Abstract
Sarcoglycanopathies include four subtypes of autosomal recessive limb-girdle muscular dystrophies (LGMDR3, LGMDR4, LGMDR5 and LGMDR6) that are caused, respectively, by mutations in the SGCA, SGCB, SGCG and SGCD genes. Delta-sarcoglycanopathy (LGMDR6) is the least frequent and is considered an ultra-rare disease. Our aim was to characterize the clinical and genetic spectrum of a large international cohort of LGMDR6 patients and to investigate whether or not genetic or protein expression data could predict a disease’s severity.
This is a retrospective study collecting demographic, genetic, clinical and histological data of patients with genetically confirmed LGMDR6 including protein expression data from muscle biopsies.
We contacted 128 paediatric and adult neuromuscular units around the world that reviewed genetic data of patients with a clinical diagnosis of a neuromuscular disorder. We identified 30 patients with a confirmed diagnosis of LGMDR6 of which 23 patients were included in this study. Eighty-seven per cent of the patients had consanguineous parents. Ninety-one per cent of the patients were symptomatic at the time of the analysis. Proximal muscle weakness of the upper and lower limbs was the most common presenting symptom. Distal muscle weakness was observed early over the course of the disease in 56.5% of the patients. Cardiac involvement was reported in five patients (21.7%) and four patients (17.4%) required non-invasive ventilation. Sixty per cent of patients were wheelchair-bound since early teens (median age of 12.0 years). Patients with absent expression of the sarcoglycan complex on muscle biopsy had a significant earlier onset of symptoms and an earlier age of loss of ambulation compared to patients with residual protein expression.
This study confirmed that delta-sarcoglycanopathy is an ultra-rare neuromuscular condition and described the clinical and molecular characteristics of the largest yet-reported collected cohort of patients. Our results showed that this is a very severe and quickly progressive disease characterized by generalized muscle weakness affecting predominantly proximal and distal muscles of the limbs. Similar to other forms of sarcoglycanopathies, the severity and rate of progressive weakness correlates inversely with the abundance of protein on muscle biopsy.
Keywords: muscular dystrophies, delta-sarcoglycan, SGCD, LGMD-R6/2F, registries
Alonso-Pérez et al. describe the largest cohort to date of patients with the ultra-rare neuromuscular disorder delta-sarcoglycanopathy, and show that the severity and rate of progressive weakness correlates inversely with the abundance of protein expression on muscle biopsy.
Introduction
Limb-girdle muscular dystrophies (LGMD) are a heterogeneous group of genetic diseases that affect skeletal muscle causing progressive loss of muscle fibres leading to muscle weakness predominantly affecting the pelvic and shoulder girdle.1 More than 30 genes causing different types of LGMD have been described so far. Among them, sarcoglycanopathies are one of the most frequent forms especially when symptoms onset during childhood.2-7 There are four sarcoglycan genes (SGCA, SGCB, SGCG and SGCD) causing autosomal recessive LGMD (LGMDR3–6 previously known as LGMD2C, D, E and F). The frequency of each type of sarcoglycanopathy varies depending on the studied population, although LGMDR3/LGMD2D and LGMDR5/LGMD2C are the two most frequent forms.8
Delta-sarcoglycanopathy (LGMDR6/LGMD2F) is caused by recessive mutations in the SGCD gene and was originally described in 1996,9–11 and is thought to be the least common type of sarcoglycanopathy although the number of existing cases is not known. There are only a few reports describing the clinical features of single cases or short cohorts and therefore neither the clinical features nor the disease progression over time is well known.12–14
Sarcoglycanopathies (LGMDR3–6) are in general severe disorders characterized by weakness onset at first decade of life leading to loss of ambulation during adolescence or early adulthood. However, patients with a milder phenotype with onset of symptoms after the second decade of life, a slowly progressive course and still ambulant after the age of 50 or 60 years have also been described.15–18 We have recently observed that symptoms’ onset before the age of 10 years, protein expression in the muscle biopsy of <30% or mutations leading to absence of protein expression were independent risk factors associated with a more severe phenotype characterized by early loss of ambulation.8 However, it is not known whether these are also risk factors of quick progression in patients with LGMD-R6.
To answer these questions, we collected demographic, genetic, clinical and muscle protein data of a large international cohort of patients with mutations in SGCD. Our aims were to describe the main clinical and genetic features, investigate potential genotype–phenotype correlations and identify factors influencing the progression of the disease.
Materials and methods
Study design
This is an observational retrospective cross-sectional study reviewing clinical and genetic data of patients with confirmed pathogenic mutations in the SGCD gene. Anonymized data from clinical reports were collected in a survey and stored in a secure server in the Hospital de la Santa Creu i San Pau (HSCSP). The study was approved by the Ethics Committee of the HSCSP. To identify patients for the study, we used two different strategies. On the one hand we contacted 128 paediatric and/or adult hospitals from 38 countries. Only 11 neuromuscular centres from nine different countries had patients with delta-sarcoglycanopathy and agreed to participate in the study. On the other hand, we also contacted colleagues that lead next-generation sequencing (NGS) studies in large cohorts of neuromuscular disease patients without diagnosis. These studies actually included 9610 patients. None of these patients had mutations in the SGCD gene.
Patient cohort
The inclusion criteria for the study were: (i) genetically confirmed diagnosis of delta-sarcoglycanopathy by identification of two heterozygous or one homozygous pathogenic mutation in the SGCD gene; and (ii) enough data available in the clinical records to answer ∼70% of the variables of the survey about disease onset and progression, presence or absence of cardiac and respiratory involvement and muscle function status at last clinical assessment.
Data sources
Participating centres completed a survey for each of the patients included in the study. The survey collected demographic, clinical and genetic data of all patients. We also collected information about the muscle biopsy if performed, including morphological features and levels of protein expression measured by immunohistochemistry or immunofluorescence. Cardiac involvement was defined according to international guidelines based on the presence of left ventricular ejection fraction lower than 50% and/or fractional shortening lower than 25% and/or the existence of morphological abnormalities in the ventricular walls evaluated by echocardiography or the existence of cardiac conduction defects identified using ECG or Holter monitoring.19,20 The need for ventilatory support was also collected as well as the age at which it was started. A copy of the survey is provided in the Supplementary material.
Mutations in the SGCD gene were centrally reviewed by experienced geneticists from HSCSP (L.G.-Q. and P.G.) to confirm pathogenicity and predict protein expression. We considered nonsense, frameshift and canonical splice site mutations as non-protein producing since these mutations will cause a disruption of the reading frame or a considerable shortening of the transcript resulting in mRNA elimination by nonsense-mediated decay or aberrant protein degradation. A sequence-based prediction of impact the of the missense mutations on protein function and/or expression was performed using different computational approaches assessing sequence conservation; for example, SIFT21 and PANTHER-PSEP,22 as well as sequence and structural features such as PolyPhen-223 and MutationTaster.24,25 Deep intronic variants were considered compatible with protein production since these variants even when resulting in abnormal splicing they usually produce a residual amount of wild-type transcript.26,27
Statistical analysis
Quantitative variables were analysed using the Shapiro–Wilk test to verify the normal distribution. Comparison between the different subgroups of patients was performed using the Chi-squared test for categorical variables and the Student's t-test or Mann–Whitney test for quantitative variables. We used a Cox proportional hazard regression model for the analyses of time to wheelchair. P was considered significant if lower than 0.05. Hierarchical analysis and graphical representation as a heatmap of muscle strength measured using the MRC scale was performed using R software v.3.1.3 as previously described.28 Statistical analysis was performed by J.D.-M. and J.A.-P. using SPSS® Statistics software v.21 from IBM®.
Data availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
Results
Patient cohort
We contacted 128 paediatric and adult neuromuscular units and/or neurology departments around the world and identified a total of 30 patients with a confirmed genetic diagnosis of LGMDR6. We excluded seven patients because clinical data were not available or updated. The 23 patients included were from nine different countries: Spain, UK, Italy, Hungary, Turkey, Iran, Pakistan, Brazil and Canada. Additionally, we also contacted international centres that have lead research projects using NGS to sequence large cohorts of patients with neuromuscular diseases.4,29–34 After reviewing the results of 9610 patients, no patients with delta-sarcoglicanopathy were found.
Among the 23 identified patients with confirmed diagnosis of LGMDR6, 10 were males and 13 were females from 18 different families. Consanguinity was present in 20 patients (87%) and 12 patients (52.2%) had another relative affected by the disease.
Most of the patients (n = 21, 91.3%) were symptomatic at the time of data collection. However, two patients were considered presymptomatic as they had neither symptoms nor signs of neuromuscular involvement on clinical examination. These two patients were 4 and 5 years old, respectively, and relatives of other affected patients. Among the 21 symptomatic patients, median age at onset of symptoms was 5.0 years (range: 2–24 years). Diagnostic delay, defined as the time from onset of symptoms to the confirmed genetic diagnosis of the disease range from 1 to 37 years (median: 6.5 years). Median diagnostic delay in patients with onset of symptoms before 2010, when genetic testing was done using Sanger sequencing was of 10.0 years (range: 4–37, n = 13), while diagnostic delay in patients with onset of symptoms after 2010, when NGS studies started to be used in the diagnosis process of LGMD was of 2.0 years (range: 0–7, n = 10). These differences were statistically significant (P = 0.001, Mann–Whitney test).
Table 1 shows the most common symptoms at onset. In summary, proximal lower limb weakness was the most frequent symptom seen in 12 of 21 patients. Five patients (23.8%) complained of muscle pain associated with muscle weakness at the onset of the disease. Seven out of the 21 symptomatic patients were ambulant at their last visit. Median age at loss of ambulation was 12.0 years (range 9–37). Median time from onset of symptoms to loss of ambulation was 7.0 years (range 4–10). We characterized disease progression based on the data collected from the clinical reports and observed that 18 patients (85.7%) were not able to run since a median age of 8.0 years old, 16 patients (76.2%) were not able to stand up from a chair since a median age of 9.5 years and nine patients (42.9%) needed aids to walk since a median age of 10.0 years (Table 1).
Table 1.
Demographic and clinical features
| LGMDR6/2F | |
|---|---|
| No. of patients | 23 |
| Sex, male/female | 10/13 |
| Consanguinity, n (%) | 21 (87) |
| Age onset, median ± SD (range) | 5.0 ± 6.8 (2–24) |
| Diagnostic delay (onset–genetic), median ± SD (range) | 6.5 ± 7 (1–37) |
| Age at last evaluation, median ± SD (range) | 17.0 ± 12.3 (4–50) |
| Evolution of the disease, years, median ± SD (range) | 11.5 ± 8.9 (3–33) |
| Symptom onset, n (%) | |
| Proximal lower limb weakness | 12 (57.1) |
| Proximal upper limb weakness | 2 (9.5) |
| Gait disturbance | 11 (52.4) |
| Muscle pain | 5 (23.8) |
| Motor function, n (%); median age ± SD (range) | |
| Stop running | 18 (85.7); 8.0 ± 6.8 (3–32) |
| Impossibility to stand from a chair | 16 (76.2); 9.5 ± 7.3 (7–34) |
| Walking with aids | 9 (42.8); 10.0 ± 1.8 (9–14) |
| Wheelchair-bound | 14 (66.7); 12.0 ± 7.1 (9–37) |
| Cardiac involvement, n (%); median age ± SD (range) | 5 (23.8); 13.0 ± 2.7 (11–17) |
| Respiratory support, n (%); median age ± SD (range) | 4 (19.0); 20.5 ± 5.1 (13–24) |
| Death, n (%) | 0 (0) |
n = number of families; SD = standard deviation.
At last clinical evaluation, clinical examination showed proximal muscle weakness in all symptomatic patients. Axial (n = 13, 61.9%) and distal (n = 12, 57.1%) weakness was also present from early stages of the disease, especially in patients with no remaining protein expression (Fig. 1). Scoliosis was observed in 12 patients (57.1%) at a median age of 11.5 years (range 7–15). Scapular winging (n = 10, 47.6%), calf hypertrophy (n = 12, 57.1%) and generalized muscle atrophy (n = 13, 61.9%) were also described. Tiptoe walking due to Achilles tendon contractures was reported in 15 patients (71.4%) at a median age of 9.5 years (range 3–16). Foot deformities, mainly pes cavus due to early lower limb distal muscle weakness was also observed (Fig. 2).
Figure 1.
Heat map showing an unsupervised hierarchical clustering of MRC values of patients included in the study. The heat map shows the MRC value for all muscles studied. Patients and muscles were ordered automatically by the software in an unsupervised manner. Hip extension, flexion, abduction and adduction were the weakest movements followed by arm abduction, elbow flexion and extension, and knee flexion and extension. Muscle strength was measured using the MRC scale that scores muscle function from 0 to 5. We observed a correlation between the degree of muscle weakness and time from onset of symptoms and also between MRC and absence of protein expression on the muscle biopsy.
Figure 2.
Foot deformities. We identified weakness of the distal muscles of the lower limbs in some of the patients in the cohort that lead to the frequent presentation of deformities as the two shown here. (A) A 9-year-old patient with bilateral clubfeet. (B) A pes cavus in an 18-year-old patient.
Cardiac involvement, defined as cardiomyopathy or heart rhythm abnormalities, was reported in five patients (23.8%) with a median age of diagnosis of 13.0 years (range 11–17). Dilated cardiomyopathies were present in three patients (60%) and heart rhythm abnormalities were present in two patients. The last cardiac assessment showed median left ventricle ejection fraction of 40% (range 30–58). Four out of these five patients were under treatment because of the cardiac involvement. Median forced vital capacity at last assessment was of 62.0% (range 44–84). Four patients (19%) required ventilatory support from a median age of 20.5 years (range 13–24) and were using it a median of 9 h per night (range 6–15). None of the patients had a tracheostomy.
A muscle biopsy was performed in 14 patients (60.9%) at a median age of 10.0 years (range 2–34) and a median time of disease duration of 4.5 years (range 1–30). Increase in the amount of fibrotic tissue and presence of necrotic muscle fibres were the most frequent features (64.3 and 50%, respectively). Inflammatory infiltrates were observed in 28.6% of biopsies (Fig. 3A and B). Residual expression of sarcoglycan proteins was studied using immunohistochemistry or immunofluorescence in all patients (Fig. 3C). Six patients had no protein expression of any sarcoglycan subunit while eight patients had some amount of residual expression. There were no differences in the age when biopsy was obtained (P = 0.295, Mann–Whitney test) or in the time of progression of the disease (P = 0.180, Mann–Whitney test) between patients with residual protein expression and those with no protein expression.
Figure 3.
Muscle biopsy. (A and B) Haematoxylin-eosin staining. Necrotic fibres (asterisk), inflammatory infiltrate (arrow), increased fibrosis tissue (arrowhead), hypercontracted fibres (plus symbol). Magnification: ×20 (left) and ×40 (right). (C) Immunofluorescence of sarcoglycan subunits. SG = sarcoglycan.
Table 2 shows the clinical, genetic, muscle biopsy features and protein expression of the patients included in this study.
Table 2.
Clinical and genetic features
c.699 + 1G>T
| Pt | Mutation | Age at last evaluation | Age of onset | WCB, age | Distal weakness | Cardiopathy, age | NIV, age | Muscle biopsy | IHQ/IF | Protein expression |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | c.657delC, p.(Thr220Profs*6) | 21 | 7 | Yes, 15 | Yes | No | No | No | ↓↓ | Yes |
| 2 | c.657delC, p.(Thr220Profs*6) | 8 | 5 | No | No | No | No | No | – | YESa |
| 3 | c.657delC, p.(Thr220Profs*6) | 27 | 8 | Yes, 15 | Yes | No | No | Yes | ↓↓ | Yes |
| 4 | c.657delC, p.(Thr220Profs*6) | 17 | 6 | Yes, 10 | Yes | No | No | Yes | ↓↓ | Yes |
| 5 | c.657delC, p.(Thr220Profs*6) | 22 | 5 | Yes, 14 | Yes | Yes, 17 | Yes, 18 | Yes | ↓↓ | Yes |
| 6 | c.1_3del, p.(Met1del) | 5 | – | – | – | – | – | – | – | Yesa |
| 7 | c.1_3del, p.(Met1del) | 25 | 23 | No | No | No | No | No | – | Yesa |
| 8 | c.1_3del, p.(Met1del) | 11 | 7 | No | Yes | No | No | No | – | Yesa |
| 9 | c.1_3del, p.(Met1del) | 37 | 24 | No | No | No | No | Yes | ↓ | Yes |
| 10 | c.(3_4–52)_(187_193-1)del), p.(Met2_Ile64del) | 5 | 3 | No | No | No | No | Yes | ↓↓ | Yes |
| 11 | c.(3_4-52)_(187_193-1)del), p.(Met2_Ile64del) | 4 | – | – | – | – | – | Yes | ↓↓ | Yes |
| 12 | c.353-357del, p.(Thr119Serfs*17) | 19 | 2 | Yes, 11 | Unknown | Yes, 12 | No | Yes | Absent | No |
| 13 | c.353-357del, p.(Thr119Serfs*17) | 24 | 9 | Yes, 15 | Yes | No | Yes, 23 | Yes | Absent | No |
| 14 | c.568G>T, p.(Glu190*) | 11 | 4 | Yes, 9 | Yes | No | No | No | – | Noa |
| 15 | c.568G>T, p.(Glu190*) | 16 | 5 | Yes, 10 | Yes | No | No | Yes | Absent | No |
| 16 | c.422dupT, p.(Thr143Asnfs*13) | 10 | 5 | No | Yes | No | No | No | – | Noa |
| 17 | c.289C>T, p.(Arg97*) | 13 | 3 | Yes, 10 | Yes | No | No | No | – | Noa |
| 18 | c.248-249delCT, p.(Ser83*) | 11 | 4 | Yes, 10 | Yes | Yes, unknown | No | Yes | Absent | No |
| 19 | c.89G>A, p.(Trp30*) | 29 | 2 | Yes, 9 | Unknown | Yes, 19 | Yes, 13 | Yes | Absent | No |
| 20 | 43 | 3 | Yes, 12 | Yes | Yes, 11 | Yes, 24 | Yes | Absent | No | |
| 21 | c.593G>C, p.(Arg198Pro) | – | 4 | Yes, 15 | Unknown | Unknown | Unknown | Yes | ↓↓ | Yes |
| 22 | c.575+1G>T | 50 | 22 | Yes, 37 | Unknown | Unknown | Unknown | Yes | ↓↓ | Yes |
| 23 | c.-519_502del), p.0 | 17 | 9 | No | Yes | No | No | Yes | ↓↓ | Yes |
IHQ/IF = Immunohistochemistry or immunofluorescence detection of protein expression of the whole sarcolgycan complex on muscle biopsies; NIV = non-invasive ventilation required and age at which was started; Pt = patient; WCB = age at which patients were wheelchair-bound. Patients 6 and 11 were asymptomatic at the last assessment.
Identifies patients whose protein expression in the muscle was predicted.
Genetics
Thirteen different pathogenic variants were identified in the SGCD gene (Fig. 4), six of them not previously described. All patients were homozygous for a single variant. The variants identified differed depending of the country of origin, with variant c.657delC, p.(Thr220Profs*6) being the most commonly detected in patients of Brazilian origin (21.7% of patients). Most of the pathogenic variants (65.2%) affected the extracellular domain of the protein. Four frameshift, three nonsense and two splicing mutations are reported in the present work, assuming that they will cause a premature termination of translation in case that the mRNA generated is not rapidly degraded by mRNA nonsense-mediated decay. In addition, three deletions are reported. The mutation c.1_3del;p.Met1del eliminates the translation initiation methionine (M1) located at the very end of exon 2. Protein production could still be possible since the first codon of exon 3 is also a methionine (M2) that could act as a starting point for the polypeptide, as is the case in the alternative transcript ENST00000435422.7. However, expression of the whole sarcoglycan complex in the muscle biopsy of the cases harbouring this mutation was identified (Table 2). The deletion c.4_192del eliminates 63 amino acids (p.Met2_Ile64del) yet keeps the reading frame so it is difficult to predict its effect on protein structure. This mutation was present in two patients of the cohort with a muscle biopsy that showed severe reduction in protein expression. The deletion c.-519_502del from exon 1 to 6 removes first half of the protein and all protein domains. We identified one missense mutation, p.(Arg198Pro) that has been previously reported as pathogenic.35 Assessing the pathogenic effect of a missense variant requires an understanding of its impact on the gene expression and protein structure and function. Missense mutations can affect protein function not only by disrupting their structure and conformation but also influencing its interaction with other proteins or molecules. Thus, the effect of missense mutations is not always easy to predict. This missense variant was neither found in gnomAD nor the 1000 Genomes databases, and its pathogenicity is supported by different prediction tools’ scores (Supplementary Table 1).
Figure 4.
Distribution of the pathogenic variants found in our cohort of patients in the SGCD gene. The graph shows the distribution of the pathogenic variants identified in the patients that participated in the study. Novel variants not previously described are underlined. Bottom right: The legend describes the type of mutation.
Genotype–protein expression–phenotype correlation
To study the potential genotype–phenotype correlation in delta-sarcoglycanopathies, we studied the impact of the mutations on the residual protein expression in 14 muscle biopsies. Based on these data we estimated the residual protein expression in the nine remaining patients without muscle biopsy but who shared the same variants in the SGCD gene with the patients who had a muscle biopsy. Patients were classified in two groups: (i) no protein expression if patients had an undetectable expression of the sarcoglycan complex measured by muscle immunohistochemistry or immunofluorescence or carried two frameshift or nonsense mutations; and (ii) residual protein expression detected by immunohistochemistry or immunofluorescence.
We did not observe differences in the duration of the disease, defined as the time from onset of symptoms to last clinical evaluation, between both groups of patients (P = 0.49, Student’s t-test).
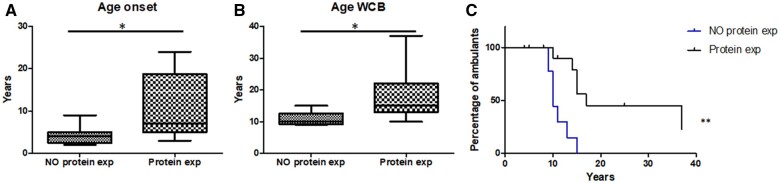
Patients with no protein expression had an earlier disease onset (4.1 versus 10.3 years; P = 0.01, Mann–Whitney test) (Fig. 5A) and lost ambulation earlier (10.9 versus 17.3 years; P = 0.03, Mann–Whitney test) than patients with residual protein expression (Fig. 5B). Consequently, patients with residual protein expression maintained ambulation for a longer period of time (P = 0.001, Mantel–Cox test) (Fig. 5C). The correlation between the age of onset and progression of the disease remains if we analyse the data independently, depending on whether the protein level was obtained from muscle biopsy or by prediction (data not shown).
Figure 5.
Influence of remaining protein expression. (A) Age of onset of patients with residual protein expression and patient with no protein expression. (B) Age of loss of ambulation of patients with residual protein expression and patient with no protein expression. Mann–Whitney test, *P < 0.05. (C) Kaplan–Meier estimates influence of remaining protein expression in age at wheelchair for patients with residual protein expression and patient with no protein expression. Mantel–Cox, **P < 0.001. exp = expression; WCB = wheelchair-bound.
There were not significant differences in the prevalence of cardiac involvement (Chi-squared test, P = 0.09) or the needed for respiratory support (Chi-squared test, P = 0.21) between both groups.
Discussion
We report the clinical, genetic and natural history data of the largest series of patients with delta-sarcoglycanopathy described so far. International collaboration has been crucial to gather this number of patients and to obtain information about clinical features and prognosis of the disease. As described in other types of sarcoglycanopathy, patients with residual protein expression had a later onset of disease symptoms and a milder clinical course characterized by later loss of ambulation than patients with no remaining protein expression. These data are especially relevant at present, as genetic therapies are under development and interventional clinical trials in patients with other types of sarcoglycanopathy are being designed.36–38
Sarcoglycanopathies, as a group, are one of the most common recessive LGMD6,39,40; however, delta-sarcoglycanopathy (LGMDR6) was supposed to be a very rare disease. In recent years, a series of studies have analysed large cohorts of patients with undiagnosed genetic muscle diseases using genes panels and/or whole exome sequencing. The frequency of LGMDR6 in these studies is very low or absent.3,8,34,40 Furthermore, published LGMDR6 reports so far have just included a few cases or isolated patients.12,13,41 A recent publication analysing a large cohort of European sarcoglycanopathies showed that only 1.5% of the cases were delta-sarcoglycanopathy.8 This previous evidence confirms that delta-sarcoglycanopathy can be considered an ultra-rare disease that it is defined as having a prevalence lower than 20 patients per million of population or one case per 50 000 habitants. In the case of delta-sarcoglycanopathy, the prevalence could be much lower probably in the order of one case per million of habitants. We do not have a clear explanation why patients with delta-sarcoglycanopathy are so infrequent compared to patients with other forms of sarcoglycanopathy. One potential explanation could be that mutations in this gene may be lethal during foetal development. If that were the case, this could be more probably related to a cardiac problem than to a skeletal muscle problem as there are no patients with congenital myopathy described so far. On the contrary, it is well known that mutations in the SGCD gene can be associated to developmental problems of the heart in preclinical animal models, such as zebrafish, and also to progressive cardiomyopathy both in humans and animal models.42–45 From a functional point of view, both the beta and the delta subunits are the core of the sarcoglycan complex suggesting than lack of beta-sarcoglycan should be as deleterious as the delta subunit for development, but patients with mutations in the SGCB are much more frequent than patients with mutations in the SGCD gene.46–48 Another reason could be that SCGD is devoid of sequence more prone to mutate compared to other sarcoglycan genes, but we observed that mutations in the SGCD were located in all the exons, same as the other sarcoglycan genes, suggesting that no particular exons are lethal if mutated.8
LGMDs produced by mutations in SGCA, SGCB and SGCG genes have been classically associated with a rapid progression of muscle weakness leading to severe disability, loss of ambulation in the second decade of life and frequently presenting cardiac and respiratory involvement.8,49 However, disease progression can be more heterogeneous as several patients, especially those with mutations in the SGCA gene, have a later onset of the disease and can loss ambulation later in life or even remain ambulant.8,16,18 To broaden the clinical presentation, patients with alpha-sarcoglycanopathy and isolated hyperCKaemia have been described.50 In contrast, patients with beta and gamma sarcoglycanopathies have in general a more homogeneous clinical picture with an early loss of ambulation.8,18,49,51 Our data shows that delta-sarcoglycanopathy is similar to other sarcoglycanopathies, with early onset in the first decade of life, rapid progression and loss of ambulation in the second decade of life in a high percentage of cases. However, we have observed that residual protein expression could influence clinical progression and severity of the disease. Indeed, 77.8% of patients with no remaining protein expression and 87.5% of patients with symptoms onset before the age of 5 years were wheelchair-bound at the age of 15 years. On the other hand, 21.4% of patients with residual protein expression and 25% of patients with symptom onset after the age of 5 years lose ambulation before adolescence. Moreover, only 42.9% of patients with residual protein expression ended up in a wheelchair and in all cases after the age of 10 years. These data support that residual protein expression is associated with a later onset of the disease and better prognosis as is shown in Fig. 3. This finding was also described for alpha, beta and gamma sarcoglycanopathies. However, despite the fact that patients with residual protein expression can have a milder disease progression, this disease is extremely severe as 91.3% of the patients were wheelchair-bound before the age of 18 years.
Beta and delta-sarcoglycan subunits form the core of the sarcoglycan complex and therefore are essential for its assembly.47 Mutations in the SGCD gene should therefore translate into a complete disruption of the complex leading to absence of expression of all sarcoglycan subunits in the muscle membrane and, consequently, to a more fragile membrane suffering more damage after every contraction, as has been described in the murine model of the disease.48,52 In fact, the delta-sarcoglycan murine model is characterized by severe muscle weakness and cardiac involvement.13,53,54 However, in this cohort some of the patients for whom a muscle biopsy was available (8 of 14) did have residual protein expression of the sarcoglycan subunits identified by immunohistochemistry. These patients presented with a later onset and less severe disease, suggesting that residual expression of the different components of the complex was enough to produce a better prognosis.
All patients were homozygous for one pathogenic variant and 87% had a history of consanguinity in the family. The most frequent types of mutation were frameshift mutations (up to 39.1% of the patients); however, 30.4% of patients carried a large deletion affecting one or more exons, which might be missed by Sanger sequencing and could only be confirmed by a quantitative technique such as multiplex ligation-dependent probe amplification (MLPA). It is therefore important to use MLPA in those cases with a high suspicion of the disease based on clinical and/or muscle biopsy data if Sanger sequencing does not identify any pathogenic mutation.55,56
Cardiac and respiratory involvement requiring ventilatory support is frequent in patients with sarcoglycanopathy.8,57,58 However, there are differences in the frequency and the type of cardiac or respiratory involvement depending on the mutated gene. For example, it is known that cardiac involvement in the form of a cardiomyopathy is more frequent in patients with beta-sarcoglycanopathy, than in patients with alpha or gamma-sarcoglycanopathy.8,49,57,59 Loss of delta-sarcoglycan is associated with cardiomyopathy is several preclinical animal models. For example, the different BIO14.6 hamster strains that harbour a deletion in the SCGD gene can develop either dilated or hypertrophic cardiomyopathy.60 In mice, delta-sarcoglycan deficiency leads to cardiomyopathy that is aggravated as a result of coronary artery vascular irregularities probably as a consequence of the absence of delta-sarcoglycan in the smooth muscle.61 Delta-sarcoglycan is key for normal cardiac development in zebrafish, leading to left–right asymmetry of the heart and disorganization of the intracellular myofibrils.42,62 There has been a long discussion related with the existence of isolated dominant cardiomyopathy produced by heterozygous mutations in the SGCD gene. Tsubata et al.63 found a single missense mutation (c.451T>G, p.Ser151Ala) associated with severe dominant cardiomyopathy in a family with three patients carrying this mutation. However, the pathogenic relevance of the p.Ser151Ala variant was challenged when this mutation was discovered in a large consanguineous family homozygous for p.Ala131Pro suffering from LGMDR6/2F but lacking any signs of cardiac disease in family members carrying the p.Ser151Ala mutation.64 Additionally, the knock-in of p.Ser151Ala caused a rather mild phenotype of cardiomyopathy in mice.45 Based on these data, the pathogenic potential of the p.Ser151Ala missense mutation to cause familial cardiomyopathy is unlikely. Surprisingly, we have only observed cardiac involvement in five patients in our cohort (23.8%), which was not related to a longer disease duration or with the level of residual protein expression. None of the patients had previous history of coronary heart disease. These data indicate that, in humans, delta-sarcoglycan defiency is not always associated to cardiac problems, although a longer follow-up of the patients included here is needed to confirm that they do not develop cardiac problems with age. Respiratory insufficiency requiring ventilatory support was only observed in four patients (19.0%) and was also not related with a longer disease duration or remaining protein expression. These data confirm that periodic assessment of cardiac and respiratory involvement is needed in all cases regardless of a patient’s age and the clinical status.
Our study has some limitations. First, data were collected retrospectively and there were some missing data in all cases. Second, quantification of protein expression was carried out through immunofluorescence performed at each centre with different antibodies and techniques including immunohistochemistry or immunofluorescence. Quantification of protein expression using western blot would have been better and could have helped us to identify a cut-off point predicting a more severe progression, as has been described in other types of sarcoglycanopathy.8 Moreover, for some patients, protein expression was estimated based on the results observed in patients sharing the same mutations in whom protein expression was studied in muscle biopsy. Although it is probable that protein expression is similar between patients with the same mutations, this is speculative and therefore these results should be interpreted cautiously. Despite our effort in contacting as many neuromuscular units as possible, we could have missed some delta-sarcoglycanopathy patients for inclusion into our study. An international registry for patients with mutations in the sarcoglycan genes, specifically in this case for patients with mutations in the SGCD gene could be useful to identify more patients. However, our series is the largest described so far and includes patients from different countries and ethnicities although there were no patients from Africa or South-East Asia.
In conclusion, our study provides new and relevant information that widens the knowledge on the clinical and genetic features of patients with mutations in the SGCD gene. We have identified that residual protein expression is associated with a later onset of disease and milder phenotype with later loss of ambulation. These data should be useful for the design of natural history studies and for clinical trial design, including gene replacement therapies, that are currently under development for other types of sarcoglycanopathy.
Funding
This investigation was sponsored by a grant from the Spanish Ministry of Health, Fondos FEDER-ISCIII PI18/01525 to J.D.-M. J.A.P. was supported by the ‘Rio Hortega’ grant (CM19/00178), Acción Estratégica de Salud (EAS), Instituto de Salud Carlos III (Spain). B.M. and K.H. were supported by grants NKFIH 119540 and EFOP-3.6.1–16-2016–00004. L.G.-Q. and P.G. received funding from FIS PI8/01585, funded by ISCIII and FEDER, ‘Una manera de hacer Europa’.
Competing interests
All authors report no competing interests.
Supplementary material
Supplementary material is available at Brain online.
Supplementary Material
Glossary
- LGMD
limb-girdle muscular dystrophies
References
- 1. Straub V, Murphy A, Udd B; LGMD workshop study group . 229th ENMC international workshop: Limb girdle muscular dystrophies – Nomenclature and reformed classification Naarden, the Netherlands, 17–19 March 2017. Neuromuscul Disord. 2018;28(8):702–710. [DOI] [PubMed] [Google Scholar]
- 2. Winckler PB, da Silva AMS, Coimbra-Neto AR, et al. Clinicogenetic lessons from 370 patients with autosomal recessive limb-girdle muscular dystrophy. Clin Genet. 2019;96(4):341–353. [DOI] [PubMed] [Google Scholar]
- 3. Ten Dam L, Frankhuizen WS, Linssen WHJP, et al. Autosomal recessive limb-girdle and Miyoshi muscular dystrophies in the Netherlands: The clinical and molecular spectrum of 244 patients. Clin Genet. 2019;96(2):126–133. [DOI] [PubMed] [Google Scholar]
- 4. Xie Z, Hou Y, Yu M, et al. Clinical and genetic spectrum of sarcoglycanopathies in a large cohort of Chinese patients. Orphanet J Rare Dis. 2019;14(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chakravorty S, Nallamilli BRR, Khadilkar SV, et al. Clinical and genomic evaluation of 207 genetic myopathies in the Indian subcontinent. Front Neurol. 2020;11:559327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vainzof M, Passos-Bueno MR, Pavanello RCM, Marie SK, Oliveira ASB, Zatz M.. Sarcoglycanopathies are responsible for 68% of severe autosomal recessive limb-girdle muscular dystrophy in the Brazilian population. J Neurol Sci. 1999;164(1):44–49. [DOI] [PubMed] [Google Scholar]
- 7. Ginjaar HB, Van Der Kooi AJ, Ceelie H, et al. Sarcoglycanopathies in Dutch patients with autosomal recessive limb girdle muscular dystrophy. J Neurol. 2000;247(7):524–529. [DOI] [PubMed] [Google Scholar]
- 8. Alonso-Pérez J, González-Quereda L, Bello L, et al. New genotype-phenotype correlations in a large European cohort of patients with sarcoglycanopathy. Brain. 2020;143(9):2696–2708. [DOI] [PubMed] [Google Scholar]
- 9. Nigro V, Piluso G, Belsito A, et al. Identification of a novel sarcoglycan gene at 5q33 encoding a sarcolemmal 35 kDa glycoprotein. Hum Mol Genet. 1996;5(8):1179–1186. [DOI] [PubMed] [Google Scholar]
- 10. Jung D, Duclos F, Apostol B, et al. Characterization of δ-sarcoglycan, a novel component of the oligomeric sarcoglycan complex involved in limb-girdle muscular dystrophy. J Biol Chem. 1996;271(50):32321–32329. [DOI] [PubMed] [Google Scholar]
- 11. Nigro V, de Sá Moreira E, Piluso G, et al. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the delta-sarcoglycan gene. Nat Genet. 1996;14(2):195–198. [DOI] [PubMed] [Google Scholar]
- 12. Younus M, Ahmad F, Malik E, et al. SGCD homozygous nonsense mutation (p.Arg97∗) causing limb-girdle muscular dystrophy type 2F (LGMD2F) in a consanguineous family, a case report. Front Genet. 2018;9:727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dinçer P, Bönnemann CG, Erdir Aker Ö, et al. A homozygous nonsense mutation in δ-sarcoglycan exon 3 in a case of LGMD2F. Neuromuscul Disord. 2000;10(4-5):247–250. [DOI] [PubMed] [Google Scholar]
- 14. Bevilacqua JA, Guecaimburu Ehuletche M. D R, Perna A, et al. The Latin American experience with a next generation sequencing genetic panel for recessive limb-girdle muscular weakness and Pompe disease. Orphanet J Rare Dis. 2020;15(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trabelsi M, Kavian N, Daoud F, et al. Revised spectrum of mutations in sarcoglycanopathies. Eur J Hum Genet. 2008;16(7):793–803. [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez-Quereda L, Gallardo E, Töpf A, et al. A new mutation of the SCGA gene is the cause of a late onset mild phenotype limb girdle muscular dystrophy type 2D with axial involvement. Neuromuscul Disord. 2018;28(8):633–638. [DOI] [PubMed] [Google Scholar]
- 17. Oliveira Santos M, Coelho P, Roque R, Conceição I.. Very late-onset limb-girdle muscular dystrophy type 2D: A milder form with a normal muscle biopsy. J Clin Neurosci. 2020;72:471–473. [DOI] [PubMed] [Google Scholar]
- 18. Tarnopolsky M, Hoffman E, Giri M, Shoffner J, Brady L.. Alpha-sarcoglycanopathy presenting as exercise intolerance and rhabdomyolysis in two adults. Neuromuscul Disord. 2015;25(12):952–954. [DOI] [PubMed] [Google Scholar]
- 19. Ponikowski P, Voors AA, Anker SD, et al. ; ESC Scientific Document Group . ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J. 2016;37(27):2129–2200m. [DOI] [PubMed] [Google Scholar]
- 20. Lipshultz SE, Law YM, Asante-Korang A, et al. Cardiomyopathy in children: Classification and diagnosis: A scientific statement from the American Heart Association. 2019;140(1). [DOI] [PubMed] [Google Scholar]
- 21. Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC.. SIFT missense predictions for genomes. Nat Protoc. 2016;11(1):1–9. [DOI] [PubMed] [Google Scholar]
- 22. Tang H, Thomas PD.. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016;32(14):2230–2232. [DOI] [PubMed] [Google Scholar]
- 23. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarz JM, Cooper DN, Schuelke M, Seelow D.. Mutationtaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. [DOI] [PubMed] [Google Scholar]
- 25. Steinhaus R, Proft S, Schuelke M, Schwarz JM, Seelow D, Cooper DN.. OUP accepted manuscript. Nucleic Acids Res. 2021;49(W1):W446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Møller LB, Tümer Z, Lund C, et al. Similar splice-site mutations of the ATP7a gene lead to different phenotypes: Classical Menkes disease or occipital horn syndrome. Am J Hum Genet. 2000;66(4):1211–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ward AJ, Cooper TA.. The pathobiology of splicing. J Pathol. 2010;220(2):152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Díaz-Manera J, Alejaldre A, González L, et al. Muscle imaging in muscle dystrophies produced by mutations in the EMD and LMNA genes. Neuromuscul Disord. 2016;26(1):33–40. [DOI] [PubMed] [Google Scholar]
- 29. Ghaoui R, Cooper ST, Lek M, et al. Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: Outcomes and lessons learned. JAMA Neurol. 2015;72(12):1424–1432. [DOI] [PubMed] [Google Scholar]
- 30. Nallamilli BRR, Chakravorty S, Kesari A, et al. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol. 2018;5(12):1574–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Savarese M, Fruscio GD, Magri F, et al. The genetic basis of undiagnosed muscular dystrophies and myopathies: Results from 504 patients. Neurology. 2016; 87(1):71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beecroft SJ, Yau KS, Allcock RJN, et al. Targeted gene panel use in 2249 neuromuscular patients: The Australasian referral center experience. Ann Clin Transl Neurol. 2020;7(3):353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gonzalez-Quereda L, Rodriguez MJ, Diaz-Manera J, et al. Targeted next-generation sequencing in a large cohort of genetically undiagnosed patients with neuromuscular disorders in Spain. 2020;11(5):539–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Töpf A, Johnson K, Bates A, et al. ; MYO-SEQ consortium . Sequential targeted exome sequencing of 1001 patients affected by unexplained limb-girdle weakness. Genet Med. 2020;22(9):1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boito C, Fanin M, Siciliano G, Angelini C, Pegoraro E.. Novel sarcoglycan gene mutations in a large cohort of Italian patients. J Med Genet. 2003;40(5):e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Allamand V, Donahue KM, Straub V, Davisson RL, Davidson BL, Campbell KP.. Early adenovirus-mediated gene transfer effectively prevents muscular dystrophy in alpha-sarcoglycan-deficient mice. Gene Ther. 2000;7(16):1385–1391. [DOI] [PubMed] [Google Scholar]
- 37. Pozsgai ER, Griffin DA, Heller KN, Mendell JR, Rodino-Klapac LR.. Systemic AAV-mediated β-sarcoglycan delivery targeting cardiac and skeletal muscle ameliorates histological and functional deficits in LGMD2E mice. Mol Ther. 2017;25(4):855–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Israeli D, Cosette J, Corre G, et al. An AAV-SGCG dose-response study in a γ-sarcoglycanopathy mouse model in the context of mechanical stress. Mol Ther - Methods Clin Dev. 2019;13:494–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu W, Pajusalu S, Lake NJ, et al. Estimating prevalence for limb-girdle muscular dystrophy based on public sequencing databases. Genet Med. 2019;21(11):2512–2520. [DOI] [PubMed] [Google Scholar]
- 40. Fanin M, Nascimbeni AC, Aurino S, et al. Frequency of LGMD gene mutations in Italian patients with distinct clinical phenotypes. Neurology. 2009;72(16):1432–1435. [DOI] [PubMed] [Google Scholar]
- 41. Passos-Bueno MR, Vainzof M, Moreira ES, Zatz M.. Seven autosomal recessive limb-girdle muscular dystrophies in the Brazilian population: From LGMD2A to LGMD2G. Am J Med Genet. 1999;82(5):392–398. [DOI] [PubMed] [Google Scholar]
- 42. Cheng L, Guo XF, Yang XY, et al. Δ-Sarcoglycan is necessary for early heart and muscle development in Zebrafish. Biochem Biophys Res Commun. 2006;344(4):1290–1299. [DOI] [PubMed] [Google Scholar]
- 43. Matsunari H, Honda M, Watanabe M, et al. Pigs with δ-sarcoglycan deficiency exhibit traits of genetic cardiomyopathy. Lab Investig. 2020;100(6):887–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sandonà D, Betto R.. Sarcoglycanopathies: Molecular pathogenesis and therapeutic prospects. Expert Rev Mol Med. 2009;11:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rutschow D, Bauer R, Göhringer C, et al. S151A δ-sarcoglycan mutation causes a mild phenotype of cardiomyopathy in mice. Eur J Hum Genet. 2014;22(1):119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tarakci H, Berger J.. The sarcoglycan complex in skeletal muscle. Front Biosci - Landmark. 2016;21(4):744–756. [DOI] [PubMed] [Google Scholar]
- 47. Draviam RA, Shand SH, Watkins SC.. The β-δ-core of sarcoglycan is essential for deposition at the plasma membrane. Muscle and Nerve. 2006;34(6):691–701. [DOI] [PubMed] [Google Scholar]
- 48. Shi W, Chen Z, Schottenfeld J, Stahl RC, Kunkel LM, Chan Y-M.. Specific assembly pathway of sarcoglycans is dependent on beta- and delta-sarcoglycan. Muscle Nerve. 2004;29(3):409–419. [DOI] [PubMed] [Google Scholar]
- 49. Semplicini C, Vissing J, Dahlqvist JR, et al. Clinical and genetic spectrum in limb-girdle muscular dystrophy type 2E. Neurology. 2015;84(17):1772–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dosi C, Rubegni A, Cassandrini D, et al. Alpha-sarcoglycanopathy presenting as myalgia and hyperCKemia in two adults with a long-term follow-up. Case reports. Acta Myol. 2020;39(4):218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cantero D, Hernández-Laín A, Martínez JFG, et al. Milder forms of α-sarcoglicanopathies diagnosed in adulthood by NGS analysis. J Neurol Sci. 2018;394:63–67. [DOI] [PubMed] [Google Scholar]
- 52. Ozawa E, Mizuno Y, Hagiwara Y, Sasaoka T, Yoshida M.. Molecular and cell biology of the sarcoglycan complex. Muscle and Nerve. 2005;32(5):563–576. [DOI] [PubMed] [Google Scholar]
- 53. van Putten M, Lloyd EM, de Greef JC, Raz V, Willmann R, Grounds MD.. Mouse models for muscular dystrophies: An overview. Dis Model Mech. 2020;13(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Straub V, Duclos F, Venzke DP, et al. Molecular pathogenesis of muscle degeneration in the δ-sarcoglycan-deficient hamster. Am J Pathol. 1998;153(5):1623–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. White SJ, Uitte de Willige S, Verbove D, et al. Sarcoglycanopathies and the risk of undetected deletion alleles in diagnosis. Hum Mutat. 2005;26(1):59. [DOI] [PubMed] [Google Scholar]
- 56. Wildförster V, Dekomien G.. Detecting copy number variations in autosomal recessive limb-girdle muscular dystrophies using a multiplex ligation-dependent probe amplification (MLPA) assay. Mol Cell Probes. 2009;23(1):55–59. [DOI] [PubMed] [Google Scholar]
- 57. Politano L, Nigro V, Passamano L, et al. Evaluation of cardiac and respiratory involvement in sarcoglycanopathies. Neuromuscul Disord. 2001;11(2):178–185. [DOI] [PubMed] [Google Scholar]
- 58. Schade van Westrum SM, Dekker LRC, De Voogt WG, et al. Cardiac involvement in Dutch patients with sarcoglycanopathy: A cross-sectional cohort and follow-up study. Muscle and Nerve. 2014;50(6):909–913. [DOI] [PubMed] [Google Scholar]
- 59. Melacini P, Fanin M, Duggan DJ, et al. Heart involvement in muscular dystrophies due to sarcoglycan gene mutations. Muscle Nerve. 1999;22(4):473–479. [DOI] [PubMed] [Google Scholar]
- 60. Sakamoto A, Ono K, Abe M, et al. Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, δ-sarcoglycan, in hamster: An animal model of disrupted dystrophin-associated glycoprotein complex. Proc Natl Acad Sci U S A. 1997;94(25):13873–13878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Coral-Vazquez R, Cohn RD, Moore SA, et al. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: A novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98(4):465–474. [DOI] [PubMed] [Google Scholar]
- 62. Guyon JR, Mosley AN, Jun SJ, et al. δ-Sarcoglycan is required for early zebrafish muscle organization. Exp Cell Res. 2005;304(1):105–115. [DOI] [PubMed] [Google Scholar]
- 63. Tsubata S, Bowles KR, Vatta M, et al. Mutations in the human δ-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest. 2000;106(5):655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goehringer C, Rutschow D, Bauer R, et al. Prevention of cardiomyopathy in δ-sarcoglycan knockout mice after systemic transfer of targeted adeno-associated viral vectors. Cardiovasc Res. 2009;82(3):404–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.