Shkarina et al. report the development and characterization of a new optogenetic toolset for apoptosis, necroptosis, and pyroptosis induction in humans, mice, and zebrafish. They demonstrate its application for studying the extrusion and efferocytosis of dead cells in 2D, 3D cultures, and in vivo.
Abstract
Targeted and specific induction of cell death in an individual or groups of cells hold the potential for new insights into the response of tissues or organisms to different forms of death. Here, we report the development of optogenetically controlled cell death effectors (optoCDEs), a novel class of optogenetic tools that enables light-mediated induction of three types of programmed cell death (PCD)—apoptosis, pyroptosis, and necroptosis—using Arabidopsis thaliana photosensitive protein Cryptochrome-2. OptoCDEs enable a rapid and highly specific induction of PCD in human, mouse, and zebrafish cells and are suitable for a wide range of applications, such as sub-lethal cell death induction or precise elimination of single cells or cell populations in vitro and in vivo. As the proof-of-concept, we utilize optoCDEs to assess the differences in neighboring cell responses to apoptotic or necrotic PCD, revealing a new role for shingosine-1-phosphate signaling in regulating the efferocytosis of the apoptotic cell by epithelia.
Introduction
Programmed cell death (PCD) controls the elimination of unnecessary, damaged, malignant, or infected cells during development and tissue homeostasis (Green, 2011). Apoptosis—the best-studied form of PCD—leads to the disintegration of dying cells into apoptotic bodies that maintain membrane integrity and are removed by phagocytes in an immunologically silent manner (Galluzzi et al., 2018). Mechanistically, apoptosis is initiated either by the extrinsic/death-receptor pathway or the intrinsic/mitochondrial pathway that controls the activation of apoptotic initiator and executioner caspases (caspase-8/-9, caspase-3/-7), a family of cysteine-aspartic proteases that cleave substrate proteins to promote cell death (Taylor et al., 2008). In contrast to apoptosis, programmed necrotic cell death, such as pyroptosis and necroptosis, results in cell lysis, which drives inflammation and immune responses (Galluzzi et al., 2018). Tumor necrosis factor receptor 1 (TNFR1) can initiate necroptosis if the activity of caspase-8 is inhibited by forming the necrosome, a signaling complex that contains the receptor-interacting kinase (RIPK)1 and RIPK3 (Vandenabeele et al., 2010). Within this complex, RIPK1 phosphorylates RIPK3, which in turn phosphorylates and activates the pseudokinase MLKL that translocates to the plasma membrane to form oligomeric ion channels that drive necroptosis. Pyroptosis is initiated by inflammasome complexes (Broz and Dixit, 2016), which assemble upon detection of pathogens or endogenous danger signals by cytosolic pattern recognition receptors, resulting in the activation of inflammatory caspases (e.g., caspase-1, -11, -4, and -5). These caspases process gasdermin D (GSDMD), thereby releasing the pore-forming GSDMDNT that permeabilizes the plasma membrane to trigger pyroptosis.
While the pathways and checkpoints that control PCD are well understood, comparably little is known about how distinct forms of PCD differ in their outcome for the dying cell, its neighbors, and the organism as a whole (Green, 2019). The effects of cell death are highly context-specific and can either induce or dampen immune responses, or induce cell proliferation and wound repair. It is generally assumed that necrotic cell death causes inflammation, while apoptosis is anti-inflammatory or even tolerogenic (Galluzzi et al., 2018). Yet studies that systematically compare and contrast different forms of PCD and the respective response of neighboring cells remain rare, particularly in an in vivo setting. Experimentally, such analyses are hindered by the low specificity of cell death-inducing stimuli, which affect both the targeted cell and its neighbors, the crosstalk between different PCD pathways, and the difficulty to induce death in a spatially and temporally controlled manner. Laser-induced ablation of single or multiple cells (Okano et al., 2020; Tirlapur et al., 2001; Uchugonova et al., 2008) has been used to overcome these obstacles, as it allows to study the signaling response of direct neighbors and dead cell extrusion. However, it is unclear what type of death is caused by laser ablation since dying cells often display features of both apoptosis (caspase activation) and necrosis (membrane permeabilization; Tirlapur et al., 2001). Another approach is to force-oligomerize cell death executors using fusions with a chemically-dimerizable domain (DmrB/FKBP; Oberst et al., 2010). Limits however are that reversion needs excess concentrations of washout ligands and the inability to selectively target and kill single cells in vitro or in vivo.
Here, we present a novel set of “clean cell death” tools for three major types of PCD—pyroptosis, necroptosis, and apoptosis—which are based on optogenetics (illumination with light), and hence named optogenetically controlled cell death effectors (optoCDEs). Light-mediated cell death induction offers several significant advantages: faster and easier signal delivery; ability to precisely control the intensity and duration of the cell death stimulus by varying illumination dose and duration; and the ability to restrict cell death induction to selected cells or tissues of interest. Different approaches exist to use optogenetics to activate proteins. Recently, light-sensitive protein domain (LOV2)-based tools to induce the separation of the catalytic subunits of caspase-3 and -7, and thus activation were reported (Mills et al., 2012; Smart et al., 2017). Our toolset on the other hand utilizes Cryptochrome 2 E490G (Cry2olig), an Arabidopsis thaliana–derived photosensitive protein, that undergoes rapid homooligomerization upon exposure to blue (450–488 nm) light (Taslimi et al., 2014). By fusing Cry2olig to the effector domains of human, mouse, or zebrafish inflammatory caspases, we designed light-activatable caspases (optoCaspases), which induce rapid and efficient pyroptosis within minutes post illumination. OptoCaspases are functional in various model systems, including multiple human and mouse cell lines, organotypic 3D cell cultures, and live zebrafish larvae, and can be utilized to precisely control, at a spatiotemporal level, the degree of caspase activation to drive sub-lethal pyroptosis activation or single-cell ablation. We extend this approach to induce apoptosis and necroptosis, by generating light-activated apoptotic effectors optoCaspase-8 and -9, and necroptotic effectors optoRIPK3 and optoMLKL. Using the optoCDEs, we then demonstrate how optogenetic induction of cell death can be used to study bystander cell responses and the extrusion of dying cells from epithelia, revealing fundamental differences in the fate of cells dying by apoptosis and programmed necrosis.
Results
Design and characterization of optogenetically controlled human inflammatory caspases
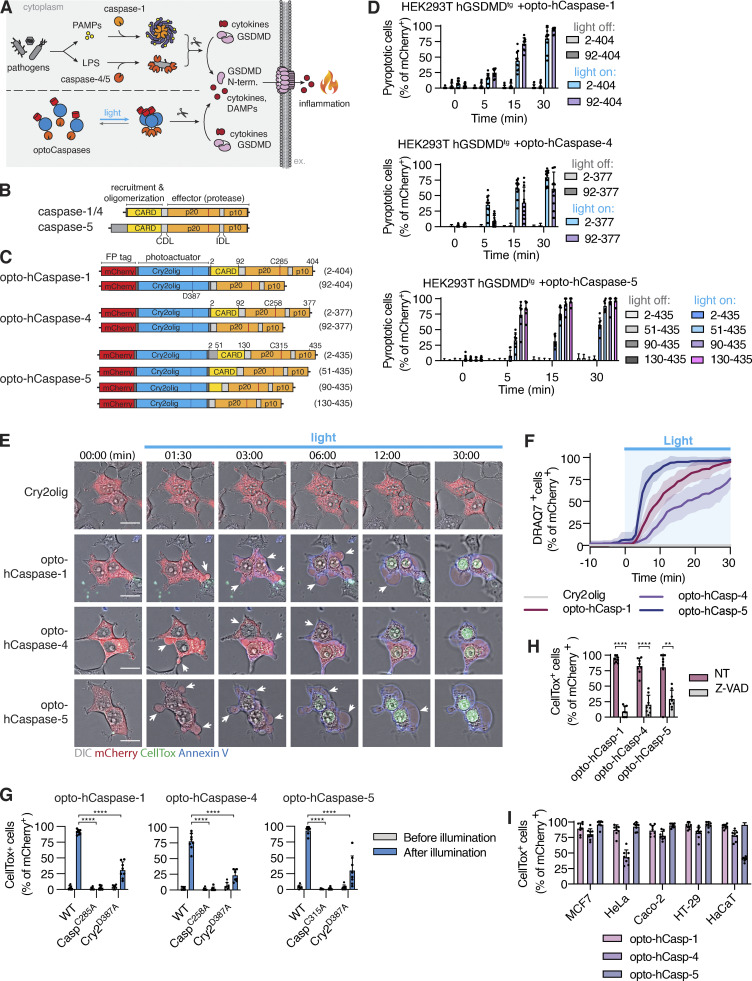
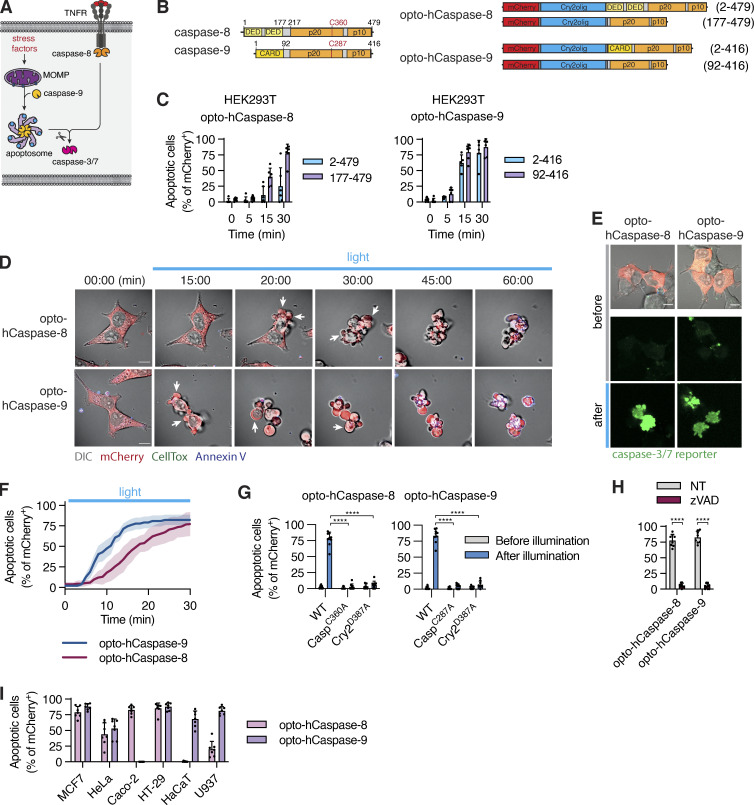
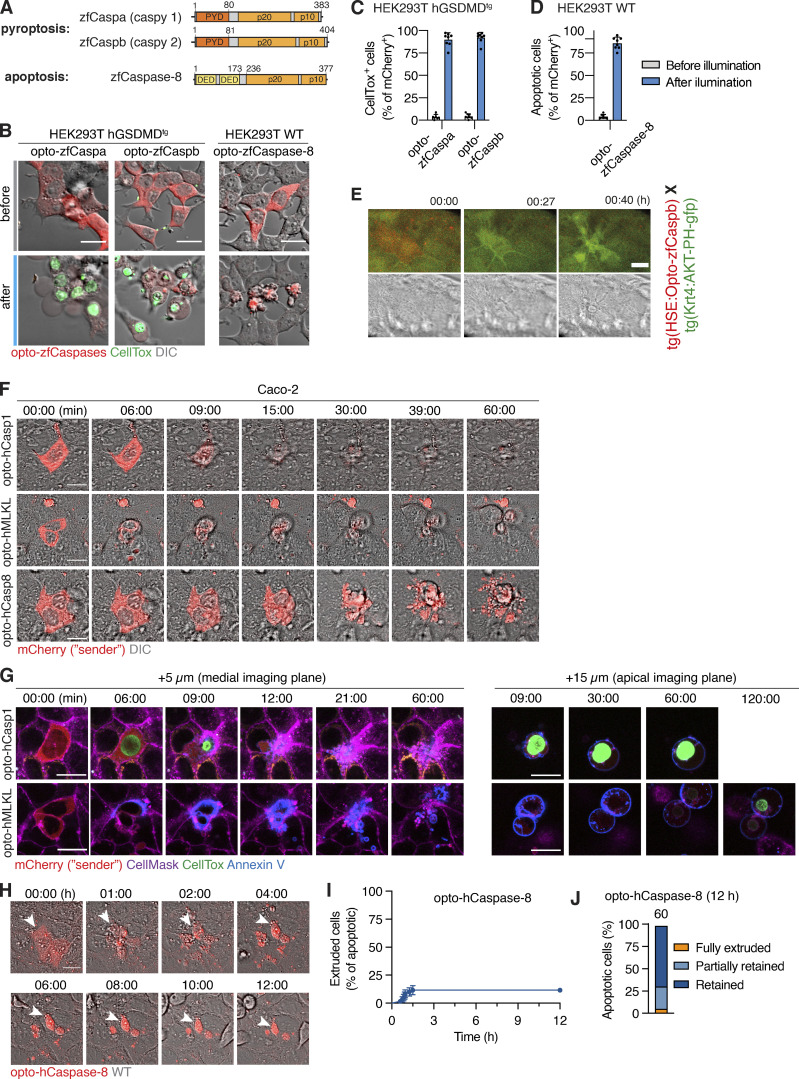
In humans, pyroptosis is induced by inflammatory caspases-1, -4, or -5 that cleave the pore-forming cell death executor GSDMD (Fig. 1 A; Broz et al., 2020). To generate light-activatable human caspases (hereafter termed opto-hCaspases), we fused the mCherry-tagged A. thaliana photosensitive protein Cry2olig N-terminally to full-length or CARD-deficient human caspase-1, -4, or -5 (incl. splice variants of caspase-5; Fig. 1, B and C) and transiently expressed the constructs in GSDMD-transgenic (GSDMDtg) HEK293T cells (lack endogenous GSDMD) to assess their ability to induce pyroptosis upon blue light illumination. For all experiments where photoactivation was done by microscopy, we used 488 nm light at below 26.3 mW/cm2, as this allowed rapid clustering of Cry2olig in HEK293T cells (Fig. S1, A and B) without detectable phototoxicity. By contrast, opto-hCaspase-expressing (mCherry+) cells responded to blue light illumination by membrane blebbing, followed by cell swelling and nuclear rounding (Figs. 1, D and E, and S1 C; and Video 1), all being hallmarks of pyroptosis. Cells also rapidly lost membrane integrity (acquisition of the membrane-impermeable DNA dye CellTox Green), acquired Annexin-V staining (measures phosphatidylserine [PS] exposure), and progressively lost cytoplasmic mCherry signal (Figs. 1 E and S1 D; and Video 1). While all opto-hCaspase constructs induced death of more than 50% of mCherry+ cells within 30 min, full-length opto-hCaspase-1 and -5 were slightly less efficient. To minimize potential endogenous regulatory interactions via the CARD, we decided thus to use CARD-deficient opto-hCaspase-1 (aa 92–404), opto-hCaspase-4 (aa 92–377), and partially CARD-deficient opto-hCaspase-5 (aa 90–435) for all further experiments (of note, fully CARD-deficient opto-hCaspase-5 showed increased background cytotoxicity). The selected constructs induced death with comparable speed and reached similar levels of pyroptosis at later timepoints, with the majority of opto-hCaspase-expressing cells (i.e. mCherry-positive) dying within 10–15 min after illumination (Fig. 1, E and F).
Figure 1.
Development and validation of optogenetically activated human inflammatory caspases. (A) Inflammasome pathway, Cry2-mediated clustering of inflammatory caspases. (B and C) Domain architecture of human inflammatory caspases and opto-hCaspase constructs. (D) Quantification of CellTox-positive (permeabilized) human GSDMD-transgenic (hGSDMDtg) HEK293T cells expressing opto-hCaspases untreated or illuminated with blue light every 30 s. Here and after, only cells expressing detectable levels (mCherry+) of optoCDEs at t = 0 were quantified, and unless stated otherwise 488 nm light at 5 mW/cm2 was used for all assays. (E) Time-lapse images of hGSDMDtg HEK293T cells after blue-light activation of Cry2olig or opto-hCaspase-1, -4, or -5 showing PS exposure (Annexin V) and membrane permeabilization (CellTox Green). Scale bar, 20 µm. (F) DRAQ7 uptake over time (quantified in 1-min intervals) in hGSDMDtg HEK293T cells expressing indicated constructs. (G and H) Percentage of CellTox+ hGSDMDtg HEK23T cells expressing WT or mutant opto-hCaspases before and after illumination; or after mock (NT) or Z-VAD-fmk treatment. (I) Percentage of CellTox+ cells in cell lines expressing indicated constructs at 1 h post illumination. All data are shown as mean ± SD (D and G–I) or mean ± SEM (F) and are representative of three independent experiments (E) or pooled from three independent experiments (D, F, G, H, and I), each with three technical replicates per experiment (n = 9 in total). Each dot represents an independently stimulated field of view. ****, P < 0.0001; **, P < 0.01 (two-tailed t test). PAMPs, pathogen-associated molecular patterns; LPS, lipopolysaccharide; DAMPs, danger-associated molecular patterns; CARD, Caspase recruitment domain; p20, catalytic p20 domain; p10, catalytic p10 domain.
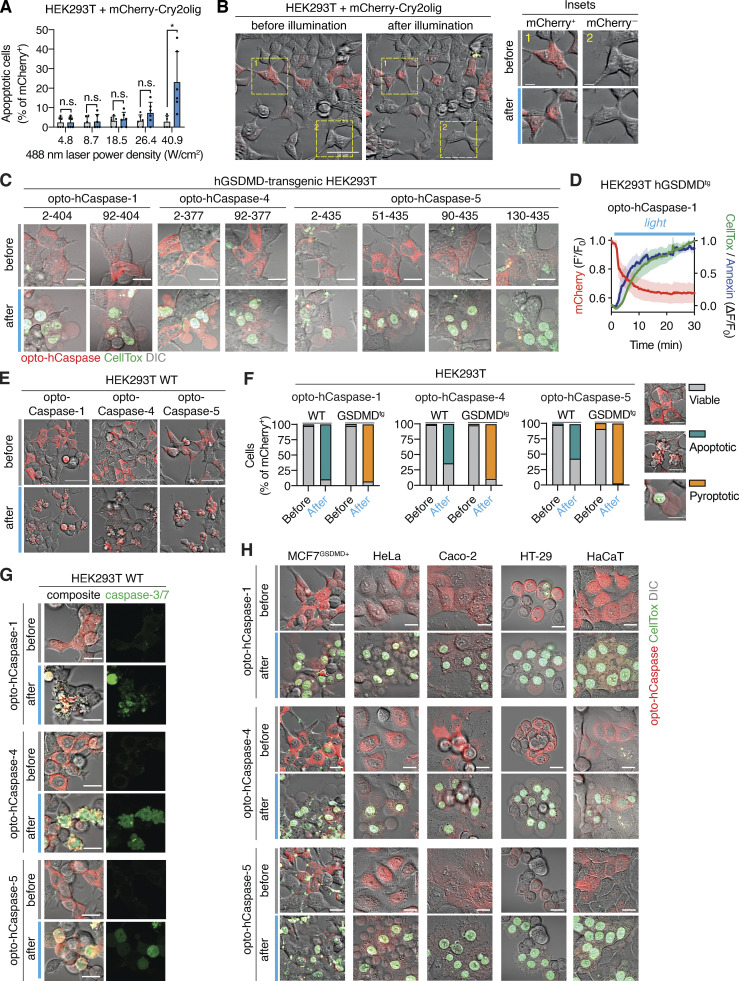
Figure S1.
Activation of optogenetic inflammatory caspases in human cells. (A) Western blot analysis of the expression of inflammatory (hCaspase-1, hCaspase-4, and hCaspase-5) and apoptotic (hCaspase-8 and hCaspase-9) caspases and hGSDMD in the cell lines used in the study. (B) To determine the phototoxicity threshold, HEK293T cells expressing mCherry-Cry2olig were illuminated with blue lights of different power density every 15 s for 1 h, and percentage of cells with altered morphology was quantified before and after illumination. (C) Representative images of cells from A before and after 1 h after illumination with blue light (5 mW/cm2). Insets show the morphology of transfected (mCherry+) and non-transfected (mCherry−) cells. (C) Representative images of HEK293T cells expressing different opto-hCaspase-1, -4, and -5 versions before and after illumination (488 nm, 5 mW/cm2). (D) Normalized fluorescence intensity of the mCherry, Annexin-V, and CellTox signals in cells expressing CARD-deficient opto-hCaspase-1 and undergoing light-induced pyroptosis. Mean ±SEM, n = 5 cells. (E) Representative images of wild-type (i.e., naturally GSDMD-deficient) HEK293T cells expressing opto-hCaspase-1, -4 or -5 (red) before and after illumination (488 nm, 5 mW/cm2). (F) Quantification of apoptotic, pyroptotic, or viable cells before and after illumination among wild-type or hGSDMDtg HEK293T cells expressing opto-hCaspase-1, -4, or -5. Images show morphological features and CellTox staining of the three categories of cells (viable, apoptotic, and pyroptotic). (G) Representative images of wild-type HEK293T cells expressing opto-hCaspase-1/-4/-5 and the genetically encoded caspase-3/7 activity reporter VC3AI (green) before and after illumination (488 nm, 5 mW/cm2). (H) Representative images of different human epithelial cell lines expressing opto-hCaspase-1/-4/-5 before and after 1 h of illumination (488 nm, 5 mW/cm2). Scale bars, 20 µm. All data are representative of at least two (A) or three (C–H) independent experiments, each done with triplicate technical replicates (n = 6 and n = 9).
Video 1.
Induction of pyroptosis by light-induced activation of optoCaspase-1, optoCaspase-4, or optoCaspase-5. Time-lapse fluorescence confocal microscopy of GSDMD-transgenic HEK293T cells transiently transfected with indicated constructs and stimulated with blue light (5 mW/cm2) for 30 min. Composite of DIC, mCherry (marking optoCaspase or Cry2olig used as a control, red), CellTox (membrane permeabilization marker, green), and Annexin V (PS exposure marker, blue) is shown. Pyroptotic cells display characteristic morphological features, such as early membrane blebbing, swelling, and ballooning, as well as nuclear rounding and condensation and acquisition of CellTox and Annexin staining. Cry2olig-expressing cells remain morphologically unchanged over the course of experiment. Scale bar, 20 µm.
Mutation of the catalytic cysteines in opto-hCaspase-1, -4, or -5 completely abrogated cell death induction upon illumination (Fig. 1 G), similarly to treatment with the pan-caspase inhibitor Z-VAD (Fig. 1 H), validating the specificity of the tools. Mutating D387 of Cry2olig, which strongly decreases its light-mediated clustering (Kennedy et al., 2010), also led to the reduction in pyroptosis levels (Fig. 1 G). Light-induced activation of opto-hCaspase-1, -4, or -5 in wild-type (WT) HEK293T cells, which cannot undergo pyroptosis due to the lack of endogenous GSDMD expression, did not induce membrane permeabilization and CellTox influx (Fig. S1, E and F). Instead, we observed a typical apoptotic morphology that correlated with the activation of a genetically encoded caspase-3/7 reporter VC3AI (Zhang et al., 2013; Fig. S1 G). This is in line with reports showing that caspase-1 activation in Gsdmd-deficient mouse macrophages engages rapid apoptosis via Bid cleavage (de Vasconcelos et al., 2020; Heilig et al., 2020; Tsuchiya et al., 2019). While mouse caspase-11 is not able to induce this type of apoptosis (Heilig et al., 2020), we found that opto-hCaspase-4 and -5 caused GSDMD-independent apoptosis, possibly due to their wider substrate profile (Bibo-Verdugo et al., 2020). Opto-hCaspase activation efficiently induced pyroptosis in commonly used cell lines, such as MCF7, HeLa, Caco-2, HT-29, and HaCaT (Figs. 1 I and S1 H). In summary, we demonstrate that fusion with the photoresponsive protein Cry2 can drive inflammatory caspase activation and that optogenetically activatable inflammatory caspases induce GSDMD-dependent pyroptosis across different cell types.
Inflammatory optoCaspases induce efficient pyroptosis and downstream substrate processing in macrophage-like cell lines
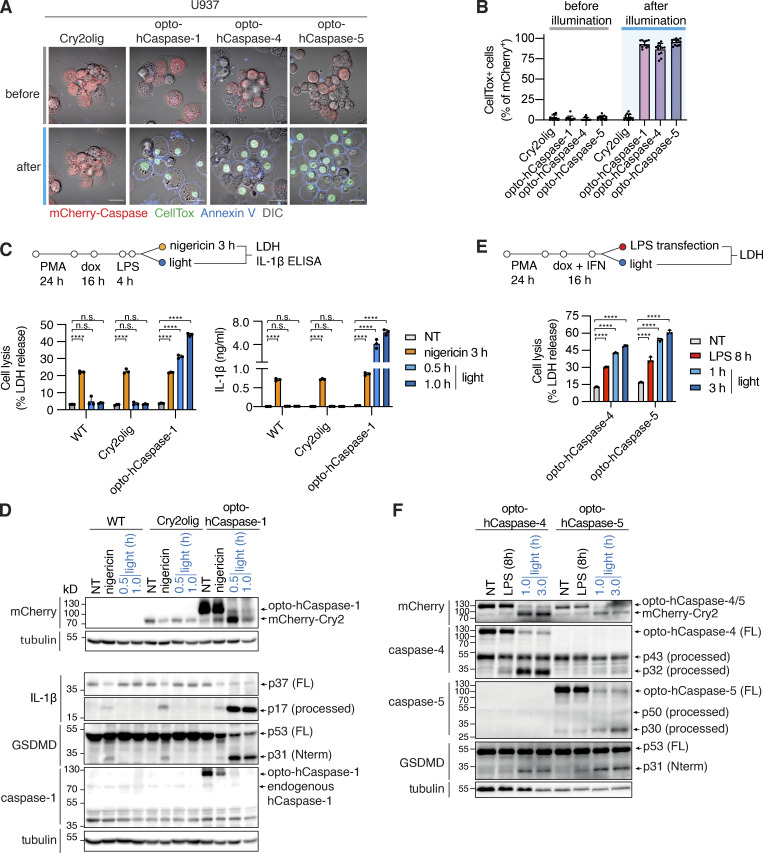
We next validated opto-hCaspases in PMA-differentiated U937 cells, as macrophages remain the most commonly used in vitro model to study pyroptosis. Blue light stimulation of U937 stably expressing optoCaspases induced rapid membrane permeabilization, Annexin-V acquisition, and pyroptotic morphology in the majority of mCherry-positive cells, while Cry2-expressing controls remained unaffected (Fig. 2, A and B). To compare the efficacy of optogenetically-induced pyroptosis to pyroptosis caused by regular inflammasome triggers, we assessed cell lysis, cytokine secretion, and substrate processing in cells stimulated by “classical” canonical and non-canonical inflammasome activation versus bulk cell blue light illumination in a LED light plate apparatus (Gerhardt et al., 2016). Nigericin (NLRP3 activator) induced around 20% of LDH release and IL-1β secretion in WT (non-transduced), mCherry-Cry2olig-, and opto-hCaspase-expressing macrophages (Fig. 2 C). By contrast, blue light illumination efficiently killed ∼50% of opto-hCaspase-1-expressing cells within 1 h but did not induce LDH or IL-1β release from WT or Cry2olig-expressing cells. Cell lysis correlated with the percentage of mCherry-positive cells in the population (Fig. S2, A and B), indicating that most opto-hCaspase-1-expressing cells had died. Immunoblot analysis confirmed opto-hCaspase-1 processing after blue light illumination, but not after Nigericin treatment (Fig. 2 D), validating that the fusion protein was functioning orthogonally to the endogenous inflammasome pathway. In line with the LDH and IL-1β data, opto-hCaspase-1 efficiently processed pro-IL-1β and GSDMD (Fig. 2 D). Consistent with the rapid opto-hCaspase-1 activation, we found that also ∼50% of opto-hCaspase-4 or -5-expressing cells died within 1–3 h of blue light illumination (Fig. 2 E), whereas activation of the non-canonical inflammasome by LPS transfection required 8 h to induce even moderate levels of cell death. Light-induced activation of opto-hCaspase-4/5 also resulted in caspase auto-processing and GSDMD cleavage (Fig. 2 F).
Figure 2.
Optogenetic pyroptosis induction in human macrophage-like cell lines. (A and B) Representative images and quantification of CellTox+ PMA-differentiated U937 cells expressing the indicated constructs before and after 1 h of blue light illumination. Scale bar, 20 µm. (C and E) LDH and IL-1β release from WT U937 or U937 cells expressing the indicated constructs, treated with nigericin (C), transfected with LPS (E), or illuminated with blue light. NT, non-treated. (D and F) Immunoblots of combined cell lysates and supernatants from C or E, respectively. Anti-mCherry is used for full-length and processed opto-hCaspase-1/-4/-5 and Cry2olig. A and C–F are representative of at least three independent experiments, each with three technical replicates per experiment (at least n = 9), B is pooled from three experiments, each with quadruplicate technical replicates (n = 12). Mean ± SD, ****, P < 0.0001 (one-way ANOVA). Source data are available for this figure: SourceData F2.
Figure S2.
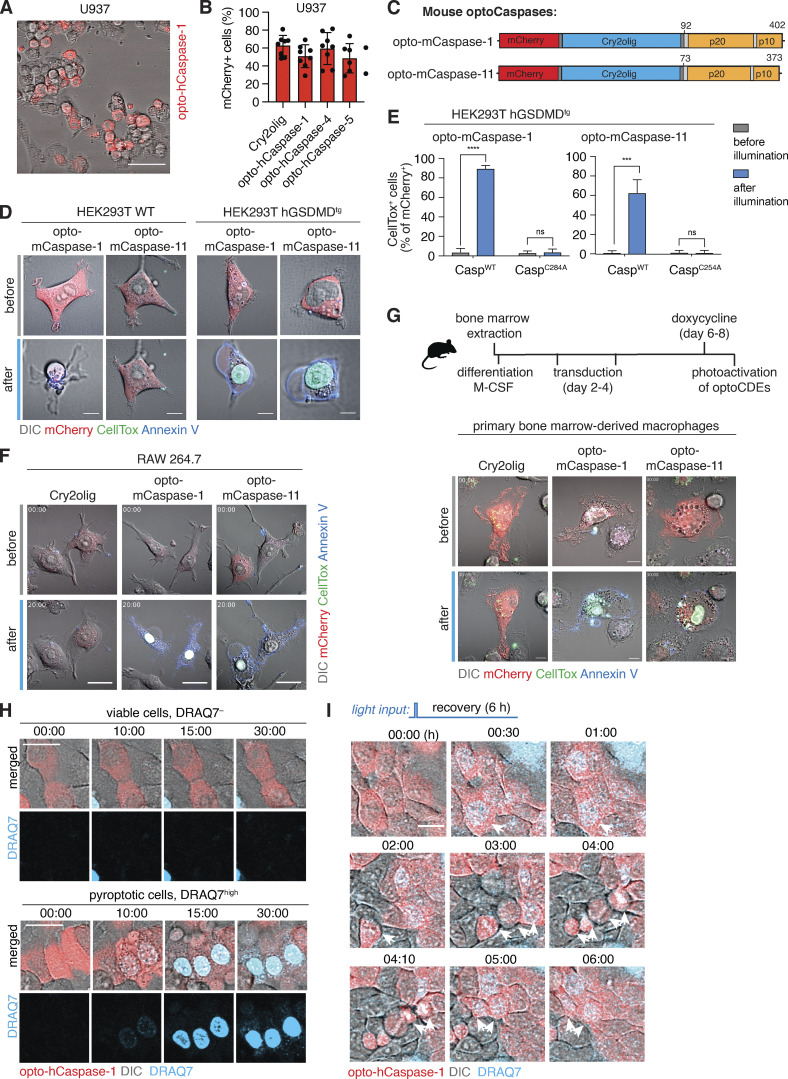
Optogenetic induction of pyroptosis in human and mouse macrophage-like cells, and sublethal induction of pyroptosis. (A) Representative image showing heterogenous mCherry expression levels in U937 cells stably transduced with opto-hCaspase-1 after doxycycline (dox) treatment. Scale bar, 50 µm. (B) Percentage of mCherry-positive cells in differentiated dox-treated U937 cell lines expressing indicated constructs. Data is pooled from two independent experiments, four fields of view per experiment, mean ± SD. (C) Schematic presentation of mouse opto-mCaspase-1 and opto-mCaspase-11 constructs. Both constructs contain CARD-deficient mouse caspase-1 or 11 proteins N-terminally fused to mCherry-tagged Cry2olig via a GGGS linker. (D) Representative images of wild-type or GSDMD-transgenic HEK293T cells expressing opto-mCaspase-1 or opto-mCaspase-11 before or 30 min after illumination (488 nm, 5 mW/cm2 every 15 s). Cells were imaged in presence of CellTox (green) and Annexin-V (blue) to visualize membrane permeabilization and PS exposure. Scale bars, 10 µm. (E) Percentage of CellTox-positive hGSDMDtg HEK293T cells expressing wild-type or catalytically-deficient opto-mCaspase-1 or opto-mCaspase-11 before and after illumination (488 nm, 5 mW/cm2 every 15 s). Mean ± SD, pooled from three independent experiments. ****, P < 0.0001; ***, P < 0.001 (two-tailed t test). (F and G) Representative images of RAW 264.7 cells and primary murine bone marrow derived macrophages (BMDMs) expressing Cry2olig, opto-mCaspase-1 or opto-mCaspase-11 before and after illumination (488 nm, 5 mW/cm2) in presence of CellTox and Annexin V. Scale bars, 20 µm (F) and 10 µm (G). (H) Representative images of cells not affected by the first illumination pulse (viable, DRAQ7−) or undergoing complete pyroptosis (pyroptotic, DRAQ7high). Related to Fig. 3 E. Scale bar, 20 µm. (I) HaCaT cells expressing opto-hCaspase-1 were transiently illuminated with low-intensity 488 nm light (3 × 0.2 mW/cm2) at t = 3min and imaged for 6 h. DRAQ7 influx (turquoise) marks the cells with initial sub-lethal membrane permeabilization, which was then repaired. Arrows indicate dividing parent cell and its daughters. Scale bar, 20 µm. (D–G) Data are representative of or combined from three independent experiments, each done with triplicate technical replicates (n = 9). H and I are representative of six independent experiments (n = 6).
As mice are commonly used to study inflammasomes both in vitro and in vivo, we also generated optogenetically activatable mouse inflammatory caspases, e.g., opto-mCaspase-1 and opto-mCaspase-11 (Fig. S2 C). Like human optoCaspases, opto-mCaspase-1 and opto-mCaspase-11 induced high levels of pyroptosis in hGSDMDtg HEK293T cells (Fig. S2, D and E), and pyroptosis induction was completely abrogated by mutating the catalytic cysteines. Opto-mCaspase-1 and opto-mCaspase-11 activation induced pyroptotic morphology and membrane permeabilization (Annexin V and CellTox co-staining) in mouse macrophage-like RAW264.5 cells and primary bone marrow–derived macrophages (BMDMs; Fig. S2, F and G). Thus, optoCaspases efficiently function in human and mouse myeloid cell models and yield a downstream response comparable with or stronger than the regular inflammasome activators.
Sub-lethal activation of optoCaspases by limited illumination
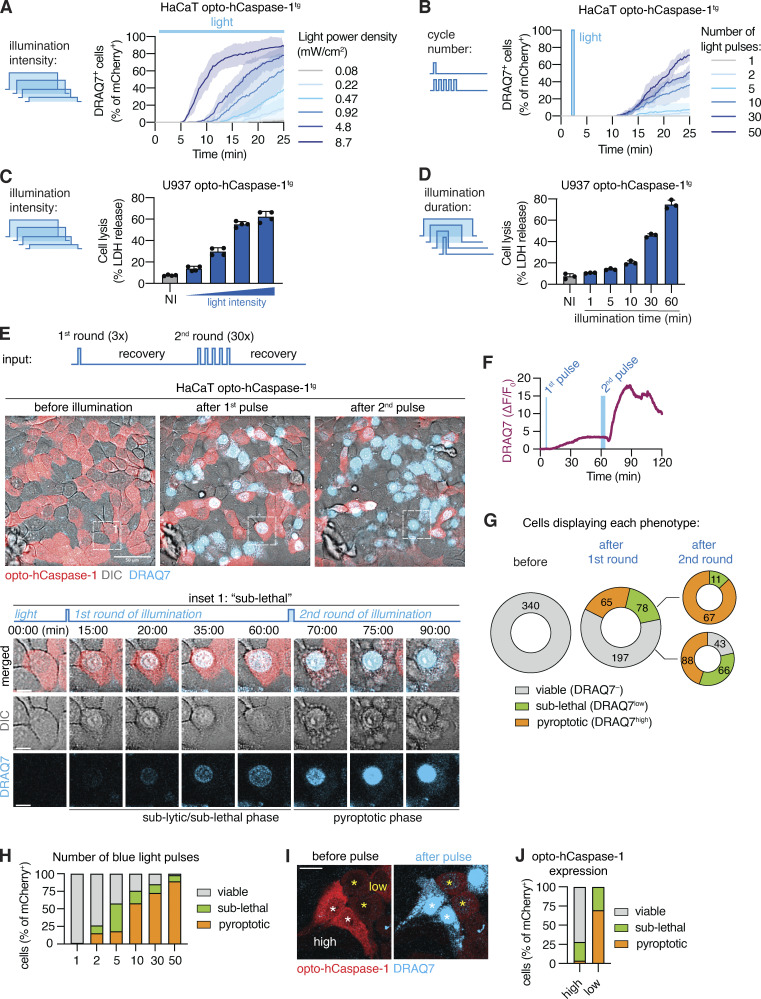
A major advantage of optogenetics is the ability to precisely control the level of protein activation or localization by changing spatial and temporal illumination parameters (i.e., illumination intensity or duration, number of light pulses, or the size of the illuminated region). We thus next assessed how these parameters affected optogenetic pyroptosis induction. Gradual increase in 488 nm light intensity resulted in a concomitant increase in the speed by which cells underwent pyroptosis, as well as the total number of pyroptotic cells at 25 min after the start of illumination (Fig. 3 A). A similar dose-dependent response was observed in cells transiently illuminated with increasing numbers of light pulses at a single timepoint (Fig. 3 B), and in opto-hCaspase-1tg U937 macrophages in which increasing blue light intensity or illumination time increased the levels of pyroptosis induction (Fig. 3, C and D).
Figure 3.
Titrating illumination dose allows for sub-lethal opto-hCaspase-1 activation. (A and B) Percentage of DRAQ7-positive nuclei in opto-hCaspase-1tg HaCaT cells illuminated with (A) varying intensity blue light every 15 min, or (B) transiently illuminated at t = 3 min with an increasing number of light pulses (0.2 mW/cm2/pulse). Mean ± SEM, representative of three independent experiments, each done with triplicate technical replicates (n = 9). (C and D) LDH release from PMA-differentiated opto-hCaspase-1tg U937 cells illuminated either (C) continuously with different intensity light (0.1–4 mW/cm2) or (D) with a defined light intensity (0.9 mW/cm2). Representative of three independent experiments, each done with triplicate (C) or quadruplicate (D) technical replicates (n = 9, n = 12), mean ± SD. (E–J) Opto-hCaspase-1tg HaCaT cells were subjected to a first round of low-intensity (3 × 0.2 mW/cm2) blue light illumination and left to recover for 60 min, followed by a second round of illumination (variable number × 0.2 mW/cm2). (E) Representative confocal images before , after first (t = 60 min), and after second (t = 120 min) round of illumination. Inset shows cell undergoing sub-lethal membrane permeabilization and reversion after first pulse and complete pyroptosis after second pulse. Scale bar: 50 µm; inset scale bar, 10 µm. (F) Normalized nuclear DRAQ7 intensity of the cell from inset in E. (G) Quantification of the cells according to phenotypes before and after illumination. Cells were classified as: “viable” = no DRAQ7 influx and normal morphology; “sub-lethal” = low DRAQ7 influx, early pyroptotic features (nuclear rounding, membrane blebbing) but reversion to normal morphology within 15–30 min; “pyroptotic” – high DRAQ7, pyroptotic morphology. (H) Percentage of viable, sub-lethal and pyroptotic cells depending on the initial number of light pulses (t = 30 min after illumination). (I) Images of cells with high (white *) and low (yellow *) opto-hCaspase-1 expression before and after pulsed illumination, respectively. Scale bar, 20 µm. (J) Percentage of cells with high or low optoCaspase-1 expression displaying each phenotype after a transient low-intensity blue light stimulation. (E–J) Data are representative of six independent experiments or pooled from six independent experiments (n = 340 cells in total).
Interestingly, we noticed that upon illumination with a single pulse of moderate-intensity light, a fraction of cells displayed signs of early pyroptosis (e.g., membrane blebbing) and moderate DRAQ7 influx (DRAQ7low), but nevertheless survived and reverted to normal morphology within 30–40 min post illumination (Fig. 3 E and Video 2). By contrast, other cells showed either no membrane permeabilization (viable nonresponding cells, DRAQ7−) or complete permeabilization (fully pyroptotic cells, DRAQ7high), the latter being followed by the acquisition of pyroptotic morphology (Fig. S2 H). Since the DRAQ7low cells remained viable (at least up to 6 h after illumination), they were able to undergo normal cell division (with both daughter cells retaining slight DRAQ7 staining; Fig. S2 I). Limited DRAQ7 uptake suggests that the plasma membrane of these cells was temporarily permeabilized by GSDMD pores, but that they afterward repaired the damage as observed for necroptosis and pyroptosis previously (Gong et al., 2017; Rühl et al., 2018). Illumination with a second round of light that was 10 times more intense induced death in the majority of these recovered cells (Fig. 3, E and F), confirming that they retain the ability to transition into the lytic stage and that their initial survival was not due to intrinsic defects in pyroptosis execution.
Video 2.
Sub-lethal induction of pyroptosis on a single-cell level. Time-lapse fluorescence confocal microscopy showing HaCaT cells stably expressing optoCaspase-1 (red) that were transiently stimulated with the low dose of blue light at 3 min and left to recover for 1 h, after which a second round of 10 times more intensive light stimulation was performed. Inset (left) shows a cell displaying a sub-lethal membrane damage (as judged by the acquisition of low-intensity DRAQ7 staining, blue) after the first pulse. The cell transitions into “full” pyroptosis (gain of strong DRAQ7 signal, loss of cytosolic mCherry and membrane blebbing) after the second pulse of illumination. Scale bar, 20 µm.
To quantitatively assess the ability of these cells to survive the transient opto-hCaspase-1 activation, we determined the percentage of DRAQ7-negative (live), DRAQ7low (“sub-lethal”), and DRAQ7high (pyroptotic) cells. Around 40% of opto-hCaspase-1 cells responded to the first pulse by gaining nuclear DRAQ7 staining, and among them around 50% underwent pyroptosis while 50% survived this low-level illumination (Fig. 3 G). A second round of longer illumination induced permeabilization in 80% of the cells, and among them 75% cells underwent pyroptosis and 25% survived. The second laser pulse affected DRAQ7low cells more strongly than the DRAQ7− cells since more DRAQ7low cells underwent pyroptosis after illumination. Consistent with the amount of caspase activation determining whether a cell survived or died, we observed that the fraction of “sub-lethal/DRAQ7low” cells was more prominent under low stimulation conditions (1–5 light pulses), yet some were present even after high-intensity stimulation (30–50 pulses; Fig. 3 H). In line with this observation, opto-Caspase-1 expression levels also correlated with the probability to induce sub-lethal pyroptosis (Fig. 3, I and J).
In summary, caspase activation can lead to either sub-lethal or lethal GSDMD pore formation, as hypothesized before (Broz et al., 2020), and the optogenetic control of caspase activity yields a unique approach to study the features and mechanisms that allow cells to avoid or revert from cell death induction, such as the expression of pro-survival factors or plasma membrane repair (Gong et al., 2017; Rühl et al., 2018).
Optogenetic pyroptosis induction enables precise single-cell ablation in 2D and 3D cell culture
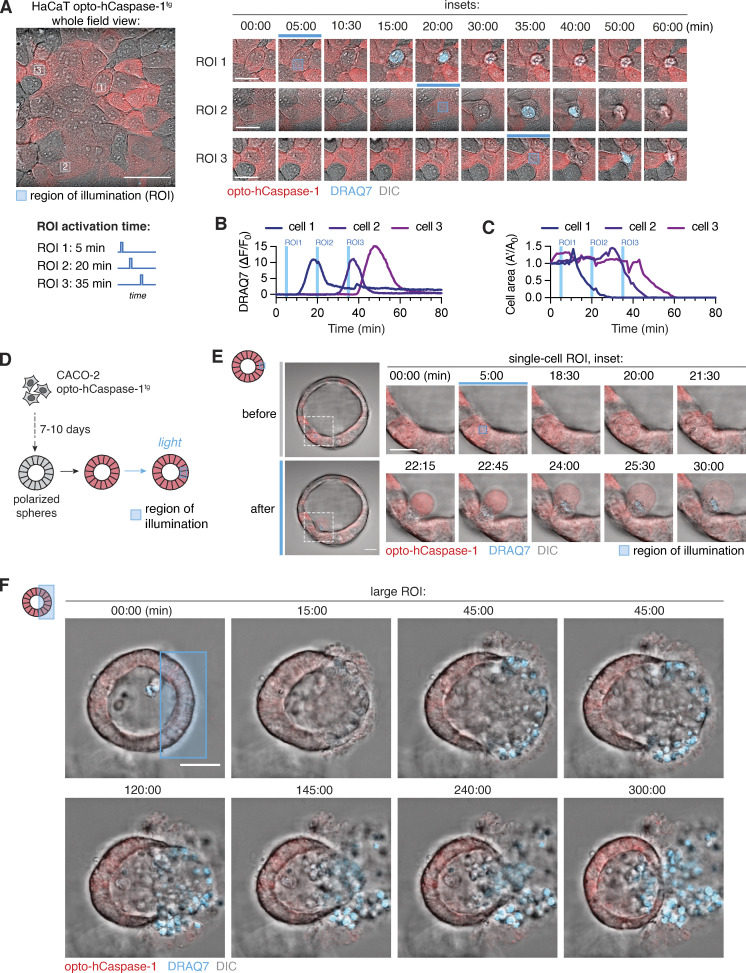
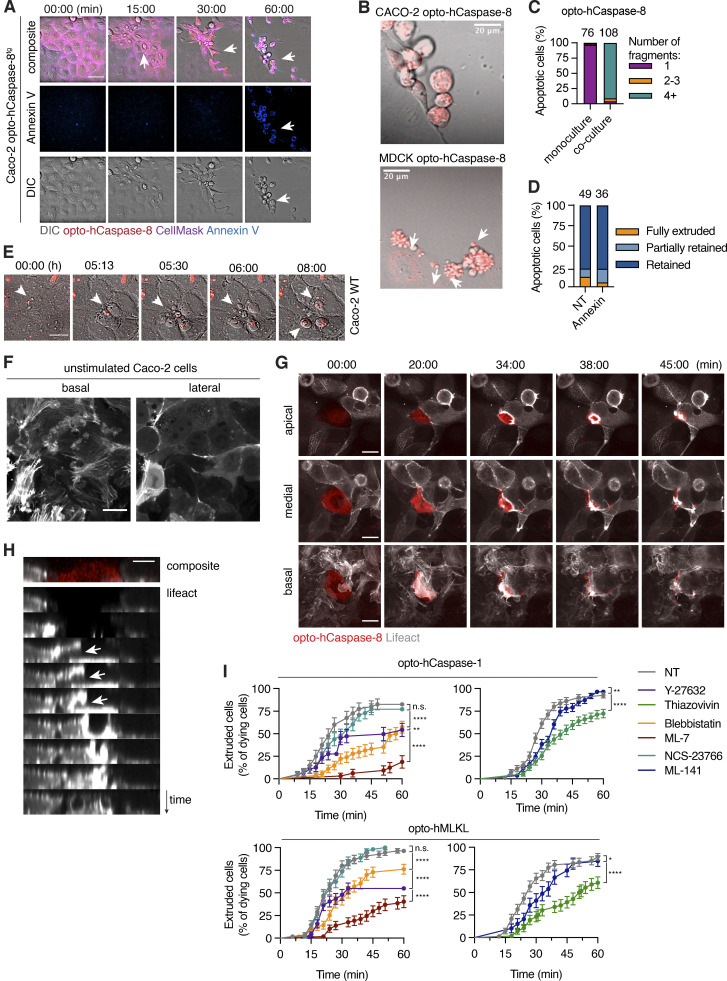
Laser-induced single cell ablation has been previously used to study the response of direct neighbors to dying epithelial cells (Kuipers et al., 2014). To test whether optoCaspase activation enabled precise single-cell pyroptosis induction and cell ablation in populations of closely attached epithelial cells, we transiently illuminated 10–15 µm2 regions of individual opto-hCaspase-1tg HaCaT cells within confluent monolayers in 15-min intervals (Fig. 4 A). Illuminated cells underwent pyroptosis within minutes (Fig. 4, A and B and Video 3), while neighboring cells remained viable and maintained membrane integrity (Fig. S3 A) despite similar opto-hCaspase-1 expression levels. The neighboring cells responded to the pyroptotic events by rapidly migrating toward the pyroptotic cell, extruding it from the monolayer and resealing the gap (Fig. 4, A–C).
Figure 4.
Optogenetic single-cell ablation in 2D and 3D cell cultures. (A) Time-lapse images of single-cell ablation in confluent monolayer of opto-hCaspase-1tg HaCaT cells. Left, whole field of view. Right, inset images of three selectively ablated cells; illuminated at 5, 20, and 35 min. Blue squares indicate the regions of interest (ROI) which were stimulated with blue light. DRAQ7 = membrane permeabilization. Scale bar, 50 µm; inset scale bar, 20 µm. (B and C) Normalized DRAQ7 intensity (B) and loss of cell area (C). Blue lines indicate timing of ROI stimulation for each cell. (D) Schematic representation of experimental setup for spatially resolved cell ablation in 3D Caco-2 spheres. (E and F) Time-lapse images showing selective ablation of a single cell (E) or large group of cells (F) in Caco-2 spheres. ROIs (blue square) were stimulated with blue light (3.4 mW/cm2) at t = 5 min, and pyroptosis measured by DRAQ7 influx. Data are representative of n = 5 (A and B), 7 (D), and 4 (E) independent experiments. Scale bars, 50 µm; inset scale bar (E), 20 µm.
Video 3.
Optogenetic pyroptosis induction in single cells. Time-lapse fluorescence confocal microscopy of HaCaT cells stably expressing optoCaspase-1 (red). Blue squares mark the regions which are transiently stimulated with the blue light at indicated timepoints. Pyroptotic cells rapidly gain DRAQ7 signal (blue) and are extruded from the monolayer by neighbors. Scale bar, 20 µm.
Figure S3.
Optogenetic activation of apoptosis, necroptosis and pyroptosis in individual cells in 2D and 3D. (A) Normalized nuclear DRAQ7 intensity of eight unstimulated directly adjacent neighbors of the cells selectively stimulated with blue light. Related to Fig. 4 A. Grey lines indicate individual cell traces. Vertical blue lines indicate the timing of each of the three local blue light stimulations. (B) Representative time-lapse images of partial cell ablation in non-lumenized Caco-2 spheroid. Blue sphere indicates the region where the stimulation was performed at t = 5 min. Scale bar, 50 µm. Representative of three independent experiments. (C) Representative images of HEK293T cells expressing full-length or CARD/DED-deficient opto-hCaspase-8 or opto-hCaspase-9 before and after illumination (488 nm, 5 mW/cm2 every 15 s). Arrows indicate apoptotic blebbing. Scale bars, 20 µm. (D) Quantification of cells from C expressing opto-hCaspase-8 or opto-hCaspase-9 and displaying apoptotic blebbing alone, or blebbing and Annexin V staining. Cells were stimulated with blue light every 3 min, and images were acquired for 3 h. (E) U937 or HaCaT cells expressing opto-hCaspase-8, or Caco-2 cells expressing opto-hCaspase-9 were treated with the SMAC mimetic AZD5582 and stimulated with blue light for 1 h, and percentage of cells displaying apoptotic morphology was quantified before and after illumination. Mean ± SD. Data in C–E are pooled from three independent experiments. ***, P < 0.001 (two-tailed t test). (F) Representative images of HEK293T cells expressing wild-type or RHIM-deficient opto-hRIPK3 (C-terminally fused to Cry2olig) in absence of blue light illumination (left). Percentage of cells with mCherry clusters in HEK293T cells expressing different opto-hRIPK3 versions (right). Scale bars, 10 μm. (G) Representative images of cells expressing wild-type C-terminal opto-hRIPK3 and displaying apoptotic morphology in absence of blue light (left). Percentage of spontaneously apoptotic cells in HEK293T cells transfected with different opto-hRIPK3 versions (right). Scale bars, 10 µm. (H) Representative images of HEK293T cells expressing different opto-hRIPK3 versions (co-transfected with human hMLKL) or opto-hMLKL before and after stimulation with blue light (5 mW/cm2). Cells were imaged in presence of Annexin V (blue) and CellTox (green) to visualize early (pre-lytic) and late (lytic) necroptosis. Scale bars, 20 µm. (I) Percentage of Annexin-V-positive HEK293T cells expressing opto-hRIPK3 versions before and after illumination, in absence of hMLKL co-expression. Data in F–I are pooled from two (E) or three (B–H) independent experiments, performed with triplicate technical replicates (D, E, and I) or quadruplicate technical replicates (F–H). n = 6 (E), n = 9 (D and I), and n = 12 (F–H). Mean ± SD (two-tailed t test). Scale bars, 10 µm (A and B) or 20 µm (C).
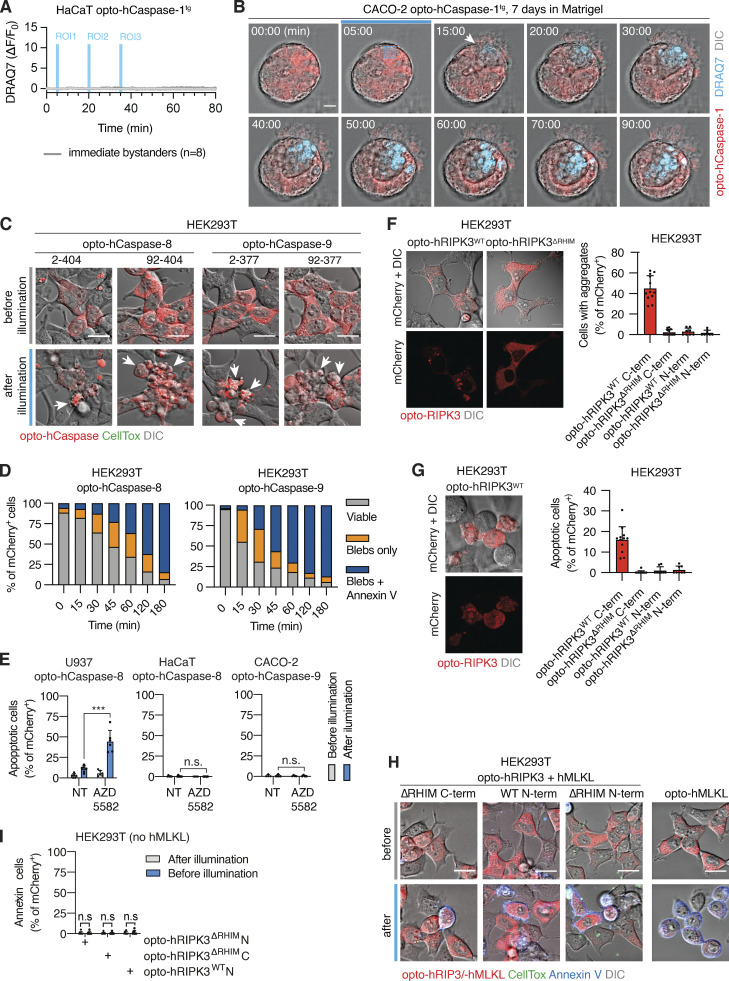
We next tested optogenetic pyroptosis induction in opto-hCaspase-1tg Caco-2 cells cultured to form polarized acini-like structures (spheres). We selectively illuminated ROIs within the sphere wall (Fig. 4 D) and monitored cell death and neighboring cell response by time-lapse microscopy. As in 2D cell cultures, illumination of single cells within the sphere wall induced their pyroptosis, highlighted by membrane blebbing and DRAQ7 influx, and the extrusion of illuminated cells into the sphere lumen (apical side; Fig. 4 E and Video 4), followed by gap closure by neighboring cells. A defined luminal space was not required for extrusion to the apical side, since pyroptotic cells were still extruded toward the spheroid center in non-lumenized spheroids, causing the formation of a lumen around them (Fig. S3 B). When larger subpopulations of the cells were killed simultaneously (Fig. 4 F), pyroptotic cell extrusion and resealing still occurred, and involved a directed migration of the viable neighboring cells toward the lesion, which physically pushed out the dead cells into the lumen. Lumen size was a limiting factor as once the lumen was completely filled with dead cells, extrusion and gap closure could not proceed properly (Fig. 4 F), resulting in a breach in the basal membrane and the spilling of dead cells from the sphere.
Video 4.
Optogenetic pyroptosis induction in 3D organotypic cell cultures. Time-lapse fluorescence confocal microscopy of Caco-2 cells grown in 3D cultures (spheres) in Matrigel and locally stimulated with the blue light at 5 min. Inset (left) shows a stimulated cell displaying membrane blebbing and swelling, gaining DRAQ7 signal (blue) and being extruded from the sphere wall into the lumen. Scale bar, 50 µm.
In summary, these data demonstrate that optogenetic caspase activation can be used for a precise single-cell ablation in 2D and 3D cell culture models and highlight the immense potential of the new toolset for the study of bystander cell responses and cell extrusion in these models.
Optogenetic induction of apoptosis
To expand our toolset to optogenetic apoptosis induction (Fig. 5 A), we fused Cry2olig N-terminally to either full-length or ΔCARD/ΔDED hCaspase-8/-9 (Fig. 5 B). Illumination of opto-hCaspase-8- or opto-hCaspase-9-expressing HEK293T cells induced apoptotic features, such as cell shrinking, detachment, and membrane blebbing in the majority of mCherry-positive cells within 30 min (Figs. 5, B and C, and S3, C and D; and Video 5). Since the DEDs of opto-hCaspase-8 and to a lesser extent the CARD of opto-hCaspase-9 delayed apoptosis induction (Fig. 5 C), we used ΔCARD/ΔDED opto-hCaspase-8/-9 for further characterization.
Figure 5.
Optogenetic activation of apoptotic initiator caspases. (A and B) Schematic representations of intrinsic and extrinsic apoptosis induction and full-length or truncated opto-hCaspase-8 and opto-hCaspase-9. (C) HEK293T cells expressing opto-hCaspase-8/9 versions were illumination with blue light and percentage of apoptotic cells over time determined based on morphology (cell shrinking, blebbing, nuclear fragmentation). (D) Time-lapse images and Annexin V acquisition of HEK293T cells expressing DED-deficient opto-hCaspase-8 and CARD-deficient opto-hCaspase-9 after illumination. Scale bar, 10 µm. (E) Images activation of caspase-3/7 reporter in opto-hCaspase-8/9-expressing HEK293T cells before and after 1 h of blue light illumination. Scale bar, 10 µm. (F) Quantification of apoptotic cells from D over time (in 1-min intervals). (G–I) Quantification of apoptosis induction in cells expressing WT (G and I) or mutated (G) opto-hCaspases or treated with Z-VAD (H) 1 h after blue light illumination. Data are representative of (D) or pooled from (C and E–I) three independent experiments, each performed with triplicate technical replicates (n = 9). Each data point corresponds to independent technical replicate. Mean ± SD; P < 0.0001 (two-tailed t test). TNFR, TNF-receptor; MOMP, mitochondrial outer membrane permeabilization; DED, death-effector domain.
Video 5.
Optogenetic induction of apoptosis via direct activation of optoCaspase-8 and optoCaspase-9. Time-lapse fluorescence confocal microscopy of HEK293T cells transiently expressing optoCaspase-8 (red, left) and optoCaspase-9 (red, right) and stimulated with blue light. The appearance of early apoptotic features (cell shrinking and blebbing) precedes the PS exposure (Annexin V staining, blue). Scale bar, 20 µm.
Validation of opto-hCaspase-8/-9 showed rapid activation of a genetically encoded caspase-3/-7 activity reporter confirming efficient activation of executioner caspases (Fig. 5, D–F). Catalytic dead or dimerization-deficient mutants failed to induce cell death (Fig. 5 G), and the pan-caspase inhibitor Z-VAD abrogated opto-hCaspase-8/9-induced apoptosis (Fig. 5 H). Unexpectedly, we observed a strong variation in opto-hCaspase-8- and opto-hCaspase-9-induced apoptosis levels between different tested cell lines (Fig. 5 I), potentially due to variable levels of endogenous caspase inhibitors (cIAPs/XIAP) or incorrect folding of the construct. Consistent with the former, treatment with the SMAC mimetic AZD5582 facilitated apoptosis induction in at least some but not all cell lines (Fig. S3 E). In summary, optogenetically activatable caspase-8/-9 can induce apoptosis in a variety of cell lines and might provide a new approach to study endogenous regulatory mechanisms that control apoptosis execution.
Optogenetic activation of necroptosis by RIPK3 and MLKL
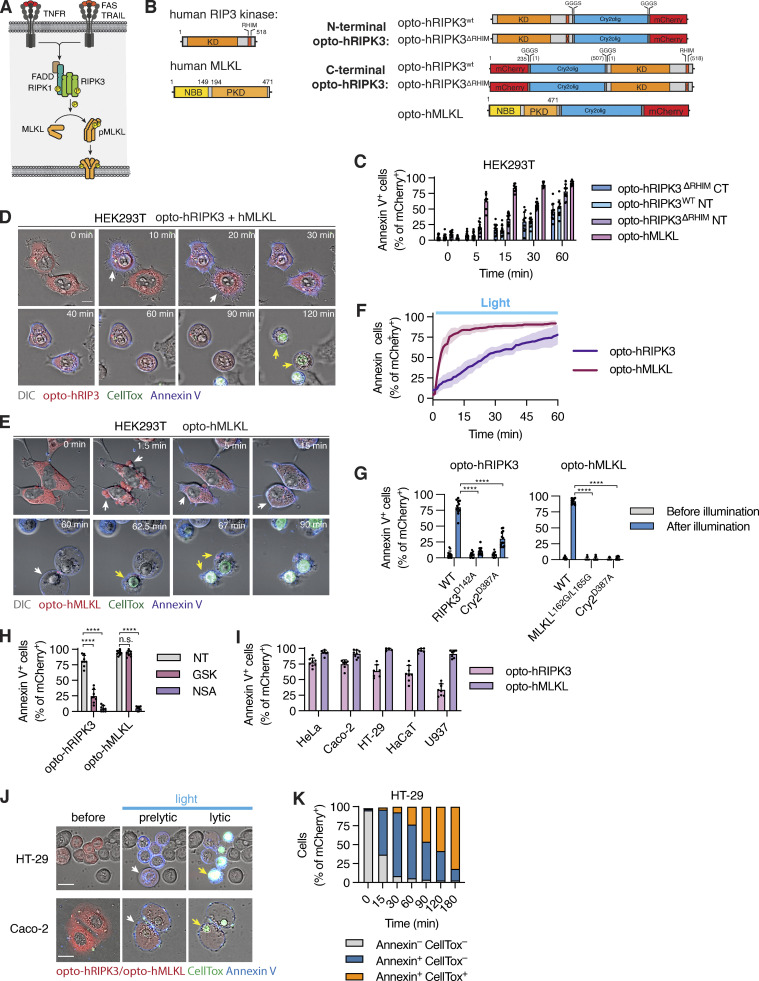
To complete the toolbox of optogenetic cell death inducers, we next constructed optogenetic inducers of necroptosis (Fig. 6 A) by C- or N-terminal fusing mCherry-tagged Cry2olig to WT or RIP homotypic interaction motif (RHIM)-deficient (QIG449–451AAA mutation) hRIPK3 or the N-terminal bundle and brace-protein kinase domain (NBB-PKD) of hMLKL (Fig. 6 B). Illumination of opto-hRIPK3 or opto-hMLKL-expressing HEK293T cells resulted in cell rounding and PS exposure (Annexin-V staining), indicating that cell death had been induced (Fig. 6, C and D). Among the four hRIPK3 constructs, the N-terminal Cry2 fusion to opto-hRIPK3ΔRHIM (in the following termed opto-hRIPK3) proved the most efficient in inducing cell death (Fig. 6 C), while the other constructs proved less efficient or led to dark-state protein aggregation and cell death in a significant fraction of mCherry-positive cells (Fig. S3, F–H).
Figure 6.
Optogenetic activation of necroptotic effectors necroptosis. (A and B) Illustration of the necroptosis pathway and opto-hRIPK3 and opto-hMLKL constructs. (C) Quantification of necroptosis based on Annexin V acquisition in HEK293T cells expressing different opto-hRIPK3 versions or opto-hMLKL from 0 to 30 min after blue light illumination. (D and E) Time-lapse images of HEK293T cells undergoing light-induced necroptosis upon opto-RIPK3 (N-terminal, RHIM-deficient) or opto-hMLKL activation. Scale bar, 10 µm. (F) Dynamics of necroptosis induction via opto-hRIPK3 and opto-hMLKL activation. Scale bar, 10 μm. (G–I) Quantification of necroptosis in cells expressing WT, catalytically deficient or oligomerization-deficient opto-hRIPK3/opto-hMLKL or treated with 1 µM GSK’872 (RIPK3 inhibitor) or 5 µM necrosulfonamide (MLKL inhibitor) after 1 h of blue light illumination. (J) Confocal images of non-stimulated, early (pre-lytic) and late (permeabilized) necroptotic Caco-2 and HT-29 cells after activation of opto-hMLKL. (K) Quantification of Annexin V or CellTox staining in opto-hMLKL-expressing HT-29 cells after illumination with blue light. Scale bar, 20 µm. Data are representative (D, E, and J) or pooled from (C, F–I, and K) three independent experiments, each performed with triplicate technical replicates (n = 9), mean ± SD. ****, P < 0.0001 (two-tailed t test). TRAIL, TNF-Related Apoptosis Inducing Ligand; FADD, Fas Associated Via Death Domain; KD, kinase domain; PKD, pseudo kinase domain; NBB, N-terminal bundle and brace.
Time-lapse microscopy of HEK293T cells expressing either opto-hRIPK3 (co-expressed with hMLKL) or opto-hMLKL revealed rapid induction of necrotic cell death upon blue light illumination, which was characterized by cell rounding, Annexin-V staining, and at later stages by CellTox acquisition (Fig. 6, D and E and Video 6). Also, opto-hRIPK3-induced necroptosis proceeded with slower kinetics than opto-hMLKL (Fig. 6 F). Opto-hRIPK3 did not induce any phenotypical changes in the absence of hMLKL co-expression (Fig. S3 I), confirming that morphological changes and cell death resulted from RIPK3-dependent MLKL phosphorylation. The mutation of hRIPK3 D142 or hMLKL L162/L165 significantly decreased the proportion of necroptotic cells upon illumination (Fig. 6 G), as did RIPK3 or MLKL inhibitors GSK’872 and necrosulfonamide (NSA; Fig. 6 H). Both constructs were highly active in a range of cell types commonly used for necroptosis studies (Fig. 6 I).
Video 6.
Optogenetic induction of necroptosis via direct activation of RIP3 and MLKL. Time-lapse fluorescence confocal microscopy showing HEK293T cells expressing optoRIP3 (left, co-transfected with MLKL) or optoMLKL (right) and stimulated with blue light. Early necroptotic cells display only Annexin V staining (blue), while late necroptotic cells lose membrane integrity and gain CellTox signal (green). Scale bar, 20 µm.
Interestingly, necroptosis proceeded through two distinct phases: (1) A “sub-lytic” phase of circa 1 h, during which cells displayed membrane blebs, acquired Annexin-V staining, and rounded up, but maintained membrane integrity, and (2) a lytic phase characterized by the sudden and complete rupture and disappearance of the cell membrane and rapid CellTox acquisition (Fig. 6, D, E, J, and K and Video 6). This differed strikingly from the pyroptotic cell, which acquired CellTox during blebbing and ballooning but never displayed a complete plasma membrane rupture and the disappearance of the plasma membrane. Thus during pyroptosis, membrane permeabilization is an early event, while it is a late event during necroptosis, and it is preceded by a sub-lytic phase during which MLKL channels do not yet reach the threshold to induce full plasma membrane rupture (Samson et al., 2020).
Optogenetic activation of zebrafish caspases allows spatial and temporal controlled induction of pyroptosis and apoptosis in vivo
Zebrafish (Danio rerio) embryos are a powerful model to visualize biological processes like cell differentiation and death in real-time and in vivo. Zebrafish feature homologs of mammalian apoptotic and inflammatory caspases, such as zf(zebrafish)Caspa which induces pyroptosis in zebrafish skin cells downstream of zfASC-dependent inflammasomes (Forn-Cuní et al., 2019) and zfCaspb that forms a non-canonical inflammasome together with zfNLRP3 (Li et al., 2020; Fig. S4 A). We designed optogenetically activatable opto-zfCaspa, opto-zfCaspb, and opto-zfCaspase-8 by replacing their PYD or DED by Cry2olig and evaluated these constructs in GSDMDtg or WT HEK293T cells (Figs. 7 A and S4 B). Activation of opto-zfCaspa/b in HEK293T cells induced rapid pyroptosis and CellTox influx, while opto-zfCaspase-8 activation caused a typical apoptotic morphology (Fig. S4, B–D), confirming both functionality and evolutionary conservation of substrate specificity of these proteins.
Figure S4.
Activation of zebrafish caspases by optogenetics and differential responses of apoptotic and necrotic cell death in epithelia. (A) Domain organization of zebrafish inflammatory (caspa and caspb) or apoptotic (caspase-8) caspases. (B) Representative images of hGSDMDtg HEK293T cells expressing opto-zfCaspa, opto-zfCaspb or opto-zfCaspase-8 before and after 30 min of blue light illumination (5 mW/cm2). CellTox staining (green) indicates membrane permeabilization. (C and D) Percentage of pyroptotic (C) or apoptotic (D) opto-zfCaspa or opto-zfCaspb-expressing (C) or opto-zfCaspase-8-expressing (D) hGSDMDtg HEK293T cells before and after blue light illumination (5 mW/cm2). Data is pooled from three independent experiments, mean ± SD. (E) Representative time-lapse images of a keratinocyte after activation of opto-zfCaspb. (F) Representative time-lapse images (showing DIC and mCherry channels) of Caco-2 cells undergoing light-induced pyroptosis, apoptosis and necroptosis in co-cultures with wild-type cells (related to Fig. 8, A and B). (G) Representative time-lapse images showing CellMask (purple), Annexin-V (blue) and CellTox (green) staining in pyroptotic (opto-hCaspase-1) and necroptotic (opto-hMLKL) cells being extruded from monolayer of WT cells. Left, images of the cells at the medial plane (5 µm above basal surface), right, apical plane (15 µm above basal surface, showing extruded cells). For opto-hMLKL cells, imaging was performed for 120 min to show CellTox influx during lytic necroptotic stage. (H) Time-lapse images of apoptotic cell (induced by opto-hCaspase-8 activation) in co-culture. Imaging was performed every 3 min for 12 h in total. Arrows indicate persisting apoptotic bodies in neighbors. (I and J) Percentage of extruded apoptotic cells during 12 h post apoptosis induction. (I) Cells were classified as “extruded,” if more than 50% of cell body or apoptotic fragments were expulsed from the monolayer. (J) Full and partial extrusion were defined as in Fig. 8. Data are pooled from two (I and J) independent experiments (n as indicated). All images are representative of at least three independent experiments (n = 3). Scale bars, 20 µm.
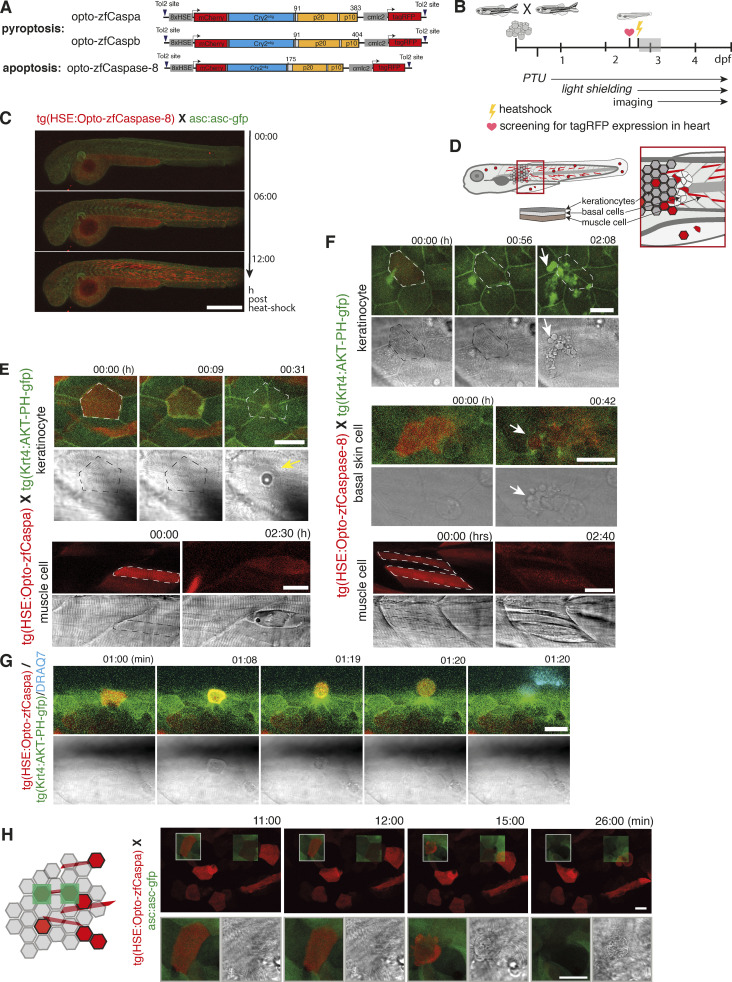
Figure 7.
Spatial and temporal control of PCD by opto-caspases in zebrafish. (A) Construct design for heat-shock-induced expression of mCherry-Cry2olig fusion with zfCaspa, zfCaspb, or zfCaspase-8. (B) Experimental setup for expression, activation, and imaging of optogenetic induction of PCD. Grey shaded region is the time frame in which C was acquired. (C) Time-lapse imaging of 2,5-dpf Tg(Opto-zfCaspa)×asc:asc-gfp larvae at 0, 6, and 12 hphs (hours post heat shock). Scale bar, 500 µm. (D) Schematic depiction of a zebrafish larvae showing the location of keratinocytes, basal cells and muscle cells. (E–H) The timecourse of light-induced activation of Caspa or Caspase-8 in Tg(Opto-zf-Caspa) or Tg(Opto-Caspase-8) lines crossed with tg(Krt4:AKT-PH-GFP). Dying cells are outlined by dashed lines. Scale bar, 20 µm. (E) Keratinocyte pyroptosis and muscle cell death after activation of opto-zfCaspa. (F) Keratinocyte, basal cell and muscle cells death after activation of opto-zfCaspase-8. (G) Keratinocyte extrusion after activation of opto-zfCaspa. (H) Two photon-activation of opto-zfCaspa (red) in single cells. Data are representative of n = 3 independent repeats.
We next generated an inducible system for the expression of optogenetic constructs under the control of a heat shock responsive element (HSE; Bajoghli et al., 2004), and created stable zebrafish lines by tol2-based transgenesis. A cardiac myosin light chain 2 (cmlc2) promoter driving tagRFP expression in the heart was used to identify transgenic larvae (Fig. 7 B). Heat-shock-induced efficient opto-caspase expression, yielding a typical mosaic expression pattern in different tissues, without any detectable spontaneous cell death induction (Fig. 7 C). While skin cells (keratinocytes and basal cells) form inflammasomes and die in response, muscle cells do not express zfASC, inflammatory caspases, or gasdermins. The mosaic expression of opto-zfCaspases in the different tissues (depicted in Fig. 7 D) allowed us to compare the effect of different optoCDEs in inflammasome-responsive and unresponsive tissues. Blue light illumination of opto-zfCaspa-expressing keratinocytes (Fig. 7, D and E and Video 7) rapidly induced pyroptosis, as judged by the appearance of a characteristic pyroptotic morphology highlighted by shrinking and eventually membrane ballooning (yellow arrow). By contrast, opto-zfCaspase-8 activation in keratinocytes and muscle cells induced apoptosis, which was highlighted by the formation of apoptotic bodies (Fig. 7 F white arrows, and Video 8). While effects of opto-caspase activation in skin cells were visible within ∼40 min, it only became apparent in muscle cells after several hours (Fig. 7, E and F). Unexpectedly, opto-zfCaspb also induced apoptosis in keratinocytes (Fig. S4 E) despite the ability of Caspb to cleave hGSDMD in HEK293 cells. Both opto-zfCaspa as well as opto-zfCaspase-8 induced apoptotic cell death in muscle cells (Fig. 7, E and F; visible as a contraction without ballooning of the cell), likely due to the absence of gasdermins in these cells (Kuri et al., 2017). In line with the observations in the mammalian cells, induction of pyroptosis in zebrafish skin triggered the rapid extrusion of the pyroptotic cells from the monolayer, followed by acquisition (Fig. 7 G and Video 9). Finally, we also used a 2-photon laser to stimulate ROIs (Fig. 7 H, light blue squares), demonstrating that we can restrict the activation of cell death to single cells. In summary, optoCDEs allow both temporal and spatial activation of cell death within living tissue down to a single-cell level.
Video 7.
Optogenetic induction of pyroptosis in zebrafish larvae. Time-lapse fluorescence confocal microscopy showing a zebrafish keratinocyte expressing opto-zfCaspa (red) and stimulated with blue light. Left, the brightfield channel. Pyroptotic cell can be seen extruded from the monolayer, followed by the gap closure. Scale bar, 20 µm.
Video 8.
Optogenetic induction of apoptosis in zebrafish larvae. Time-lapse fluorescence confocal microscopy showing a zebrafish keratinocyte expressing opto-zfCaspase-8 (red) and stimulated with blue light. Left, the brightfield channel. Apoptotic cell is fragmented into apoptotic bodies, which are retained within the monolayer. Scale bar, 20 µm.
Video 9.
Pyroptotic cell permeabilization in zebrafish. Time-lapse fluorescence confocal microscopy showing a zebrafish keratinocyte expressing opto-zfCaspa (red) and stimulated with blue light. Left, the brightfield channel. Pyroptotic cell is rapidly extruded from the monolayer, which is followed by the rapid membrane permeabilization, loss of cellular content and gain of DRAQ7 signal (turquoise). Scale bar, 20 µm.
Distinct cell death modes induce differential fates of epithelial cells
To highlight the usefulness and versatility of the novel tools for the study of complex biological questions, we compared the fates of cells dying by lytic (pyroptosis or necroptosis) versus non-lytic (apoptotic) cell death within confluent epithelial monolayers. Previous studies reported that apoptosis is followed by the extrusion of dead cells from monolayers, but death was usually induced non-specifically by laser ablation that causes a mixed apoptotic/necrotic phenotype (Okano et al., 2020; Tirlapur et al., 2001; Uchugonova et al., 2008), and the fate of necroptotic/pyroptotic cells were never studied on a single cell level before.
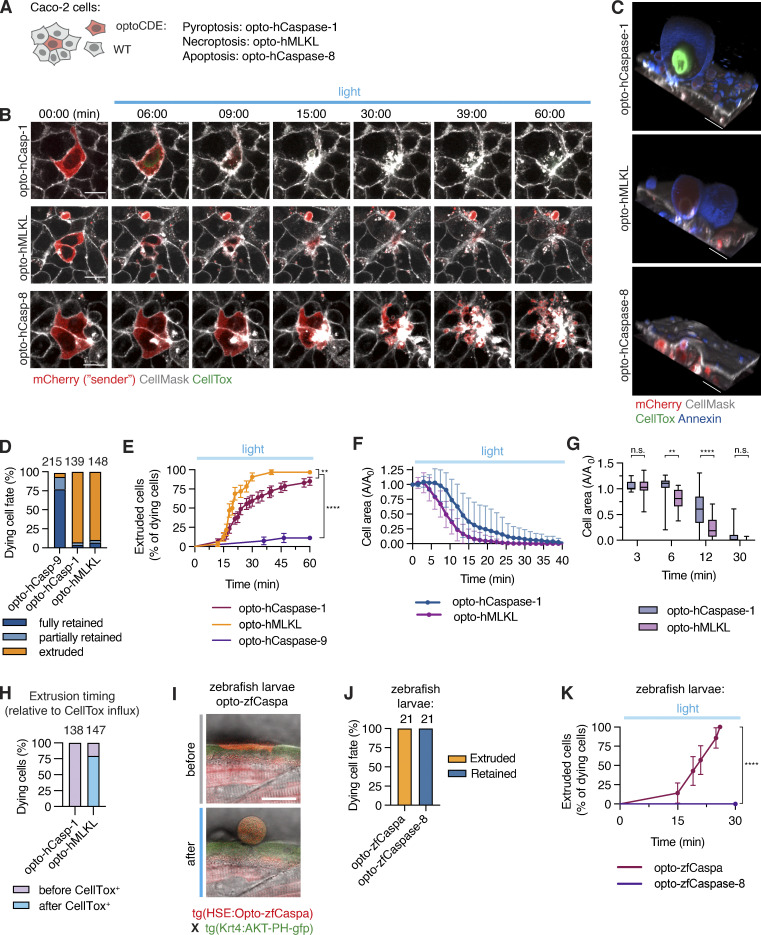
To avoid confounding effects of low-level cell death activation in neighboring cells due to light diffusion, we used confluent mosaic monolayers of opto-hCaspase-1-, opto-hCaspase-8-, or opto-hMLKL-expressing Caco-2 cells co-cultured with 20–50-fold excess of WT cells (Fig. 8 A). Illumination of optoCDE-expressing cells initiated the respective cell death programs, which was followed by a rapid rearrangement of neighboring cells and the re-establishment of epithelial integrity (Fig. 8 B and Video 10). As observed before (Fig. 3), both pyroptotic and necroptotic cells were rapidly extruded within minutes after the appearance of the first morphological signs of cell death, Annexin-V staining and, in case of pyroptosis, CellTox influx (Figs. 8, B–E; and S4, F and G), with some Annexin-V-positive membrane remnants being left behind and taken up by the neighboring cells.
Figure 8.
Response of neighboring cells to PCD. (A) Co-culture system consisting of Caco-2 cells expressing mCherry-tagged optoCDEs and 50× excess of WT Caco-2 neighbors. (B–H) Clusters of one or several optoCDE-expressing cells surrounded by WT cells were stimulated with 488 nm light (8.7 mW/cm2) every 180 s for 90 min of total time. (B) Timelapse images showing cell boundaries (CellMask) and membrane permeabilization (CellTox). (C) 3D reconstructions showing extruded pyroptotic and necroptotic cells (top and middle panels) or efferocytosed apoptotic bodies (bottom panel) at 1 h. Annexin V labels necrotic cell membranes. (D) Quantification of dying cell fates as described in the material and method section. Total number of cells quantified per condition is indicated. (E) Pyroptotic, necroptotic and apoptotic cell extrusion over time. n = 56, 32, and 27 cells. **, P < 0.01; ****, P < 0.0001 (Mantel-Cox test). (F and G) Decrease in pyroptotic and necroptotic cell area over time (normalized to t = 0). Mean n = 20 and 18 cells. *, P < 0.05; **, P < 0.01 (two-tailed t test). (H) Percentage of pyroptotic and necroptotic cells fully extruded before or after gain of CellTox signal. (I) Images of opto-zfCaspa-expressing zebrafish keratinocytes before and after illumination (488 nm). (J) Quantification of extruded versus retained cells during opto-zfCaspa-induced pyroptosis and opto-zfCaspase-8-induced apoptosis in zebrafish larvae. (K) Analysis of pyroptotic and apoptotic cell extrusion in zebrafish larvae. n = 21 events for each type of cell death; ****, P < 0.0001 (Mantel-Cox test). Mean ± SD (F and G) or mean ± SEM (E and K). Data are representative (B, C, and I) or pooled from (D–H and J–K) of three independent experiments, each done with triplicate technical replicates (n = 9). Scale bars, 20 µm.
Video 10.
Necrotic cell extrusion and apoptotic cell efferocytosis in human epithelial cells. Time-lapse fluorescence confocal microscopy showing cells (red) undergoing optoCaspase-1-induced pyroptosis, optoMLKL-induced necroptosis or optoCaspase-8-induced apoptosis in presence of WT neighbors. The upper panels show CellMask (grey) and mCherry (red) signal, bottom panels show DIC and mCherry. Scale bar, 20 µm.
By contrast, the majority of apoptotic Caco-2 cells were retained within the monolayer, rapidly fragmented into apoptotic bodies and either partially or fully engulfed by their neighbors (Fig. 8, B–E). Apoptotic bodies persisted within neighbors for up to 12 h before being degraded (Fig. S4, H–J). Unexpectedly, the fragmentation of apoptotic Caco-2 cells was only observed in the presence of neighboring cells, as when isolated Caco-2 cells were induced to die by apoptosis they did not form apoptotic bodies, despite displaying other apoptotic features, i.e., cell shrinking and Annexin-V acquisition (Fig. S5, A–C). Apoptotic cell fragmentation and efferocytosis preceded PS exposure and were not blocked by incubation with Annexin-V (Fig. S5, A–D), suggesting that, unlike in the case of efferocytosis by professional phagocytes, PS sensing was not a major driver of apoptotic cell engulfment by epithelial cells. Importantly, WT cells that spontaneously underwent apoptosis were also retained, fragmented, and efferocytosed (Fig. S4 E), confirming that engulfment was not due to laser illumination or the lack of natural apoptotic signals.
Figure S5.
Elimination of epithelial cells dying by apoptosis, pyroptosis, or necroptosis. (A) Representative time-lapse images of opto-hCaspase-8-transgenic Caco-2 cells undergoing apoptosis in absence of neighboring cells. Annexin V staining (blue) marks PS exposure during late timepoint. Arrows indicate characteristic apoptotic features: initial cell contraction (15 min), cell shrinking and detachment (30 min), rounding up and PS exposure (60 min). (B) Absence of apoptotic body formation in isolated Caco-2 cells versus formation of apoptotic bodies in MDCK cells. (C) Percentage of fragmented apoptotic cells in opto-hCaspase-8-transgenic Caco-2 cells cultured either alone (monoculture) or together with WT Caco-2 and stimulated with blue light for 1 h. (D) Percentage of extruded, partially extruded apoptotic cells in co-cultures treated with Annexin-V (10 µg/ml). (E) Representative time-lapse images of spontaneous apoptosis and apoptotic corpse efferocytosis in wild-type Caco-2. Scale bars, 20 µm. (F) Representative images of lifeact-GFP distribution in non-stimulated Caco-2 cells. Basal plane (0 µm) shows randomly oriented lamellipodia, and medial plane (+5 µm) shows cortical actin enrichment at the cell junctions). (G) Representative time-lapse images of actin rearrangement in neighboring cells during apoptotic cell extrusion. Scale bar, 20 µm. Cells were stimulated and images were acquired as in Fig. 8, A–C. (H) Reconstructed side view (xz) of time-lapse series from B. Arrows indicate incomplete phagocytic cup formation during failed corpse engulfment. (I) Percentage of extruded cells over time. Cell death was induced by illumination as described previously in the co-cultures treated with the indicated inhibitors. The time of extrusion was determined by complete gap closure in the imaging plane, determined by the CellMask signal. Data are pooled from three (C) or two (D) independent experiments (total number of cell death events as indicated). Data in I are pooled from two independent experiments (total number of cell death events as indicated in Fig. 9 H. Mean ± SEM. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. Scale bars, 20 µm.
Since a hallmark of programmed necrosis is the loss of membrane integrity, we investigated if extrusion of pyroptotic and necroptotic Caco-2 cells correlated with membrane permeabilization. While membrane permeabilization and CellTox influx always preceded extrusion of pyroptotic cells, the majority of necroptotic cells were fully extruded even before acquiring CellTox signal, and marginally faster than pyroptotic cells (20 vs. 26 min on average; Figs. 8, F–H, and S4 G), implying that, cell lysis cannot be the only factor determining cell extrusion. Apoptotic cells were also efferocytosed in zebrafish larvae, while optogenetic induction of pyroptosis resulted in cell extrusion (Figs. 7 G and 8, I–K). In summary, our data demonstrate that unlike unspecific induction of mixed apoptotic/necrotic cell death modes by laser ablation, the specific cell death induction using optoCDEs allows us to identify previously unknown differences in the ways the cells respond to apoptotic and necrotic cells.
Extrusion and uptake of dying cells require active cytoskeleton remodeling
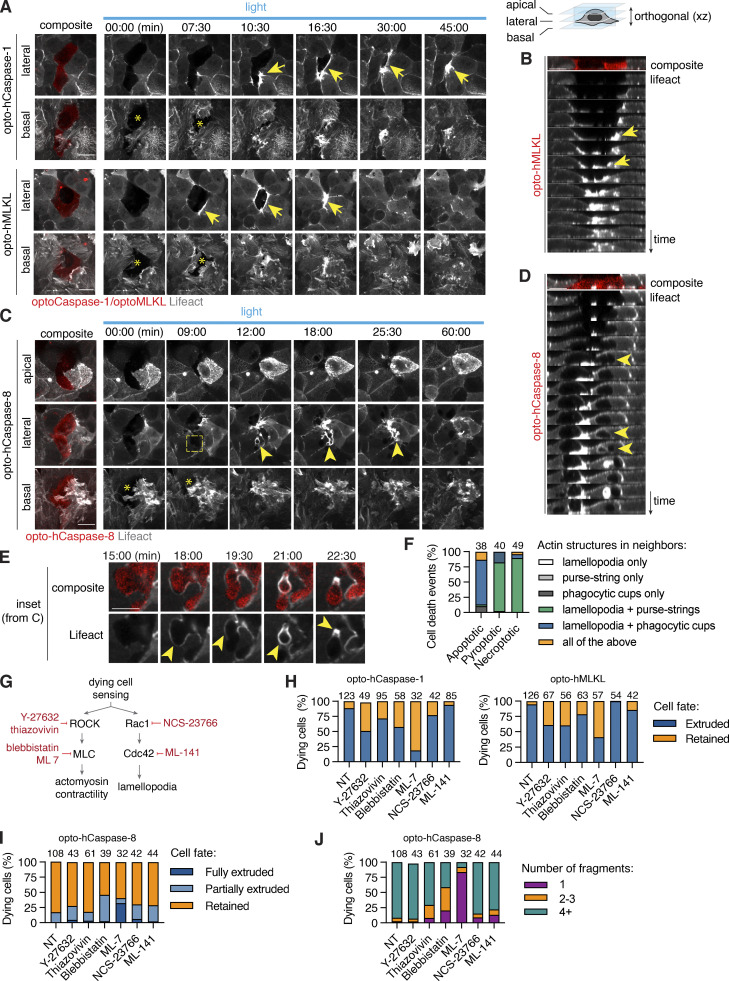
Both efferocytosis and extrusion of dead cells involve extensive cytoskeletons and the formation of characteristic structures like phagocytic cups or contractile actomyosin rings (Green et al., 2016; Kuipers et al., 2014; Le et al., 2021; Rosenblatt et al., 2001) to visualize actin dynamics in the neighbors of apoptotic or pyroptotic/necroptotic cells; we thus used Lifeact-GFP Caco-2 cells. Resting neighbors showed an actin-rich cortex and dynamic randomly oriented lamellipodial protrusions at the basal side (Fig. S5 F). By contrast, neighbors involved in the extrusion of pyroptotic or necroptotic cells formed polarized lamellipodia at the basal cell surface (0 µm, asterisk) as well as contractile purse strings, visible as thick Lifeact-positive structures around the dying cell at the apico-lateral side (arrowheads; Fig. 9, A, B, and F), consistent with cytoskeletal structures previously implicated in the extrusion of laser-ablated dead cells (Duszyc et al., 2021; Le et al., 2021).
Figure 9.
Necrotic cell extrusion and apoptotic cell efferocytosis require differential cytoskeletal rearrangement in neighbors. (A–D) Time-lapse images and reconstructed side-views showing actin dynamics in neighboring cells during pyroptotic and necroptotic Caco-2 cell extrusion, or apoptotic Caco-2 cell engulfment. (A and B) White arrows indicate lamellipodia at the basal (0 µm) plane, yellow arrows show actin cables at the medial (+5 µm) plane. Photostimulation (2 mW/cm2) and z-stack acquisition were performed every 90 s. (C and D) Images show apical closure (apical plane), efferocytic cup-like structure formation (medial plane, yellow arrows) and lamellopodia (basal plane) around apoptotic cell. Apical plane is the maximum intensity projection of four planes acquired at 0.6 µm intervals. Data are representative of at least n = 3 independent experiments. (E) Inset images from C showing actin polymerization in neighbors during apoptotic corpse fragmentation and uptake. (F) Quantification of actin structures formed in neighboring cells in response to different types of cell death. Numbers indicate cell death events per conditions, pooled from three independent experiments. (G) Major regulators of actin remodeling during lamellipodia and purse-string formation, and inhibitors targeting them. (H–J) Quantification of pyroptotic, necroptotic and apoptotic cell extrusion (at t = 60 min) and apoptotic cell fragmentation in co-cultures treated with the different cytoskeletal inhibitors, pooled from six independent experiments (n as indicated). Cells were photostimulated with blue light 8.7 mW/cm2 every 180 s to activate optoCDEs. Scale bars, 20 µm.
Apoptosis induction, on the other hand, led to a rapid polarized apical movement of the neighbors on top of the apoptotic cell, starting immediately after the appearance of the first signs of apoptosis. This was accompanied by the simultaneous engagement of basal lamellipodia (asterisk), which dynamically extended below the apoptotic cell, sealing the basal gap. Neighboring cells polymerized actin from the apico-lateral to the basal side, leading to the formation of structures resembling phagocytic cups all around the apoptotic cell (arrows), followed by the engulfment of apoptotic fragments (Fig. 9, C–E). Occasionally, we observed the formation contractile rings around apoptotic cells, which typically resulted in full or partial corpse extrusion and was associated with a lack of full apical closure or failed corpse engulfment (Figs. 9 F and S5, G and H).
To mechanistically assess the contribution of lamellipodia-based cell motility versus actomyosin contractility in extrusion or efferocytosis, we used the well-characterized Myosin II inhibitors, blebbistatin and ML-7; the ROCK inhibitors, Y-27632 and thiazovivin; the Rac1 inhibitor, NSC-23766; and the CDC42 inhibitor ML-141 (Fig. 9 G). Consistent with previous work on cell extrusion, both myosin II and ROCK inhibition significantly delayed necrotic cell extrusion and increased the fraction of retained necrotic corpses 1 h post-death induction (Figs. 9 H and S5 I). Blocking lamellipodia formation by Rac1 and CDC42 inhibitors only weakly affected necrotic cell extrusion. Blebbistatin and ML-7 treatment also affected apoptotic cell efferocytosis by reducing the number of apoptotic cell fragments and moderately increasing the fraction of fully or partially extruded apoptotic cells (Fig. 9, I and J). Together, these data suggest that in Caco-2 cells, actomyosin contractility plays a dominant role in both necrotic cell extrusion and apoptotic cell fragmentation.
Sphingosine-1-phosphate signaling is needed for efferocytosis of apoptotic cells in monolayers
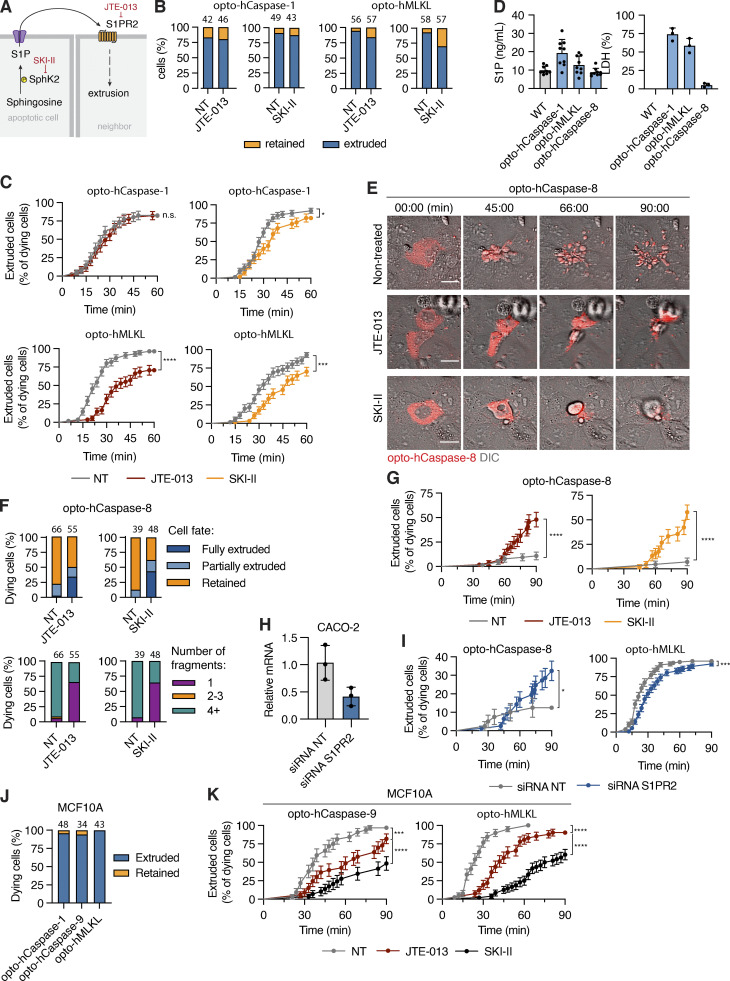
The release of sphingosines-1-phosphate from apoptotic cells and its sensing through S1P receptor 2 (S1PR2) by neighboring cells has been proposed to be a major driver of apoptotic cell extrusion (Atieh et al., 2021; Gu et al., 2011; Santacreu et al., 2021; Fig. 10 A). We thus tested whether inhibition of this pathway also blocks pyroptotic or necroptotic cell extrusion or apoptotic cell efferocytosis. Although treatment with JTE-013 (S1PR2 inhibitor) or SKI-II (Sphingosine kinase 2 inhibitor) delayed the extrusion of necroptotic cells (Fig. 10, B and C) from epithelia, it had no significant effect on the extrusion of the pyroptotic cells, even though pyroptotic cells released the highest levels of S1P (Fig. 10 D). By contrast, both JTE-013 and SKI-II treatment strongly reduced the levels of apoptotic cell efferocytosis (highlighted by the reduced numbers of apoptotic cell bodies being formed). This loss in cell fragmentation correlated with the reduced uptake of apoptotic cells by neighbors and an increase in the number of extruded cells (Fig. 10, E–G and Video 11), suggesting that in the absence apoptotic cell engulfment, other apoptotic cell-derived signals may trigger cell extrusion. The phenotypes for necroptotic cell extrusion and apoptotic cell efferocytosis were recapitulated upon knocking down S1PR2 expression in the neighboring cells (Fig. 10, H and I).
Figure 10.
S1P signaling regulates apoptotic cell efferocytosis and necroptotic cell extrusion. (A) Illustration of S1P signaling, and inhibitor targets. (B) Quantification of extruded versus retained pyroptotic and necroptotic cells in co-cultures treated with SKI-II or JTE-013. (C) Extrusion of pyroptotic and necroptotic cells over time in presence and absence of inhibitors. n = 3 independent experiments, number of cell death events is indicated above the bars. (D) S1P and LDH release from Caco-2 cells upon light-induced activation of optoCDEs. (E) Time-lapse images of opto-hCaspase-8-expressing Caco-2 cells undergoing apoptosis in presence of SKI-II or JTE-013. Scale bar, 20 µm. (F and G) Quantification of apoptotic cell retention versus extrusion in Caco-2 co-cultures treated with SKI-II and JTE013. (H) S1PR2 expression in Caco-2 cells transfected with non-targeting (NT) or anti-S1PR2 siRNA. (I) Extrusion of necroptotic and apoptotic cells over time co-cultured with bystander cells transfected with NT or S1PR2 siRNA. (J) Quantification of dying cell fates in MCF10A co-cultures. (K) Cell extrusion over time in MCF10A co-cultures treated with JTE-013 or SKI-II. Cells were photostimulated with blue light 8.7 mW/cm2 every 180 s to activate optoCDEs. Mean ± SEM (C, G, I, and K) or mean ± SD (D and H). *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (Mantel-Cox test). All data are representative of (E) or pooled from (B–D and F–K) three independent experiments, each done with triplicate technical replicates (n = 9). (D) Data points represent technical replicates.
Video 11.
Sphingosine-1-pathway regulates apoptotic cell efferocytosis by neighbors. Time-lapse fluorescence confocal microscopy showing cells (red) undergoing optoCaspase-8-induced apoptosis in co-cultures which are either non-treated (left), treated with JTE-013 (S1PR2 inhibitor, middle) or SKI-II (Sphk2 inhibitor, right). Upper panels show CellMask (grey) and mCherry (red) signal, bottom panels show DIC and mCherry. Scale bar, 20 µm.
As the finding that apoptotic cells can be efferocytosed in an S1P-dependent manner contradicts previous work that reported the extrusion of apoptotic MDCK and MCF10A cells (Gagliardi et al., 2018; Gu et al., 2011; Rosenblatt et al., 2001), we tested our optoCDE approach in these cell lines. Unlike in zebrafish keratinocytes or Caco-2 cells, we observed that both MCF10A monolayers extruded apoptotic, pyroptotic, and necroptotic cells and that S1P signaling was required for this process (Fig. 10, J and K). Similar data were obtained for MDCK cells (data not shown). These findings are in line with previous works and imply that the fate of a dying cell is highly cell-line specific. Nevertheless, S1P signaling appears to play an important role both for efferoctosis and extrusion of apoptotic cells.
Discussion
Here, we report the development, optimization, and characterization of a new toolset of optogenetically controlled cell death effectors (optoCDEs) for the induction of three major PCD types—apoptosis, necroptosis, and pyroptosis—in human and mouse cells and zebrafish larvae. OptoCDEs allow the specific induction of these forms of cell death orthogonally to and with faster kinetics and higher efficiency than the endogenous cell death pathways, offering several advantages over other currently used methods of “clean” cell death induction such as chemical activators or laser ablation. Compared with forced dimerization and the activation of caspases (Gargett and Brown, 2014; Oberst et al., 2010; Straathof et al., 2005) and RIPK3/MLKL (Hu et al., 2021; Wang et al., 2014; Wu et al., 2014) by modified FKBP domains, optoCDE excel not only in their rapid kinetics of activation and the possibility to exactly titrate the amount of stimulus, but above all in their use for single-cell manipulation and imaging in 2D, 3D, or in vivo settings. Furthermore, as Cry2 is known to become inactive within minutes of ceasing illumination, they also offer the promise of fast inactivation of signal. As we show, focused laser illumination yields superior spatiotemporal resolution that allows to rapidly and selectively kill individual cells without directly affecting the neighbors or the organism, even if these also express optoCDEs. Although other laser-based methods (such as laser ablation, laser-induced DNA-damage, or photosensitizers, such as KillerRed) can induce cell death rapidly in selected cells and in vivo (Jewhurst et al., 2014; Riani et al., 2018; Teh et al., 2010; Williams et al., 2013; Xu and Chisholm, 2016), they often fail to elicit specific cell death modes, rather relying on less specific types of cellular damage (Wang et al., 2021). Laser ablation for example induces mixed apoptotic and necrotic phenotypes (Okano et al., 2020; Uchugonova et al., 2008), and thus cannot be used to study cellular or tissue responses to specific PCD types. By contrast, the optoCDE approach allows highly specific activation of selected PCD programs directly at the effector protein level, increasing the specificity of cell death programs and reducing the impact of endogenous regulation and inter-pathway crosstalk. In contrast to optogenetic strategies for apoptosis induction via clustering of death receptors (Kim et al., 2020), recruitment of Bax (Hughes et al., 2015) to mitochondria or allosteric non-proteolytic activation of effector caspases via interdomain linker extension (Mills et al., 2012; Smart et al., 2017) allows our system to utilize a similar molecular approach and illumination parameters for different death effectors, enabling the comparison of signaling events induced by different types of cell death both in vitro and in vivo. Finally, the Cry2-based activation of cell death could easily be expanded to other death pathways that involve oligomerization of signaling components, such as lysosomal cell death or autophagy-driven cell death, and recently identified light-cleavable proteins allow to extend this principle to cleavage-activated cell death executors, such as Bid and Gasdermin D (He et al., 2021; Lu et al., 2020 Preprint).
Although “classical” methods for PCD induction might take hours and require co-stimulation with multiple ligands, direct optogenetic activation of cell death on the effector level takes just a few minutes, allowing for mechanistic studies of cells undergoing cell death with extremely high temporal resolution and independently of confounding effects associated with regularly used activators (e.g., TNFa + SMAC or LPS + nigericin). Combined with advanced live imaging techniques, this approach could offer unprecedented insights into the early cellular and metabolic events occurring during PCD; and careful titration of illumination offers a new way to study cellular mechanisms used to avoid or revert from cell death (anastasis; Tang et al., 2012; Tang and Tang, 2018), or the effects of sub-lytic/sub-lethal activation of cell death effectors like caspases and gasdermins. Such sub-lethal cell death induction recently gained attention as low-level caspase activation results in “minority mitochondrial outer membrane permeabilization” (MiniMOMP) that can drive DNA damage and genome instability (Ichim et al., 2015) or is required for differentiation (Oberst et al., 2016). Finally, the most valuable application and the largest advantage over current methods lies in the use of optoCDEs in specific single-cell death induction in 2D epithelia, 3D organoids, and live animals, and in the study of how these multicellular systems respond to distinct forms of PCD.
To highlight this application of optoCDEs, we studied the differential fates of individual epithelial cells upon apoptosis, necroptosis, or pyroptosis induction. Dying cells release a number of “find-me” and “eat-me” signals that allow professional phagocytes to migrate toward and engulf dead cells (efferocytosis; Ravichandran, 2011). Efferocytosis by non-professional phagocytes, including epithelial cells, was reported as well (Davies et al., 2018), but is less well understood as most epithelia, such as the gut epithelium, are thought to react to cell death only by extruding dying cells (Ohsawa et al., 2018). For example, multiple in vitro studies show extrusion of dying cells in response to different proapoptotic triggers, such as etoposide treatment (Andrade and Rosenblatt, 2011; Teo et al., 2020), UV illumination (Andrade and Rosenblatt, 2011; Gu et al., 2011; Rosenblatt et al., 2001), or starvation (Gagliardi et al., 2021). While we found that apoptotic MCF10A and MDCKO cells are extruded as reported before (Andrade and Rosenblatt, 2011; Atieh et al., 2021; Duszyc et al., 2021; Eisenhoffer et al., 2012; Gu et al., 2011; Le et al., 2021; Rosenblatt et al., 2001; Teo et al., 2020), our results show that apoptosis induction in Caco-2 cells and zebrafish keratinocytes results in efferocytosis by their neighbors. This implies that the decision to extrude or engulf apoptotic cells is highly cell line–specific. By contrast, we found that necroptotic and pyroptotic cells are generally extruded from monolayers, in line with previous study. showing extrusion of epithelial cells after inflammasome activation (Rauch et al., 2017; Sellin et al., 2014) and that this extrusion involves the simultaneous lamellipodia-based neighbor motility and contractile purse-string actin structures, as reported previously for laser-ablated cells (Duszyc et al., 2021; Kuipers et al., 2014; Le et al., 2021). Additional studies will be required to determine which pathway controls efferocytosis or extrusion of epithelial cells, but S1P release and signaling appear to be key signaling pathways determining both cell fates.
In summary, optoCDEs provide a versatile and specific approach to investigate PCD pathways at a spatiotemporal level, not only in individual cells but also in their neighbors in a multicellular setting, and their application might allow the identification and study new biological processes during PCD and a better understanding of associated phenomena such as membrane repair, sub-lethal cell death, or cell extrusion and migration.
Materials and methods
Plasmids
The following constructs were obtained from Addgene: Cry2olig-mCherry (plasmid #60032), mouse RIPK3-GFP (plasmid #41382), and Lifeact-miRFP703 (plasmid #79993) and used as the cloning templates. The vector containing human caspase-5 was obtained from Sino Biological (cat. #HG11152-M). The genes encoding human caspase-1, -4, -5, -8 or -9, RIPK3 and MLKL, mouse caspase-1 and caspase-11 were amplified from human or mouse cDNA. Amplified fragments were fused C- or N-terminally to Cry2olig (as indicated in the text and figures) using overlap-extension PCR and subcloned into NheI/BstBI cloning sites of pLJM1 (plasmid 19319; Addgene) for constitutive expression or EcoRI site of pLVX (cat. #632164; TaKaRa) for doxycycline-inducible expression using InFusion HD cloning kit (TaKaRa). An additional GGGS linker was introduced between the Cry2olig and caspase/kinase domains or Cry2olig and mCherry to reduce the potential sterical interference between the domains. The exact construct design is indicated in the corresponding figures. For in vivo studies in zebrafish, the catalytic domains of zf caspa, caspb, and caspase-8 were fused N-terminally to Cry2olig and subcloned into the pTH2 vector backbone containing a bidirectional heat-shock element (HSE) as a promoter (Bajoghli et al., 2004), allowing heat-shock-induced expression of the cassette. The plasmids also contain the cmlc2:tagRFP as a transgenic marker and the insertion cassette is flanked by Tol2 sites for transgenesis (Dick et al., 2016; Fig. 7 A). All inserts were verified by sequencing to ensure the absence of unwanted mutations.
Cell lines and tissue culture
HaCaT cell line was a generous gift from Gian-Paolo Dotto (University of Lausanne, Lausanne, France), Caco-2 cell line was a gift of Shaynoor Dramsi (Institut Pasteur, Paris, France), and MCF7 cell line was a gift from Nouria Hernandez (University of Lausanne). HT-29 cell line was purchased from Sigma-Aldrich (cat. #91072201). The human GSDMD-transgenic cell line was described previously (Rühl et al., 2018). HEK293T, HeLa, Caco-2, HT-29, MCF7, 3T3, and RAW264.7 cells were maintained in DMEM (Gibco) supplemented with the 10% FCS (Bioconcept), 200 U/ml penicillin, and 200 μg/ml streptomycin (Bioconcept). HaCaT, U937, and THP-1 cells were maintained in RPMI medium supplemented with the 10% FCS and penicillin-streptomycin mix. Cells were maintained at 37°C, 5% CO2.
Generation of stable cell lines
All stable cell lines were generated using an optimized lentiviral transduction protocol. For the production of lentiviral particles, 1 × 106 HEK293T cells were transiently transfected using 1.9 μg of lentiviral expression vector (pLJM1 or pLVX), 1.9 μg of third-generation packaging vector PsPax2, 0.2 μg of VSVg, and 5 μl of JetPRIME transfection reagent. The supernatants were collected 24–48 h after transfection, filtered, and either used immediately or preserved at −80°C for long-term storage. For transduction, approximately 1 × 106 cells of each cell line were spin-infected for 1 h at 1.9 × 103 g (3,000 rpm) in the presence of 10 μg/ml polybrene (Merck) to facilitate the infection. The virus-containing medium was replaced 24 h later, and the cells were left to recover for an additional 24–48 h in fresh medium. The stably transduced population of cells was selected using appropriate antibiotics (5 µg/ml puromycin or 50–100 μg/ml hygromycin Gold, both from Invivogen) for at least 5 d. To maintain stable expression, cell lines were subjected to the regular rounds of antibiotic treatment, and expression was monitored using fluorescence microscopy. Lifeact-GFP-expressing Caco-2 cells were additionally sorted based on GFP fluorescence to achieve a more uniform transgene expression level.
Generation and lentiviral transduction of primary murine bone marrow–derived macrophages
The primary bone marrow–derived macrophages were isolated from 6- to 8-wk-old wild-type C57BL/6 mice and differentiated in DMEM supplemented with 20% conditioned 3T3 cell supernatant (as a source of macrophage colony-stimulating factor M-CSF), 10% FCS, 10 mM Hepes (BioConcept), penicillin/streptomycin (Gibco), and non-essential aminoacids (Gibco). To express the opto-hCaspase-1/11, the macrophages were transduced with the lentiviral particles after 48 and 72 h of culture, as described previously (Dick et al., 2016), and kept in the dark afterward to avoid spontaneous construct activation. Photoactivation and imaging were performed on days 6–8.
3D cell culture
Single-cell Caco-2 suspension was mixed with 1.5–2 volumes of ice-cold Phenol Red Free Matrigel (Cornig) and a 40 μl drop of the resulting mixture was placed in the middle of each well of a preheated four-well μ-Slide (Ibidi), followed by immediate transfer at 37°C to induce Matrigel polymerization. After 10 min, 250–300 μl of complete medium was added to each well and the cells were maintained for 7–10 d to induce sphere formation. The medium was replaced every 2–3 d. To induce the transgene expression, the medium was supplemented with 1 μg/ml doxycycline approximately for 16 h before the experiment. For visualization of pyroptotic cells, the imaging medium was supplemented with DRAQ7 for at least 1 h before imaging to allow for dye diffusion in Matrigel.
Co-culture experiments
To generate mosaic co-cultures for cell extrusion studies, the optoCDE-expressing Caco-2 cells were trypsinized, mixed 1:20 with the wild-type Lifeact-GFP-expressing “neighbor” cells, and plated on the tissue culture–treated eight-well µ-chambers (ibidi) at a concentration of 2 × 105 cells/well to form a confluent monolayer. OptoCDE expression was induced using 1 µg/ml doxycyclin treatment overnight and imaging/photostimulation were performed the following day. To accurately determine the dying cell and neighboring cell borders, the cell membranes were pre-stained with 5 ng/ml CellMask DeepRed (Invitrogen) for 60 min.
Live cell imaging
All in vitro imaging experiments were performed using LSM800 point-scanning confocal microscope (Zeiss) equipped with 20× air immersion objective (for sphere imaging) and 63 × oil immersion Plan Apo objective (for all other experiments), as well as 405, 488, 563, and 630 nm lasers. Temperature, CO2, and humidity were controlled throughout live imaging using an automated temperature control system and a gas mixer. For time-lapse imaging, Zeiss Definite Focus.2 system was used to ensure image stabilization. Images were acquired using Zen 2 software (Zeiss) at 16-bit depth, and the acquisition settings were kept constant for the same type of experiments to aid statistical analysis.
Photoactivation of optoCDE constructs
The photoactivation experiments were performed using 488 nm laser. The laser intensity was determined using FieldMaxII laser power meter (Coherent), and the light power density was calculated as follows (Dessauges et al., 2021 Preprint). Light power density (W/cm2) = Laser peak power [W]/effective focal spot area [cm2]−1. The illumination settings were kept constant for the experiments of the same type, unless indicated otherwise.
For transient stimulation (Figs. 3 and 4), cells were illuminated at the selected timepoint with the indicated number of pulses using Zen “experimental regions” and “bleaching” functions. For sustained activation, the stimulation was performed repeatedly every 15 s (unless otherwise indicated in figure legends) or every 90–180 s (cell extrusion experiments). Initial testing and validation of the optoCDE constructs was performed in HEK293T cells. Briefly, 4 × 104 cells were seeded in each well of collagen-coated eight-well µ-chamber (ibidi) and transfected with 150–300 ng of indicated construct using XtremeGene 9 transfection reagent (Roche) according to the manufacturer’s recommendation, and photostimulation/imaging was performed 24 h later. Prior to imaging, the cells were briefly washed and the cell culture medium was replaced with the pre-warmed optiMEM. To visualize PS exposure and membrane permeabilization, the imaging medium was supplemented with CellTox Green (50,000 × dilution; Promega), 1 µM DRAQ7, and/or 1 µg/ml PacificBlue-conjugated Annexin V (BioLegend). For cell death induction in transgenic cell lines, the cells were plated at a concentration of 1.5–2.0 × 105 cells/well ∼24 h before the experiment, and optoCDE expression was induced by overnight treatment with 1 µg/ml doxycycline. To avoid cell death due to spontaneous construct activation, cells were protected from light following transfection or expression induction, and all manipulations were performed under dim light.
Activation and live imaging of optogenetic caspase variants in zebrafish
Selected larvae were heat-shocked at 2.5 dpf by incubation at 39°C for 30 min and kept under light-shielding conditions. For imaging, larvae were anesthetized by adding 40 µg/ml ethyl m-aminobenzoate methanesulfonate (tricaine; Sigma-Aldrich) into the medium. They were then mounted in 1% low-melting-point agarose (PEQ LAB Biotechnologie) on glass-bottom dishes (MaTek). A Zeiss Biosystems 780 inverted confocal microscope was used for live imaging at RT. For skin and muscles cells, we used a 40× water objective (LD C-Apochromat 40×/1.1 W Corr M27 or C-Apochromat 40×/1.2 W Corr M27; Zeiss Bio-systems). For whole larvae and overnight imaging, we acquired tile scans using a 20×air objective (Plan-Apochromat; Zeiss Biosystems) and stitching. The 488-laser was used at a power between 3% and 5% to induce optogenetic activation and excitation of GFP. The 2-photon laser was used at 950 nm wavelength and 20% laser intensity. For visualization of pyroptotic cells, the anesthetizing medium and mounting agarose solution was supplemented with DRAQ7 (1:100).
Phototoxicity and Cry2olig-induced cytotoxicity assessment
To ensure the absence of phototoxicity and Cry2olig-induced toxicity, the cells were transiently transfected with Cry2olig-mCherry plasmid and whole-imaging field was repeatedly scanned with 488 nm laser of varying intensity (0.5–4.5% laser power, 4.8–40.9 mW/cm2) every 15 s for at least 1 h. The cell viability was assessed at the end timepoint based on cell morphology and the presence of Annexin V and CellTox staining. Based on these data, the illumination intensity range between 4.8 and 25 mW/cm2 was defined as non-toxic for the cells and used for all following experiments. To assess Cry2olig-induced toxicity, the percentage of mCherry-positive cells displaying altered morphology, Annexin V staining, or CellTox influx was compared with the non-transfected (mCherry-negative) cells in the same field of view.
Generation and maintenance of Zebrafish strains
Experimental animals were cared for in accordance with EMBL guidelines and regulations and according to standard procedures (Westerfield, 2000). The following transgenic lines were created by co-injecting embryos at the one-cell stage with Opto-caspase plasmids with transposase mRNA (100 ng/μl). Transgenic larvae were selected based on heart specific expression of tagRFP under the cardiac myosin light chain (cmlc2) promoter. The following lines were generated: tg(mCherry-Cry2olig-caspa), tg(mCherry-Cry2olig-caspb), and tg(mCherry-Cry2olig-zfCaspase-8). Additionally, we used the asc:asc-gfp (Forn-Cuní et al., 2019) for skin cell labeling. For visualizing cell boundaries the Opto-lines were crossed with tg(Krt4:AKT-PH-GFP; Gault et al., 2014). To follow actin dynamics in basal retaining cells, we used the tg(krt4:Gal4) line (Wada et al., 2013) crossed to tg(6xUAS:mneonGreen-UtrCH) generated in the lab of D. Gilmour by Jonas Hartmann. To inhibit pigmentation in larvae for imaging, we treated embryos with 0.2 mM 1-phenyl-2-thiourea (PTU; Sigma-Aldrich) in E3 medium.
Drug treatment
The following inhibitors were used in the study: 20 μM Z-VAD-FMK (Invivogen), 1 μM GSK’872 (Selleck Chemicals), 5 μM Necrosulfonamide (Calbiochem), 50 µM blebbistatin (Sigma-Aldrich), 30 µM Y-27632 (StemCell), 15 µM thiazovivin (StemCell), 50 µM ML-141 (Sigma-Aldrich), 30 µM ML-7 (Tocris), 200 µM NSC-23766 (Tocris), 20 µM JTE-013 (Sigma-Aldrich), and 30 µM SKI-II (Tocris). All inhibitors were dissolved in DMSO and diluted to 10× concentration in OptiMEM, then added directly to the cells to achieve 1× concentration at least 1 h before imaging.
Canonical and non-canonical inflammasome activation in U937 cells
For canonical inflammasome activation, PMA-differentiated U937 cells grown in 24-well plates were primed with LPS for 2 h, followed by the treatment with the 15 μM Nigericin for 3 h to induce NLRP3 inflammasome activation. For non-canonical inflammasome activation, the cells were primed with human IFNγ (10 ng/ml) overnight and LPS transfection was performed as described previously (Heilig et al., 2020). Briefly, 15 μg of smooth LPS from Escherichia coli O111:B4 (Invivogen) was diluted in 200 μl of OptiMEM and incubated with Lipofectamine 2000 (5 μl per well) for 20 min at room temperature. Transfection complexes were then added directly on top of the cells in 200 μl of OptiMEM medium. To facilitate the LPS uptake, the cells were centrifuged at 500 g for 15 min, followed by incubation at 37°C for 8 h.
Optogenetic cell death induction in cell populations
The U937 cell were seeded onto black tissue culture–treated 24-well plates (4titude) at a concentration of 4 × 105 cells/well. The differentiation toward the macrophage-like phenotype was induced using PMA treatment (5 ng/ml) for 24 h, after which the cells were washed once and left to recover for additional 24 h prior to induction of optoCDE expression. The HaCaT cells were seeded at a concentration of 4 × 105 cells/well ∼24 h before an experiment. Expression of the constructs was induced in both cell lines by treatment with the 1 μg/ml doxycycline overnight. To induce cell death, each of the wells was illuminated with blue-light LEDs (450 nm) using a custom-built Light Plate Apparatus device (Gerhardt et al., 2016). To manipulate light intensity and illumination duration, custom illumination programs were created using Iris software (http://taborlab.github.io/Iris/).
Western blotting
U937 were seeded at a density of 4 × 105 cells/well and stimulated as indicated in the figure legends. After stimulation, the cells were lysed using pre-heated NuPage LDS sample buffer (Thermo Fisher Scientific) supplemented with 66 mM tris-Cl (pH 7.4), 2% SDS, and 10 mM dithiothreitol (DTT). Cell supernatants were also collected, precipitated using methanol and chloroform, and combined with cell lysates. Proteins were separated using 12% polyacrylamide gels and transferred onto a PVDF membrane using Trans-Blot Turbo (Bio-Rad). The following primary antibodies were used: rabbit anti-cleaved IL-1β (83186, CST; 1:1,000), mouse anti-IL-1β (12242, CST; 1:1,000), rabbit anti-GSDMD (ab210070; 1:1,000; Abcam), rabbit anti-cleaved N-terminal GSDMD (ab215203; 1:1,000; Abcam), mouse anti-caspase-1 (clone Bally-1 AG-20B-0048-C100; 1:1,000; AdipoGen), mouse anti-mCherry (ab125096; 1:2,000; Abcam), and HRP-conjugated mouse anti-tubulin (ab40742; 1:5,000; Abcam). Secondary antibodies conjugated to HRP (1:10,000; Southern Biotech) were used for the chemiluminescent detection.
Evaluation of cell lysis and IL-1β release
Cell lysis was assessed by measuring lactate dehydrogenase (LDH) activity in cell supernatants using the LDH cytotoxicity detection kit (TaKaRa). To obtain the positive control, the cells were fully lysed using 1% Tryton. To normalize for spontaneous cell lysis, the percentage of cell death was calculated as follows: (LDHsample − LDHnegative control)/(LDHpositive control − LDHnegative control) × 100. The level of IL-1β in cell culture supernatants was measured using ELISA (R&D Systems) according to the manufacturer’s protocol.
Image analysis of in vitro data
All image analyses were performed using FiJi (https://imagej.net/Fiji) and Zen 2 (Zeiss) software. To aid visualization, the brightness and contrast of the representative images of the same panels were adjusted and set to similar values, and illumination correction for the DIC images was performed using the Bandpass filter option (FiJi). For Figs. 1, 2, 4, and 6, the quantification of CellTox-positive and Annexin-positive cells at selected timepoints was performed manually due to the detachment and loss of late pyroptotic and necroptotic cells from the imaging plane. Apoptotic cells were defined based on morphology (cell shrinking and blebbing) and Annexin V staining (Figs. 1, 4, and S4). Quantification of DRAQ7-positive nuclei per field (Fig. 3, B and C) was performed automatically using a particle analysis tool and custom-written FiJi Macro, and the total number of DRAQ7+ nuclei per frame was divided by the total number of the mCherry-positive cells. The quantification of single-cell DRAQ7 intensities (Figs. 3 and 7) was performed by manually segmenting cell area and measuring the intensity density of selected regions over time, and intensity was normalized to the region intensity at t = 0. Pyroptotic and necroptotic cell area was quantified by the manual segmentation and tracking of dying cell borders based on CellMask and mCherry signals and additionally validated using DIC channel. The extrusion time for Figs. 8, 9, and 10 was defined based on complete closure of the area previously occupied by the mCherry-positive cell, and for all experiments the t = 0 was defined as the beginning of 488 laser stimulation. Pyroptotic and necroptotic cells were quantified as “extruded,” when the cell body and nucleus were fully removed from the plane of imaging and the remaining gap was completely closed by the neighbors (determined by membrane staining) and “retained” if the nucleus remained in the imaging plane at the end of the experiment. Apoptotic cells were quantified as “fully extruded,” if the majority (over 80%) of apoptotic bodies were extruded by the end of the experiment, “retained” if less than 20% of the cell body was extruded, and “partially extruded,” if at least 20%, but not more than 80% of cell body remained within the monolayer.
Quantification of apoptotic and pyroptotic events in vivo
Quantification of in vivo optoCaspase induced apoptotic and pyroptotic events in zebrafish keratinocytes was performed manually in several independent experiments for each of the three lines (tg(mCherry-Cry2olig-caspa), tg(mCherry-Cry2olig-caspb), and tg(mCherry-Cry2olig-zf_caspase-8), and cells were classified as pyroptotic or apoptotic based on the appearance of dead cells/cell debris. For the quantification of epithelial closure of either retention or extrusion of cells, we counted the time starting at the frame before cell death–related cell shape changes that occurred to the timepoint at which surrounding cells have closed the gap completely at the apical site.
Statistical analysis
All quantitative data analysis was performed using Microsoft Excel and GraphPad Prism 8. Statistical significances are referred as * (P < 0.05), ** (P < 0.01), *** (P < 0.001), and **** (P < 0.0001). Data distribution was assumed to be normal but was not formally tested. For comparison of the two groups, a two-tailed Student t test was used. For comparison of three or more groups P values were determined using the one- way analysis of variance (ANOVA) for multiple comparisons, and correction for the multiple comparisons was performed using Dunnett’s method. Comparison of extrusion probabilities of apoptotic, pyroptotic, and necroptotic cells was performed using a Mantel-Cox test.
Online supplemental material
Fig. S1 contains additional information on optoCaspase-1/-4 and -5 activation in human cells. Fig. S2 shows the testing of human and mouse optoCDEs in macrophages. Fig. S3 shows additional information on the activation of optoCaspase-1 in 2D and 3D cell culture systems, and additional information on the generation and testing of light-activatable apoptosis and necroptosis systems. Fig. S4 describes the generation and testing of zebrafish optoCDEs and contains additional information on the elimination of pyroptotic, apoptotic, or necroptotic cells. Fig. S5 contains additional information on the elimination of pyroptotic, apoptotic, or necroptotic cells. Video 1 shows the induction of pyroptosis by light-induced activation inflammatory caspases in HEK293T. Video 2 shows sub-lethal induction of pyroptosis on a single-cell level. Video 3 shows optogenetic pyroptosis induction in single cells. Video 4 shows optogenetic pyroptosis induction in 3D organotypic cell cultures. Video 5 shows optogenetic induction of apoptosis in HEK293T cells. Video 6 shows optogenetic induction of necroptosis in HEK293T cells. Video 7 shows optogenetic induction of pyroptosis in zebrafish larvae. Video 8 shows optogenetic induction of apoptosis in zebrafish larvae. Video 9 shows pyroptotic cell permeabilization and extrusion in zebrafish. Video 10 shows necrotic cell extrusion and apoptotic cell efferocytosis in human epithelial cells. Video 11 shows that the sphingosine-1-pathway regulates apoptotic cell efferocytosis by neighbors.
Supplementary Material
is the source file for Fig. 2.
Acknowledgments
We thank the UNIL Cellular imaging facility, UNIL Flow Cytometry Facility and EMBL Advanced Light Microscopy Facility for technical assistance and to D. Gilmour and J. Hartmann for the tg(6xUAS:mneonGreen-UtrCH) zebrafish line.
This work was supported by grants from the ERC (ERC2017-CoG-770988-InflamCellDeath), the Swiss National Science Foundation (175576 and 198005), the the OPO Stiftung and Novartis to P. Broz.
The authors declare no competing financial interests.
Author contributions: K. Shkarina and P. Broz conceptualized the study. K. Shkarina, J.C. Santos, and S. Ramos performed in vitro experiments; M. Leptin and E. Hasel de Carvalho designed and performed in vivo experiments. All authors contributed to data analysis and manuscript writing.
References
- Andrade, D., and Rosenblatt J.. 2011. Apoptotic regulation of epithelial cellular extrusion. Apoptosis. 16:491–501. 10.1007/s10495-011-0587-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atieh, Y., Wyatt T., Zaske A.M., and Eisenhoffer G.T.. 2021. Pulsatile contractions promote apoptotic cell extrusion in epithelial tissues. Curr. Biol. 31:1129–1140.e4. 10.1016/j.cub.2020.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajoghli, B., Aghaallaei N., Heimbucher T., and Czerny T.. 2004. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev. Biol. 271:416–430. 10.1016/j.ydbio.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Bibo-Verdugo, B., Snipas S.J., Kolt S., Poreba M., and Salvesen G.S.. 2020. Extended subsite profiling of the pyroptosis effector protein gasdermin D reveals a region recognized by inflammatory caspase-11. J. Biol. Chem. 295:11292–11302. 10.1074/jbc.RA120.014259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz, P., and Dixit V.M.. 2016. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 16:407–420. 10.1038/nri.2016.58 [DOI] [PubMed] [Google Scholar]
- Broz, P., Pelegrín P., and Shao F.. 2020. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 20:143–157. 10.1038/s41577-019-0228-2 [DOI] [PubMed] [Google Scholar]
- Davies, S.P., Reynolds G.M., and Stamataki Z.. 2018. Clearance of apoptotic cells by tissue Epithelia: a Putative role for hepatocytes in liver efferocytosis. Front. Immunol. 9:44. 10.3389/fimmu.2018.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vasconcelos, N.M., Van Opdenbosch N., Van Gorp H., Martín-Pérez R., Zecchin A., Vandenabeele P., and Lamkanfi M.. 2020. An apoptotic caspase network safeguards cell death induction in pyroptotic macrophages. Cell Rep. 32:107959. 10.1016/j.celrep.2020.107959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessauges, C., Mikelson J., Dobrzyński M., Jacques M.-A., Frismantiene A., Gagliardi P.A., Khammash M., and Pertz O.. 2021. Optogenetic actuator/biosensor circuits for large-scale interrogation of ERK dynamics identify sources of MAPK signaling robustness. Syst. Biol. 10.1101/2021.07.27.453955(PreprintpostedJuly272021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick, M.S., Sborgi L., Ruhl S., Hiller S., and Broz P.. 2016. ASC filament formation serves as a signal amplification mechanism for inflammasomes. Nat. Commun. 7:11929. 10.1038/ncomms11929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszyc, K., Gomez G.A., Lagendijk A.K., Yau M.-K., Nanavati B.N., Gliddon B.L., Hall T.E., Verma S., Hogan B.M., Pitson S.M., et al. 2021. Mechanotransduction activates RhoA in the neighbors of apoptotic epithelial cells to engage apical extrusion. Curr. Biol. 31:1326–1336.e5. 10.1016/j.cub.2021.01.003 [DOI] [PubMed] [Google Scholar]
- Eisenhoffer, G.T., Loftus P.D., Yoshigi M., Otsuna H., Chien C.-B., Morcos P.A., and Rosenblatt J.. 2012. Crowding induces live cell extrusion to maintain homeostatic cell numbers in epithelia. Nature. 484:546–549. 10.1038/nature10999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forn-Cuní, G., Meijer A.H., and Varela M.. 2019. Zebrafish in inflammasome research. Cells. 8:901. 10.3390/cells8080901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, P.A., Dobrzyński M., Jacques M.-A., Dessauges C., Ender P., Blum Y., Hughes R.M., Cohen A.R., and Pertz O.. 2021. Collective ERK/Akt activity waves orchestrate epithelial homeostasis by driving apoptosis-induced survival. Dev. Cell. 56:1712–1726.e6. 10.1016/j.devcel.2021.05.007 [DOI] [PubMed] [Google Scholar]
- Gagliardi, P.A., Somale D., Puliafito A., Chiaverina G., di Blasio L., Oneto M., Bianchini P., Bussolino F., and Primo L.. 2018. MRCKα is activated by caspase cleavage to assemble an apical actin ring for epithelial cell extrusion. J. Cell Biol. 217:231–249. 10.1083/jcb.201703044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galluzzi, L., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Agostinis P., Alnemri E.S., Altucci L., Amelio I., Andrews D.W., et al. 2018. Molecular mechanisms of cell death: recommendations of the nomenclature committee on cell death 2018. Cell Death Differ. 25:486–541. 10.1038/s41418-017-0012-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargett, T., and Brown M.P.. 2014. The inducible caspase-9 suicide gene system as a “safety switch” to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 5:235. 10.3389/fphar.2014.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault, W.J., Enyedi B., and Niethammer P.. 2014. Osmotic surveillance mediates rapid wound closure through nucleotide release. J. Cell Biol. 207:767–782. 10.1083/jcb.201408049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardt, K.P., Olson E.J., Castillo-Hair S.M, Hartsough L.A., Landry B.P., Ekness F., Yokoo R., Gomez E.J., Ramakrishnan P., Suh J., et al. 2016. An open-hardware platform for optogenetics and photobiology. Sci. Rep. 6:35363. 10.1038/srep35363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Y.N., Guy C., Olauson H., Becker J.U., Yang M., Fitzgerald P., Linkermann A., and Green D.R.. 2017. ESCRT-III acts downstream of MLKL to regulate necroptotic cell death and its consequences. Cell. 169:286–300 e16. 10.1016/j.cell.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R. 2019. The coming decade of cell death research: five riddles. Cell. 177:1094–1107. 10.1016/j.cell.2019.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D.R. 2011. Means to an End: Apoptosis and other Cell Death Mechanisms. Cold Spring Harbor Laboratory Pres, Cold Spring Harbor, NY. [Google Scholar]
- Green, D.R., Oguin T.H., and Martinez J.. 2016. The clearance of dying cells: table for two. Cell Death Differ. 23:915–926. 10.1038/cdd.2015.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y., Forostyan T., Sabbadini R., and Rosenblatt J.. 2011. Epithelial cell extrusion requires the sphingosine-1-phosphate receptor 2 pathway. J. Cell Biol. 193:667–676. 10.1083/jcb.201010075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., Huang Z., Huang K., Chen R., Nguyen N.T., Wang R., Cai X., Huang Z., Siwko S., Walker J.R., et al. 2021. Optogenetic control of non-apoptotic cell death. Adv. Sci. (Weinh). 8:2100424. 10.1002/advs.202100424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilig, R., Dilucca M., Boucher D., Chen K.W., Hancz D., Demarco B., Shkarina K., and Broz P.. 2020. Caspase-1 cleaves Bid to release mitochondrial SMAC and drive secondary necrosis in the absence of GSDMD. Life Sci. Alliance. 3:e202000735. 10.26508/lsa.202000735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, H., Wu X., Wu G., Nan N., Zhang J., Zhu X., Zhang Y., Shu Z., Liu J., Liu X., et al. 2021. RIP3-mediated necroptosis is regulated by inter-filament assembly of RIP homotypic interaction motif. Cell Death Differ. 28:251–266. 10.1038/s41418-020-0598-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, R.M., Freeman D.J., Lamb K.N., Pollet R.M., Smith W.J., and Lawrence D.S.. 2015. Optogenetic apoptosis: light-triggered cell death. Angew. Chem. Int. Ed. Engl. 54:12064–12068. 10.1002/anie.201506346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichim, G., Lopez J., Ahmed S.U., Muthalagu N., Giampazolias E., Delgado M.E., Haller M., Riley J.S., Mason S.M., Athineos D., et al. 2015. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol. Cell. 57:860–872. 10.1016/j.molcel.2015.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewhurst, K., Levin M., and McLaughlin K.A.. 2014. Optogenetic control of apoptosis in targeted tissues of Xenopus laevis embryos. J. Cell Death. 7:25–31. 10.4137/JCD.S18368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., and Tucker C.L.. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 7:973–975. 10.1038/nmeth.1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S., Kim N., Lee J., Kim S., Hong J., Son S., and Heo W.D.. 2020. Dynamic Fas signaling network regulates neural stem cell proliferation and memory enhancement. Sci. Adv. 6:eaaz9691. 10.1126/sciadv.aaz9691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuipers, D., Mehonic A., Kajita M., Peter L., Fujita Y., Duke T., Charras G., and Gale J.E.. 2014. Epithelial repair is a two-stage process driven first by dying cells and then by their neighbours. J. Cell Sci. 127:1229–1241. 10.1242/jcs.138289 [DOI] [PubMed] [Google Scholar]
- Kuri, P., Schieber N.L., Thumberger T., Wittbrodt J., Schwab Y., and Leptin M.. 2017. Dynamics of in vivo ASC speck formation. J. Cell Biol. 216:2891–2909. 10.1083/jcb.201703103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, A.P., Rupprecht J.-F., Mège R.-M., Toyama Y., Lim C.T., and Ladoux B.. 2021. Adhesion-mediated heterogeneous actin organization governs apoptotic cell extrusion. Nat. Commun. 12:397. 10.1038/s41467-020-20563-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.-Y., Wang Y.-Y., Shao T., Fan D.-D., Lin A.-F., Xiang L.-X., and Shao J.-Z.. 2020. The zebrafish NLRP3 inflammasome has functional roles in ASC-dependent interleukin-1β maturation and gasdermin E-mediated pyroptosis. J. Biol. Chem. 295:1120–1141. 10.1074/jbc.RA119.011751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X., Wen Y., Zhang S., Zhang W., Chen Y., Shen Y., Lemieux M.J., and Campbell R.E.. 2020. Improved Photocleavable proteins with faster and more Efficient dissociation. Synth. Biol. 10.1101/2020.12.10.419556 (Preprint posted December 10, 2020) [DOI] [Google Scholar]
- Mills, E., Chen X., Pham E., Wong S., and Truong K.. 2012. Engineering a photoactivated caspase-7 for rapid induction of apoptosis. ACS Synth. Biol. 1:75–82. 10.1021/sb200008j [DOI] [PubMed] [Google Scholar]
- Oberst, A., Ichim G., and Tait S.W.G.. 2016. Mitochondrial permeabilization: from lethality to vitality. In Mitochondria and Cell Death. Hockenbery, D.M., editor. Springer New York, New York, NY. 213–226. 10.1007/978-1-4939-3612-0_11 [DOI] [Google Scholar]
- Oberst, A., Pop C., Tremblay A.G., Blais V., Denault J.-B., Salvesen G.S., and Green D.R.. 2010. Inducible dimerization and inducible cleavage reveal a requirement for both processes in caspase-8 activation. J. Biol. Chem. 285:16632–16642. 10.1074/jbc.M109.095083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa, S., Vaughen J., and Igaki T.. 2018. Cell extrusion: a stress-responsive force for good or evil in epithelial homeostasis. Dev. Cell. 44:284–296. 10.1016/j.devcel.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano, K., Wang C.-H., Hong Z.-Y., Hosokawa Y., and Liau I.. 2020. Selective induction of targeted cell death and elimination by near-infrared femtosecond laser ablation. Biochem. Biophys. Rep. 24:100818. 10.1016/j.bbrep.2020.100818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch, I., Deets K.A., Ji D.X., von Moltke J., Tenthorey J.L., Lee A.Y., Philip N.H., Ayres J.S., Brodsky I.E., Gronert K., and Vance R.E. 2017. NAIP-NLRC4 inflammasomes coordinate intestinal epithelial cell expulsion with eicosanoid and IL-18 release via activation of caspase-1 and -8. Immunity. 46:649–659. 10.1016/j.immuni.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravichandran, K.S. 2011. Beginnings of a good apoptotic meal: the find-me and eat-me signaling pathways. Immunity. 35:445–455. 10.1016/j.immuni.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riani, Y.D., Matsuda T., Takemoto K., and Nagai T.. 2018. Green monomeric photosensitizing fluorescent protein for photo-inducible protein inactivation and cell ablation. BMC Biol. 16:50. 10.1186/s12915-018-0514-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt, J., Raff M.C., and Cramer L.P.. 2001. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11:1847–1857. 10.1016/s0960-9822(01)00587-5 [DOI] [PubMed] [Google Scholar]
- Rühl, S., Shkarina K., Demarco B., Heilig R., Santos J.C., and Broz P.. 2018. ESCRT-dependent membrane repair negatively regulates pyroptosis downstream of GSDMD activation. Science. 362:956–960. 10.1126/science.aar7607 [DOI] [PubMed] [Google Scholar]
- Samson, A.L., Zhang Y., Geoghegan N.D., Gavin X.J., Davies K.A., Mlodzianoski M.J., Whitehead L.W., Frank D., Garnish S.E., Fitzgibbon C., et al. 2020. MLKL trafficking and accumulation at the plasma membrane control the kinetics and threshold for necroptosis. Nat. Commun. 11:3151. 10.1038/s41467-020-16887-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santacreu, B.J., Romero D.J., Pescio L.G., Tarallo E., Sterin-Speziale N.B., and Favale N.O.. 2021. Apoptotic cell extrusion depends on single-cell synthesis of sphingosine-1-phosphate by sphingosine kinase 2. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1866:158888. 10.1016/j.bbalip.2021.158888 [DOI] [PubMed] [Google Scholar]
- Sellin, M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives expulsion of infected enterocytes to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Smart, A.D., Pache R.A., Thomsen N.D., Kortemme T., Davis G.W., and Wells J.A.. 2017. Engineering a light-activated caspase-3 for precise ablation of neurons in vivo. Proc. Natl. Acad. Sci. USA. 114:E8174–E8183. 10.1073/pnas.1705064114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straathof, K.C., Pulè M.A., Yotnda P., Dotti G., Vanin E.F., Brenner M.K., Heslop H.E., Spencer D.M., and Rooney C.M.. 2005. An inducible caspase 9 safety switch for T-cell therapy. Blood. 105:4247–4254. 10.1182/blood-2004-11-4564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.L., Tang H.M., Mak K.H., Hu S., Wang S.S., Wong K.M., Wong C.S.T., Wu H.Y., Law H.T., Liu K., et al. 2012. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol. Biol. Cell. 23:2240–2252. 10.1091/mbc.E11-11-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.M., and Tang H.L.. 2018. Anastasis: recovery from the brink of cell death. R. Soc. Open Sci. 5:180442. 10.1098/rsos.180442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taslimi, A., Vrana J.D., Chen D., Borinskaya S., Mayer B.J., Kennedy M.J., and Tucker C.L.. 2014. An optimized optogenetic clustering tool for probing protein interaction and function. Nat. Commun. 5:4925. 10.1038/ncomms5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R.C., Cullen S.P., and Martin S.J.. 2008. Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 9:231–241. 10.1038/nrm2312 [DOI] [PubMed] [Google Scholar]
- Teh, C., Chudakov D.M., Poon K.-L., Mamedov I.Z., Sek J.-Y., Shidlovsky K., Lukyanov S., and Korzh V.. 2010. Optogenetic in vivo cell manipulation in KillerRed-expressing zebrafish transgenics. BMC Dev. Biol. 10:110. 10.1186/1471-213X-10-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo, J.L., Tomatis V.M., Coburn L., Lagendijk A.K., Schouwenaar I.-M., Budnar S., Hall T.E., Verma S., McLachlan R.W., Hogan B.M., et al. 2020. Src kinases relax adherens junctions between the neighbors of apoptotic cells to permit apical extrusion. Mol. Biol. Cell. 31:2557–2569. 10.1091/mbc.E20-01-0084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirlapur, U.K., König K., Peuckert C., Krieg R., and Halbhuber K.J.. 2001. Femtosecond near-infrared laser pulses elicit generation of reactive oxygen species in mammalian cells leading to apoptosis-like death. Exp. Cell Res. 263:88–97. 10.1006/excr.2000.5082 [DOI] [PubMed] [Google Scholar]
- Tsuchiya, K., Nakajima S., Hosojima S., Thi Nguyen D., Hattori T., Manh Le T., Hori O., Mahib M.R., Yamaguchi Y., Miura M., et al. 2019. Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 10:2091. 10.1038/s41467-019-09753-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchugonova, A., Isemann A., Gorjup E., Tempea G., Bückle R., Watanabe W., and König K.. 2008. Optical knock out of stem cells with extremely ultrashort femtosecond laser pulses. J. Biophotonics. 1:463–469. 10.1002/jbio.200810047 [DOI] [PubMed] [Google Scholar]
- Vandenabeele, P., Galluzzi L., Vanden Berghe T., and Kroemer G.. 2010. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 11:700–714. 10.1038/nrm2970 [DOI] [PubMed] [Google Scholar]
- Wada, H., Ghysen A., Asakawa K., Abe G., Ishitani T., and Kawakami K.. 2013. Wnt/Dkk negative feedback regulates sensory organ size in zebrafish. Curr. Biol. 23:1559–1565. 10.1016/j.cub.2013.06.035 [DOI] [PubMed] [Google Scholar]
- Wang, H., Sun L., Su L., Rizo J., Liu L., Wang L.F., Wang F.S., and Wang X.. 2014. Mixed lineage kinase domain-like protein MLKL causes necrotic membrane disruption upon phosphorylation by RIP3. Mol. Cell. 54:133–146. 10.1016/j.molcel.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Wang, K.-N., Liu L.-Y., Qi G., Chao X.-J., Ma W., Yu Z., Pan Q., Mao Z.-W., and Liu B.. 2021. Light-driven cascade mitochondria-to-nucleus photosensitization in cancer cell ablation. Adv. Sci. (Weinh). 8:2004379. 10.1002/advs.202004379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield, M. 2000. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio). Fourth edition. University of Oregon Press, Eugene. [Google Scholar]
- Williams, D.C., Bejjani R.E., Ramirez P.M., Coakley S., Kim S.A., Lee H., Wen Q., Samuel A., Lu H., Hilliard M.A., and Hammarlund M.. 2013. Rapid and permanent neuronal inactivation in vivo via subcellular generation of reactive oxygen with the use of KillerRed. Cell Rep. 5:553–563. 10.1016/j.celrep.2013.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, X.-N., Yang Z.-H., Wang X.-K., Zhang Y., Wan H., Song Y., Chen X., Shao J., and Han J.. 2014. Distinct roles of RIP1-RIP3 hetero- and RIP3-RIP3 homo-interaction in mediating necroptosis. Cell Death Differ. 21:1709–1720. 10.1038/cdd.2014.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S., and Chisholm A.D.. 2016. Highly efficient optogenetic cell ablation in C. elegans using membrane-targeted miniSOG. Sci. Rep. 6:21271. 10.1038/srep21271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Wang X., Cui W., Wang W., Zhang H., Liu L., Zhang Z., Li Z., Ying G., Zhang N., and Li B.. 2013. Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nat. Commun. 4:2157. 10.1038/ncomms3157 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 2.