Abstract
The Parkinson Progression Marker Initiative (PPMI) is a comprehensive observational, international, multicenter study designed to identify PD progression biomarkers both to improve understanding of disease etiology and course and to provide crucial tools to enhance the likelihood of success of PD modifying therapeutic trials. The PPMI cohort will comprise 400 recently diagnosed PD and 200 healthy subjects followed longitudinally for clinical, imaging and biospecimen biomarker assessment using standardized data acquisition protocols at twenty-one clinical sites. All study data will be integrated in the PPMI study database and will be rapidly and publically available through the PPMI web site- www.ppmi-info.org. Biological samples including longitudinal collection of blood, cerebrospinal fluid (CSF) and urine will be available to scientists by application to an independent PPMI biospecimen review committee also through the PPMI web site. PPMI will rely on a partnership of government, PD foundations, industry and academics working cooperatively. This approach is crucial to enhance the potential for success of this ambitious strategy to develop PD progression biomarkers that will accelerate research in disease modifying therapeutics.
Keywords: biomarker, Dopamine transporter imaging, Diffusion tensor imaging, cerebrospinal fluid, alpha synuclein
Reliable and well-validated biomarkers for Parkinson Disease (PD) progression would dramatically accelerate research into both PD etiology and therapeutics. During the past two decades much progress has been made in identifying and assessing PD biomarkers, but as yet no fully validated biomarker for PD is currently available (Maetzler et al., 2009; Marek et al., 2008; Schapira, 2004; Olanow et al., 2008; Gerlach et al., 2008; Shi et al., 2009; Scherzer et al., 2007; Ravina et al., 2009). Given the recent advances in molecular genetics, neurobiology, and imaging technology and the recognition that the lack of PD biomarkers has created a roadblock for further studies of disease modifying therapies, a major initiative to develop PD biomarkers is both timely and necessary. Therefore, the Parkinson Progression Marker Initiative (PPMI), a collaborative effort of PD researchers with expertise in biomarker development, PD clinical study design and implementation, and bioinformatics, statistics, and data management, was developed to identify and validate PD progression markers (The Lancet, 2010). PPMI is a public–private partnership, largely sponsored by the Michael J Fox Foundation with substantial industry partnership (GE Healthcare, Pfizer, Roche, Genentech, Merck, Abbott, Biogen, Covance, Glaxo-Smith-Kline).
PPMI is a five-year observational, international, multi-center study designed to identify PD progression biomarkers both to improve understanding of disease etiology and course and to provide crucial tools to enhance the likelihood of success of PD disease modifying therapeutic trials. The specific goals of PPMI are to:
Establish standardized protocols for acquisition, transfer and analysis of clinical, imaging and biospecimen data that can be used by the PD research community.
Investigate existing and identify novel clinical, imaging and biospecimen PD progression markers that individually or in combination will rapidly demonstrate interval change in PD patients in comparison to healthy controls or in sub-sets of PD patients defined by baseline assessments, progression milestones and/or rate of clinical, imaging or biospecimen change.
Optimize bioassays and conduct preliminary verification studies on promising biological markers using stored biospecimen.
The PPMI study was planned during a two-year period in a series of workshops with input from academic PD experts, Michael J Fox Foundation scientific staff, and government and industry partners. The study was launched in June 2010 and enrollment is underway. The overall study design and key study features are detailed below.
1. PPMI study design
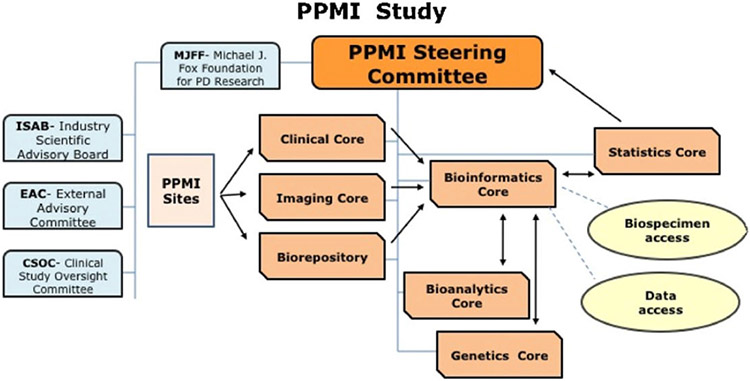
The PPMI cohort will comprise 400 recently diagnosed PD and 200 healthy subjects followed longitudinally and comprehensively for biomarker assessment using standardized data acquisition protocols at twenty-one clinical sites with expertise in subject recruitment and biomarker assessment. The PPMI steering committee directs the study through the clinical, imaging, genetics, bioanalytic, biorepository, statistics, and bioinformatics cores (Fig. 1). The steering committee includes PD biomarker experts, study core leaders, and MJFF and industry scientists. All study data will be integrated in the PPMI study database and will be rapidly and publically available through the PPMI web site – http://www.ppmi-info.org/. Biological samples including longitudinal collection of blood, cerebrospinal fluid (CSF) and urine will be stored at the PPMI biorepository and will be available to scientists by application to an independent PPMI biospecimen review committee through the PPMI web site. In addition the PPMI website will serve as a source of ongoing information about standardized study procedures, study progress, and study results.
Fig. 1.
PPMI study organization and governance.
1.1. Study cohort and subject selection criteria
PD subjects will be recruited at disease threshold. They will be required to have an asymmetric resting tremor or asymmetric bradykinesia or two of bradykinesia, resting tremor and rigidity with diagnosis within two years and to be untreated for PD. All subjects will undergo dopamine transporter (DAT) imaging and DAT deficit will be required for PD subject eligibility. Healthy subjects must have no significant neurologic dysfunction, no first degree family member with PD and Montreal Cognitive Assessment MoCA > 26. This first use of an imaging biomarker for eligibility in a PD biomarker study offers three major advantages. First, use of DAT imaging will enhance the accuracy of diagnosis given increasing evidence that subjects enrolled in PD clinical trials with scans without DAT deficit are unlikely to have PD (Parkinson Study Group, 2004, 2007; Whone et al., 2003). Second PD subjects can be enrolled in the study earlier in their disease course with only a single asymmetric sign. There is growing consensus that enrolling PD subjects at the earliest detectable stage of disease would likely enhance the potential to both identify a progression biomarker and provide a better population for eventual disease modifying drug trials. Third, enrolling PD subjects earlier would likely prolong the duration of biomarker evaluation prior to initiating symptomatic treatment. This strategy would allow more complete assessment and comparison of PD biomarkers prior to symptomatic medications, while still allowing assessment during the transition period when symptomatic medications are needed, and after longitudinal follow-up while on symptomatic medications. PD subjects enrolled in PPMI should not require symptomatic PD medications for at least six months (subjects will be treated and retained in the study if treatment is required before six months). PD subjects will be allowed to participate in clinical trials after 12 months participation in PPMI to encourage retention in PPMI.
1.2. Subject recruitment
Given the acknowledged difficulty in identifying early-untreated PD study subjects and healthy subjects compounded by the comprehensive longitudinal PPMI assessment requiring substantial subject time and commitment, it is anticipated that study recruitment will be a major challenge for PPMI. Within the PPMI study we have piloted several novel recruitment strategies. Working closely with the Michael J Fox Foundation we have developed a template of recruitment material and an ongoing series of focused local events at each site to introduce the study to both referral physicians and potential study candidates. PPMI will also be publicized more globally with an international publicity campaign to highlight the need for participation by both PD subjects and healthy subjects. An important strategy has been to engage referral physicians, a key source of early-untreated PD subjects, to assist with recruitment by fully educating them about the critical need for biomarkers. We anticipate that sites can recruit one early-untreated PD subject each month and one healthy subject every two months. We therefore expect recruitment to be completed within 24 months. Subject retention strategies are also a key to study success given that the goal of PPMI is to identify biomarkers of disease progression. We plan to provide information to subjects throughout the study through websites and newsletters and other outreach. We plan to establish a PPMI community (within the confines of confidentiality) for interested participants and will continue to work closely with the Michael J Fox Foundation and other public relations resources to promote study retention.
1.3. Site selection
Twenty-one clinical study sites (sixteen US and five European) with expertise and experience in PD research have been selected for the PPMI study. These clinical sites have undergone a two stage assessment by the PPMI steering committee beginning with a site questionnaire followed by an in person visit by a steering committee member to assess site resources and capabilities for PD and healthy subject recruitment and retention and to conduct all study clinical assessments, for collection of biospecimens and for acquisition of positron emission tomography (PET), single photon emission computerized tomography (SPECT) and MRI. All sites have undergone and will undergo ongoing training to ensure standardization of data acquisition and biospecimen collection. PPMI has included sites in both the US and Europe to enhance the generalizability of the study, to broaden the recruitment pool, and to encourage wide reaching collaboration among the PD research community.
1.4. Study assessments
Both the PD and healthy subjects in PPMI will undergo a comprehensive longitudinal schedule of clinical and imaging assessments and biosampling (Table 1). Evaluations will occur at screening/baseline and at 3 month intervals during the first year of participation and then every 6 months thereafter. In general the PD and healthy subjects will be assessed identically. PPMI will provide both extensive motor and non-motor longitudinal clinical data. Subjects will be assessed with the MDS-UPDRS (Colosimo et al., 2010) and with additional clinical tests evaluating cognition, depression, anxiety, autonomic function, sleep and olfaction. All PD subjects will undergo longitudinal DAT imaging to monitor the change in DAT density during the study. A subset of subjects will undergo longitudinal MRI DTI building on recent data suggesting that DTI measures may distinguish early PD from healthy subjects (Peran et al., 2010; Vaillancourt et al., 2009). Based on extensive discussion within the PPMI imaging core DTI MRI will be restricted to those sites with a uniform camera and image acquisition system to best acquire poolable longitudinal data.
Table 1.
| Clinical assessments | Imaging assessments | ||
|---|---|---|---|
| Motor | MDS-UPDRS | Dopamine transporter imaging | DaTSCAN |
| NeuroBehavior | Geriatric depression scale (GDS) State – trait anxiety inventory (STAI) |
MRI | Volumetric, DTI, resting state |
| Questionnaire for impulsive-compulsive disorders (QUIP) | Biospecimen collection | ||
| Cognitive testing | MoCA total score | ||
| Hopkins verbal learning test – revised | Blood | Alpha synuclein, DJ1, amyloid, tau, urate | |
| Benton judgment of line orientation | CSF | ||
| Semantic fluency | Urine | ||
| Letter number sequencing Symbol digit modalities test | Genetics | DNA, RNA | |
| Autonomic testing | SCOPA-AUT total autonomic score SCOPA-AUT sub scores | ||
| Sleep disorders | Epworth sleepiness scale total score REM sleep disorder | ||
| Olfactory testing | UPSIT |
Longitudinal collection of biospecimens including blood, CSF and urine is a key component of PPMI. For example, CSF samples will be collected every six months the first year and every twelve months thereafter. Samples will be collected according to standardized procedures and will be stored in the PPMI biorepositories in the US (Coriell, Camden, NJ) and Europe (BioRep, Milan, Italy). The focus on CSF collection in PPMI is both novel and ambitious. Recent data from both the Alzheimer Disease Neuroimaging initiative (ADNI) and from single site studies of PD subjects demonstrating that CSF biomarkers such as alpha synuclein, tau, and ß-amyloid can potentially provide useful and novel insights for PD provide the scientific rationale for CSF collection (Montine et al., 2010; Shaw et al., 2009; Siderowf et al., 2010; El-Agnaf et al., 2006; Mollenhauer and Trenkwalder, 2009). In addition the recent availability of non-traumatic spinal needles reducing discomfort and shortening the time of collection of CSF provide the practical justification for requiring longitudinal CSF in PPMI (Peskind et al., 2009).
It is anticipated based on previous clinical trials that most PD subjects enrolled in PPMI will require symptomatic treatment within six-twelve months of enrollment (Parkinson Study Group, 2007; Olanow et al., 2009). The timing of the PPMI study assessments will be adjusted as needed to ensure that subjects are fully assessed just prior to initiation of symptomatic medications. The goal of PPMI is to identify biomarkers that track progression both before and after initiation of commonly used symptomatic PD medications.
1.5. Scheme for incorporating additional PPMI assessments and biospecimen analyses
PPMI is designed to be a flexible and adaptive biomarker identification consortium. Biomarker development is an iterative process and as novel biomarkers are identified by the research community they can be tested and further validated in PPMI with its robust clinical, biospecimen, imaging and genetic data. Working groups within PPMI will continue to assess novel clinical, imaging and biospecimen biomarkers throughout the study. The PPMI Biospecimen Review Advisory Committee (BRC) formed by experts within and independent of PPMI will evaluate proposals for additional clinical and imaging studies utilizing existing and/or adding additional assessments and biosampling.
The imaging working group has identified several potential imaging biomarkers for consideration for inclusion in PPMI (Table 2). The inclusion of these imaging markers in PPMI is dependent on the available data supporting their use as PD progression markers, the ease of incorporating these markers in a multi-center study and the subject burden associated with implementing these imaging assessments. Similarly the biologic working group has established a scheme for identifying CSF, blood and urine biomarkers for inclusion in PPMI as additional data is available. The biologic working group will be charged with identifying biomarkers with adequate assay systems that could be PD progression markers. Proposals to assess PPMI biospecimen generated from either the PPMI biologic working group or those unaffiliated with PPMI in the scientific community will be reviewed by the PPMI BRC. In general, PPMI biospecimen will be used to verify or help to validate biomarkers rather than for biomarker discovery. The use of PPMI samples to study the same analytes already under investigation in PPMI with different assay platforms or methods will be discouraged in favor of proposals that focus on new, albeit validated biomarker assays. All assessment, biosampling, and biomarker data will be rapidly made public on the PPMI website.
Table 2.
Scheme for incorporating imagine and biospecimen biomarkers.
| Modality | Target | Ligand | |
|---|---|---|---|
| A. Imaging markers | |||
| TIER 1 – in use in PPMI in all or sub set of sites | |||
| SPECT | Dopamine transporter | DaTSCAN | |
| MRI | DTI/resting state | ||
| TIER II – supportive data and clear rationale for biomarker for PD, but limited data in disease progression | |||
| PET | VMAT2 | DTZB, AV133 | |
| PET | Glucose metabolism | FDG | |
| Ultrasound | Substantia nigra | ||
| PET | Amyloid | AV45, BAY94, PIB | |
| SPECT | Cardiac sympathetic function | MIBG | |
| TIER III – limited data in PD but rationale for biomarker for PD | |||
| PET/SPECT | PBR – inflammation | PK11195, PBR06, PBR111 | |
| PET/SPECT | Striatal function – MGluR5, CB1, A2a, GlyT1 | FPEB, MK9640, FCPyPB | |
| PET/SPECT | DASB, MZIENT, INER, MPPF | SERT, NET, 5HT1a, 5HT2a | |
| Optical coherence tomography (OCT) | Retinal morphology | ||
| TIER IV – strong rationale for biomarker for PD, no data for PD | |||
| PET/SPECT | Alpha-synuclein | ||
| PET/SPECT | Tau | ||
| B. Biospedmen biomarkers | |||
| TIER I – plans for use in PPMI. Markers for which there is some evidence for a disease association, preliminary data around the detection of the marker in a biochemical assay exist | CSF alpha synuclein, DJ1, amyloid, tau, blood urate | ||
| TIER II – putative markers with weak data correlating to PD, standardized assays exist and are straightforward to study in PD subjects | EGF, cytokines, inflammatory markers, glutamine/glutamate | ||
| TIER III – minimal data available, relationship to PD hypotheses and mechanisms of disease exist | Proteomics, glutathione, 8-OHdG | ||
1.6. Standardization of data acquisition/training
PPMI has developed and implemented standardized procedures for acquisition of all study data. The goal is that these procedures will set standards for future multi-center PD studies. The PPMI study cores have established study manuals detailing the collection of data. All sites undergo extensive training in the MDS-UPDRS, neuropsych and neurobehavioral testing, electronic data capture, collection and handling of biospecimen, and acquisition of imaging outcomes before enrolling study subjects. This includes web based and in person training. Retraining will be implemented when quality control issues occur and/or staff and site resources change during the study. Comprehensive quality control of data provides rapid feedback to sites to ensure that standard procedures remain fully implemented and to optimize data quality. For example, biospecimen collection parameters (such as volume collected, aliquoting, time to freezing) are monitored and quality control of specimens received at the biorepository (such as number of samples received, visual inspection of samples, hemoglobin in CSF samples) are assessed and reported to sites as necessary. Imaging acquisition has also been standardized at each site and customized study phantoms are acquired to enable ongoing assessment of camera performance (Seibyl et al., 2010). PPMI will continue to harmonize with the common data elements establish by the NIH taskforce (http://www.commondataelements.ninds.-nih.gov/PD) in selecting tools for data acquisition.
1.7. Database/statistical analysis
PPMI is fully committed to rapidly making study data publically available to the PD research community through a web portal. PPMI has developed a comprehensive yet flexible, user friendly, secure and easily accessible database. The database has been developed and will be maintained by the Laboratory of NeuroImaging (LONI) at UCLA. The bioinformatics core will receive diverse datasets from several sources including the clinical, imaging, bioanalytic and genetics cores and the biorepository. Clinical core data are transferred to the PPMI database through nightly transfers including the multi-site clinical assessments and reconciled biospecimen collection, and inventory and QC data from the biorepository. Similarly raw imaging data and PPMI analyzed imaging data and PPMI biospecimen analyses will be transferred from the imaging core and integrated into the PPMI database.
The database will allow a coordinated assessment of clinical, imaging and biospecimen data. While the specific analysis plans and sample size estimates to assess biospecimen and imaging progression biomarkers remain for the most part exploratory, it will be possible to examine the data front subject assessments in PPMI to compare healthy controls to PD subjects and/or subsets of PD subjects defined by clinical, imaging, genetic, and/or biochemical characteristics. The first set of analyses will compare baseline characteristics among PD subjects and healthy controls. The second set will use backwards selection to build a model for examining short-term change during the first six months for each progression endpoint of interest. The third set of analyses will examine whether the short-term changes in the progression endpoints are predictive of changes in long-term endpoints, such as the MDS-UPDRS score. A ten-fold cross validation procedure will be used to test the predictive validity of each model. If successful, the final model will provide a subset of one or more short-term progression endpoints that are predictive of the change in one or more of the long-term endpoints. This would suggest that these short-term progression endpoints ate valid biomarkers for future studies of interventions in PD patient populations. Finally, each of the first three sets of analyses will be repeated comparing subsets of PD subjects. If successful, the final model from these subset comparisons will determine whether some of the short-term progression endpoints are more predictive of long-term change in the MDS-UPDRS score for only some subsets of PD subjects.
While formal sample size estimation is difficult for these potential biomarkers we can examine the ability of the proposed sample size to detect meaningful effects of interest for the preliminary comparisons of baseline characteristics and univariate assessments of progression markers across the groups of interest. For all calculations, we assume a two-sided alpha level of 0.05 and a target power of 80%. With 600 total subjects (400 PD patients and 200 healthy controls), the PPMI study is adequately powered for a detectable difference in prevalence of 13% (for a dichotomous endpoint) and a detectable standardized mean difference of 0.24 (for a continuous endpoint). This suggests that the PPMI study is adequately powered to detect effects that would generally be of clinical interest. Given that PPMI is designed to identify biomarkers of change that would be useful in clinical studies of disease modifying drugs, it is reasonable to focus first on those biomarkers that demonstrate detectable change during a six-month interval. The analysis plan and sample size estimate will continue to be refined as additional biomarker data becomes available.
1.8. PPMI website
The PPMI website, http://www.ppmi-info.org/ is an essential component of the study design. The website will provide access to both PPMI data and biospecimen and to PPMI protocols, standard operating procedures, recruitment and retention materials and study presentations and publications.
The PPMI leadership is committed to rapid data sharing of the PPMI data. Individuals requesting data access will simply need to submit brief identifying information and agree that data analyses would be provided to the PPMI database upon completion. The PPMI data committee will be informed of any request for data access and would be expected to respond with approval within twenty-four hours. The database will provide for data access at several levels of expertise with simple data queries possible. The goal would be to stimulate data analyses and data mining and to incorporate some of these additional data analyses into the PPMI database.
All requests for biospecimen access would also flow through the PPMI website. The inventory of biosamples available on the PPMI website will be updated though nightly transfer of these data from the biorepository. There is a two-step request process for biosample access including a brief request outlining the research question, the samples requested and the associated PPMI data requested followed by a mote detailed proposal. The application will be reviewed by the biospecimen review committee (BRC), an independent group of biomarker experts. Successful applicants will be requited to provide their data for entry into the PPMI database to further enhance the PPMI data set.
2. Discussion
While PPMI seeks to discover and validate PD biomarkers for all phases of PD, the most critical need is to identify one or mote biomarkers to monitor disease progression. During the past two decades numerous studies have investigated potential disease modifying therapies (Parkinson Study Group, 1993, 2004, 2007; Olanow et al., 2006, 2009; Vornov et al., 2006; Schapira et al., 2010). Much has been learned from these studies, but all have failed to clearly demonstrate slowing of disease. While there are many challenges to developing a disease-modifying drug including limited animal models and uncertain clinical designs, the lack of validated biomarkers remains a major barrier to success. Current disease modifying drug development is limited to large long duration very costly studies. Even when the study subjects are enrolled early in disease, substantial neurodegeneration has already occurred (Braak and Del Tredici, 2008; Eckert et al., 2007; Fearnley and Lees, 1990; Langston, 2006; Marek and Jennings, 2009). Furthermore, the test drug may have symptomatic benefit and/or study subjects are typically treated with available symptomatic medications during the study further confounding the study outcomes (Parkinson Study Group, 1993, 2004; Ravina et al., 2005; Shults et al., 2002). Identifying PD biomarkers enabling both accurate early detection and objective monitoring of PD would transform disease modifying drug development. The primary goal of PPMI is to identify markers that would provide more rapid and sensitive demonstration of change. This might allow a biomarker signal to be assessed in a relatively small sample size short duration Phase 2 study rather the large Phase3 studies that have been unsuccessful. The biomarker driven Phase 2 studies might provide a rationale strategy to identify promising drugs to move to the costly Phase 3. These studies would also enable more rational drug dosage determination in Phase 2 studies, another major impediment to disease modifying drug development. Finally, PD biomarkers would also enable more accurate early detection of study subjects both ensuring that the study population actually has early PD and enabling study subjects to be assessed for a longer period before requiring potentially confounding symptomatic drugs.
PPMI directly addresses yet another potential flaw in current disease modifying trials – that medications tested may be of benefit to specific sub-groups of PD patients and given our inability to define these subgroups effectively the potential drug effect may be washed out and overlooked. For example, biomarkers defining genetic risk and/or change in biochemical or imaging outcomes may identify PD subgroups that are rapid or slow progressors and at whom specific therapies might be directed and assessed. In these cases medications that might be focused on specific disease mechanisms may slow disease for PD subgroups and may ultimately provide clues for more generalizable PD therapeutics. In addition, PPMI would allow us to combine biomarkers to better define subgroups to more effectively assess potential disease modifying drugs. Given the multiple genetic etiologies for PD already identified, the marked variability in the loss of dopaminergic markers measured by imaging at motor symptom onset and the well-known heterogeneity of clinical symptoms in PD onset and clinical progression, it is clear that many biomarkers with a focus ranging from clinical symptoms to PD pathobiology to molecular genetic mechanisms will be necessary to fully map PD progression (Jankovic, 1990; Klein and Schlossmacher, 2007; Beyer et al., 2007). Finally, the longitudinal design of PPMI will further allow us to assess the temporal pattern of biomarker change. Recent data from the ADNI study has developed a working hypothesis for the timing of AD biomarkers that has informed clinical AD studies (Jack et al., 2010). Likely PD biomarkers also have a temporal pattern that may be dependent on PD clinical and/or pathological stage. Of particular interest in PPMI is the opportunity to compare progression biomarkers prior to symptomatic treatment, at the time symptomatic treatment is required and following long-term symptomatic treatment. Finally the longitudinal study design will enable PPMI to flexibly assess biomarkers ‘on the fly’ and discard putative markers that are not useful while adding other promising markers that may be newly available. The accessibility of PPMI data and biospecimen will enable the PD research community to both learn from and contribute to PPMI accelerating this adaptive approach.
It is crucial to develop PD progression biomarkers to speed development of PD treatments. PPMI is an ambitious, but essential approach for biomarker development and will rely on a partnership of government, PD foundations, industry and academics working cooperatively. As in the highly successful ADNI consortium for Alzheimer disease, this collaborative and cooperative strategy is necessary to make progress in developing biomarkers that would enhance the potential for success of PD modifying therapeutics (Trojanowski et al., 2010; Aisen et al., 2010; Schmidt et al., 2010; Weiner et al., 2010).
Appendix A. The Parkinson Progression Marker Initiative (PPMI)
Executive Steering Committee:
Kenneth Marek, MD, Principal Investigator, Danna Jennings, MD, Shirley Lasch, Institute for Neurodegenerative Disorders, New Haven, CT; Andrew Siderowf, MD, University of Pennsylvania, Philadelphia, PA; Caroline Tanner, MD, PhD (Site Investigator), The Parkinson’s Institute, Sunnyvale, CA; Tanya Simuni, MD (Site Investigator), Northwestern University, Chicago, IL; Chris Coffey, PhD, (Statistics Core, PI), University of Iowa, Iowa City, IA, Karl Kieburtz, MD, MPH (Clinical Core, PI), Emily Flagg, (Clinical Core, Project Manager), Clinical Trials Coordination Center, University of Rochester, NY; Sohini Chowdhury, Michael J. Fox Foundation, New York, NY
Steering Committee/Cores:
Werner Poewe, MD (Site Investigator), Innsbruck Medical University, Innsbruck, Austria; Brit Mollenhauer, MD (Site Investigator), Paracelsus-Elena Klinik, Kassel, Germany; Todd Sherer, PhD, Mark Frasier, PhD, Claire Meunier, Michael J. Fox Foundation, New York, NY
Michael J. Fox Foundation, New York, NY
Study Cores:
Clinical Coordination Core:
Alice Rudolph, PhD, Cindy Casaceli, Clinical Trials Coordination Center, University of Rochester, NY
Imaging Core:
John Seibyl, MD, Principal Investigator, Susan Mendick, MPH, Institute for Neurodegenerative Disorders, New Haven, CT; Norbert Schuff, PhD, University of California, San Francisco
Statistics Core:
Ying Zhang, University of Iowa, Iowa City, IA
Bioinformatics Core:
Arthur Toga, PhD, Principal Investigator, Karen Crawford, Laboratory of Neuroimaging (LONI), University of California, Los Angeles
BioRepository:
Alison Ansbach, MS, Principal Investigator, Coriell Institute for Medical Research, Camden, NJ; Pasquale De Blasio, Michele Piovella, BioRep, Milan, Italy
Bioanalytics Core:
John Trojanowski, MD, PhD, Principal Investigator, Les Shaw, PhD, Principal Investigator, University of Pennsylvania, Philadelphia, PA
Genetics Core:
Andrew Singleton, PhD, Principal Investigator, National Institute on Aging, NIH, Bethesda, MD
Neuropsychological and Cognitive Assessments:
Keith Hawkins, PsyD, Yale University, New Haven, CT
Michael J Fox Foundation
Jamie Eberling, PhD, Deborah Brooks
Site Investigators and Coordinators:
David Russell, MD, PhD, Laura Leary, BS, Institute for Neurodegenerative Disorders, New Haven, CT; Stewart Factor, DO, Barbara Sommerfeld, RN, MSN, Emory University of Medicine, Atlanta, GA; Penelope Hogarth, MD, Emily Pighetti, Oregon Health and Science University, Portland, OR; Karen Williams, Northwestern University, Chicago, IL; David Standaert, MD, PhD, Stephanie Guthrie, University of Alabama at Birmingham; Robert Hauser, MD, Holly Delgado, RN, University of South Florida, Tampa, FL; Joseph Jankovic, MD, Christine Hunter, RN, CCRC, Baylor College of Medicine, Houston, TX; Matthew Stern, MD, Baochan Tran, University of Pennsylvania, Philadelphia, PA; Jim Leverenz, MD, Marne Baca, University of Washington, Seattle, WA; Sam Frank, MD, Cathi-Ann Thomas, RN, MS, Boston University, Boston, MA; Irene Richard, MD, Cheryl Deeley, MS, RNC, University of Rochester, Rochester, NY; Linda Rees, The Parkinson’s Institute, Sunnyvale, CA; Fabienne Sprenger, Innsbruck Medical University, Innsbruck, Austria; Elisabeth Lang, Paracelsus-Elena Klinik, Kassel, Germany; Holly Shill, MD, Sanja Obradov, BA, Banner Research Institute, Sun City, AZ; Hubert Fernandez, MD, Adrienna Winters, BS, Cleveland Clinic, Cleveland, OH; Daniela Berg, MD, Katharina Gauss, University of Tuebingen, Germany; Douglas Galasko, MD, Deborah Fontaine, RNCS, MS, University of California, San Diego; Zoltan Mari, MD, Melissa Gerstenhaber, RNC, MSN, Johns Hopkins University, Baltimore, MD; David Brooks, MD, Sophie Malloy, MD, Imperial College London, UK; Paolo Barone, MD, PhD, Katia Longo, MD, Universita Federico II, Naples, Italy
ISAB (Industry Scientific Advisory Board):
Tom Comery, PhD, Pfizer, Inc., Groton, CT; Bernard Ravina, MD, MSCE, Biogen Idee, Cambridge, MA; Igor Grachev, MD, PhD, Kim Gallagher, PhD, GE Healthcare, Princeton, NJ; Michelle Collins, PhD, Abbott Laboratories, Abbott Park, IL; Katherine L. Widnell, MD, PhD, Abbott Neuroscience Research & Development, Abbott Park, IL; Suzanne Ostrowizki, MD, PhD, Paulo Fontoura, MD, PhD, F.Hoffmann La-Roche, Basel, Switzerland; Tony Ho, MD, Johan Luthman, DDS, PhD, Merck & Co., North Wales, PA; Marcel van der Brug, PhD, Genentech, Inc., South San Francisco, CA; Alastair D. Reith, PhD, GlaxoSmithKline, Stevenage, United Kingdom; Peggy Taylor, ScD, Covance, Dedham, MA
Footnotes
Please see Appendix A for authors list with affiliations.
References
- Aisen PS, et al. , 2010. Clinical core of the Alzheimer’s Disease Neuroimaging Initiative: progress and plans. Alzheimers Dement. 6 (3), 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer MK, Larsen JP, Aarsland D, 2007. Gray matter atrophy in Parkinson disease with dementia and dementia with Lewy bodies. Neurology 69 (8) 747–754. [DOI] [PubMed] [Google Scholar]
- Braak H, Del Tredici K, 2008. Invited Article: nervous system pathology in sporadic Parkinson disease. Neurology 70 (20), 1916–1925. [DOI] [PubMed] [Google Scholar]
- Colosimo C, et al. , 2010. Task force report on scales to assess dyskinesia in Parkinson’s disease: critique and recommendations. Mov. Disord 25 (9), 1131–1142. [DOI] [PubMed] [Google Scholar]
- Eckert T, Tang C, Eidelberg D, 2007. Assessment of the progression of Parkinson’s disease: a metabolic network approach. Lancet Neurol. 6 (10), 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Agnaf OM, et al. , 2006. Detection of oligomeric forms of alpha-synuclein protein in human plasma as a potential biomarker for Parkinson’s disease. FASEB J. 20 (3), 419–425. [DOI] [PubMed] [Google Scholar]
- Fearnley J, Lees A, 1990. Striatonigral degeneration: a clinico-pathological study. Brain 113, 1823–1842. [DOI] [PubMed] [Google Scholar]
- Gerlach M, et al. , 2008. Early detection of Parkinson’s disease: unmet needs. Neurodegener. Dis 5 (3–4), 137–139. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr., et al. , 2010. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol. 9 (1), 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic J, 1990. Lower body (vascular) parkinsonism. Arch. Neurol 47 (7), 728. [DOI] [PubMed] [Google Scholar]
- Klein C, Schlossmacher MG, 2007. Parkinson disease, 10 years after its genetic revolution: multiple clues to a complex disorder. Neurology 69 (22), 2093–2104. [DOI] [PubMed] [Google Scholar]
- Langston JW, 2006. The Parkinson’s complex: parkinsonism is just the tip of the iceberg. Ann. Neurol 59 (4), 591–596. [DOI] [PubMed] [Google Scholar]
- Maetzler W, Liepelt I, Berg D, 2009. Progression of Parkinson’s disease in the clinical phase: potential markers. Lancet Neurol. 8 (12), 1158–1171. [DOI] [PubMed] [Google Scholar]
- Marek K Jennings D, 2009. Can we image premotor Parkinson disease? Neurology 72 (Suppl. 7), S21–S26. [DOI] [PubMed] [Google Scholar]
- Marek K, et al. , 2008. Biomarkers for Parkinson’s disease: tools to assess Parkinson’s disease onset and progression. Ann. Neurol 64 (Suppl. 2), S111–S121. [DOI] [PubMed] [Google Scholar]
- Mollenhauer B, Trenkwalder C, 2009. Neurochemical biomarkers in the differential diagnosis of movement disorders. Mov. Disord 24 (10), 1411–1426. [DOI] [PubMed] [Google Scholar]
- Montine TJ, et al. , 2010. CSF Abeta(42) and tau in Parkinson’s disease with cognitive impairment. Mov. Disord 25 (15), 2682–2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olanow CW, et al. , 2006. TCH346 as a neuroprotective drug in Parkinson’s disease: a double-blind, randomised, controlled trial. Lancet Neurol. 5 (12), 1013–1020. [DOI] [PubMed] [Google Scholar]
- Olanow CW, Kieburtz K, Schapira AH, 2008. Why have we failed to achieve neuroprotection in Parkinson’s disease? Ann. Neurol 64 (Suppl. 2), S101–S110. [DOI] [PubMed] [Google Scholar]
- Olanow CW, et al. , 2009. A double-blind, delayed-start trial of rasagiline in Parkinson’s disease. N. Engl. J. Med 361 (13), 1268–1278. [DOI] [PubMed] [Google Scholar]
- Parkinson Study Group, 1993. Effects of tocopherol and deprenyl on the progression of disability in early Parkinson’s disease. N. Engl. J. Med 328, 176–183. [DOI] [PubMed] [Google Scholar]
- Parkinson Study Group, 2004. Levodopa and the progression of Parkinson disease. NEMJ 351, 18–28. [DOI] [PubMed] [Google Scholar]
- Parkinson Study Group, 2007. Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease. Neurology 69 (15), 1480–1490. [DOI] [PubMed] [Google Scholar]
- Peran P, et al. , 2010. Magnetic resonance imaging markers of Parkinson’s disease nigrostriatal signature. Brain 133 (11), 3423–3433. [DOI] [PubMed] [Google Scholar]
- Peskind E, et al. , 2009. Safety of lumbar puncture procedures in patients with Alzheimer’s disease. Curr. Alzheimer Res 6 (3), 290–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravina B, et al. , 2005. The role of radiotracer imaging in Parkinson disease. Neurology 64 (2), 208–215. [DOI] [PubMed] [Google Scholar]
- Ravina B, et al. , 2009. A longitudinal program for biomarker development in Parkinson’s disease: a feasibility study. Mov. Disord 24 (14), 2081–2090. [DOI] [PubMed] [Google Scholar]
- Schapira AH, 2004. Disease modification in Parkinson’s disease. Lancet Neurol. 3 (6), 362–368. [DOI] [PubMed] [Google Scholar]
- Schapira AH, et al. , 2010. Rationale for delayed-start study of pramipexole in Parkinson’s disease: the PROUD study. Mov. Disord 25 (11), 1627–1632. [DOI] [PubMed] [Google Scholar]
- Scherzer CR, et al. , 2007. Molecular markers of early Parkinson’s disease based on gene expression in blood. Proc. Natl. Acad. Sci. U.S.A 104 (3), 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt ME, et al. , 2010. The Alzheimer’s Disease Neuroimaging Initiative: perspectives of the Industry Scientific Advisory Board. Alzheimers Dement. 6 (3), 286–290. [DOI] [PubMed] [Google Scholar]
- Seibyl J, Marek K, Zubal IG, 2010. The role of the core imaging laboratory in multicenter trials. Semin. Nucl. Med 40 (5), 338–346. [DOI] [PubMed] [Google Scholar]
- Shaw LM, et al. , 2009. Cerebrospinal fluid biomarker signature in Alzheimer’s Disease Neuroimaging Initiative subjects. Ann. Neurol 65 (4), 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M, Caudle WM, Zhang J, 2009. Biomarker discovery in neurodegenerative diseases: a proteomic approach. Neurobiol. Dis 35 (2), 157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults CW, et al. , 2002. Effects of coenzyme Q10 in early Parkinson disease: evidence of slowing of the functional decline. Arch. Neurol 59 (10), 1541–1550. [DOI] [PubMed] [Google Scholar]
- Siderowf A, et al. , 2010. CSF amyloid β 1–42 predicts cognitive decline in Parkinson disease. Neurology 75 (12), 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Lancet, N., 2010. Biomarker promise for Parkinson’s disease. Lancet Neurol. 9 (12), 1139. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, et al. , 2010. Update on the biomarker core of the Alzheimer’s Disease Neuroimaging Initiative subjects. Alzheimers Dement. 6 (3), 230–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaillancourt DE, et al. , 2009. High-resolution diffusion tensor imaging in the substantia nigra of de novo Parkinson disease. Neurology 72 (16), 1378–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vornov J, et al. , 2006. GPI1485, a neuroimmunophilin ligand, fails to alter disease progression in mild to moderate Parkinson’s disease. Mov. Disord. [Google Scholar]
- Weiner MW, et al. 2010. The Alzheimer’s Disease Neuroimaging Initiative: progress report and future plans. Alzheimers Dement. 6 (3), 202 e7–211 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whone AL, et al. , 2003. Slower progression of Parkinson’s disease with ropinirole versus levodopa: the REAL-PET study. Ann. Neurol 54 (1), 93–101. [DOI] [PubMed] [Google Scholar]