Abstract
The aim of this study was to determine the impact of local muscle heating during endurance exercise on human skeletal muscle mitochondrial-related gene expression. Twelve subjects (25 ± 6 yr, 177 ± 8 cm, 78 ± 16 kg, and peak aerobic capacity 45 ± 8 mL·kg−1·min−1) cycled with one leg heated (HOT) and the other serving as a control (CON). Skin and intramuscular temperatures were taken before temperature intervention (Pre), after 30 minutes (Pre30), after exercise (Post) and four hours after exercise (4Post). Muscle biopsies were taken from each leg at Pre and 4Post. Intramuscular temperature increased within HOT (34.4 ± 0.7 °C to 36.1 ± 0.5 °C, p < 0.001) and was higher than CON at Pre30 (34.0 ± 0.7 °C, p < 0.001). However, temperatures at POST were similar (HOT 38.4 ± 0.7 °C, CON 38.3 ± 0.5 °C, p = 0.661). Skin temperature was higher than CON at Post30 (30.3 ± 1.0 °C, p < 0.001) and Post (HOT 34.6 ± 0.9 °C, CON 32.3 ± 1.6 °C, p < 0.001). PGC-1α, VEGF and NRF2 mRNA increased with exercise (p < 0.05) but was not altered with heating (p > 0.05). TFAM increased after exercise with heat application (HOT, p = 0.019) but not with exercise alone (CON, p = 0.422). There was no difference in NRF1, ESRRα, or any of the mitophagy related genes in response to exercise or temperature (p > 0.05). In conclusion, TFAM is enhanced by local heat application during endurance exercise, whereas other genes related to mitochondrial homeostasis are unaffected.
Résumé :
Cette étude se propose de déterminer l’impact de l’échauffement musculaire local pendant l’exercice d’endurance sur l’expression des gènes liés aux mitochondries des muscles squelettiques humains. Douze sujets (25 ± 6 ans, 177 ± 8 cm, 78 ± 16 kg et consommation d’oxygène de pointe de 45 ± 8 mL·kg−1·min−1) font du vélo avec une jambe chauffée (HOT) et l’autre servant de contrôle (CON). Les températures cutanées et intramusculaires sont prises avant l’échauffement musculaire (Pre), après 30 minutes (Pre30), après l’exercice (Post) et quatre heures après l’exercice (4Post). Des biopsies musculaires sont prélevées sur chaque jambe à Pre et 4Post. La température intramusculaire augmente dans la condition HOT (de 34,4 ± 0,7 °C à 36,1 ± 0,5 °C, p < 0,001) et est supérieure à la condition CON à Pre30 (34,0 ± 0,7 °C, p < 0,001). Cependant, les températures à POST sont similaires (HOT 38,4 ± 0,7 °C, CON 38,3 ± 0,5 °C, p = 0,661). La température de la peau est plus élevée que dans la condition CON à Post30 (30,3 ± 1,0 °C, p < 0,001) et à Post (HOT 34,6 ± 0,9 °C, CON 32,3 ± 1,6 °C, p < 0,001). L’ARNm de PGC-1±, VEGF et NRF2 augmente avec l’exercice (p < 0,05) mais n’est pas modifié avec l’échauffement musculaire (p > 0,05). TFAM augmente après l’exercice avec l’échauffement musculaire (HOT, p = 0,019) mais pas avec l’exercice seul (CON, p = 0,422). Il n’y a aucune différence de NRF1, ESRRα et l’un des gènes liés à la mitophagie en réponse à l’exercice ou à la température (p > 0,05). En conclusion, TFAM est amélioré par l’échauffement musculaire local pendant l’exercice d’endurance tandis que les autres gènes liés à l’homéostasie mitochondriale ne sont pas affectés. [Traduit par la Rédaction]
Mots-clés : TFAM, PGC-1±, ARNm, muscle squelettique, échauffement local, exercice d’endurance, TFAM, PGC-1α, mRNA, skeletal muscle, local heating, endurance exercise
Introduction
Mitochondrial function is essential to overall health and well-being. Links between mitochondrial dysfunction and aging (Derbré et al. 2012), obesity (Bournat and Brown 2010), metabolic diseases (Bullon et al. 2014), inflammatory, and neurodegenerative diseases (Witte et al. 2010) highlight the importance of healthy mitochondrial function. To avoid these health-related problems, mitochondrial health is essential. Mitochondrial health is characterized by mitochondrial growth (biogenesis) and mitochondrial breakdown (mitophagy). This constant cycle of growth and breakdown allows for optimum function by the breakdown and replacement of damaged mitochondria. Mitochondrial growth is largely controlled by the gene peroxisome proliferator activated receptor gamma coactivator 1 alpha (PGC-1α), and thus known as the master regulator of mitochondrial biogenesis (Hood et al. 2006). Increased expression of PGC-1α and the genes associated with its downstream transcription factors (i.e., estrogen related receptor α (ESRRα), nuclear respiratory factor 1 (NRF1), nuclear respiratory factor 2 (NRF2), mitochondrial transcription factor A (TFAM), and vascular endothelial growth factor (VEGF)) indicate enhanced potential for mitochondrial development (Wu et al. 1999). Further understanding how these mechanisms interact with specific interventions will help advance the practices to enhance mitochondrial function.
Exercise is a potent stimulator of increased mitochondrial biogenesis-related gene expression (Seebacher and Glanville 2010; Slivka et al. 2012, 2013; Zak et al. 2017; Opichka et al. 2019). In addition to exercise, there may be other important stimuli that influence mitochondrial gene expression (Puigserver et al. 1998). Previous work from our lab suggests temperature, when paired with exercise, can alter gene expression related to mitochondrial development (Slivka et al. 2012, 2013; Heesch et al. 2016; Zak et al. 2017; Opichka et al. 2019). More specifically, exercise in cold ambient temperatures has demonstrated increased expression of PGC-1α messenger RNA (mRNA) compared to exercise alone, yet there is an attenuated expression when exercising in hot temperatures (Slivka et al. 2012; Heesch et al. 2016). However, these studies investigated the effect of environmental temperature on gene expression rather than local temperature application to a specific muscle. More recent work in the cold suggests exercise paired with ambient and local temperatures affect core, muscle, and skin temperature differently. Local cold application during exercise results in lower PGC-1α expression when compared to exercise alone (Meister et al. 2021). However, the effect of local heat application on exercise-induced gene expression is not known.
The purpose of this study is to determine the effect of localized heat on skeletal muscle gene expression related to mitochondrial development during aerobic exercise. By adding additional heat to the active muscle of one leg during exercise, we can observe the impact of increased muscle temperature without differentially altering whole body core temperature. This will allow us to isolate muscle temperature’s effect on aerobic exercise, specifically how it is related to gene expression for mitochondrial development.
Materials and methods
Study design and participants
Participants were informed of experimental procedures, consented to the risks and benefits of their participation, and all procedures were in accordance with the Declaration of Helsinki and approved by the University Institutional Review Board (Protocol no. 491-19-FB). This study recruited 12 recreationally trained, healthy individuals between the ages of 19 and 45 years (11 males and 1 female). To be considered healthy, subjects were free from any signs or symptoms of cardiovascular disease or orthopedic limitations that would impact their ability to exercise. Recreationally active was defined as participating in regular exercise three times per week for at least the last 8 weeks. Participants completed two visits to the laboratory (the initial visit and the experimental trial). The initial visit and the experimental trial were separated by at least 48 hours to allow for recovery. Participants were instructed to eat a light meal before arriving and to avoid strenuous exercise 24 hours before the experimental visit. In addition, participants were asked to avoid consuming alcohol, tobacco, or other drugs for at least 24 hours before the experimental visit.
Initial visit
Participants were instructed to come to the lab in a 3-hour fasted state before the initial visit. During the initial visit height (Seca Standometer; Hamburg, Germany), weight (Befour Digital Scale; Soukvill Wisconsin), percent body fat (InBody Body Composition Analyzer; Seoul, Korea), and peak aerobic capacity () were measured. Subjects rested for 15 minutes in the supine position prior to analyzing body composition via bioelectrical impedance analysis to allow for fluid shifts. was conducted on a Lode Excalibur Sport cycle ergometer (Lode Medical Technology, Groningen, the Netherlands) using a graded exercise protocol until volitional fatigue. Stage one of the graded protocol was set to 95 W of resistance for 3 minutes and then increased by 35 W every 3 minutes until the subject could no longer continue. The highest oxygen consumption recorded via the metabolic measurement system (Parvo Medics TrueOne 2400 System; Sandy, UT) was defined as the . Maximum workload was calculated as the highest completed stage (in watts) + the proportion of time in the last stage multiplied by the 35 W per stage increment. The workload for the experimental trial was set at 65% of the maximum workload.
Experimental trial
The experimental trial consisted of four muscle biopsies, eight skin temperature measurements, and eight intramuscular temperature measurements. Upon arrival to the lab, the subjects had their legs shaved around the vastus lateralis and rested for 30 minutes in the supine position. Skin temperatures were then measured on each leg and then a muscle biopsy and intramuscular temperature measurement was taken. Thermal pads (ThermaZone, Innovative Medical Equipment LLC, Cleveland, OH) were applied around each thigh for 30 minutes. One leg was heated to 40 °C (HOT), the other leg served as the control (CON) and was not heated (randomized). Once 30 minutes was complete, skin temperatures and intramuscular temperatures were again measured. Subjects then cycled for 1 hour at 65% of the power output associated with with the thermal pads still in place. Immediately following the cycling protocol thermal pads were removed and skin temperatures and intramuscular temperatures were recorded (Post; within 92 s ± 4 s of pad removal). Thirty minutes after exercise subjects were provided a small, standardized meal of commercially pre-packaged food (721 ± 43 kcal, 76 ± 5 g carbohydrates, 25 ± 3 g protein, 38 ± 3 g fat) and instructed to consume the meal within 30 minutes in order to ensure that all participants were in a similarly fed state. Participants then recovered in standard laboratory conditions for four hours. At four hours after cycling (4Post) skin temperatures were taken on each leg. Muscle biopsies were obtained before the exercise session (Pre) and 4Post from each leg using the same incision site, followed by intramuscular temperatures.
Biopsies
The skin was cleaned with an alcohol wipe and ~2 mL of 1% lidocaine was administered to the subcutaneous tissue and fascia adjacent to the vastus lateralis via a 25-gauge needle to numb the area. A sterile field was created with the use of an iodine sponge and covered with a fenestrated drape. A small puncture incision was made in the skin over the belly of the vastus lateralis (~10 cm proximal to the patella and ~3 cm from the middle of the thigh). A 14-gauge micro-biopsy needle with a trigger-based system (Argon Medical Devices, Athens, TX) was inserted through the incision about 3–4 cm into the muscle and the biopsy was obtained. Four muscle biopsies (2 per leg) were taken in total. Two biopsy samples were obtained at Pre (one from each leg) and two more biopsy samples were obtained following 4Post (one from each leg). This time point was based on the peak gene expression of our target genes (Leick et al. 2010). The time between biopsies of the HOT and CON legs was 108 ± 12 s. The delay in biopsy time allowed time for the investigator to move from one leg to the other. The second biopsy on a given leg was obtained from the same incision as the initial biopsy but the needle was inserted angled proximal to the previous biopsy. After removal of excess blood, connective tissue and fat, tissue samples (9.41 ± 2.66 mg) were immersed in All-Protect (All-Protect, Qiagen, Hilden, North Rhine-Westphalia, Germany) and stored at 4 °C overnight, followed by storage at −30 °C for later analysis. The muscle biopsies were used to evaluate alterations in specific mitochondrial genes related to biogenesis and mitophagy.
Intramuscular temperature
Immediately after the initial muscle biopsies a hypodermic needle (~26 g) thermocouple, (Physitemp, Physitemp Instruments LLC, Clifton, NJ) was inserted 4 cm into the initial biopsy site. Intramuscular temperatures were recorded on a Thermometer Datalogger (Extech Instruments, Nashua, NH) at four time periods: Pre, after one leg has been heated for 30 minutes (Pre30), Post and at 4Post.
Skin temperature
Skin temperatures were measured using a skin surface probe (Physitemp, Physitemp LLC, Clifton, NJ) and Thermometer/Datalogger (Extech Instruments, Nashua, NH). Skin temperature was measured on the skin surface 3–4 cm distal from where the biopsy was taken at the Pre, Pre30, Post, and 4Post time periods immediately before the biopsies and intramuscular temperature procedures. The thermistor was held on the surface of the skin proximal to the site of the biopsy for 60 s simultaneously for each leg which allowed for stable temperature measurements.
Exercise trial data
In order to characterize the intensity of the exercise bout, rating of perceived exertion (RPE; Noble et al. 1983) was recorded in the last 5 minutes of the exercise bout. Heart rate was monitored continuously during the cycling exercise using a HRM Dual heart rate monitor (Garmin International Inc., Garmin Ltd.). The pedals (Vector 3, Garmin Intern. Inc. Olathe, KS) measured the right and left power independently to determine the relative amount of work done by each leg.
Gene expression
Muscle samples were analyzed using the real-time reverse transcriptase polymerase chain reaction (real-time RT-qPCR) to analyze relative mRNA content. Skeletal muscle tissue (9.4 ± 2.6 mg) was homogenized in 500 μL of Trizol (invitrogen, Carlsbad, CA) using an electric homogenizer (Homogenizer 150, Fisher Scientific, Atlanta, GA). Samples were then incubated at room temperature for 5 minutes, 100 μL of chloroform per 500 μL of Trizol was added to the Trizol solution, and the tubes were shaken vigorously by hand for 15 s. After another incubation at room temperature for 3 minutes, the samples were centrifuged at 12 000g for 15 minutes and the aqueous layer was transferred to a fresh tube. Next, 250 μL of isopropyl alcohol was added and incubated overnight at −20 °C to precipitate mRNA. The next day samples were centrifuged at 12 000g for 10 minutes at 4 °C and the mRNA was washed by removing the supernatant, adding 500 μL of 75% ethanol, and the mRNA pellet was dried. Finally, mRNA was dissolved in 30 μL RNA storage buffer and RNA was quantified (160 ± 39 ng/μL) using a spectrophotometer (Nanodrop, Thermo Fisher Scientific, MA, Wilmington, DE). An Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA) was used to inspect the RNA quality (RIN: 8.5 ± 0.4).
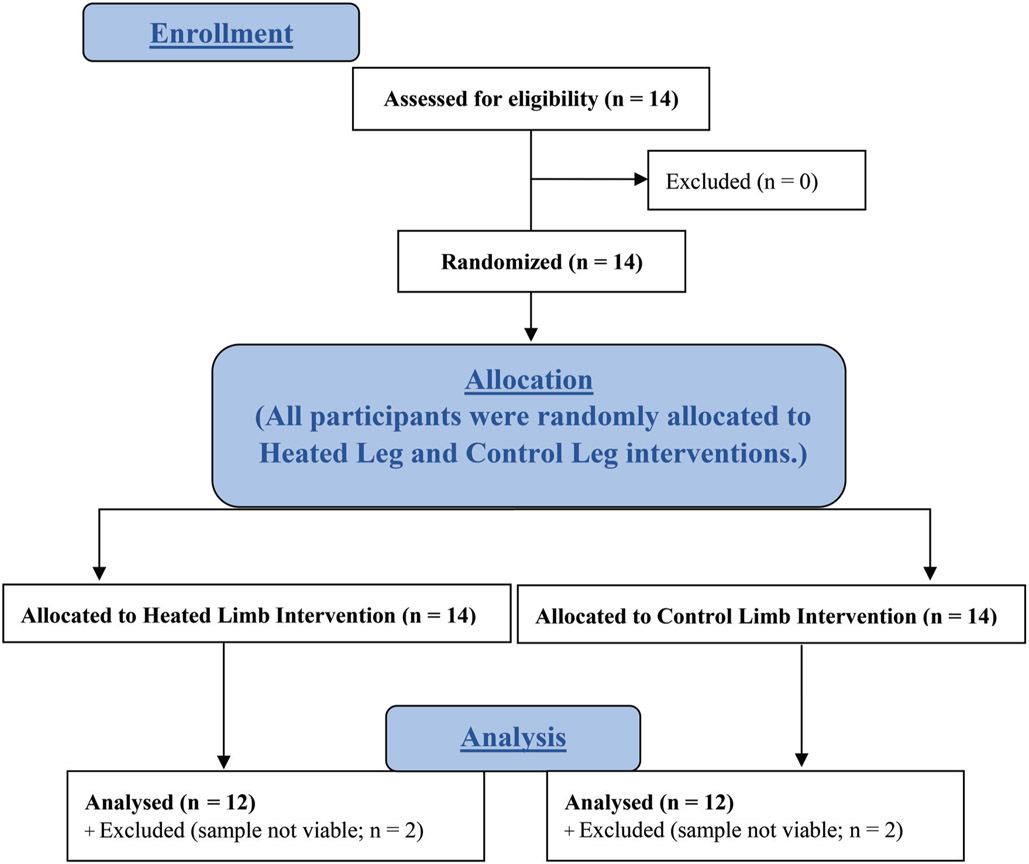
First-strand synthesis system for real time RT-qPCR (Superscript IV, Invitrogen, Carlsbad, CA) was used to synthesize RNA to complimentary DNA (cDNA). Each sample within a subject was adjusted to contain a standard RNA concentration (3 ng/μL) by dilution using RNase free water. Each 20 μL PCR reaction volume contained 1 μL of probe and primer mix (PrimeTime qPCR assay, Integrated DNA technologies), 10 μL Brilliant III Ultra-Fast qPCR master mix (Agilent Technologies Inc., Santa Clara, CA), 0.3 μL of dye, 4.2 μL of water, and 4.5 μL of sample (13.5 ng cDNA). PCR was run in triplicate samples using an Agilent Technologies Aria Mx real time PCR detection system (Agilent Technologies Inc.) running 1 cycle at 95 °C for 3 minutes, then 40 cycles of 95 °C for 5 s and 60 °C for 10 s. Quantification of mRNA for our genes of interest in the experimental leg was calculated relative to the control leg and relative to stable reference genes using the 2−ΔΔCT method (Livak and Schmittgen 2001). After the samples of 2 subjects were deemed not viable, these subjects were removed from all analyses. See Fig. 1.
Fig. 1.
Subject reporting, CONSORT 2010 flow diagram.
The most stable reference genes were determined via Normfinder Software (Andersen et al. 2004) for each subject. The reference genes used were beta actin (ACTB), ribosomal protein S18 (RPS-18), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and beta-2-Microglobulin (B2M). The genes of interest related to mitochondrial biogenesis were PGC-1α, NRF1, NRF2, TFAM, estrogen related receptor α (ERRα), and VEGF. The genes related to mitophagy were PTEN induced kinase 1 (PINK-1), Parkin RBR E3 ubiquitin protein ligase (PARK-2), BCL2 interacting protein 3 (BNIP-3) and BCL2 interacting protein 3 like (BNIP-3L) (Integrated DNA Technologies, Coralville, IA).
Statistical analysis
A repeated-measures two-way ANOVA (time × condition) was used for the dependent variables. If the F-ratio was found to be significant then a Fisher’s protected Least Significant Difference post hoc was performed to evaluate where differences occurred. The probability of type I error less than 5% was considered significant (p < 0.05). All statistical data was analyzed using the Statistical Package for Social Sciences software SPSS (version 26; IBM Corp., Armonk, NY). Previous data from our lab with exercise and temperature has demonstrated that at 80% power, a sample size of n = 12 is necessary to observe a treatment effect.
Results
Subject descriptive data
Twelve recreationally active college aged males (n = 11) and females (n = 1) completed the procedures associated with this study (height, 177.2 ± 1.9 cm; weight, 78.0 ± 3.9 kg; and body composition, 13.5 ± 2.4% fat; , 45.1 ± 2.3 mL·kg−1·min−1).
Exercise trial data
During the one-hour cycling exercise bout, rating of perceived exertion (RPE) was 15 ± 1, average heart rate was 160 ± 9 bpm, and average power was 150 ± 28 W. Temperature had no effect on the percent power contribution between limbs (CON 51 ± 3% and HOT 49 ± 3%, p = 0.420).
Skin temperature
Skin temperatures were similar between groups prior to heating (p = 0.805). Skin temperature increased from Pre to Pre30 in the heated leg (p < 0.001), whereas there was no change in CON (p = 0.912). Skin temperature increased in both legs from Pre to Post (p < 0.001) and was higher in HOT than CON at the Post time point (p < 0.001). Skin temperature increased from Pre30 to Post in CON (p < 0.001), but not in HOT (p = 0.958). Skin temperatures returned to baseline 4Post (HOT, p = 0.920; CON, p = 0.622) and were similar between groups (p = 0.667). See Fig. 2A.
Fig. 2.
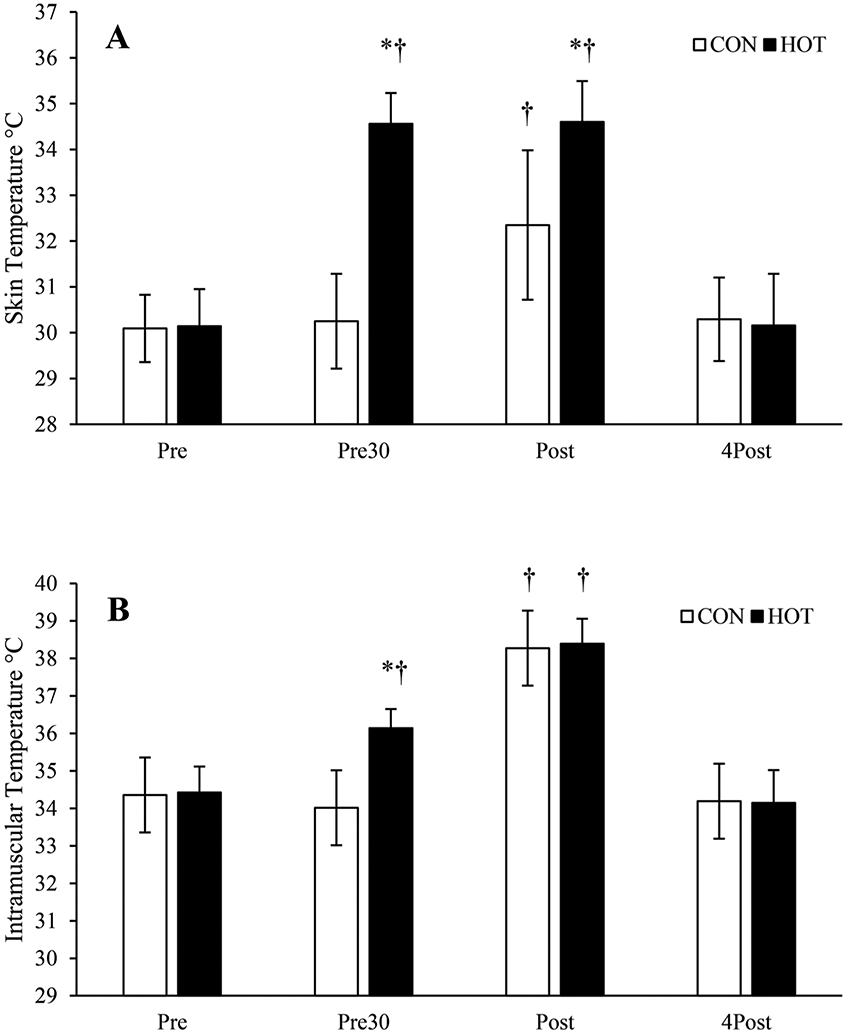
Exercise trial temperature data. (A) Skin temperatures for heated (HOT) and control (CON) legs during the experimental trial. (B) Intramuscular temperatures of the vastus lateralis in control (CON) and heated (HOT) legs during the experimental trial. 4Post, after 4 hours of recovery; Post, immediately after exercise; Pre, before any treatment; Pre30, after one leg has been heated for 30 minutes. Data are means ± SD; *, p < 0.05 from CON; †, p < 0.05 from Pre of the same limb.
Intramuscular temperature
Intramuscular temperature was not different between legs prior to heating (p = 0.668). Intramuscular temperature increased in HOT after heat treatment (p < 0.001) while CON did not change (p = 0.912). Intramuscular temperature increased in both legs from Pre to Post (p < 0.001) and was similar between legs Post (p = 0.661). Intramuscular temperature increased in both CON (p < 0.001) and HOT (p < 0.001) from Pre30 to Post. Intramuscular temperature returned to baseline at 4Post for HOT (p = 0.198) and CON (p = 0.425) and were not different between legs (p = 0.840). See Fig. 2B.
Gene expression
PGC-1α, VEGF, and NRF2 mRNA expression increased with exercise regardless of temperature (PGC-1α, p < 0.001; VEGF, p < 0.001; NRF2, p = 0.016). There were no differences between HOT and CON (PGC-1α, p = 0.512; VEGF, p = 0.187; NRF2, p = 0.298). TFAM increased after exercise in HOT (p = 0.019), but not in CON (p = 0.422). There were no changes in NRF1 or ESRRα in response to exercise (NRF1, p = 0.069; ESRRα, p = 0.650) or temperature (NRF1, p = 0.121; ESRRα, p = 0.443). Gene expression data related to mitochondrial biogenesis are presented in Fig. 3.
Fig. 3.
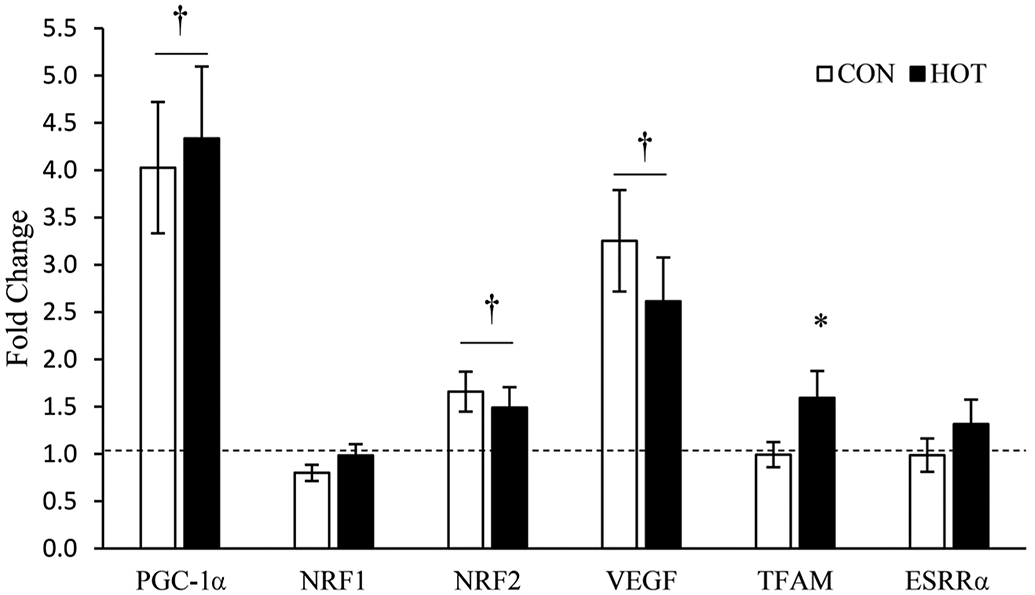
Genes related to mitochondrial biogenesis messenger RNA expression after 4 hours of recovery (4Post). All 4Post samples were normalized to the pre-treatment (Pre) samples, represented as the dashed line at 1.0. Data are means ± SEM; *, p < 0.05 from CON; †, p < 0.05 from Pre.
Mitophagy related genes did not change due to exercise (BNIP3, p = 0.113; BNIP3L, p = 0.760; PINK1, p = 0.113; PARK, p = 0.086) or in response to temperature (BNIP3, p = 0.098; BNIP3L, p = 0.157; PINK1, p = 0.894; PARK, p = 0.927). Gene expression data related to mitophagy are presented in Fig. 4.
Fig. 4.
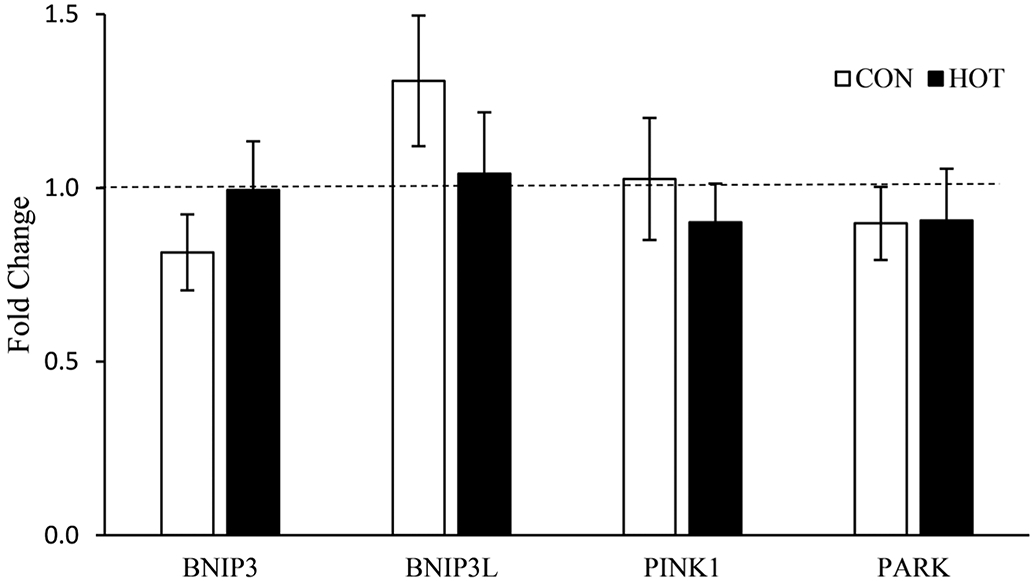
Genes related to mitophagy mRNA expression after 4 hours of recovery (4Post). All 4Post samples were normalized to the pre-treatment samples, represented as the dashed line at 1.0. Data are means ± SEM.
Discussion
The present study aimed to determine the effect of localized heat application on skeletal muscle during endurance exercise on gene expression related to mitochondrial homeostasis (mitophagy and biogenesis). The main finding of this study was that only TFAM expression was differentially expressed with local heating after endurance exercise. Other genes related to mitochondrial homeostasis were unaffected by the temperature intervention. As expected, local heat application before exercise only altered skin and intramuscular temperature in the limb that received the treatment. However, heat production of the contracting muscle over-whelmed the external input of the localized heating resulting in similar temperatures between limbs after exercise. This strong thermoregulatory response to exercise is a likely explanation for similar intramuscular temperatures between legs after exercise.
Exercising in hot environments increases whole-body core temperature (Slivka et al. 2012; Heesch et al. 2016), whereas it is unlikely that the local heating of the small area of application (25 cm × 70 cm) would alter whole-body temperature outside of the expected increase observed from exercise. By applying additional heat to the active muscle of one leg, we observed the impact of increased muscle temperature without significantly altering core temperature and/or whole-body skin temperature. This bilateral leg model allowed us to isolate muscle temperature in the exercise-induced response of aerobic exercise as well as accounting for variation within day-to-day gene expression. Hence, we have demonstrated a methodology that may take advantage of the benefits of local heat application, while removing the influence of core temperature regulation.
Environmental temperature and local temperature application affect muscle and skin temperature differently. Local temperature application has a greater impact on local skin and intramuscular temperature but does not have a large impact on whole-body skin or core temperature (Shute et al. 2017; Kim et al. 2020). Exercising in cold environments has been shown to increase mitochondria-related gene expression, (Slivka et al. 2012, 2013; Opichka et al. 2019) while local cold application had a blunting effect (Meister et al. 2021). Exercise in hot environments lead to a suppression of many genes related to mitochondrial development (Slivka et al. 2012; Heesch et al. 2016), while we demonstrate that this suppression in not present when only local active musculature is heated. Collectively, the results of this project, and that of our previous work (Slivka et al. 2012; Heesch et al. 2016) indicate environmental heat and local heat application effect gene expression in different ways.
TFAM is essential for mitochondrial DNA (mtDNA) maintenance (Larsson et al. 1998; Kang and Hamasaki 2005; Kang et al. 2018) and when disrupted can cause major mitochondrial dysfunction (Larsson et al. 1998). Although muscle temperatures after endurance exercise were similar between legs, there was an increase in transcription of TFAM in the HOT. Unlike some other genes related to mitochondrial biogenesis, TFAM has not been previously shown to be stimulated with a similar exercise and experimental protocol in cold environments, at room temperature, or in the heat (Slivka et al. 2012, 2013; Heesch et al. 2016; Meister et al. 2021). However, a recent publication investigating the influence of exercise on mtDNA copy number suggests that ambient heat may induce greater mtDNA damage, i.e., a reduced copy number (McGlynn et al. 2021). This may suggest an increased mtDNA turnover in heat and thus, increased TFAM transcription. In this investigation, TFAM did not increase with exercise alone, but when exercise was paired with local heat application TFAM was stimulated despite only a short period of time when muscle temperatures were different between limbs. Only after the 30 minutes of pre-heating (Pre30) were muscle temperatures different. The current study cannot determine if these TFAM alterations were induced by temperature alone or if exercise is a critical component to enhanced TFAM expression. The applied implications (i.e., the actual amount of protein translated and the resulting adaptive response) of this magnitude of change in TFAM expression is not clear. Nonetheless, these findings suggest that by locally preheating muscle, one may increase their mitochondrial adaptive potential, while avoiding the blunted gene expression response of acute exercise within environmental heat (Slivka et al. 2012; Heesch et al. 2016).
Mitophagy targets dysfunctional mitochondria for degradation by modulating several genes including PINK1, PARK2, BNIP3, and BNIP3-L. Given the small changes in mitophagy-related mRNA expression, there was no detectable influence after exercise or with local heat application. Previous investigations using cold environmental temperature and local cold application also reported no alterations in these genes (Opichka et al. 2019; Meister et al. 2021). These genes do not appear to be robustly altered with acute exercise or temperature interventions within the common 4-hour post-exercise time-point. However, the time course for expression of these genes could be different than the 4Post time-point. Mitophagy related gene expression for BNIP3 and BNIP3-L has been shown to increase immediately post exercise (Schwalm et al. 2017), while PARK2 and PINK1 were shown to decrease 3 hours post exercise (Opichka et al. 2019). It is possible that this investigation missed the window of gene expression for these mitophagy genes. However, it appears that these mitophagy related genes are not robustly altered with exercise at 3–4 hours after exercise.
In conclusion, local heat application during exercise appears to enhance TFAM expression whereas other genes related to mitochondrial homeostasis are unaffected. These findings paired with previous research on environmental heat (Slivka et al. 2012; Heesch et al. 2016) suggest that localized heat application does not elicit the same blunting response to mitochondrial related gene expression as environmental heat when coupled with endurance exercise.
Novelty:
The main finding of this study is that localized heating increased TFAM mRNA expression.
The normal exercise-induced increased PGC-1α gene expression was unaltered by local muscle heating.
Les nouveautés :
La principale observation de cette étude est que l’échauffement musculaire local augmente l’expression de l’ARNm de TFAM.
L’augmentation normale de l’expression du gène PGC-1α induite par l’exercice n’est pas altérée par l’échauffement musculaire local.
Acknowledgements
We thank Ben Meister and Camille Larson for assistance with data collection for this study. This research was funded by Nebraska IDeA Networks of Biomedical Research Excellence (INBRE), National Institute for General Medical Science (NIGMS P20GM103427) and a Graduate Research and Creative Activity Grant.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- Andersen CL, Jensen JL, and Ørntoft TF 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Bournat JC, and Brown CW 2010. Mitochondrial dysfunction in obesity. Curr. Opin. Endocrinol. Diabetes. Obes 17(5): 446–452. doi: 10.1097/MED.0b013e32833c3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullon P, Newman HN, and Battino M 2014. Obesity, diabetes mellitus, atherosclerosis and chronic periodontitis: a shared pathology via oxidative stress and mitochondrial dysfunction? Periodontol. 2000. 64(1): 139–153. doi: 10.1111/j.1600-0757.2012.00455.x. [DOI] [PubMed] [Google Scholar]
- Derbré F, Gomez-Cabrera MC, Nascimento AL, Sanchis-Gomar F, Martinez-Bello VE, Tresguerres JAF, et al. 2012. Age associated low mitochondrial biogenesis may be explained by lack of response of PGC-1α to exercise training. Age, 34(3): 669–679. doi: 10.1007/s11357-011-9264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesch MW, Shute RJ, Kreiling JL, and Slivka DR 2016. Transcriptional control, but not subcellular location, of PGC-1α is altered following exercise in a hot environment. J. Appl. Physiol 121(3): 741–749. doi: 10.1152/japplphysiol.01065.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood DA, Irrcher I, Ljubicic V, and Joseph AM 2006. Coordination of metabolic plasticity in skeletal muscle. J. Exp. Biol 209(12): 2265–2275. doi: 10.1242/jeb.02182. [DOI] [PubMed] [Google Scholar]
- Kang D, and Hamasaki N 2005. Mitochondrial transcription factor A in the maintenance of mitochondrial DNA. Ann. N.Y. Acad. Sci 1042(1): 101–108. doi: 10.1196/annals.1338.010. [DOI] [PubMed] [Google Scholar]
- Kang I, Chu CT, and Kaufman BA 2018. The mitochondrial transcription factor TFAM in neurodegeneration: emerging evidence and mechanisms. FEBS Lett. 592(5): 793–811. doi: 10.1002/1873-3468.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Reid BA, Casey CA, Bender BE, Ro B, Song Q, et al. 2020. Effects of repeated local heat therapy on skeletal muscle structure and function in humans. J. Appl. Physiol 128(3): 483–492. doi: 10.1152/japplphysiol.00701.2019. [DOI] [PubMed] [Google Scholar]
- Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, et al. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet 18(3): 231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Leick L, Plomgaard P, Grønlykke L, Al-Abaiji F, Wojtaszewski JFP, and Pilegaard H 2010. Endurance exercise induces mRNA expression of oxidative enzymes in human skeletal muscle late in recovery. Scand. J. Med. Sci. Sports, 20(4): 593–599. doi: 10.1111/j.1600-0838.2009.00988.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25(4): 402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- McGlynn ML, Schnitzler H, Shute R, Ruby B, and Slivka D 2021. The acute effects of exercise and temperature on regional mtDNA. Int. J. Environ. Res. Public Health, 18(12): 6382. doi: 10.3390/ijerph18126382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister B, Collins C, McGlynn M, and Slivka D 2021. Effect of local cold application during exercise on gene expression related to mitochondrial homeostasis. Appl. Physiol. Nutr. Metab 46(4): 318–324. doi: 10.1139/apnm-2020-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble BJ, Borg GAV, Jacobs I, Ceci R, and Kaiser P 1983. A category-ratio perceived exertion scale: relationship to blood and muscle lactates and heart rate. Med. Sci. Sports Exerc 15(6): 523–528. [PubMed] [Google Scholar]
- Opichka M, Shute R, Marshall K, and Slivka D 2019. Effects of exercise in a cold environment on gene expression for mitochondrial biogenesis and mitophagy. Cryobiology, 90: 47–53. doi: 10.1016/j.cryobiol.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, and Spiegelman BM 1998. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell, 92(6): 829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Schwalm C, Deldicque L, and Francaux M 2017. Lack of activation of mitophagy during endurance exercise in human. Med. Sci. Sports Exerc 49(8): 1552–1561. doi: 10.1249/MSS.0000000000001256. [DOI] [PubMed] [Google Scholar]
- Seebacher F, and Glanville EJ 2010. Low levels of physical activity increase metabolic responsiveness to cold in a rat (Rattus fuscipes). PLoS One, 5(9): e13022. doi: 10.1371/journal.pone.0013022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shute R, Heesch M, Laursen T, and Slivka D 2017. Local muscle cooling does not impact expression of mitochondrial-related genes. J. Therm. Biol 67: 35–39. doi: 10.1016/j.jtherbio.2017.04.008. [DOI] [PubMed] [Google Scholar]
- Slivka D, Tucker TJ, Slivka D, Dumke C, Cuddy J, and Ruby B 2012. Human mRNA response to exercise and temperature. Int. J. Sports Med 33(2): 94–100. doi: 10.1055/s-0031-1287799. [DOI] [PubMed] [Google Scholar]
- Slivka D, Dumke C, Hailes W, Cuddy J, Heesch M, and Ruby B 2013. Effects of post-exercise recovery in a cold environment on muscle glycogen, PGC-1α, and downstream transcription factors. Cryobiology, 66: 250–255. doi: 10.1016/j.cryobiol.2013.02.005. [DOI] [PubMed] [Google Scholar]
- Witte ME, Geurts JJ, de Vries HE, van der Valk P, and van Horssen J 2010. Mitochondrial dysfunction: A potential link between neuroinflammation and neurodegeneration? Mitochondrion, 10(5): 411–418. doi: 10.1016/j.mito.2010.05.014. [DOI] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. 1999. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivato r PGC-1. Cell, 98(1): 115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- Zak RB, Shute RJ, Heesch MWS, La Salle DT, Bubak MP, Dinan NE, et al. 2017. Impact of hot and cold exposure on human skeletal muscle gene expression. Appl. Physiol. Nutr. Metab 42(3): 319–325. doi: 10.1139/apnm-2016-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]