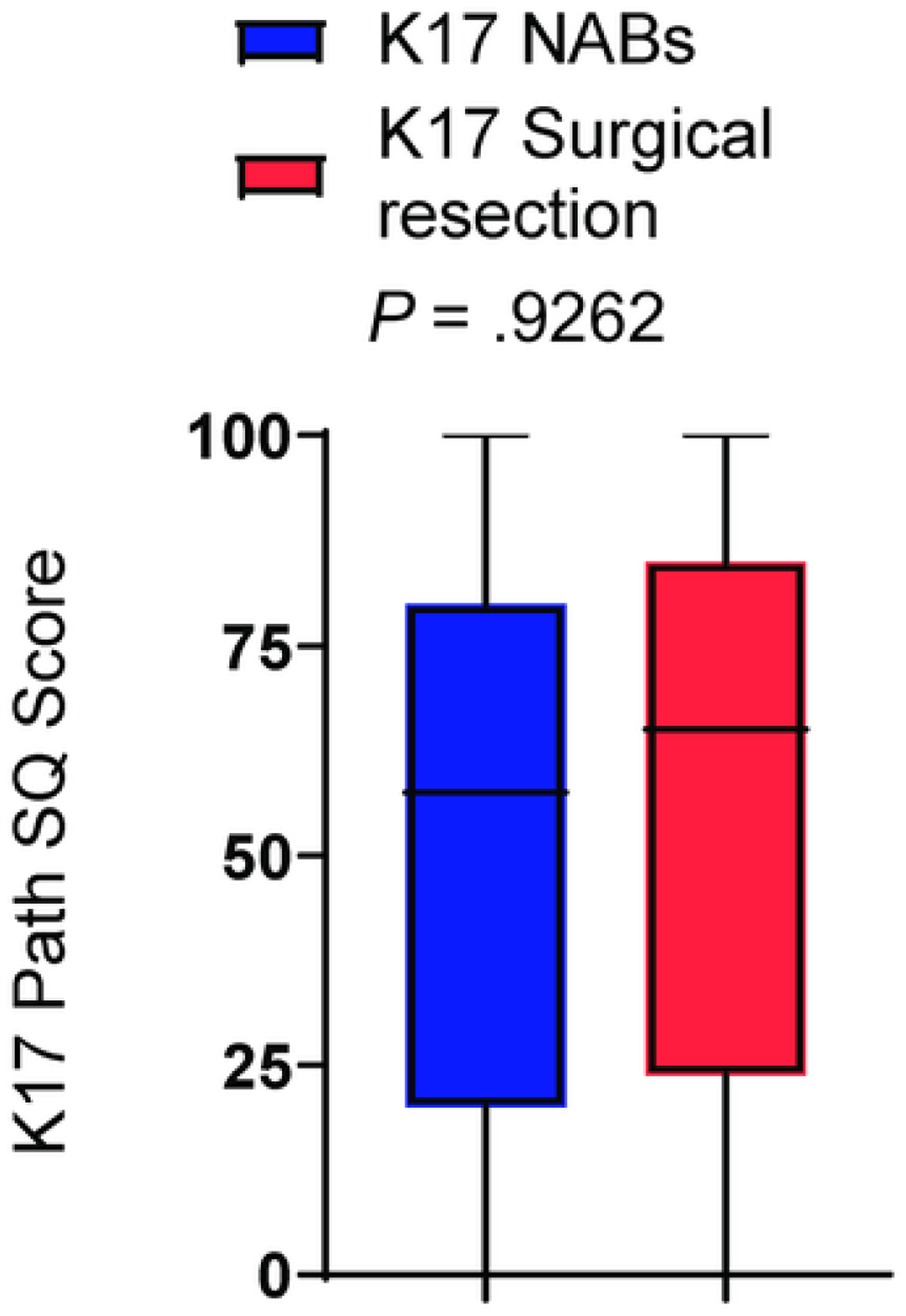
Graph illustrating K17 IHC PathSQ in NAB and matched resection tissues. P-value was calculated using the Mann Whitney test.
K17 IHC Testing in PDAC NAB Predicts Patient Outcomes
K17 IHC was evaluated in cell block samples from NABs of 211 patients at the time of diagnosis from discovery and validation cohorts (Table 1).
K17 IHC was performed in a discovery cohort consisting of 106 cell block samples from PDAC NABs. The K17 IHC level of expression was unrelated to the morphologic features of the tumors (Fig. 2A–D). The mean PathSQ score for K17 was 67.5%. Samples were classified on the basis of the maximum likelihood fit of a Cox proportional hazards model to identify the best threshold for stratifying low- or high-K17 case survival probability. The best cutoff point for stratifying cases was identified as a PathSQ score ≥ 80. High-K17 cases were classified as having a score of ≥80%, which represented 37% of cases (n = 39), whereas low-K17 cases had a score of <80% and accounted for 63% of cases (n = 67; Fig. 2E). The mean K17 PathSQ score was 30% for cases categorized as low-K17 cases and 95% for the group of high-K17 cases (Supporting Fig. 2A).
Figure 2.
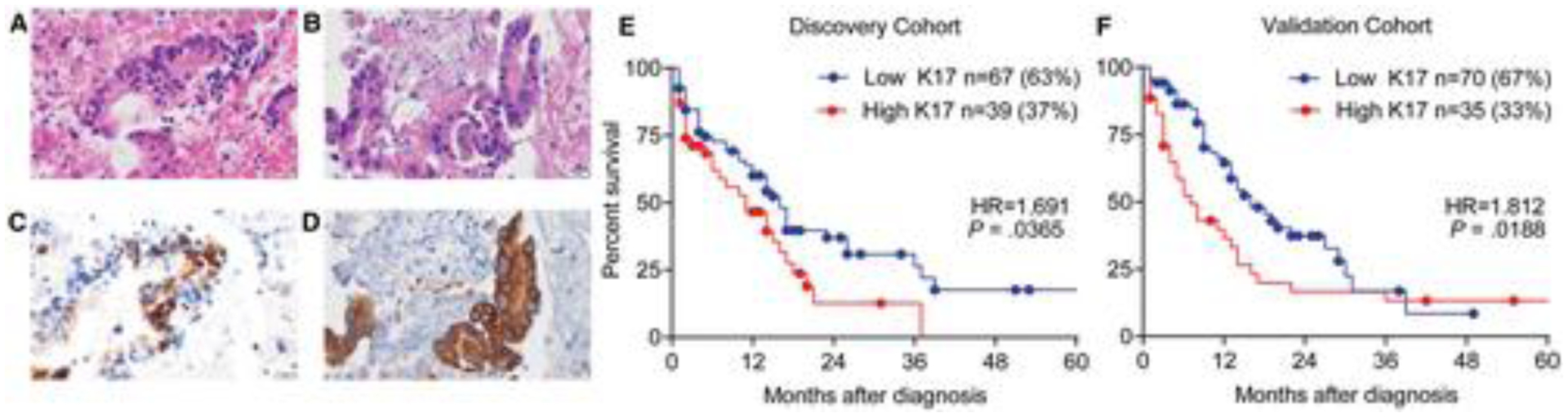
PDAC cases with high K17 protein expression have shortened survival. (A-D) Representative images from 2 PDAC NAB samples with similar morphologic features: (A,B) H & E-stained sections and (C,D) corresponding sections processed for immunohistochemistry showing low and high K17 expression, respectively (scale bar = 20 µm). (E,F) Kaplan-Meier curves for the overall survival analysis of K17 from PDAC NAB in the discovery and validation cohorts, respectively. The P values were calculated with the log-rank test. HRs and P values are shown. HR indicates hazard ratio; K17, keratin 17; NAB, needle aspiration biopsy; PDAC, pancreatic ductal adenocarcinoma.
To determine whether K17 is a prognostic biomarker, we performed a Kaplan-Meier and Cox proportional hazards regression model of survival. We found that patients with high-K17 PDACs had a median survival of 11 months, whereas patients with low-K17 PDACs had a median survival of 16 months (hazard ratio [HR], 1.7; P = .0365; Fig. 2E).
To evaluate whether the K17 status is independent of other clinicopathologic features, univariate and multivariate analyses using Cox proportional hazards regression of individual risk factors were performed. The univariate analysis (Fig. 3A) and the multivariate analysis (Fig. 3B) showed that K17 was an independent negative prognostic biomarker in the discovery cohort.
Figure 3. Keratin 17 is an independent negative prognostic biomarker in PDAC NAB.
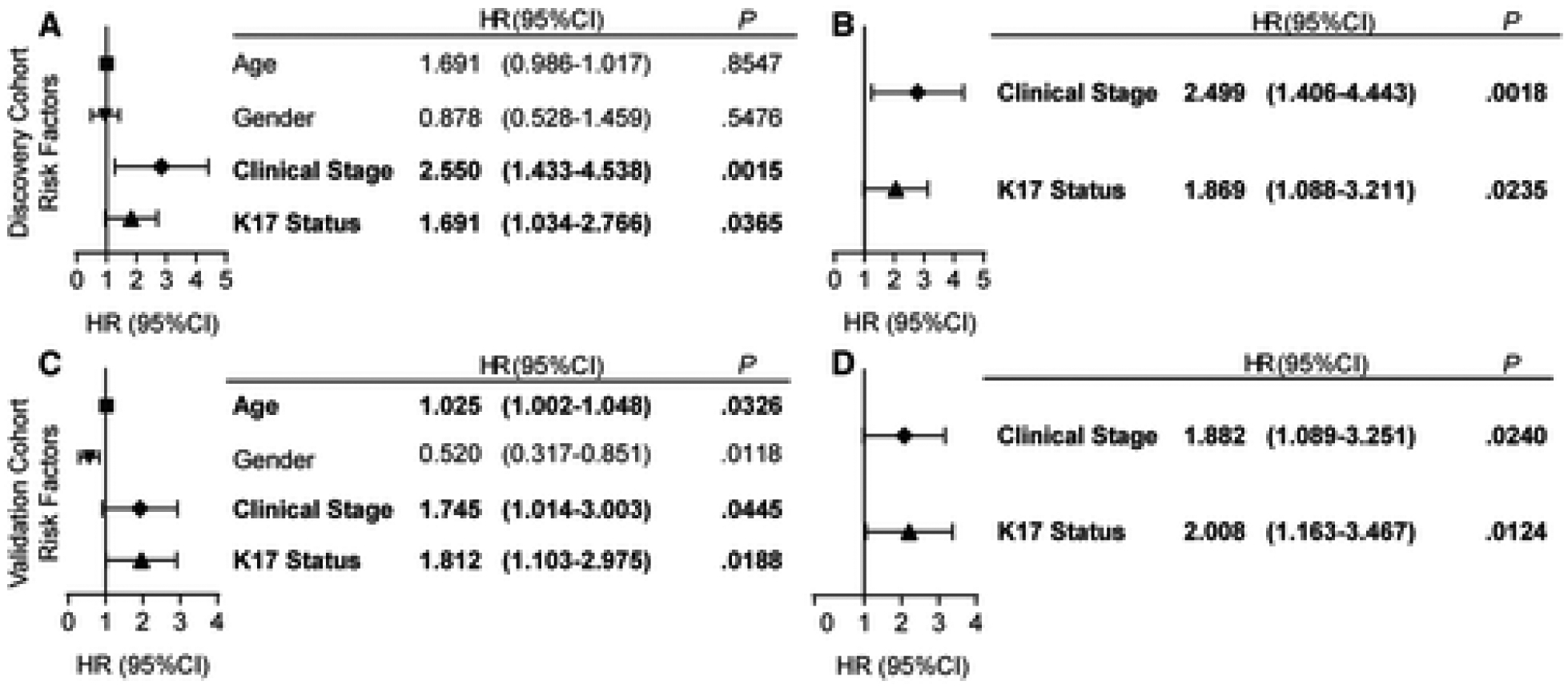
Forest plots showing the (A and C) univariate and (B and D) multivariate analyses using Cox proportional hazards regression for K17 as a binary variable and other PDAC risk factors in the discovery (A and B) and validation (C and D) cohorts. Hazard ratios (HR), confidence intervals (CI) and p-values are shown.
To corroborate the findings from the discovery cohort, we used a validation cohort of 105 PDAC NAB cases. K17 IHC was performed, and the same cutoff value used in the discovery cohort was applied to the validation cohort to stratify samples on the basis of the PathSQ score. The mean PathSQ score was 70%, and again, the level of expression was not related to the histologic characteristics of the tumor. Thirty-three percent of the cases (n = 35) were classified as low K17, whereas 67% (n = 70) were high K17 (Fig. 2F). The mean expression for K17 IHC was 40% for low K17 and 95% for high K17 in the validation cohort (Supporting Fig. 2B).
To validate the accuracy of K17 IHC as a negative prognostic biomarker in PDAC NAB, we applied the same maximum likelihood fit of a Cox proportional hazards model threshold to the validation cohort. We found that high levels of expression of K17 were again a negative prognostic biomarker, with a median survival of 7 months for high-K17 cases and a median survival of 16 months for low-K17 cases (HR, 1.8; P = .0188; Fig. 2F). The univariate analysis (Fig. 3C) and the multivariate analysis (Fig. 3D) showed that K17 was also an independent negative prognostic biomarker in the validation cohort. In summary, results from the discovery and validation cohorts show that K17 in NAB is an independent prognostic biomarker for a combined group of PDAC patients with a resectable or unresectable disease status.
The analysis of the combined discovery and validation cohorts (n = 211) showed that 35% of the cases (n = 74) were classified as low K17, whereas 65% (n = 137) were high K17. The mean PathSQ score was 40% for low K17 and 95% for high K17 (Supporting Fig. 2C). The Kaplan-Meier and Cox proportional hazards regression model analyses showed that K17 was a negative prognostic biomarker with a median survival of 7 months for high K17 and 16 months for low K17 (HR, 1.7; P = .0017; Supporting Fig. 3A).
To determine whether K17 is also prognostic in patients who are not candidates for surgical resection, we analyzed this population from the combined discovery and validation cohorts as 1 group. The Kaplan-Meier and Cox proportional hazards regression model showed a longer median survival of 11 months for low K17 in comparison with the 6-month median survival for high K17 (HR, 1.5; P = .0676; Supporting Fig. 3B). The mean PathSQ score of the combined cohort was 40% for low K17 and 95% for high K17 (Supporting Fig. 2D).
K17 Provides Additional Prognostic Information in Cases at an Advanced Clinical Stage
To establish whether K17 provides additional prognostic information independently of the tumor stage, the interactions between K17 and clinical stage were analyzed in the combined discovery and validation cohorts. Clinical information on stage was available for 86% of the cases (stages I-IIA, n = 78; stages IIB-IV, n = 103). Data on tumor stage were not available for patients who received treatment outside the institution where the original diagnosis had been established.
The Kaplan-Meier and Cox proportional hazards regression model showed that K17 stratified survival differences within patients with advanced disease (Fig. 4B) but did not correlate with localized disease (Fig. 4A). The median survival at an advanced stage was 14 months for low-K17 PDACs and only 5 months for high-K17 PDACs (HR, 1.9; P = .0061).
Figure 4. Keratin 17 provides additional prognostic information in advanced clinical stage.
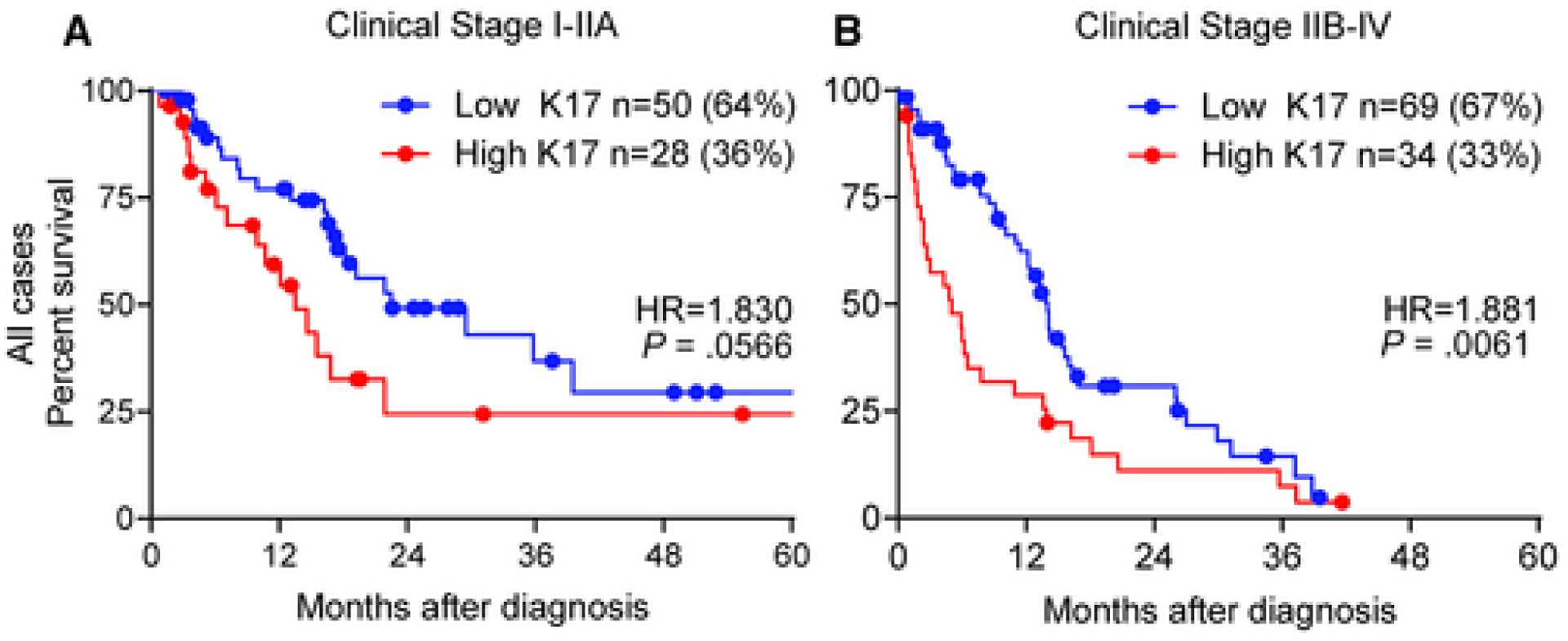
Kaplan–Meier curves depicting the overall survival integrating K17 status and clinical stage for the combined discovery and validation cohorts (A: clinical stage I-IIA, B: clinical stage IIB-IV). P-values were calculated using the log-rank test. Hazard ratios (HR) and p-values are shown.
Given that approximately 50% of patients with PDAC die within the first year after diagnosis and our prior finding that K17 identifies the most aggressive form of PDAC, we performed Kaplan-Meier and Cox proportional hazards regression model analyses to correlate survival with the K17 status at localized (Fig. 5A) or advanced clinical stages (Fig. 5B) only for cases that died within 12 months of the diagnosis (n = 95). The median survival during the first year at an advanced clinical stage for all cases was 8 months with low-K17 PDACs and only 3 months with high-K17 PDACs (HR, 2.5; P = .0011). The HR was higher for the first year of survival than for 5-year survival with an advanced clinical stage.
Figure 5. High Keratin 17 determines shorter survival during the first year in advanced clinical stage cases.
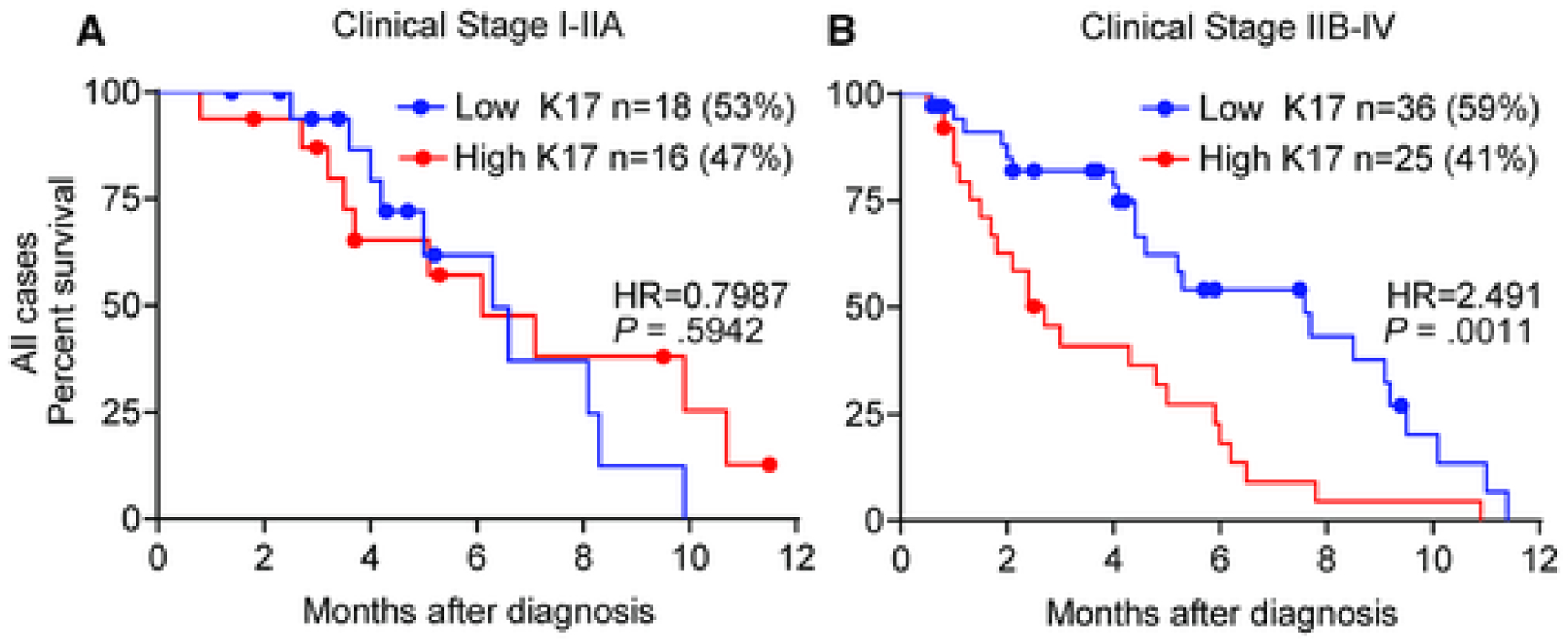
Kaplan–Meier curves depicting the overall survival for the first year integrating K17 status and clinical stage for all cases (combined discovery and validation cohorts) (A: clinical stage I-IIA, B: clinical stage IIB-IV). Hazard ratios (HR) and p-values are shown.
These findings demonstrate that K17 provides prognostic information for cases at an advanced clinical stage, especially during the first year after diagnosis.
DISCUSSION
In our prior study, we showed that a high level of K17 determined by IHC in surgical resection tissues was a negative prognostic biomarker that also identified the subgroup of patients with an advanced disease stage and a negative margin status who had the shortest survival.4 The potential value of K17 as a prognostic biomarker was initially limited to cases that were candidates for surgical excision. The current study, however, supports the conclusion that K17 IHC can also be used to predict the survival of PDAC patients with unresectable disease. Although the 5-year relative survival rate for pancreatic cancer is only 9%,1 we have found that patient survival during the first year is dramatically reduced in cases with high K17. By testing K17 at the time of diagnosis, we can identify patients at higher risk of mortality in order to intervene at an earlier time.
NAB has been used not only to improve the diagnostic efficacy12 but also to evaluate molecular modifications in DNA, RNA, and protein as prognostic and predictive biomarkers.13–16 The major advantage of using NAB for these tests is that a cytologic diagnosis can be provided even for patients with nonresectable tumors to inform therapeutic interventions.
We have demonstrated that K17 IHC scores in resected tissues generally correlate with those of their matched NABs. Therefore, K17 testing of NAB is as accurate as performing the assay on surgical resection specimens. A possible limitation in the use of NAB to determine the expression of a protein by IHC is that an NAB sample represents a very small fragment of the tumoral tissue, and the level of expression of this protein cannot represent accurately the protein expression in a tumor resection. Previous studies, however, have shown that the expression of different proteins evaluated by IHC corresponds between surgical resection specimens and NAB samples.16–18 Given that relatively few cases had both NAB and resected PDACs, however, we were not able to statistically address the minimal threshold NAB tumor cellularity. Thus, our threshold of at least 100 viable cells in the sample to be included in the study was empirically determined but has not yet been rigorously defined.
K17 in PDAC NAB from the discovery, validation, and combined cohorts was a robust, clinically relevant, independent prognostic biomarker that stratified clinical outcomes. As in our previous publication, in the combined cohort, K17 independently identified a subset of patients at an advanced stage with the shortest survival. Although there was a trend, K17 was not significant in the group of patients who did not undergo surgery. Our clinical data were incomplete for patients who were treated at institutions different from the health care institution at which the diagnosis was first established. Thus, obtaining follow-up information on the treatment and outcome of the patients limited our analysis of clinical outcome correlation in 14% of cases.
Although we have not yet used K17 IHC in a diagnostic setting, the development of this test in NAB and tissue resection as a clinically deployable prognostic biomarker is supported by our previous observation that K17 detection by IHC is superior to messenger RNA–based tests, which may be affected by the variability of cellularity in surgically resected whole-tumor specimens.4 K17 IHC in surgical resections and NAB enables a precise evaluation of tumor cells. Furthermore, K17 IHC could be readily incorporated into the workflow of pathology diagnostic laboratories, and test results can be scored with the same general approaches that surgical pathologists use in routine diagnostic practices.
Our laboratory is currently attempting to determine how testing the K17 status could serve as a predictive biomarker for currently available therapeutic agents for the most aggressive forms of PDAC. In a recent study,19 we demonstrated that K17 expression correlates with the resistance of PDAC to gemcitabine, and using both in vitro and in vivo models with human and murine PDAC cells and orthotopic xenografts, we found that K17 expression drives increased resistance to gemcitabine and 5-fluorouracil, agents that are currently included in first-line chemotherapy for PDAC. Thus, determining the K17 status at the time of diagnosis could one day help to optimize therapeutic efficacy and to minimize the side effects attributable to agents that are unlikely to provide a survival advantage.
In summary, our study has demonstrated that K17 IHC of NAB enables prognostic stratification of PDAC patients. Thus, K17 IHC testing in NAB should be further validated and tested in clinical trials to determine whether it also has predictive value that could affect clinical opportunities for chemotherapeutic intervention.
Supplementary Material
FUNDING SUPPORT
This work was supported by a Pancreatic Cancer Action Network Translational Research Grant (grant 18-65-SHRO to Kenneth R. Shroyer), the National Cancer Institute (K99-R00 CA226342-01 to Luisa F. Escobar-Hoyos), the Hirshberg Foundation (to Luisa F. Escobar-Hoyos), the Damon Runyon Foundation (Innovator Award to Luisa F. Escobar-Hoyos), and the National Pancreas Foundation (grant to Luisa F. Escobar-Hoyos). Cindy V. Leiton was partly supported by a postdoctoral fellowship awarded by the National Science Foundation’s workforce development program, the Alliance for Graduate Education in the Professoriate (transformation grant HRD-1311318), through the Stony Brook University Center for Inclusive Education (Stony Brook, New York).
We thank the Stony Brook Cancer Center Biorepository for access to tissue specimens (Dr. R. Kew) and the cancer registries of the UMass Memorial Medical Center and Stony Brook University (X. Barzilay).
Footnotes
Additional supporting information may be found in the online version of this article.
CONFLICT OF INTEREST DISCLOSURES
Cindy V. Leiton reports other from the Department of Therapeutic Radiology of Yale University outside the submitted work. Jonathan Buscaglia reports personal fees from AbbVie outside the submitted work. Luisa F. Escobar-Hoyos and Kenneth R. Shroyer report a pending provisional patent application; they are also consultants for KDx Diagnostics, Inc. The other authors made no disclosures.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551 [DOI] [PubMed] [Google Scholar]
- 2.Martens S, Lefesvre P, Nicolle R, et al. Different shades of pancreatic ductal adenocarcinoma, different paths towards precision therapeutic applications. Ann Oncol. 2019;30:1428–1436. doi: 10.1093/annonc/mdz181 [DOI] [PubMed] [Google Scholar]
- 3.Kamarajah SK. Validation of the American Joint Commission on Cancer (AJCC) 8th edition staging system for patients with pancreatic adenocarcinoma: a Surveillance, Epidemiology and End Results (SEER) analysis. Ann Surg Oncol. 2017;24:2023–2030. doi: 10.1245/s10434-017-5810-x [DOI] [PubMed] [Google Scholar]
- 4.Roa-Peña L, Leiton CV, Babu S, et al. Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer. Sci Rep. 2019;9:11239. doi: 10.1038/s41598-019-47519-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kommalapati A, Tella SH, Goyal G, Ma WW, Mahipal A. Contemporary management of localized resectable pancreatic cancer. Cancers (Basel). 2018;10:24. doi: 10.3390/cancers10010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soweid AM. The borderline resectable and locally advanced pancreatic ductal adenocarcinoma: definition. Endosc Ultrasound. 2017;6(suppl 3):S76–S78. doi: 10.4103/eus.eus_66_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Escobar-Hoyos LF, Shah R, Roa-Peña L, et al. Keratin-17 promotes p27KIP1 nuclear export and degradation and offers potential prognostic utility. Cancer Res. 2015;75:3650–3662. doi: 10.1158/0008-5472.CAN-15-0293 [DOI] [PubMed] [Google Scholar]
- 8.Escobar-Hoyos LF, Yang J, Zhu J, et al. Keratin 17 in premalignant and malignant squamous lesions of the cervix: proteomic discovery and immunohistochemical validation as a diagnostic and prognostic biomarker. Mod Pathol. 2014;27:621–630. doi: 10.1038/modpathol.2013.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merkin RD, Vanner EA, Romeiser JL, et al. Keratin 17 is overexpressed and predicts poor survival in estrogen receptor–negative/human epidermal growth factor receptor-2–negative breast cancer. Hum Pathol. 2017;62:23–32. doi: 10.1016/j.humpath.2016.10.006 [DOI] [PubMed] [Google Scholar]
- 10.Bai JDK, Babu S, Roa-Peña L, et al. Keratin 17 is a negative prognostic biomarker in high-grade endometrial carcinomas. Hum Pathol. 2019;94:40–50. doi: 10.1016/j.humpath.2019.09.005 [DOI] [PubMed] [Google Scholar]
- 11.Babu S, Mockler DC, Roa-Peña L, et al. Keratin 17 is a sensitive and specific biomarker of urothelial neoplasia. Mod Pathol. 2019;32:717–724. doi: 10.1038/s41379-018-0177-5 [DOI] [PubMed] [Google Scholar]
- 12.Nigam N, Rastogi A, Bhatia V, Sureka B, Jain P, Bihari C. EUS-guided FNA in diagnosing pancreatic lesions: strength and cytological spectrum. J Cytol. 2019;36:189–195. doi: 10.4103/joc.Joc_5_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada R, Mizuno S, Uchida K, et al. Human equilibrative nucleoside transporter 1 expression in endoscopic ultrasonography–guided fine-needle aspiration biopsy samples is a strong predictor of clinical response and survival in the patients with pancreatic ductal adenocarcinoma undergoing gemcitabine-based chemoradiotherapy. Pancreas. 2016;45:761–771. doi: 10.1097/mpa.0000000000000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elhanafi S, Mahmud N, Vergara N, et al. Comparison of endoscopic ultrasound tissue acquisition methods for genomic analysis of pancreatic cancer. J Gastroenterol Hepatol. 2019;34:907–913. doi: 10.1111/jgh.14540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishiwada S, Sho M, Cui Y, et al. A gene expression signature for predicting response to neoadjuvant chemoradiotherapy in pancreatic ductal adenocarcinoma. Int J Cancer. 2021;148:769–779. doi: 10.1002/ijc.33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C, Wallace MB, Yang J, et al. ZIP4 is a novel diagnostic and prognostic marker in human pancreatic cancer: a systemic comparison between EUS-FNA and surgical specimens. Curr Mol Med. 2014;14:309–315. doi: 10.2174/1566524013666131217112921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senoo J, Mikata R, Kishimoto T, et al. Immunohistochemical analysis of IMP3 and p53 expression in endoscopic ultrasound–guided fine needle aspiration and resected specimens of pancreatic diseases. Pancreatology. 2018;18:176–183. doi: 10.1016/j.pan.2017.12.010 [DOI] [PubMed] [Google Scholar]
- 18.Hendry S, Byrne DJ, Christie M, et al. Adequate tumour cellularity is essential for accurate PD-L1 immunohistochemistry assessment on cytology cell-block specimens. Cytopathology. 2020;31:90–95. doi: 10.1111/cyt.12795 [DOI] [PubMed] [Google Scholar]
- 19.Pan CH, Otsuka Y, Sridharan B, et al. An unbiased high-throughput drug screen reveals a potential therapeutic vulnerability in the most lethal molecular subtype of pancreatic cancer. Mol Oncol. 2020;14:1800–1816. doi: 10.1002/1878-0261.12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


