Abstract
Genetic prion diseases are a rare and diverse group of fatal neurodegenerative disorders caused by pathogenic sequence variations in the prion protein gene, PRNP. Data on CSF biomarkers in patients with genetic prion diseases are limited and conflicting results have been reported for unclear reasons.
Here, we aimed to analyse the diagnostic accuracy of CSF biomarkers currently used in prion clinical diagnosis in 302 symptomatic genetic prion disease cases from 11 prion diagnostic centres, encompassing a total of 36 different pathogenic sequence variations within the open reading frame of PRNP.
CSF samples were assessed for the surrogate markers of neurodegeneration, 14-3-3 protein (14-3-3), total-tau protein (t-tau) and α-synuclein and for prion seeding activity through the real-time quaking-induced conversion assay. Biomarker results were compared with those obtained in healthy and neurological controls. For the most prevalent PRNP pathogenic sequence variations, biomarker accuracy and associations between biomarkers, demographic and genetic determinants were assessed. Additionally, the prognostic value of biomarkers for predicting total disease duration from symptom onset to death was investigated.
High sensitivity of the four biomarkers was detected for genetic Creutzfeldt–Jakob disease associated with the E200K and V210I mutations, but low sensitivity was observed for mutations associated with Gerstmann–Sträussler–Scheinker syndrome and fatal familial insomnia. All biomarkers showed good to excellent specificity using the standard cut-offs often used for sporadic Creutzfeldt–Jakob disease. In genetic prion diseases related to octapeptide repeat insertions, the biomarker sensitivity correlated with the number of repeats. New genetic prion disease-specific cut-offs for 14-3-3, t-tau and α-synuclein were calculated. Disease duration in genetic Creutzfeldt–Jakob disease-E200K, Gerstmann–Sträussler–Scheinker-P102L and fatal familial insomnia was highly dependent on PRNP codon 129 MV polymorphism and was significantly associated with biomarker levels.
In a large cohort of genetic prion diseases, the simultaneous analysis of CSF prion disease biomarkers allowed the determination of new mutation-specific cut-offs improving the discrimination of genetic prion disease cases and unveiled genetic prion disease-specific associations with disease duration.
Keywords: biomarker, cerebrospinal fluid, diagnostic marker, genetic prion diseases
Schmitz et al. assess the diagnostic accuracy of the three CSF biomarkers and the RT-QuIC assay most commonly used for the clinical diagnosis of genetic prion diseases. They show that each disease has a characteristic marker profile, and determine new mutation-specific cut-off values that will help to improve discrimination of the diseases.
Introduction
Prion diseases are transmissible and invariably fatal diseases characterized by rapidly progressive dementia. The most prevalent form is sporadic Creutzfeldt–Jakob disease (sCJD). Genetic prion diseases (gPrD) arise due to pathogenic sequence variations (either point mutations or insertions) within the open reading frame of the prion protein gene (PRNP), accounting for 10–15% of prion diseases. Historically, gPrD have been classified into three main types based on clinicopathological observations: genetic CJD (gCJD), Gerstmann–Sträussler–Scheinker syndrome (GSS) and fatal familial insomnia (FFI).1-3 The most common gPrD is gCJD caused by the E200K mutation (gCJD-E200K), which clinically resembles sCJD.4 Many other point mutations have been described to underlie gCJD, such as V210I (gCJD-V210I). GSS is characterized by longer survival often with later cognitive impairment, compared to other prion diseases. The most prevalent GSS-associated mutation is P102L (GSS-P102L). In contrast to the other gPrD, FFI is caused by a unique PRNP single point mutation, D178N, most commonly combined with methionine in cis position at codon 129, presenting as FFI. In contrast, patients bearing the same D178N mutation when valine is present at codon 129 of the mutated allele typically develop gCJD.5 In the presence of octapeptide repeat insertions (OPRI) in PRNP, the number of repeats is a strong factor determining the disease phenotype. Although contention still exists regarding the pathogenicity of 1- and 2-OPRI, usually when the OPRI is <5 carriers manifest gCJD, whereas OPRI ≥5 cases associate with a heterogeneous presentation and often a long disease course.3,6
Clinical evaluation and differential diagnosis of prion diseases is supported by CSF biomarkers. They have been primarily established based on sCJD samples and, thus, cut-off points were determined based on maximization of combined sensitivity and specificity to be optimal for the discrimination of this disease. CSF biomarkers currently included in the sCJD diagnostic criteria are 14-3-37 and the real-time quaking-induced conversion (RT-QuIC) assay,8 although high total-tau (t-tau) levels have also traditionally been used to support diagnosis.9
Both 14-3-3 and t-tau are surrogate markers of neuro-axonal damage, being elevated in sCJD compared to controls.10 By contrast, prion protein (PrP) seeding activity detected by RT-QuIC is a direct marker of pathology and, therefore, specifically present in prion diseases.11,12 In addition, α-synuclein (α-syn) has been recently demonstrated to be highly increased in sCJD, and its diagnostic value has been interlaboratory-validated, achieving similar performance to that of 14-3-3 and t-tau.13,14 The good diagnostic accuracy of sCJD CSF biomarkers is usually translated to gCJD, which presents clinicopathological features similar to those of sCJD, but not to other gPrD such as GSS-P102L or FFI,15 which currently lack robust CSF diagnostic biomarkers.
In the present work, we analysed the CSF prion disease biomarker profiles in a large cohort composed of gPrD, healthy controls and neurological disease controls. We report data on 14-3-3 levels, obtained with both the traditional western-blot approach and enzyme-linked immunosorbent assay (ELISA), t-tau and α-syn concentration and RT-QuIC results. The diagnostic utility of these biomarkers for the most prevalent gPrD forms were calculated and new genetic disease-specific cut-off points are proposed to improve the sensitivity offered by each biomarker. Additionally, because several CSF biomarkers have recently been associated with overall disease duration or with survival from the time of lumbar puncture (biomarker measurement) in sCJD,16–18 their prognostic value in gPrD was also investigated.
Materials and methods
Study population
We retrospectively analysed a total of 302 CSF samples obtained from symptomatic gPrD patients recruited at the reference units from 11 different participant centres: (i) Clinical Dementia Center and the National Reference Center for CJD Surveillance at the University Medical Center, Göttingen, Germany; (ii) National Referral Center for CJD, Coimbra, Portugal; (iii) Alzheimer’s Disease and Other Cognitive Disorders Unit, Hospital Clínic, Barcelona, Spain; (iv) Medical University of Lodz, Poland; (v) National Center of Microbiology-Carlos III Institute of Health, Madrid, Spain; (vi) Istituto Superiore di Sanità, Rome, Italy; (vii) Slovak Medical University, Bratislava, Slovakia; (viii) Australian National CJD Registry, The Florey Institute of Neuroscience and Mental Health, Melbourne, Australia; (ix) Department of Neurology, Memory and Aging Center, University of California, San Francisco (UCSF), San Francisco, California, USA; (x) Austrian Reference Centre for Human Prion Diseases, Vienna, Austria; and (xi) Hungarian Reference Centre for Human Prion Diseases, Budapest, Hungary. The diagnoses of genetic prion diseases were carried out according to surveillance criteria after PRNP analysis. The patients were classified according to established diagnostic criteria.19,20 For comparison, 51 healthy controls and 111 neurological disease controls were included. The neurological disease control group was composed of cases diagnosed with non-primarily neurodegenerative neurological and psychiatric conditions according to acknowledged standard neurological clinical and paraclinical findings based on the ICD10 definitions cases, without cognitive impairment or dementia at the time of sampling.
CSF analyses
Lumbar punctures were performed for diagnostic purposes at the time point of first clinical workup (UCSF was at subject’s first research visit). CSF samples were stored in polypropylene tubes at −80°C until analysis. CSF was analysed in the diagnostic, research and neurochemistry laboratories of the Clinical Dementia Center, Göttingen, except 14-3-3 western blot, which was locally analysed through each national CJD surveillance unit. The presence of 14-3-3 protein was analysed by western blot as described previously.21 Quantification of 14-3-3 was performed using the CircuLex 14-3-3 gamma ELISA kit as reported before.22 T-tau was quantitatively measured using the INNOTEST® hTAU-Ag ELISA kit from Fujirebio. α-Syn was quantified using an electrochemiluminiscent ELISA platform as described before.13 RT-QuIC was performed following an established protocol.23
Statistical analyses
Statistical analyses were only performed with diagnostic groups in which n > 10: healthy controls, neurological disease controls, gCJD-E200K, gCJD-V210I, FFI and GSS-P102L.
Differences in age between groups were tested with ANOVA, and P-values were corrected with Tukey’s post-test. Differences in the sex ratio and PRNP codon 129 MV genotypes between groups were tested with the chi-squared test followed by Bonferroni P-value adjustment. The codon 129 VV genotype was excluded from analysis because of the low number of cases.
Differences in biomarker values depending on the diagnostic groups were tested with Tobit regression models, available in the AER R-package.24 These models allow the inclusion of censored dependent variables, which in this study were considered as the cases out of the quantification limits of each ELISA test of 14-3-3, t-tau and α-syn. For RT-QuIC, left-censoring was considered at 10 000 relative fluorescence units (RFU) (the definition of a negative RT-QuIC outcome) and right-censoring was considered at 65 000 RFU (the maximum fluorimetric value retrieved by the plate reader). In each model, biomarker data were log-transformed and the following parameters were included as covariates: age, sex and cohort of origin (the latter as random-effects variable). Pair-wise multiple comparisons of means were corrected with the Tukey method through the lsmeans R-package.25 Association between OPRI and biomarker data and correlations between biomarker values in the disease groups were assessed with Kendall’s tau coefficient, which measures the monotonic association between two measured parameters. We used the cenken function from the NADA R-package26 that allows inclusion of left-censored data. This was an advantage because correlations could be computed with RT-QuIC data that were negative (RFU ≤ 10 000). On the contrary, it was not possible to include right-censoring, which implied assuming the upper limit of quantification for the values above of this. Kappa index, through the vcd R-package,27–29 was used to assess agreement between the classification results with the different biomarkers in a pair-wise fashion.
Diagnostic accuracy analyses were conducted with the three ELISA-based biomarkers (14-3-3, t-tau and α-syn). Receiver operating characteristic (ROC) curves were calculated and area under the curve (AUC) values were extracted with GraphPad Prism 6.0. Optimal disease-specific cut-off points were determined by maximizing the sensitivity × specificity product using the cut-off point R package.30 The same package was used to obtain the precision-recall plots. RT-QuIC data were excluded from this analysis because all control cases had a negative outcome, thus establishing cut-off points made no sense.
Differences in disease duration (time from disease onset to death) were tested with Cox proportional hazards models for survival data, available through the coxph function from survival R-package.31,32 Proportional hazards assumptions were met in all cases. Covariates were included in the models when they were significant, and in the case of multiple comparisons, P-value Tukey corrections were conducted with the multcomp R-package.33 To evaluate the prognostic performance of the biomarkers, these were used as explanatory variables in the Cox proportional hazard models. Biomarker values were divided by 1000 to facilitate interpretation of hazard ratios (HR). In this case, due to the numerous cases with RT-QuIC negative outcome, we used RT-QuIC data as a dichotomized predictor (dummy variable).
Statistical significance was considered at P ≤ 0.05.
Ethics
This study was carried out in accordance with the Declaration of Helsinki and with informed written consent provided by patients or by their next of kin in the case of cognitive impairment. The study was approved by the ethics committees at the University of Göttingen and by local ethic committees from each participating centre:
Hospital Clínic de Barcelona Ethics Committee approval ‘Genetic dementia (autosomal dominant Alzheimer’s disease and genetic prion diseases) in preclinical and early stages of the disease: cognitive performance, neuroimaging and biochemical markers’, code HCB/2016/0329.
Ministry of Health of the Slovak Republic (No: 33-SZU-11): Genetic risk factors of prion diseases: identification, characterization, study of interactions with exogenous risks and effective prevention.
Local ethic committee of the University Medicine Goettingen, Von Siebold-Str. 3 37075 Göttingen, approval No. 24/8/12 Biomarker based diagnosis in rapid progressive dementias – optimisation of diagnostic protocols and approval No. 11/11/93 Studies on the epidemiology, early diagnosis and molecular pathology of human spongiform encephalopathies.
Ethics committee of the Medical University of Lodz approval No. RNN/282/16/KE (Collecting of biological samples from neurodegenerative diseases) and No. RNN/355/17/KE (Genetic, epigenetic and proteomic analyses in prion diseases: studies in archival samples).
Ethics committee granted by the University of Melbourne, approval No. 1648441 ‘Investigation of biomarkers for Alzheimer’s disease and neurodegenerative diseases in human cerebrospinal fluid’.
The ethics committee approval is in Portuguese HUC-43-09: ‘Early diagnosis of dementia: evaluation of classification criteria and of new research instruments’.
Committee on Human Research (CHR), University of California, San Francisco (UCSF), approval No. 10-04905.
The ethics committee approval is in Italy, No. CE/12/365 ‘Study of the genetic forms of Transmissible Spongiform Encephalopathies (TSE)’.
Samples were registered in the National Biobank Registry as collection #C.0002119. The clinical data information was associated to anonymous data according to the ISCIII Ethics Committee inform CEI 34_2017.
The ethics committee approval is at the Medical University of Vienna EK 397/2011. ‘Development of diagnostic multiparametric markers in the CSF for the early diagnosis and differentiating neurodegenerative diseases: a retrospective study’.
Role of the funding source
The funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the paper for publication.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.
Results
General description of the working cohort
A total of 464 cases were included in this study, stratified into healthy controls (n = 51), neurological disease controls (n = 111), GSS (n = 25; A117V and D202N are not shown) and gCJD (n = 181; P238S is not shown) associated mutations, FFI (n = 68), OPRI (n = 22), nonsense mutations (n = 1) and five other mutations for which a causative role for prion disease has not been validated (n = 5) (not shown). Demographic, genetic (including PRNP codon 129) and relevant biomarker information is provided in Table 1. There were considerable differences in age, with FFI cases characterized by significantly younger disease onset compared to neurological disease controls, gCJD-E200K and gCJD-V210I (P < 0.0001 in all comparisons). The gCJD-V210I cases were also significantly older than healthy controls (P = 0.0049) and GSS-P102L (P = 0.0222) cases. In all disease groups, incidence was higher in females than in males, except in the case of FFI; however, the difference in sex ratio was only significant between the gCJD-E200K and FFI group (P = 0.0017). Regarding PRNP codon 129, the MM genotype was the most frequent one across all gPrD, followed by the MV genotype, with no differences among disease groups (P = 0.4194). The exception was the 5-OPRI group, in which 6 of 10 samples were VV, although no statistical test was performed due to low number of cases (Table 1).
Table 1.
Diagnostic accuracy of CSF biomarkers in gPrD
| Demographic and genetic data | Biomarkers | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WB | ELISA | Seeding assay | |||||||||||||||
| Sex | Age | Codon 129a | 14-3-3 | 14-3-3 (AU/ml) | t-tau (pg/ml) | α-Syn (pg/ml)b | RT-QuIC (RFU) | ||||||||||
| gPrD | Mutation | n | F | M | Mean ± SD | MM | MV | VV | Sens. | Sens. | Mean ± SD | Sens. | Mean ± SD | Sens. | Mean ± SD | Sens. | Mean ± SD |
| GSS | P102L | 14 | 8 | 6 | 54 ± 11 | 12 | 2 | 0 | 14% | 43% | 37 772 ± 45 859 | 43% | 2355 ± 4000 | 43% | 1509 ± 1982 | 43% | 17 214 ± 13 473 |
| P105T | 3 | 2 | 1 | 36 ± 17 | 0 | 3 | 0 | 33% | 0% | 88 59 ± 5629 | 0% | 668 ± 523 | 33% | 466 ± 293 | 0% | 7945 | |
| A133V | 1 | 1 | 0 | 62 | 1 | 0 | 0 | 0% | 0% | 10 456 | 0% | 446 | 0% | 169 | 100% | 49 481 | |
| V176G | 1 | 1 | 0 | 61 | 0 | 0 | 1 | 100% | 0% | 13 300 | 0% | 909 | 0% | 162 | 0% | 8456 | |
| F198S | 1 | 0 | 1 | 51 | 0 | 1 | 0 | 100% | 0% | 14 155 | 100% | 1612 | 0% | 461 | 100% | 14 155 | |
| D202G | 1 | 0 | 1 | 65 | 0 | 1 | 0 | 100% | 100% | 24 553 | 0% | 678 | 100% | 1970 | 0% | 7897 | |
| Q217R | 1 | 0 | 1 | 59 | 0 | 1 | 0 | 100% | 100% | 21 578 | 100% | 2787 | 100% | 1074 | 0% | 8312 | |
| Y218N | 1 | 1 | 0 | NA | 0 | 1 | 0 | 0% | 0% | 2133 | 0% | 125 | 0% | 258 | 0% | 8123 | |
| FFI | D178N | 68 | 24 | 44 | 51 ± 10 | 45 | 23 | 0 | 9% | 13% | 11 018 ± 12 075 | 18% | 917 ± 1433 | 21% | 763 ± 1808 | 28% | 14 028 ± 8426 |
| gCJD | G114V | 1 | 0 | 1 | 20 | 0 | 1 | 0 | 0% | 100% | 64 771 | 0% | 540 | 0% | 130 | 100% | 36 987 |
| D178N(V) | 1 | 1 | 0 | 48 | 0 | 0 | 1 | 0% | 0% | 15 604 | 0% | 885 | 100% | 1045 | 100% | 25 039 | |
| V180I | 1 | 0 | 1 | 77 | 1 | 0 | 0 | 100% | 100% | 26 707 | 100% | 1730 | 100% | 884 | 0% | 7435 | |
| T188A | 1 | 1 | 0 | 82 | 1 | 0 | 0 | 100% | 100% | 69 047 | 100% | 3093 | 100% | 5003 | 0% | 7967 | |
| T188K | 2 | 2 | 0 | 63 ± 8 | 0 | 2 | 0 | 100% | 100% | 46 492 ± 5178 | 100% | 7536 ± 7432 | 100% | 2901 ± 2753 | 100% | 46 182 ± 403 | |
| K194E | 1 | 0 | 1 | 71 | 0 | 1 | 0 | 0% | 100% | 32 803 | 0% | 1116 | 100% | 852 | 100% | 31 256 | |
| E196K | 7 | 4 | 3 | 70 ± 6 | 4 | 2 | 1 | 86% | 86% | 56 499 ± 35 599 | 86% | 6524 ± 6544 | 100% | 3790 ± 2102 | 100% | 48 515 ± 12 820 | |
| E200K | 112 | 74 | 38 | 61 ± 10 | 76 | 34 | 1 | 70% | 82% | 60 356 ± 39 852 | 81% | 4851 ± 5628 | 87% | 4377 ± 5294 | 93% | 45 411 ± 14 065 | |
| V203G | 1 | 1 | 0 | 76 | 0 | 0 | 1 | 100% | NA | NA | 100% | 6332 | NA | NA | NA | NA | |
| R208H | 4 | 2 | 2 | 62 ± 7 | 3 | 1 | 0 | 100% | 100% | 101 791 ± 22 151 | 100% | 7599 ± 2321 | 100% | 3527 ± 1355 | 100% | 58 745 ± 2321 | |
| V210I | 47 | 26 | 21 | 64 ± 10 | 34 | 11 | 2 | 79% | 94% | 94 874 ± 32 428 | 96% | 10 543 ± 9998 | 96% | 8028 ± 10 861 | 87% | 47 817 ± 192 86 | |
| E211Q | 2 | 2 | 0 | 62 ± 30 | 2 | 0 | 0 | 50% | 50% | 25 193 ± 20 471 | 100% | 4281 ± 4131 | 50% | 1853 ± 1851 | 100% | 50 524 ± 20 471 | |
| Insert | OPRI | 1 | 1 | 0 | 68 | 0 | 1 | 0 | 0% | 100% | 20 786 | 100% | 1936 | 0% | 392 | 100% | 367 66 |
| 1-OPRI | 3 | 2 | 1 | 64 ± 4 | 2 | 1 | 0 | 100% | 100% | 90 422 ± 28 746 | 100% | 7095 ± 3674 | 100% | 4925 ± 2405 | 100% | 41 482 ± 13 212 | |
| 2-OPRI | 1 | 0 | 1 | 61 | 1 | 0 | 0 | 100% | 100% | 38 773 | 100% | 3454 | 100% | 1874 | 100% | 44 438 | |
| 4-OPRI | 5 | 3 | 2 | 61 ± 7 | 3 | 0 | 2 | 60% | 80% | 51 480 ± 45 801 | 60% | 1934 ± 1377 | 60% | 3025 ± 2832 | 60% | 28 295 ± 23 518 | |
| 5-OPRI | 10 | 9 | 1 | 63 ± 10 | 1 | 3 | 6 | 50% | 90% | 30 196 ± 16 730 | 80% | 2367 ± 1922 | 80% | 1531 ± 846 | 60% | 25 556 ± 14 822 | |
| 6-OPRI | 1 | 0 | 1 | 37 | 1 | 0 | 0 | 0% | 0% | 3715 | 0% | 321 | 0% | 112 | 0% | 8745 | |
| 8-OPRI | 1 | 0 | 1 | 48 | 0 | 1 | 0 | 0% | 0% | 9928 | 0% | 212 | 0% | 221 | 0% | 8453 | |
| HC | – | 51 | 28 | 23 | 56 ± 10 | NA | NA | 100% | 3064 ± 1684 | 100% | 247 ± 124 | 100% | 237 ± 73 | 100% | 7842 | ||
| ND | – | 111 | 52 | 59 | 6 0 ± 16 | NA | 89% | 91% | 7953 ± 6502 | 92% | 403 ± 433 | 93% | 326 ± 194 | 100% | 8345 | ||
Demographic (number of cases, sex and age), genetic (PRNP codon 129 MV genotype) and CSF biomarker (14-3-3, t-tau, α-syn and RT-QuIC) data in the study cohort is shown, which includes 161 controls and 302 symptomatic gPrD cases. Sensitivity and specificity are calculated based on sCJD cut-offs: 14-3-3 WB = positive/negative (inconclusive results were considered negative), 14-3-3 ELISA > 20 000 AU/ml, t-tau > 1300 pg/ml, α-syn > 680 pg/ml, RT-QuIC > 10 000 RFU. F = female; HC = healthy controls; M = male; NA = not available; ND = neurological disease controls; RFU = relative fluorescence units from positive cases; Sens. = sensitivity; WB = western blot.
The performance of prion disease biomarkers was evaluated in each disease group considering the established cut-off points for sCJD: >20 000 AU/ml for ELISA 14-3-3,22 >1300 pg/ml for t-tau,10 >680 pg/ml for aSyn13 and >10 000 RFU for RT-QuIC.23 The best sensitivities were obtained in the gCJD group, generally >80% for all tested quantitative biomarkers: western blot 14-3-3 performed worst in all diagnostic groups. Biomarker sensitivities dropped to <50% in the GSS-P102L group and were very low for FFI, in which the best performer was RT-QuIC with a sensitivity of only 28%. Specificity was 100% with all biomarkers when compared to healthy controls. When compared to neurological disease controls, only RT-QuIC exhibited 100% specificity, whereas other biomarker specificities ranged between 89% and 93% (Table 1). In the total gPrD population, ELISA 14-3-3 displayed a higher sensitivity (63%) than the western blot method (52%; Table 1).
Concentrations of different biomarkers in genetic prion diseases
CSF biomarkers in the most prevalent groups were statistically analysed controlling for the effect of age, sex and cohort of origin. Because a significant proportion of cases had results beyond the assay limits of quantification, regression models considering data censoring were applied. The four analysed biomarkers displayed the same concentration profiles across the panel of the diagnostic groups, with the following ascending order: healthy controls < neurological disease controls < FFI < GSS-P102L < gCJD-E200K < gCJD-V210I. Each major cohort presented with similar characteristic values across all four biomarkers (e.g. for gCJD, all four were very high), consistent with the fact that most pair-wise comparisons between major cohorts were statistically significant (Fig. 1E, legend statistic). The only non-significant differences in pair-wise comparisons were observed for 14-3-3 levels between gCJD-E200K and GSS-P102L (P = 0.138; Fig. 1A, legend statistic), for t-tau levels between GSS-P102L and FFI (P = 0.079) and for α-syn levels between neurological disease controls and FFI (P = 0.108; Fig. 1B and C, legend statistic). In addition, the distinction between healthy controls and neurological disease controls was not significant for t-tau (P = 0.742) and α-syn (P = 0.788). Although RT-QuIC is traditionally read-out as a binary technique (positive versus negative result), we considered it in our analysis as a quantitative method. Thus, maximum RFU values were compared among diagnostic groups. Because all healthy controls and neurological disease controls cases were negative in RT-QuIC, all comparisons involving these two groups were not analysed. Regarding the gPrD groups, we could observe two subgroups with significantly distinct mean RT-QuIC outcomes: a high RFU group composed of both gCJD forms (E200K and V210I) and a low RFU group composed of GSS-P102L and FFI (Fig. 1D, legend statistic).
Figure 1.
Evaluation of biomarker values in the different diagnostic groups. The levels of 14-3-3 (measured by ELISA) (A), t-tau (B) and α-syn (C) were measured in healthy controls (HC), neurological disease controls (ND), gCJD-E200K, gCJD-V210I, GSS-P102L and FFI cases. (D) RT-QuIC maximum RFU was also recorded in the same diagnostic groups. Data are plotted on a logarithmic scale, except RT-QuIC RFU data. Horizontal bars represent mean and standard error (SE). Differences among diagnostic groups were tested with Tobit models and Tukey contrasts after controlling the effect of age, sex and cohort, as explained in the ‘Materials and methods’ section. Resultant corrected P-values for pair-wise group comparisons: HC versus ND 14-3-3 P < 0.0001, t-tau P = 0.7421, α-syn P = 0.7880, RT-QuIC not available (NA); HC versus gCJD-E200K P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; HC versus gCJD-V210I 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; HC versus GSS-P102L 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; HC versus FFI 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P = 0.0107, RT-QuIC NA; ND versus gCJD-E200K P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; ND versus gCJD-V210I 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; ND versus GSS-P102L 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC NA; ND versus FFI 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P = 0.1084, RT-QuIC NA; gCJD-E200K versus gCJD-V210I 14-3-3 P = 0.0113, t-tau P = 0.0020, α-syn P = 0.0060, RT-QuIC P = 0.9430; gCJD-E200K versus GSS-P102L 14-3-3 P = 0.1383, t-tau P = 0.0099, α-syn P = 0.0057, RT-QuIC P < 0.0001; gCJD-E200K versus FFI 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC P < 0.0001; gCJD-V210I versus GSS-P102L 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC P < 0.0001; gCJD-V210I versus FFI 14-3-3 P < 0.0001, t-tau P < 0.0001, α-syn P < 0.0001, RT-QuIC P < 0.0001; GSS-P102L versus FFI 14-3-3 P = 0.0004, t-tau P = 0.0789, α-syn P = 0.0327, RT-QuIC P = 0.8256.
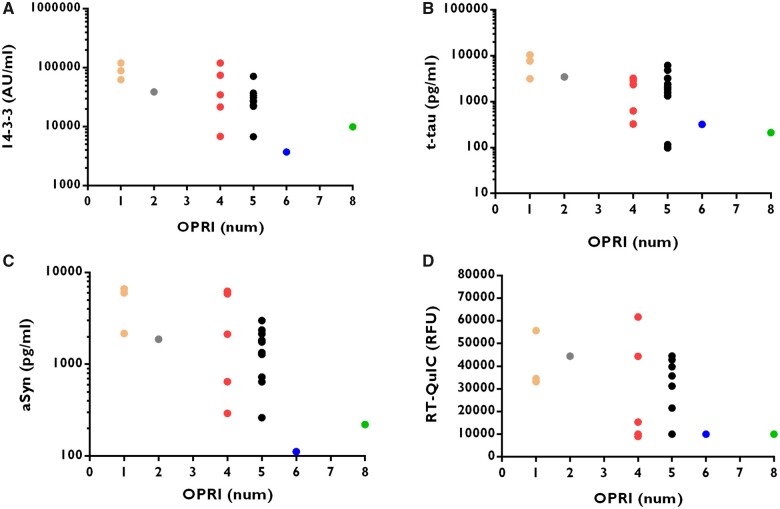
In all OPRI-related gPrD, the number of insertions was significantly inversely correlated with each biomarker concentration or maximum RFU for RT-QuIC (Fig. 2A–D).
Figure 2.
Biomarker concentration is related to the number of OPRI. The levels of 14-3-3 (A), t-tau (B) and α-syn (C) were measured in genetic OPRI cases. RT-QuIC maximum RFU was also recorded in the same cases (D). Data are plotted on a logarithmic scale, except RT-QuIC RFU data. Association between the biomarker value and number of insertions was measured with Kendall’s tau (shown together with related P-value), which is a non-parametric correlation coefficient (cc) as explained in the ‘Materials and methods’ section. For 14-3-3 we calculated cc = −0.433, P = 0.0035, for t-tau cc = −0.3714, P = 0.0125, for α-syn cc = −0.4190 P = 0.0045, and for RT-QuIC cc = −0.2810, P = 0.0486. The figure shows that the number of OPRI is indirectly related to the biomarker level—the lower the OPRI, the higher the biomarker level.
Correlation between biomarker values in genetic prion diseases
Correlations between different biomarkers or PrPSc detection via RT-QuIC were assessed with Kendall’s tau coefficient that allowed accommodation of values out of quantification limits (Table 2). Both gCJD forms had similar profiles, characterized by positive correlations between all surrogate biomarkers: 14-3-3, t-tau and α-syn. GSS-P102L data rendered significant correlations in all biomarkers, including RT-QuIC. FFI data unravelled significant correlations between all biomarkers except t-tau, although these correlations were not as strong as those found in the other gPrD groups (Table 2). Kappa index was used to evaluate agreement between diagnostic biomarker outcomes using previously established sCJD cut-off points in the classification of gPrD (Table 3). For gCJD, moderate and substantial agreement indices were found between surrogate markers of neuronal damage, but not with RT-QuIC results. In the case of GSS-P102L, although all pair-wise combinations showed agreement, highest agreement indices were observed between RT-QuIC and 14-3-3 and between RT-QuIC and t-tau. Only moderate agreement was found in the classification of FFI cases, being worse when involved with t-tau results (Table 3).
Table 2.
Correlation between different surrogate biomarker levels in CSF in the four largest mutation cohorts
| 14-3-3 | t-tau | α-Syn | RT-QuIC | |
|---|---|---|---|---|
| gCJD-E200K | ||||
| 14-3-3 | – | <0.0001 | <0.0001 | 0.2383 |
| t-tau | 0.5119 | – | <0.0001 | 0.5866 |
| α-Syn | 0.5494 | 0.5115 | – | 0.7080 |
| RT-QuIC | 0.0778 | 0.0360 | 0.0249 | – |
| gCJD-V210I | ||||
| 14-3-3 | – | <0.0001 | <0.0001 | 0.7586 |
| t-tau | 0.6087 | – | <0.0001 | 0.9554 |
| α-Syn | 0.4977 | 0.5190 | – | 0.9851 |
| RT-QuIC | −0.0305 | −0.0065 | −0.0028 | – |
| GSS-P102L | ||||
| 14-3-3 | – | 0.0035 | 0.0035 | 0.0039 |
| t-tau | 0.5934 | – | 0.0015 | 0.0034 |
| α-Syn | 0.5934 | 0.6483 | – | 0.0110 |
| RT-QuIC | 0.5055 | 0.5165 | 0.4505 | – |
| FFI | ||||
| 14-3-3 | – | 0.3661 | 0.0003 | 0.0015 |
| t-tau | 0.0760 | – | 0.1104 | 0.3168 |
| α-Syn | 0.3008 | 0.1339 | – | 0.0014 |
| RT-QuIC | 0.2013 | 0.0638 | 0.2026 | – |
The correlations between the concentration of 14-3-3, t-tau, α-syn and the RFU of RT-QuIC in the four largest single mutation groups in our cohort were assessed with Kendall’s tau, which allowed accommodation of left-censored data, as explained in the ‘Materials and methods’ section. Kendall’s tau are shown and associated P-values are shown above (P < 0.05 values are bolded). 14-3-3, t-tau and α-syn correlated strongly with each other in gCJD-E200K, gCJD-V210I, GSS-P102L, but only 14-3-3 with t-tau in FFI. RT-QuIC correlated with the three other biomarkers only in GSS-P102L and with 14-3-3 and α-syn in FFI.
Table 3.
Agreement between biomarker results among the four most common single mutations
| Biomarker pair agreement | gCJD-E200K | gCJD-V210I | GSS-P102L | FFI | ||||
|---|---|---|---|---|---|---|---|---|
| Kappa | SE | Kappa | SE | Kappa | SE | Kappa | SE | |
| 14-3-3 versus t-tau | 0.6296 | 0.0975 | 0.7892 | 0.2038 | 0.4167 | 0.2453 | 0.2687 | 0.1498 |
| 14-3-3 versus α-syn | 0.6259 | 0.1014 | 0.7892 | 0.2038 | 0.4167 | 0.2453 | 0.5677 | 0.1347 |
| 14-3-3 versus RT-QuIC | 0.0476 | 0.0916 | 0.1499 | 0.1914 | 0.7083 | 0.1907 | 0.5037 | 0.1225 |
| t-tau versus α-syn | 0.5346 | 0.1127 | 0.4778 | 0.3153 | 0.4167 | 0.2453 | 0.1643 | 0.1391 |
| t-tau versus RT-QuIC | 0.0625 | 0.0984 | −0.0682 | 0.0379 | 0.7083 | 0.1907 | 0.2357 | 0.1311 |
| α-Syn versus RT-QuIC | 0.0070 | 0.0933 | 0.1989 | 0.2018 | 0.4167 | 0.2453 | 0.4587 | 0.1256 |
Agreement in the disease state classification results among the four most common single mutations, achieved by biomarkers, was assessed in a pair-wise fashion with Kappa index using established cut-off values for sCJD: >20 000 AU/ml for ELISA 14-3-3, >1300 pg/ml for t-tau, >680 pg/ml for α-syn and >10 000 RFU for RT-QuIC. For each pair-wise comparison, Kappa statistic is shown along with its approximate standard error (SE).
Diagnostic accuracy of prion disease biomarkers in genetic prion diseases
With the concentrations obtained for each quantitative biomarker (t-tau, ELISA 14-3-3 and α-syn), we conducted ROC analyses with all pair-wise comparisons involving a disease and a control group. Because the three biomarkers fail in detecting most of GSS-P102L and FFI cases when applied using established sCJD cut-offs, we sought new disease-specific cut-off points. By maximizing the product sensitivity × specificity, we obtained cut-off points for each gPrD versus neurological disease controls and versus healthy controls that enabled an increase in the biomarker’s sensitivity while maintaining acceptable specificity values (>65%; Table 4). Because of the marked class imbalance, especially for the gCJD-V210I and GSS-P102L groups, precision–recall curves were chosen to visualize the trade-off between the true positive rate and the positive predictive value for each biomarker (Supplementary Fig. 1).
Table 4.
Diagnostic accuracy of different biomarkers for the four most common genetic prion disease mutations in our cohort
| Healthy controls | Neurological controls | |||||
|---|---|---|---|---|---|---|
| 14-3-3 | t-tau | α-Syn | 14-3-3 | t-tau | α-Syn | |
| gCJD-E200K | ||||||
| AUC (95% CI) | 0.99 (0.97–1) | 0.99 (0.98–1) | 0.96 (0.93–0.99) | 0.95 (0.92–0.97) | 0.95 (0.93–0.98) | 0.94 (0.90–0.98) |
| Cut-off | >7747 AU/ml | >434 pg/ml | >401 pg/ml | >14 426 AU/ml | >643 pg/ml | >710 pg/ml |
| Sensitivity | 96% | 94% | 93% | 89% | 90% | 87% |
| Specificity | 100% | 98% | 98% | 89% | 89% | 95% |
| gCJD-V210I | ||||||
| AUC (95% CI) | 1 | 0.99 (0.99–1) | 1 | 0.99 (0.97–1) | 0.99 (0.89–1) | 0.99 (0.99–1) |
| Cut-off | >9706 AU/ml | >431 pg/ml | >624 pg/ml | >39 695 AU/ml | >2071 pg/ml | >1389 pg/ml |
| Sensitivity | 100% | 100% | 100% | 94% | 94% | 94% |
| Specificity | 100% | 96% | 100% | 100% | 99% | 100% |
| GSS-P102L | ||||||
| AUC (95% CI) | 0.92 (0.79–1) | 0.75 (0.55–0.96) | 0.83 (0.67–0.97) | 0.80 (0.65–0.94) | 0.71 (0.52–0.90) | 0.72 (0.56–0.88) |
| Cut-off | >5550 AU/ml | >659 pg/ml | >311 pg/ml | >9891 AU/ml | >659 pg/ml | >358 pg/ml |
| Sensitivity | 93% | 64% | 71% | 79% | 64% | 64% |
| Specificity | 90% | 98% | 88% | 79% | 89% | 73% |
| FFI | ||||||
| AUC (95% CI) | 0.88 (0.82–0.94) | 0.81 (0.73–0.89) | 0.67 (0.57–0.77) | 0.58 (0.50–0.67) | 0.70 (0.62–0.78) | 0.56 (0.47–0.65) |
| Cut-off | >3855 AU/ml | >284 pg/ml | >291 pg/ml | >7465 AU/ml | >366 pg/ml | >355 pg/ml |
| Sensitivity | 87% | 78% | 51% | 49% | 62% | 44% |
| Specificity | 82% | 80% | 84% | 66% | 74% | 72% |
ROC analyses were performed with the biomarker concentration obtained for the different major mutation (diagnostic) groups. Diagnostic accuracy was assessed with ROC-derived AUC values (with 95% CI) for the discrimination of each tested genetic prion disease from healthy controls and neurological controls. Optimal cut-off points were determined based on the maximization of the sensitivity × specificity product. Resultant sensitivity and specificity in the study cohort are also shown.
With the three tested biomarkers (t-tau, ELISA 14-3-3 and α-syn), both gCJD groups could be almost perfectly discriminated from both types of controls (Table 4). In the case of GSS-P102L, biomarker performance decreased, especially in the discrimination from neurological disease controls, 14-3-3 being the biomarker displaying the best accuracy (AUC = 0.80). As expected, the worst discrimination occurred with FFI cases from neurological disease controls, in which t-tau rendered the highest but moderate AUC value (0.70). Compared to sCJD cut-off points, the newly determined gPrD-specific cut-offs points resulted in higher sensitivities in the case of gCJD-E200K, GSS-P102L and FFI (Table 4), showing considerable variability across different gPrD. Kappa index measuring the agreement between biomarkers was calculated again for each disease using the new disease-specific cut-off points (Supplementary Table 1). Improved agreement occurred among surrogate biomarkers only in the case of gCJD-V210I and GSS-P102L groups.
PRNP codon 129 MV polymorphism and disease duration in genetic prion diseases
Stratification of disease duration in gPrD diagnostic groups is shown as Kaplan–Meier curves (Fig. 3A–E). Differences in disease duration were tested using Cox proportional hazards models and controlling for the effect of age at onset, sex and PRNP codon 129 genotype. Longer survival times (from onset) were observed in the GSS-P102L group, followed by FFI, gCJD-E200K and gCJD-V210I groups [median survival time in months with interquartile ranges was 57.7 (14.9–74.8), 12.0 (9.8–15.3), 6.0 (3.8–10.3) and 4.0 (3.0–6.9), respectively] (Fig. 3A). All pair-wise comparisons were significant except in the case of gCJD-E200K versus gCJD-V210I with P = 0.528 (Fig. 3, legend statistic). Because the PRNP codon 129 MV genotype was a highly significant covariate, disease duration in each diagnostic group was also stratified depending on the genotype (the VV genotype was not considered due to the low number of cases). Kaplan–Meier curves were plotted and differences were assessed controlling for the effect of age and sex. MV cases had a significantly longer disease duration than MM cases in FFI and gCJD-E200K, whereas age and sex had no significant effect in the Cox models (Fig. 3B, E and legend statistic). By contrast, in the gCJD-V210I group, no influence of genotype was observed (P = 0.6226), but age and sex behaved as significant covariates (HR = 1.0587 and P = 0.0016 for age, and HR = 1.9972 and P = 0.0360 for male sex; Fig. 3C, legend statistic). Disease duration differences between PRNP codon 129 MM and MV genotype could not be tested in the GSS-P102L group because there were only 2 MV cases.
Figure 3.
Disease duration (and the effect of codon 129 MV genotype) in the four most common genetic prion disease mutations in our cohort. (A) Disease duration in gCJD-E200K, gCJD-V210I, GSS-P102L and FFI cases is represented as Kaplan–Meier survival curves. (B–E) Hazard ratios (HR) and associated Tukey-corrected P-values were obtained with Cox proportional hazards (PH) models for each disease group (B: gCJD-E200K; C: gCJD-V210I; D: GSS-P102L; and E: FFI).Differences in disease duration depending on PRNP codon 129 MV genotype were represented with Kaplan–Meier curves. Hazard ratios and associated P-values are shown. Because of low case numbers, the VV genotype was not included. Accommodating age, sex and PRNP codon 129 MV genotype as covariates, we obtained following pair-wise differences between diagnostic groups: gCJD-E200K versus FFI HR = 1.8239, P = 0.010; GSS-P102L versus FFI HR = 0.1469, P < 0.001; gCJD-V210I versus FFI HR 2.3580, P < 0.001; GSS-P102L versus gCJD-E200K HR = 0.0806, P < 0.001; gCJD-V210I versus gCJD-E200K HR 1.2928, P = 0.528; gCJDV210I versus GSS-P102L HR 16.0483, P < 0.001. *No statistics were computed in the GSS-P102L group due to low number of MV cases.
Following the stratification based on PRNP codon 129 MV genotype, we assessed the prognostic performance of each biomarker with Cox proportional hazards models (Table 5). Statistical analyses showed that ELISA 14-3-3 strongly trended towards association with disease duration in gCJD-E200K (P > 0.05). T-tau and α-syn showed significant prognostic value in the gCJD-E200K-MV and GSS-P102L-MM groups, whereas in FFI-MV cases only t-tau appeared associated with disease duration. In the case of RT-QuIC (here it was considered a binary technique, see the ‘Material and methods’ section), we observed a significant association with disease duration in the FFI-MM cases. In all significant cases, shorter survival was associated with higher biomarker values, except in the case of RT-QuIC in FFI-MM, for which surprisingly a negative RT-QuIC was associated with shorter survival (Table 5).
Table 5.
Prognostic values of CSF biomarkers in the four most common genetic prion disease mutations stratified by PRNP codon 129 MV genotype
| 14-3-3 | t-tau | α-Syn | RT-QuIC (binary) | |||||
|---|---|---|---|---|---|---|---|---|
| HR | P | HR | P | HR | P | HR | P | |
| gCJD-E200K-MM | 1.007 | 0.052 | 1.014 | 0.513 | 1.020 | 0.351 | 0.543 | 0.254 |
| gCJD-E200K-MV | 1.010 | 0.049 | 1.170 | 0.039 | 1.226 | 0.003 | 0.352 | 0.325 |
| gCJD-V210I-MM | 1.009 | 0.187 | 1.003 | 0.870 | 0.992 | 0.549 | 1.383 | 0.612 |
| gCJD-V210I-MV | 0.983 | 0.128 | 0.974 | 0.404 | 0.969 | 0.362 | 1.125 | 0.884 |
| GSS-P102L-MM | 0.999 | 0.903 | 1.248 | 0.045 | 2.014 | 0.025 | 1.479 | 0.566 |
| FFI-MM | 0.988 | 0.360 | 0.910 | 0.529 | 0.915 | 0.827 | 0.287 | 0.012 |
| FFI-MV | 0.966 | 0.146 | 1.639 | 0.045 | 0.501 | 0.168 | 1.155 | 0.776 |
The prognostic performance of CSF biomarkers in each gPrD group stratified by PRNP codon 129 MV genotype was assessed with Cox proportional hazards (PH) models in which disease duration was the variable dependent on the biomarker values. The associated P-values of hazard ratios (HR) are displayed and highlighted in bold when significant. Because of the low number of GSS-P102L-MV cases, this group was excluded from the statistical analysis. Biomarker values were divided by 1000 to facilitate interpretation of HR, so the following units apply: AU/1000 for ELISA 14-3-3 and ng/ml for t-tau and α-syn. RT-QuIC was considered as a binary biomarker in these analyses (see the ‘Materials and methods’ section). Higher HR indicates shorter total disease duration. Higher 14-3-3, t-tau and α-syn were associated with shorter disease duration in gCJD-E200K-MV, and t-tau and α-syn (highest HR) in GSS-P102L-MM. For FFI, a negative RT-QuIC was associated with shorter disease duration in FFI-MM, whereas an elevated t-tau indicated a shorter disease duration in FFI-MV.
Discussion
The diagnosis of gPrD may be complex due to heterogeneous clinical presentations and very low incidence, especially for non-gCJD cases. In many cases of gPrD, a positive familial history is present, but even in highly penetrant mutations a negative familial history is possible.19 Information about some family members may be unavailable or mutation carriers may have died before clinical onset of gPrD. Therefore, a familial prion disease diagnosis is not always suspected and, therefore, DNA testing for pathogenic sequence variations in PRNP can be delayed or not performed. Moreover, in known mutation carriers, the symptoms at disease onset may be quite non-specific for gPrD. In these scenarios, robust biomarkers are necessary to support reliable clinical diagnosis. This is not only important for patient and family counselling, but also because gPrD will be of major interest in future clinical trials that will likely have to enrol patients at very early disease and even presymptomatic stages.34,35
In the present work, we evaluated well-known and verified CSF prion disease biomarkers (i.e. 14-3-3 western blot and t-tau), as well as those more recently delineated (i.e. ELISA 14-3-3, α-syn and RT-QuIC) in a large cohort of 302 gPrD and 162 control cases.
To improve data analysis, quantification limits of each test were considered as data-censoring factors. Overall, the good diagnostic accuracy of the four tested biomarkers of sCJD translated to gPrD with similar clinical features and presentations, especially gCJD and OPRI variants with a low number of repeats. In the OPRI group, an inverse correlation between number of insertions and biomarker values was observed, in agreement with an association between low number of repeats and an sCJD-like phenotype. In this context, contention exists regarding 1- and 2-OPRI cases, as it is still not clear whether they are pathogenic and, thus, should be considered as gPrD or if they represent non-pathogenic polymorphic variants and are actually sCJD cases (1-OPRI has also been found in healthy individuals).3,36
In contrast, a GSS-like phenotype or atypical dementia is generally observed in cases with >5-OPRI.3 The differential disease phenotype of GSS and FFI compared to gCJD and sCJD implies distinct pathological and molecular features, including longer disease duration, that probably cause poor performance of surrogate prion disease biomarkers when applied in these hereditary disorders with sCJD-derived cut-off points. However, our data showed that GSS-P102L and FFI had elevated values of all tested prion disease biomarkers compared to healthy controls and neurological disease controls, albeit with low sensitivity when using sCJD-based cut-offs.
These findings prompted us to hypothesize that better accuracy could be possible by lowering the cut-off points in a disease-specific manner. Therefore, new cut-off points for 14-3-3, t-tau and α-syn seeking higher sensitivity at the expense of specificity are proposed. This is based on the fact that in the current clinical diagnostic process, surrogate markers of neuro-axonal damage are used as frontline tools upon suspicion of prion disease, which are then followed by the more expensive and specialized test RT-QuIC that provides very high specificity. Thus, we propose that centres analysing these biomarkers should consider adjusted cut-off values versus neurological disease controls when a gPrD is clinically suspected, so physicians do not rule out this diagnostic possibility and continue to recommend that PRNP is sequenced, particularly when a familial history is absent. In addition, adjusted cut-off values versus healthy controls might be useful for contributing to determine clinical onset in mutation carriers.
Implementation of RT-QuIC in clinical practice has revolutionized the field of prion disease fluid biomarkers, obtaining almost full discrimination of sCJD from non-CJD cases.11,37
In contrast to the surrogate markers (14-3-3-, tau and α-syn), which show a high degree of conformity, RT-QuIC results can occasionally differ with these surrogate biomarkers. A potential reason is that levels of surrogate markers become elevated in CSF as a consequence of massive neuro-axonal damage in the brain tissue, which is typical in more fulminant or faster forms of human prion diseases. In contrast, the RT-QuIC assay is the only test specifically assaying for the pathogenic PrPRes in CSF, which is released from the brain in prion disease patients. Indeed, in our series of 162 controls, none of the samples were positive for RT-QuIC. Nonetheless, RT-QuIC presents low sensitivity in some gPrDs, and it is usually interpreted in a binary manner with the impossibility of adjusting a cut-off point depending on the type of gPrD. In the present work, we also analysed RT-QuIC data in a quantitative manner considering the maximum RFU value achieved. This strategy revealed that among the cases classified as RT-QuIC-positive, statistically significant differences existed in RFU between the four most common gPrD mutations in our cohort (disease groups). Similar to the scenario observed with the other prion disease biomarkers, RT-QuIC RFU could distinguish between a gCJD group (gCJD-E200K and gCJD-V210I) and a non-gCJD group (GSS-P102L and FFI), with the latter group displaying lower RFU. This implies that RT-QuIC not only fails in detecting most GSS and FFI cases due to low intrinsic seeding capacity, but also that those detected are likely to be characterized by a lower signal intensity compared to other prion diseases. In addition to possibly low intrinsic seeding capacity, there are probably other molecular mechanistic reasons underlying this finding, but further research is necessary to elucidate them. Enhanced RT-QuIC assays (such as second generation of the RT-QuIC with a truncated recombinant PrP substrate) may be able to increase sensitivity for GSS-P102L and FFI.38
Our data on t-tau and 14-3-3 are in line with previous reports assessing a limited number of gPrD cases, with high sensitivities in gCJD and lower utility in FFI and GSS.19,39–41 With gPrD diagnostics, despite Sano et al.40 describing good sensitivities for RT-QuIC in FFI (83.3%) and GSS-P102L (90%), other studies demonstrated good sensitivity only in gCJD among the spectrum of gPrD.37,38,42 Potential reasons may be the choice of recombinant PrP substrate used for the RT-QuIC assay or the composition of the patient cohorts.
α-Syn has not been systematically analysed in gPrD, but a previous report in a limited number of cases displayed a similar profile with elevated concentrations in gCJD-E200K and gCJD-V210I but levels similar to those of controls in FFI and GSS-P102L cases.13 Importantly, the comparative analysis of western blot and ELISA 14-3-3 detection in our study validated previous reports of a superior accuracy of ELISA over the western blot method in the discrimination of sCJD cases.22,43 Moreover, one of the advantages offered by continuous markers versus binary markers is the possibility to adjust cut-off points. In the present study, we could observe that although similar accuracies were achieved by the four biomarkers when sCJD cut-offs were used (Table 1), the adjustment of cut-offs in a mutation-specific fashion enabled the three continuous markers (ELISA 14-3-3, α-syn and t-tau) to discriminate gCJD-E200K and gCJD-V210I with almost full accuracy, surpassing the accuracy displayed by RT-QuIC.
The different phenotypes associated with GSS-P102L and FFI compared to other prion diseases also includes the correlation between biomarkers. Whereas surrogate biomarkers (14-3-3, t-tau and α-syn) are highly correlated with each other in gCJD (this study), as well as in sCJD13,43 and iCJD,44 the lack of association with RT-QuIC data has been explained by the fact that the amount of pathological and seeding-competent PrP molecules circulating in the CSF were too low. For example, it is not necessarily related to the degree of neuronal damage in the brain, or is also biased by the category of PrPSc types or strains.37
However, this rationale may not apply in the case of GSS-P102L and FFI, in which we generally observed significant association between RT-QuIC data and the surrogate markers. Interestingly, we found in FFI cases that t-tau was not associated with the other studied biomarkers. Overall, despite significant correlations, the level of agreement between biomarker classification results is rather moderate, suggesting that at least two biomarkers must be undertaken for diagnostic screening upon gPrD suspicion before accepting a negative outcome. An alternative diagnostic marker (not investigated here) is the cellular prion protein (PrPC). In contrast to controls without prion disease, gPrD cases exhibit a decrease in PrPC levels in CSF. Previous ROC analysis of PrPC as a diagnostic marker for gPrD exhibited a moderate to good diagnostic accuracy for gPrD indicated by AUC values between 0.57 and 0.83. While discrimination of gCJD from controls was accurate with CSF PrPC levels, PrPC showed only a poor diagnostic accuracy for GSS P102L,45 which is in line with our observations with the four biomarkers we studied here.
In this work, we also focused on the disease duration data and found increasing survival times (total disease duration) in the following order: gCJD-V210I < gCJD-E200K < FFI < GSS-P102L. The longer survival of FFI and GSS-P102L patients was not due to younger age at onset because this variable was already included in the Cox model and, indeed, previous reports already documented longer disease duration for these two gPrD compared to gCJD.5,40,46,47 Increased survival time was associated with MV genotype in the gCJD-E200K and FFI groups, as reported before,40,46,48 but not in the gCJD-V210I group. For this mutation, younger age at onset and female sex were the best disease prognostic predictors, in agreement with previously published data.46
Regarding GSS-P102L, previous investigations did not show an association of the codon 129 polymorphism and disease duration, but reported earlier disease onset in heterozygous GSS (P102L) patients.49 In this study, the number of heterozygous GSS-P102L patients was too low for statistical computation as performed for FFI, but the few GSS-P102L cases with MV genotype (n = 2) showed a tendency towards longer survival.
When investigating the potential prognostic value of each biomarker in gPrD, we found a modest but significant effect of ELISA 14-3-3 in gCJD-E200K-MV. Although previous studies reported associations between negative western blot 14-3-3 outcome and increased survival time,46,47 we are not aware of similar reported analysis in gPrD with quantitative ELISA 14-3-3 data. The association between t-tau and total disease duration and/or survival from time of measurement has recently been reported in sCJD,16,17,50,51 but is unknown in gPrD. Here, we observed a significant association of t-tau with total disease duration in GSS-P102L-MM, FFI-MV, as well as in gCJD-E200K-MV, in agreement with previous reports in which gCJD-E200K cases showed a positive correlation between CSF t-tau, disease severity and degree of cognitive decline.52 A significant prognostic value of α-syn also appeared in gCJD-E200K-MV and GSS-P102L groups. However, considering the fact that some of the associated P-values are close to the significance threshold and that HR values are close to 1, we acknowledge the limited clinical utility of these observations.
We found a very intriguing negative association between RT-QuIC and disease duration in FFI-MM. The biological meaning of these data remains to be elucidated. Published RT-QuIC data on sCJD revealed no association with disease duration either when RT-QuIC results were used dichotomously or as RFU,11,17 confirming that despite the excellent diagnostic performance of this technique, it lacks prognostic value.
By contrast, CSF markers of synaptic/neuronal damage may help prognosticate disease duration in a mutation-specific manner. Due to their continuous nature, these surrogate biomarkers can exert a modest but significant modulation effect over the influence of the PRNP codon 129 MV genotype, which studies have found contradictory results as to whether it is a determinant of total disease duration, although most have shown an effect in several gPrDs, particularly FFI (D178N) (except in gCJD-V210I).48 Thus, our data should serve to improve development of algorithms for predicting disease duration in gPrD, similarly to what we have recently proposed for sCJD based on the contribution of CSF t-tau to the prognostic value of demographic and genetic parameters.16 In light of the results and due to the relatively long disease duration of GSS-P102L, it would be worth exploring the possibility of developing a prognostic model based on PRNP codon 129 MV genotype, α-syn and t-tau, with applicability not only in patient counselling but also in the evaluation of clinical trials.
Limitations intrinsically associated with our work include, on the one hand, the natural low incidence of some gPrD, restricting our statistical analyses to the most prevalent groups. Thus, data on other mutation groups, some of them composed of single cases, remain purely descriptive. Yet, we considered the disclosure of those data interesting to the clinical community. On the other hand, we lack longitudinal data, impeding our ability to ascertain whether the biomarker levels in gPrD are stable or manifest alterations along disease progression. Additionally, there was only a limited number of clinically well-characterized control patients available for this analysis.
Other limitations associated with data analysis include the possible bias introduced in the reported biomarker accuracies due to the fact that the new proposed cut-off points for each biomarker were evaluated in the study cohort and not a separate cohort. In addition, we must emphasize that P-value corrections were applied only in those tests involving multilevel factors to correct for multiple pair-wise comparisons. Therefore, considering the amount of statistical tests performed along this study, we cannot exclude that some P-values reached the significance threshold by chance.
The main strength of the present work is the size of our study cohort, which to the best of our knowledge is the largest ever used in a study of CSF biomarkers in gPrD. This was possible by collecting cases from 11 clinical prion disease centres worldwide. Potential bias associated with this multicentric collection of samples was avoided by including cohort as a covariate in the analyses of the biomarker values and also having all assays except 14-3-3 western blot done at a single site. Therefore, the reported results are supported by statistically appropriate models that allowed us to offer sound conclusions. We also highlight that our study included CSF new-generation biomarkers, which despite their recent development are currently in use in clinical practice; e.g. RT-QuIC was included in the new diagnostic criteria of prion diseases.8
Supplementary Material
Acknowledgements
We acknowledge the generosity of all the patients involved and their families. We thank Silja Köchy for technical assistance.
Funding
We thank CERCA Programme of the Generalitat de Catalunya for institutional support. This research was funded by the Institute of Health Carlos III (ISCIII) (grants CP/00041 and PI19/00144) to F.L. and by the Robert Koch Institute through funds from the Federal Ministry of Health (grant No. 1369–341) to I.Z. A.V.P. is supported by the Beatriu de Pinós programme (2018-BP-00129) from the Ministry of Business and Knowledge of the Government of Catalonia, co-funded by the EU Horizon 2020 programme under an MSCA grant agreement (801370). S.C. is supported in part by an NHMRC Practitioner Fellowship (#APP1105784). For this manuscript, M.D.G. was supported by the US National Institutes of Health (NIH), National Institute of Aging (NIA) grants R01AG031189, R01AG062562 and R56 AG055619. I.Z. is supported by the Robert-Koch Institute through funds of the Federal Ministry of Health (no. 1369-341) and by the CJD Foundation (Title: Implementation of a blood based biomarker test for sporadic Creutzfeldt-Jakob disease and clinical pratice).
Competing interests
The authors declare that they have no conflict of interest.
Glossary
- α-syn
α-synuclein
- ELISA
enzyme-linked immunosorbent assay
- g/sCJD
genetic/sporadic Creutzfeldt–Jakob disease
- gPrD
genetic prion diseases
- GSS
Gerstmann–Sträussler–Scheinker syndrome
- FFI
fatal familial insomnia
- OPRI
octapeptide repeat insertions
- PrP
prion protein
- RFU
relative fluorescence units
- RT-QuIC
real-time quaking-induced conversion
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Tee BL, Longoria Ibarrola EM, Geschwind MD. Prion Diseases. Neurol Clin. 2018;36(4):865–897. [DOI] [PubMed] [Google Scholar]
- 2. Schmitz M, Dittmar K, Llorens F, et al. Hereditary human prion diseases: An update. Mol Neurobiol. 2017;54(6):4138–4112. [DOI] [PubMed] [Google Scholar]
- 3. Takada LT, Kim M-O, Cleveland RW, et al. Genetic prion disease: Experience of a rapidly progressive dementia center in the United States and a review of the literature. Am J Med Genet Part B Neuropsychiatr Genet. 2017;174(1):36–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gambetti P, Kong Q, Zou W, Parchi P, Chen SG.. Sporadic and familial CJD: Classification and characterisation. Br Med Bull. 2003;66:213–239. [DOI] [PubMed] [Google Scholar]
- 5. Goldfarb LG, Petersen RB, Tabaton M, et al. Fatal familial insomnia and familial Creutzfeldt–Jakob disease: Disease phenotype determined by a DNA polymorphism. Science. 1992;258(5083):806–808. [DOI] [PubMed] [Google Scholar]
- 6. Croes EA, Theuns J, Houwing-Duistermaat JJ, et al. Octapeptide repeat insertions in the prion protein gene and early onset dementia. J Neurol Neurosurg Psychiatry. 2004;75(8):1166–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zerr I, Kallenberg K, Summers DM, et al. Updated clinical diagnostic criteria for sporadic Creutzfeldt–Jakob disease. Brain. 2009;132(Pt 10):2659–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hermann P, Laux M, Glatzel M, et al. Validation and utilization of amended diagnostic criteria in Creutzfeldt–Jakob disease surveillance. Neurology. 2018;91(4):e331–e338. [DOI] [PubMed] [Google Scholar]
- 9. Hermann P, Appleby B, Brandel JP, et al. Biomarkers and diagnostic guidelines for sporadic Creutzfeldt–Jakob disease. Lancet Neurol. 2021;20(3):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanchez-Juan P, Green A, Ladogana A, et al. CSF tests in the differential diagnosis of Creutzfeldt–Jakob disease. Neurology. 2006;67(4):637–643. [DOI] [PubMed] [Google Scholar]
- 11. McGuire LI, Peden AH, Orrú CD, et al. RT-QuIC analysis of cerebrospinal fluid in sporadic Creutzfeldt–Jakob disease. Ann Neurol. 2012;72(2):278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cramm M, Schmitz M, Karch A, et al. Stability and reproducibility underscore utility of RT-QuIC for diagnosis of Creutzfeldt–Jakob disease. Mol Neurobiol. 2016;53(3):1896–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Llorens F, Kruse N, Schmitz M, et al. Evaluation of α-synuclein as a novel cerebrospinal fluid biomarker in different forms of prion diseases. Alzheimer’s Dement. 2017;13(6):710–719. [DOI] [PubMed] [Google Scholar]
- 14. Kruse N, Heslegrave A, Gupta V, et al. Interlaboratory validation of cerebrospinal fluid α-synuclein quantification in the diagnosis of sporadic Creutzfeldt–Jakob disease. Alzheimers Dement. 2018;10:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Llorens F, Schmitz M, Karch A, et al. Comparative analysis of cerebrospinal fluid biomarkers in the differential diagnosis of neurodegenerative dementia. Alzheimer’s Dement. 2016;12(5):577–589. [DOI] [PubMed] [Google Scholar]
- 16. Llorens F, Rübsamen N, Hermann P, et al. A prognostic model for overall survival in sporadic Creutzfeldt–Jakob disease. Alzheimer’s Dement. 2020;16(10):1438–1447. [DOI] [PubMed] [Google Scholar]
- 17. Staffaroni AM, Kramer AO, Casey M, et al. Association of blood and cerebrospinal fluid tau level and other biomarkers with survival time in sporadic Creutzfeldt–Jakob disease. JAMA Neurol. 2019;76(8):969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Llorens F, Kruse N, Karch A, et al. Validation of α-Synuclein as a CSF biomarker for sporadic Creutzfeldt–Jakob disease. Mol Neurobiol. 2018;55(3):2249–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kovács GG, Puopolo M, Ladogana A, et al. ; EUROCJD . Genetic prion disease: The EUROCJD experience. Hum Genet. 2005;118(2):166–174. [DOI] [PubMed] [Google Scholar]
- 20. World Health Organisation . WHO manual for surveillance of human transmissible spongiform encephalopathies including variant Creutzfeldt–Jakob disease. WHO Man Surveill Hum Transm Spongiform Enceph, 2003:105. [Google Scholar]
- 21. Zerr I, Bodemer M, Gefeller O, et al. Detection of 14-3-3 protein in the cerebrospinal fluid supports the diagnosis of Creutzfeldt–Jakob disease. Ann Neurol. 1998;43(1):32–40. [DOI] [PubMed] [Google Scholar]
- 22. Schmitz M, Ebert E, Stoeck K, et al. Validation of 14-3-3 protein as a marker in sporadic Creutzfeldt–Jakob disease diagnostic. Mol Neurobiol. 2016;53(4):2189–2199. [DOI] [PubMed] [Google Scholar]
- 23. Schmitz M, Cramm M, Llorens F, et al. The real-time quaking-induced conversion assay for detection of human prion disease and study of other protein misfolding diseases. Nat Protoc. 2016;11(11):2233–2242. [DOI] [PubMed] [Google Scholar]
- 24. Kleiber C, Zeileis A.. Applied Econometrics with {R}. Springer-Verlag; 2008. https://cran.r-project.org/package=AER [Google Scholar]
- 25. Lenth RV. Least-squares means: The R package lsmeans. J Stat Softw. 2016;69(1):1–33. [Google Scholar]
- 26. Lee L. NADA: Nondetects and Data Analysis for Environmental Data. 2020. https://cran.r-project.org/package=NADA
- 27. Meyer D, Zeileis A, Hornik K. vcd: Visualizing Categorical Data. 2020.
- 28. Meyer D, Zeileis A, Hornik K.. The Strucplot framework: visualizing multi-way contingency tables with vcd. J Stat Softw. 2006;17(3):1–48. [Google Scholar]
- 29. Zeileis A, Meyer D, Hornik K.. Residual-based shadings for visualizing (conditional) independence. J Comput Graph Stat. 2007;16(3):507–525. [Google Scholar]
- 30. Thiele C. cutpointr: Determine and Evaluate Optimal Cutpoints in Binary Classification Tasks. 2020. https://cran.r-project.org/package=cutpointr
- 31. Therneau TM. A Package for Survival Analysis in R. 2020. https://cran.r-project.org/package=survival
- 32. Therneau TM, Grambsch PM.. Modeling Survival Data: Extending the Cox Model. Springer; 2000. [Google Scholar]
- 33. Bretz F, Hothorn T, Westfall P.. Multiple Comparisons Using R; Taylor & Francis; 2016. [Google Scholar]
- 34. Hermann P, Koch JC, Zerr I.. Genetic prion disease: Opportunities for early therapeutic intervention with rigorous pre-symptomatic trials. Expert Opin Investig Drugs. 2020;29(12):1313–1316. [DOI] [PubMed] [Google Scholar]
- 35. Minikel EV, Vallabh SM, Orseth MC, et al. Age at onset in genetic prion disease and the design of preventive clinical trials. Neurology. 2019;93(2):e125–e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beck JA, Poulter M, Campbell TA, et al. PRNP allelic series from 19 years of prion protein gene sequencing at the MRC prion unit. Hum Mutat. 2010;31(7):E1551–E1563. [DOI] [PubMed] [Google Scholar]
- 37. Cramm M, Schmitz M, Karch A, et al. Characteristic CSF prion seeding efficiency in humans with prion diseases. Mol Neurobiol. 2015;51(1):396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franceschini A, Baiardi S, Hughson AG, et al. High diagnostic value of second generation CSF RT-QuIC across the wide spectrum of CJD prions. Sci Rep. 2017;7(1):10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ladogana A, Sanchez-Juan P, Mitrova E, et al. Cerebrospinal fluid biomarkers in human genetic transmissible spongiform encephalopathies. J Neurol. 2009;256(10):1620–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sano K, Satoh K, Atarashi R, et al. Early detection of abnormal prion protein in genetic human prion diseases now possible using real-time QUIC assay. PLoS One. 2013;8(1):e54915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krasnianski A, Heinemann U, Ponto C, et al. Clinical findings and diagnosis in genetic prion diseases in Germany. Eur J Epidemiol. 2016;31(2):187–196. [DOI] [PubMed] [Google Scholar]
- 42. Lattanzio F, Abu-Rumeileh S, Franceschini A, et al. Prion-specific and surrogate CSF biomarkers in Creutzfeldt–Jakob disease: Diagnostic accuracy in relation to molecular subtypes and analysis of neuropathological correlates of p-tau and Aβ42 levels. Acta Neuropathol. 2017;133(4):559–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leitão MJ, Baldeiras I, Almeida MR, et al. Sporadic Creutzfeldt–Jakob disease diagnostic accuracy is improved by a new CSF ELISA 14-3-3γ assay. Neuroscience. 2016;322:398–407. [DOI] [PubMed] [Google Scholar]
- 44. Llorens F, Villar-Piqué A, Hermann P, et al. Diagnostic accuracy of prion disease biomarkers in iatrogenic Creutzfeldt–Jakob disease. Biomolecules. 2020;10(2):290- 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Villar-Piqué A, Schmitz M, Lachmann I, et al. Cerebrospinal fluid total prion protein in the spectrum of prion diseases. Mol Neurobiol. 2019;56(4):2811–2821. [DOI] [PubMed] [Google Scholar]
- 46. Pocchiari M, Puopolo M, Croes EA, et al. Predictors of survival in sporadic Creutzfeldt–Jakob disease and other human transmissible spongiform encephalopathies. Brain. 2004;127(Pt 10):2348–2359. [DOI] [PubMed] [Google Scholar]
- 47. Chen C, Wang JC, Shi Q, et al. Analyses of the survival time and the influencing factors of Chinese patients with prion diseases based on the surveillance data from 2008-2011. PLoS One. 2013;8(5):e62553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Minikel EV, Zerr I, Collins SJ, et al. Ascertainment bias causes false signal of anticipation in genetic prion disease. Am J Hum Genet. 2014;95(4):371–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Webb TEF, Poulter M, Beck J, et al. Phenotypic heterogeneity and genetic modification of P102L inherited prion disease in an international series. Brain. 2008;131(Pt 10):2632–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karch A, Hermann P, Ponto C, et al. Cerebrospinal fluid tau levels are a marker for molecular subtype in sporadic Creutzfeldt–Jakob disease. Neurobiol Aging. 2015;36(5):1964–1968. [DOI] [PubMed] [Google Scholar]
- 51. Li QX, Varghese S, Sarros S, et al. CSF Tau supplements 14-3-3 protein detection for sporadic Creutzfeldt–Jakob disease diagnosis while transitioning to next generation diagnostics. J Clin Neurosci. 2018;50:292–293. [DOI] [PubMed] [Google Scholar]
- 52. Cohen OS, Chapman J, Korczyn AD, et al. CSF tau correlates with CJD disease severity and cognitive decline. Acta Neurol Scand. 2016;133(2):119–123. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon reasonable request.