Abstract
SHV extended-spectrum β-lactamases (ESBLs) arise through single amino acid substitutions in the parental enzyme, SHV-1. In order to evaluate the effect of genetic dissimilarities around the structural gene on MICs, we had previously devised an isogenic system of strains. Here, we present an extended version of the system that now allows assessment of all major types of SHV β-lactamases as well as of two types of promoters of various strengths. Moreover, we devised a novel vector, pCCR9, to eliminate interference of the selection marker. A substitution within the signal sequence, I8F found in SHV-7, slightly increased MICs, suggesting more efficient transfer of enzyme precursor into the periplasmic space. We also noted that combination of G238S and E240K yielded higher resistance than G238S alone. However, the influence of the additional E240K change was more pronounced with ceftazidime and aztreonam than with cefotaxime and ceftriaxone. The SHV enzymes characterized by the single change, D179N, such as SHV-8, turned out to be the weakest SHV ESBLs. Only resistance to ceftazidime was moderately increased compared to SHV-1.
Since 1983 (15, 16), clinical isolates resistant to expanded-spectrum cephalosporins have increasingly been reported. They were derived through single amino acid substitutions from one of three parental enzymes, TEM-1, TEM-2, or SHV-1. The resulting structures were designated extended-spectrum β-lactamases (ESBLs) (13, 29), and they were classified in a new subgroup, 2be (4). Since the responsible genes are easily transferable due to frequent localization on plasmids (34) the situation has recently been called a “plague of plasmids” (6).
Phenotypic differences due to the variably mutated β-lactamases were noted early in vitro, and, although ESBL production appears to frequently lead to treatment failure, MICs for ESBL producers may be barely significantly increased compared to those for fully susceptible variants (18). Therefore, it is crucial to be able (i) to detect ESBLs or blaESBL genes easily and reliably (19) and (ii) to judge the clinical significance of given ESBLs by studying the structure-function relationships of the various ESBLs. The present study is a contribution to the second aim.
In order to examine the influence of amino acid substitutions in known SHV β-lactamases as well as of various promoter strengths on the level of resistance, we exploited a previously developed system of strains (26) which allows direct phenotypic comparison of such derivatives under isogenic conditions. To some extent, the effect of the plasmid copy number could also be estimated through introduction of a novel low-copy-number vector system.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli DH5α (10) was used as a recipient for transformation with cloned blaSHV genes based on a novel plasmid vector, pCCR9 (this study). Relevant information on recombinant strains and plasmids are given in Tables 1 and 2.
TABLE 1.
Susceptibilities of isogenic strains carrying recombinant plasmids based on the low-copy-number vector pCCR9
| Strain | Promoter | SHV | MICa (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CF | FOX | CXM | CAZ | CTX | CRO | FEP | AMC | ATM | CPX | |||
| CTA-1 | Strong | SHV-1 | 96 | 1 | 0.75 | 0.38 | <0.016 | <0.016 | 0.094 | 6 | 0.023 | 0.38 |
| CTB-1 | Weak | SHV-1 | 8 | 0.75 | 0.38 | 0.094 | <0.016 | <0.016 | 0.032 | 4 | <0.016 | 0.125 |
| CTA-2 | Strong | SHV-2 | >256 | 1 | 24 | 1 | 12 | 6 | 1 | 4 | 0.75 | >256 |
| CTB-2 | Weak | SHV-2 | >256 | 1 | 4 | 0.5 | 4 | 3 | 0.38 | 2 | 0.25 | 32 |
| CTA-2a | Strong | SHV-2a | >256 | 1 | 32 | 1 | 16 | 8 | 1 | 3 | 0.5 | >256 |
| CTB-2a | Weak | SHV-2a | >256 | 0.75 | 4 | 0.5 | 2 | 1.5 | 0.19 | 2 | 0.19 | 24 |
| CTA-3 | Strong | SHV-3 | >256 | 1 | 24 | 1.5 | 12 | 8 | 0.75 | 3 | 0.75 | >256 |
| CTB-3 | Weak | SHV-3 | >256 | 0.75 | 3 | 1 | 2 | 3 | 0.19 | 2 | 0.19 | 24 |
| CTA-4 | Strong | SHV-4 | >256 | 1 | 96 | >256 | 16 | 2 | 1 | 3 | >256 | >256 |
| CTB-4 | Weak | SHV-4 | >256 | 0.75 | 8 | 32 | 4 | 3 | 0.25 | 2 | 24 | 64 |
| CTA-5 | Strong | SHV-5 | >256 | 1 | 64 | >256 | 16 | 12 | 1.5 | 3 | >256 | >256 |
| CTB-5 | Weak | SHV-5 | >256 | 0.75 | 24 | 24 | 4 | 3 | 0.38 | 3 | 24 | 64 |
| CTA-7 | Strong | SHV-7 | >256 | 1 | 64 | >256 | 24 | 16 | 1 | 2 | 64 | >256 |
| CTB-7 | Weak | SHV-7 | >256 | 0.75 | 6 | 16 | 4 | 3 | 0.19 | 1.5 | 12 | 24 |
| CTA-8 | Strong | SHV-8 | 6 | 1 | 1 | 16 | 1 | 1 | 0.25 | 1.5 | 0.047 | 2 |
| CTB-8 | Weak | SHV-8 | 3 | 0.75 | 0.75 | 2 | 0.032 | 0.094 | 0.125 | 1.5 | 0.032 | 0.5 |
| E. coli DH5α(pCCR9) | 2 | 1 | 0.38 | 0.023 | <0.016 | <0.016 | <0.016 | 1 | <0.016 | 0.094 | ||
| E. coli DH5α | 4 | 0.75 | 0.50 | 0.023 | <0.016 | <0.016 | <0.016 | 1.5 | <0.016 | 0.094 | ||
Numbers represent mean values from five independent tests rounded up or down to the closest E test standard MIC, because reproducibility was such that many consecutive tests yielded identical results, and the maximum range was ≤2-fold.
TABLE 2.
Susceptibilities of isogenic strains carrying amino acid substitutions within the signal sequence
| Strain | Promoter | SHV | MICa (μg/ml) of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CF | FOX | CXM | CAZ | CTX | CRO | FEP | AMC | ATM | CPX | |||
| CTA-5 | Strong | SHV-5 | >256 | 1 | 64 | >256 | 16 | 12 | 1.5 | 3 | >256 | >256 |
| CTB-5 | Weak | SHV-5 | >256 | 0.75 | 24 | 24 | 4 | 3 | 0.38 | 3 | 24 | 64 |
| CTA-5F8 | Strong | SHV-5F8 | >256 | 1 | 64 | >256 | 12 | 12 | 1 | 3 | >256 | >256 |
| CTB-5F8 | Weak | SHV-5F8 | >256 | 0.75 | 24 | 24 | 4 | 3 | 0.25 | 3 | 64 | 64 |
| CTA-7I8 | Strong | SHV-7I8 | >256 | 1 | 64 | 192 | 16 | 8 | 0.75 | 2 | 32 | >256 |
| CTB-7I8 | Weak | SHV-7I8 | >256 | 0.75 | 6 | 12 | 3 | 2 | 0.19 | 1.5 | 8 | 24 |
| CTA-7 | Strong | SHV-7 | >256 | 1 | 64 | >256 | 24 | 16 | 1 | 2 | 64 | >256 |
| CTB-7 | Weak | SHV-7 | >256 | 1 | 6 | 16 | 4 | 3 | 0.19 | 1.5 | 12 | 24 |
| E. coli DH5α(pCCR9) | 2 | 1 | 0.38 | 0.023 | <0.016 | <0.016 | <0.016 | 1 | <0.016 | 0.094 | ||
| E. coli DH5α | 4 | 0.75 | 0.50 | 0.023 | <0.016 | <0.016 | <0.016 | 1.5 | <0.016 | 0.094 | ||
Numbers represent mean values from five independent tests rounded up or down to the closest E test standard MIC, because reproducibility was such that many consecutive tests yielded identical results, and the maximum range was ≤2-fold.
Antibiotics.
Ampicillin was obtained from SmithKline Beecham Pharmaceuticals (Surrey, England), and tetracycline was from Pfizer (Groton, Conn.).
Susceptibility tests.
Inhibition zone diameters were ascertained by disk testing according to the guidelines of the NCCLS (23). E tests (AB Biodisk, Solna, Sweden) were performed on Mueller-Hinton agar plates (Difco, Detroit, Mich.), according to the manufacturer's instructions.
DNA preparation.
Plasmid DNA was prepared using the Qiagen (Hilden, Germany) plasmid kit according to the manufacturer's instructions. Total DNA was extracted following standard protocols (32).
Cloning and sequencing of β-lactamase genes.
Total DNA or recombinant plasmid as well as vector DNA was digested with appropriate restriction enzymes following the supplier's protocols (Hoffmann La Roche, Basel, Switzerland). Size and orientation of inserts were determined by visualizing the restriction fragments under UV light after agarose gel electrophoresis (0.7% agarose, 1 mg of ethidium bromide per liter, 4 V/cm). The gels were photographed using a Polaroid camera (type 667 Professional). Calf intestinal phosphatase, T4 DNA ligase and ligation buffer were obtained from Hoffmann La Roche, and cloning of purified restriction fragments into vector pCCR9 followed by transformation of competent E. coli DH5α cells was performed according to the method of Sambrook et al. (32). Clones were selected and purified on Mueller-Hinton agar plates (Difco) with 10 μg of tetracycline per ml. DNA sequencing was performed by the dideoxy chain termination method (33) using the ABI Prism Big Dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Foster City, Calif.) and an ABI Prism 310 genetic analyzer.
SDM and oligonucleotides.
Single nucleotide mutations were introduced using the QuikChange site-directed mutagenesis (SDM) kit (Stratagene, La Jolla, Calif.). Introduction of each point mutation was confirmed, and the entire gene and promoter region were checked, all by sequencing. Correctly mutated genes were recloned as 3.6-kb fragments into fresh pCCR9 vector using EcoRI and HindIII restriction sites for genes with promoter A and Asp718 and SphI sites for those governed by promoter B. The recombinant plasmids were used to transform E. coli DH5α. Oligonucleotides for sequencing (20-mers) and SDM (20- and 23-mers) were custom synthesized (Microsynth, Balgach, Switzerland). Oligonucleotides for SDM are listed in Table 3.
TABLE 3.
Manipulations performed by SDM and the necessary mutagenic oligonucleotide primers
| Recombinant plasmids | Parental structure | Amino acida substitutionc | Mutagenic primersc |
|---|---|---|---|
| MPA-1,b MPB-1b | |||
| MPA-2,b MPB-2b | |||
| MPA-2a,b MPB-2ab | |||
| MPA-3,b MPB-3b | |||
| MPA-4, MPB-4 | MPA-5, MPB-5 | R205→L (CGG→CTG) | 5′TTCGCAACTGCAGCTGCTGC3′, |
| 3′AAGCGTTGACGTCGACGACG5′ | |||
| MPA-5,b MPB-5b | |||
| MPA-5F8, MPB-5F8 | MPA-5, MPB-5 | I8→F (ATT→TTT) | 5′GTGGTTATGCGTTATTTTCG3′, |
| 3′CACCAATACGCAATAAAAGC5′ | |||
| MPA-7, MPB-7 | MPA-7I8, MPB-7I8 | I8→F (ATT→TTT) | 5′GTGGTTATGCGTTATTTTCG3′, |
| 3′CACCAATACGCAATAAAAGC5′ | |||
| MPA-7I8, MPB-7I8 | MPA-5, MPB-5 | R43→S (CGC→AGC) | 5′CAGCTGTCGGGCAGCGTAGG3′, |
| 3′GTCGACAGCCCGTCGCATCC5′ | |||
| MPA-8, MPB-8 | MPA-1, MPB-1 | D179→N (GAC→AAC) | 5′GGCGACGCCCGCAACACCACTACC3′, |
| 3′CCGCTGCGGGCGTTGTGGTGATGG5′ |
Amino acids are indicated by conventional single-letter nomenclature.
Construction by Nüesch et al. (26).
Boldface type indicates mutagenized codons.
Nucleotide sequence accession number.
The complete nucleotide sequences of vectors pAW9 and pCCR9 have been deposited in the EMBL database under the accession numbers AJ289102 and AJ277764, respectively.
RESULTS
Construction of a low-copy-number plasmid vector, pCCR9.
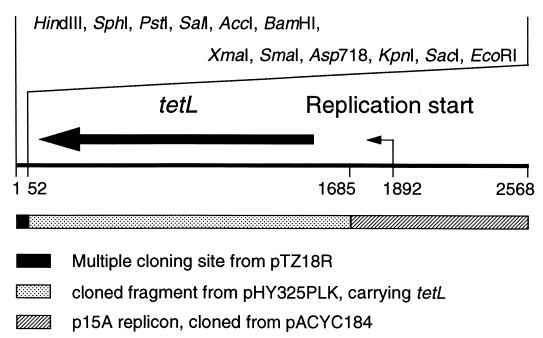
The original vector of the isogenic system, pTZ18R, is a high-copy-number system and expresses TEM-1 as a selection marker, which can interfere with part of the phenotype of the product of cloned blaSHV genes. In order to eliminate such adverse effects, an alternative vector, pAW9, was constructed, a 2,553-bp low-copy-number vector with a tetracycline selection marker. The low-copy-number replicon originated from pACYC184 (EMBL accession no. X06403), and the tetracycline resistance determinant originated from the shuttle vectors pHY300PLK (12) and pHY325PLK (EMBL accession no. D00054). The multiple cloning site (MCS) from pHY300PLK (12) is also contained in pAW9. Since, however, this MCS did not contain a KpnI/Asp718 site, we replaced it by the MCS from pTZ18R. The resulting new 2,568-bp vector was designated pCCR9, and a physical map is given in Fig. 1. The complete nucleotide sequences of both pAW9 and pCCR9 have been determined.
FIG. 1.
Physical map of the vector pCCR9. Numbers indicate nucleotide coordinates. Only unique restriction sites are depicted.
Derivation of isogenic blaSHV mutants.
The two described isogenic systems are based on a stronger promoter (PA) and a weaker promoter (PB) (26). PA originates from blaSHV-2a, and PB originates from blaSHV-2. PA leads to four to eight times more transcript (30). Introduction of point mutations into the bla genes of plasmids pMPA-1, pMPB-1, pMPA-5, and pMPB-5 resulted in further recombinants according to the genealogy shown in Table 3. Thus, the naturally occurring genes for SHV-1, SHV-2, SHV-2a, SHV-3, SHV-4, SHV-5, SHV-7, and SHV-8 were included in both systems, PA and PB. Moreover, two additional genes were constructed, coding for SHV-5F8 and SHV-7I8, which are not found in nature. Together with the producers of SHV-5 and SHV-7, these constructions provided a system for the study of a particularly interesting amino acid variation within the signal sequence. All mutations were verified by sequencing the entire blaSHV genes and flanking regions, 400 nucleotides upstream and 300 nucleotides downstream of the open reading frame, and no mistakes were found.
Since, after mutagenesis, the derived genes were all associated with the pTZ18R background (MPA-MPB system), they were recloned into the pCCR9 vector, using EcoRI/HindIII in the PA background and Asp718/SphI in the PB background to recover the desired 3.6-kb fragments for ligation. In order to indicate the promoters PA and PB, the final plasmids were designated pCTA or pCTB, respectively, followed by the number of the encoded SHV enzyme, e.g., pCTA-1 or pCTB-5F8. Correct recloning and orientation of inserts was confirmed by restriction analysis (not shown).
Resistance phenotypes of naturally occurring SHV β-lactamases.
The MICs for carriers of gene constructions cloned on vector pCCR9 are listed in Table 1. As expected, pCCR9 had barely any measurable effect on MICs as is shown by rows of data for ECDH5α and ECDH5α(pCCR9), which represent host alone and host with unmodified vector, respectively. In contrast to the ESBLs, except SHV-8, SHV-1 caused MICs of cephalothin (CF) of only 8 or 96 μg/ml depending on the promoter. The cephamycins, represented by cefoxitin (FOX), were completely unaffected by any SHV derivative, regardless of the promoter or the plasmid copy number.
The results obtained with the derivatives expressing naturally occurring SHV ESBLs (i) confirmed the importance of three substitutions at the active site, and (ii) for the first time, provided the possibility of a direct comparison of the phenotypic influence of most substitutions encountered within the SHV structure. SHV-4, SHV-5, and SHV-7 containing both substitutions G238S and E240K yielded the highest levels of resistance, significantly higher than SHV-2, SHV-2a, and SHV-3 with only G238S. SHV-8, containing D179N, turned out to be the weakest SHV ESBL, and the MICs for these were markedly lower, similar to those for SHV-1. It was the least effective β-lactamase against CF and amoxicillin-clavulanic acid (AMC). These MICs were even markedly below those of the non-ESBL SHV-1 and in the range of those reached by the non-β-lactamase control strains (Table 1). Interestingly, the only drug to which SHV-8 conferred a level of resistance, 16 μg/ml, that was intermediate according to NCCLS breakpoints (24) was ceftazidime (CAZ), even though the MICs of other oxyimino cephalosporins rose by ≥10-fold to levels still ≤1 μg/ml.
The enzymes SHV-3 and SHV-4, carrying an additional mutation, R205L (25, 27), compared to SHV-2 and SHV-5, respectively, conferred 1.5- to 2-fold less resistance than the latter. However, some exceptions to this trend were found (Table 1): looking at resistance to expanded spectrum cephalosporins, 7 of 20 producers of an enzyme with R205 gave rise to MICs higher than those reached by the respective L205 counterpart, while the opposite was true in only 4 of the 20 comparisons. The remaining nine pairs showed identical MICs (Table 1). The second non-active-site substitution, R43S, which turned SHV-5 into SHV-7 (3), also led to a general but slight decrease of resistance. Again, the results were slightly ambiguous: in a total of 20 comparisons, 11 producers of a β-lactamase with R43 were more resistant to expanded spectrum cephalosporins than those with S43, while only 2 were less resistant and 7 were equally resistant (Tables 1 and 2).
As expected, the promoter PA gave rise to higher MICs, and the effect varied from 1.5- to 16-fold. Finally, the results obtained by the disk method (data not shown) were in good agreement with the E tests.
Influence on resistance of amino acid substitution within the signal sequence.
Since one difference between SHV-7 and all other SHV ESBLs involved amino acid position 8 within the signal sequence, the influence of this change on the level of resistance was investigated. The MICs of the respective strain constructions carrying either isoleucine or phenylalanine at position 8 are listed in Table 1. With the introduction of phenylalanine at amino acid position 8 in SHV-5 we obtained SHV-5F8, which mediated increased resistance against all β-lactam antibiotics tested. Conversely, replacement of the phenylanine at position 8 within SHV-7 by isoleucine resulted in SHV-7I8, which conferred decreased resistance. This effect was very small in SHV-5 derivatives but more pronounced in those of SHV-7. In summary, judging from our two examples the substitution I8F within the SHV signal sequence was always likely to increase resistance.
DISCUSSION
A previously conceived isogenic system for accurate phenotypic comparison of SHV β-lactamases carrying single amino acid substitutions (26) has been exploited to analyze the contribution of all natural SHV sequence changes that are important to the level of resistance conferred. The system has now further been refined by recloning all inserts carrying blaSHV genes into an especially constructed low-copy-number vector, pCCR9. This allowed (i) elimination of any interference by the blaTEM-1 gene present on the previously used vector, pTZ18R, and (ii) greater sensitivity for small phenotypic changes thanks to the low gene dosage.
Substitution at position 238 (15, 37) is a hallmark of the SHV ESBLs, since, except in six of them, it is found in all 26 derivatives, as well as in some TEM ESBLs conferring high ceftazidime resistance (36). S238 causes displacement of the B3 β-strand in ESBLs, thus giving rise to a slight opening of the active site, allowing the entrance of bulkier oxyimino cephalosporins (7, 17, 21). The second-most important modification is the neighboring E240K, found in at least 8 SHV ESBLs, including uncharacterized ones such as SHV-15 (EMBL accession no. AJ011428). The effect in SHV ESBLs of E240K alone is not known because it is always associated with G238S. Together with S238, it does, however, improve interaction of the enzyme with the carboxylic acid group on oxyimino substituents of CAZ and aztreonam (ATM) (11). The effects of the two substitutions, G238S and E240K, are well documented by this study: G238S alone had a rather low to moderate impact on resistance, but caused a 50- to 100-fold increase in MICs towards cefotaxime (CTX) and ceftriaxone (CRO) while altering MICs of CAZ and ATM only two- to fivefold (Table 1). Conversely, addition of E240K to G238S boosted resistance to CAZ and ATM by another 20- to 250-fold, while increasing the MICs of CTX and CRO further by only 1.5- to 3-fold.
Concerning the three non-active-site substitutions, the effects are generally much smaller. Earlier results, e.g., the 1.5- to 3-fold increase of resistance by L35Q when governed by PA on the multicopy replicon (26), are confirmed by the present study. However, the effects are minimized under the control of the weaker promoter PB and by the low-copy-number effect. SHV-3 and SHV-4 differ from SHV-2 and SHV-5, respectively, by the substitution R205L, which is thought to cause cephalosporins with large C-3 substituents to fit better into the active-site pocket (17). Our results do not seem to support this view. Despite ambiguities in some of the tested strain pairs, the MICs for derivatives expressing arginine rather than leucine at position 205 were slightly elevated.
SHV-6 (1), SHV-8 (31), and four uncharacterized enzymes—SHV-16 (EMBL accession no. AF072684), SHV-24 (EMBL accession no. AB023477), SHV-25 (EMBL accession no. AF208796), and SHV-26 (EMBL accession no. AF227204)—are the only SHV ESBLs that do not contain the typical G238S substitution. SHV-6, SHV-8, and SHV-24 carry D179A, -N, and -G, respectively. D179 and R164 form a salt bridge and are the anchor points of the omega loop that contains the catalytically important E166 (20). Disruption of this salt bridge leads to extension of the substrate profile (2, 28). For unknown reasons, many natural TEM ESBLs are altered in position 164 (22), but none are altered in position 179, while in SHV ESBLs the opposite is true. The effects of D179A, -N, and -G had been poorly understood either for a lack of investigation, or, in the case of SHV-8, because the enzyme was masked by additional mechanisms (31). Examination of SHV-8 in our isogenic system revealed that the D179N change resulted (i) in a moderate increase in CAZ resistance, (ii) in a small but insignificant increase of resistance to CTX, CRO, cefepime (FEP), and ATM, and (iii) in a significant decrease of resistance to narrow-spectrum cephalosporins and AMC. In fact, SHV-8 conferred the lowest level of CF resistance of all derivatives tested, even with the strong promoter (Table 1). Thus, the future will show whether SHV derivatives, lacking the G238S substitution, will emerge as important ESBLs and disseminate successfully.
The precursor of SHV-7 carries a substitution, I8F (3), that can have no influence on the native β-lactamase because it is trimmed off with the signal sequence after secretion into the periplasmic space. Since testing of the I8F substitution in the SHV-7 and SHV-5 background revealed slightly higher MICs for derivatives with phenylalanine in position 8 (Table 2), we propose that the signal sequence of the original SHV-7 precursor may lead to more efficient transfer of β-lactamase into the periplasm. The effect is stronger in SHV-7 than in SHV-5. These results suggest that, apart from primary structure and expression, the level of resistance conferred by a particular β-lactamase can be modulated by the rate of transfer into the periplasm.
Increased resistance has been ascribed to hyperproduction of β-lactamases (9), and hyperproduction may be due to promoter variations (8). This effect is clearly documented by the two presented isogenic systems. In some cases, the contribution of the different promoters to the level of resistance appeared to be even greater than that of certain amino acid substitutions, as exemplified by the MICs of CTX for strains CTA-2, CTB-2, CTA-2a, and CTB-2a (Table 1). Moreover, the low-copy-number system, pCCR9, yielded generally greater promoter effects than the high-copy-number system based on vector pTZ18R (data not shown). This may be due to saturation effects in the latter system.
The detection of ESBLs is still not satisfactory (19). The data in Table 1 demonstrate that cefpodoxime (CPX) is the expanded-spectrum cephalosporin most efficiently destroyed by these ESBLs. As suggested earlier (5, 35), it is therefore the most sensitive single screening agent for SHV ESBLs, clearly superior to ceftazidime (14). This is another example for the versatility and also for the practical use of the isogenic system for the investigation of the resistance phenotype of the clinically important SHV-type ESBLs.
ACKNOWLEDGMENT
This work was supported by the Swiss National Foundation (grants 3200-39466.93/3 and 3200-52532.97).
REFERENCES
- 1.Arlet G, Rouveau M, Philippon A. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol Lett. 1997;152:163–167. doi: 10.1016/s0378-1097(97)00196-1. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee S, Pieper U, Kapadia G, Pannell L K, Herzberg O. Role of the omega-loop in the activity, substrate specificity, and structure of class A β-lactamase. Biochemistry. 1998;37:3286–3296. doi: 10.1021/bi972127f. [DOI] [PubMed] [Google Scholar]
- 3.Bradford P A, Urban C, Jaiswal A, Mariano N, Rasmussen B A, Projan S J, Rahal J J, Bush K. SHV-7, a novel cefotaxime-hydrolyzing β-lactamase, identified in Escherichia coli isolates from hospitalized nursing home patients. Antimicrob Agents Chemother. 1995;39:899–905. doi: 10.1128/aac.39.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrhardt A F, Sanders C C, Moland E S. Use of an isogenic Escherichia coli panel to design tests for discrimination of β-lactamase functional groups of Enterobacteriaceae. Antimicrob Agents Chemother. 1999;43:630–633. doi: 10.1128/aac.43.3.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fierer J, Guiney D. Extended-spectrum β-lactamases: a plague of plasmids. JAMA. 1999;281:563–564. doi: 10.1001/jama.281.6.563. [DOI] [PubMed] [Google Scholar]
- 7.Fonzé E, Charlier P, Toth Y, Vermeire M, Raquet X, Dubus A, Frere J M. Tem1 β-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallogr Sect D Biol Crystallogr. 1995;51:682–694. doi: 10.1107/S0907444994014496. [DOI] [PubMed] [Google Scholar]
- 8.Fournier B, Lagrange P H, Philippon A. β-Lactamase gene promoters of 71 clinical strains of Klebsiella oxytoca. Antimicrob Agents Chemother. 1996;40:460–463. doi: 10.1128/aac.40.2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gheorghiu R, Yuan M F, Hall L, Livermore D M. Bases of variation in resistance to β-lactams in Klebsiella oxytoca isolates hyperproducing K1 β-lactamase. J Antimicrob Chemother. 1997;40:533–541. doi: 10.1093/jac/40.4.533. [DOI] [PubMed] [Google Scholar]
- 10.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 11.Huletsky A, Knox J R, Levesque R C. Role of Ser-238 and Lys-240 in the hydrolysis of 3rd-generation cephalosporins by SHV-type β-lactamases probed by site-directed mutagenesis and 3-dimensional modeling. J Biol Chem. 1993;268:3690–3697. [PubMed] [Google Scholar]
- 12.Ishiwa H, Shibahara-Sone H. New shuttle vectors for Escherichia coli and Bacillus subtilis. IV. The nucleotide sequence of pHY300PLK and some properties in relation to transformation. Jpn J Genet. 1986;61:515–528. [Google Scholar]
- 13.Jacoby G A, Medeiros A A. More extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1991;35:1697–1704. doi: 10.1128/aac.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katsanis G P, Spargo J, Ferraro M J, Sutton L, Jacoby G A. Detection of Klebsiella pneumoniae and Escherichia coli strains producing extended-spectrum β-lactamases. J Clin Microbiol. 1994;32:691–696. doi: 10.1128/jcm.32.3.691-696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kliebe C, Nies B A, Meyer J F, Tolxdorff N R, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/aac.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 17.Knox J R. Extended-spectrum and inhibitor-resistant TEM-type β-lactamases: mutations, specificity, and three-dimensional structure. Antimicrob Agents Chemother. 1995;39:2593–2601. doi: 10.1128/aac.39.12.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livermore D M. β-Lactamases in laboratory and clinical resistance. Clin Microbiol Rev. 1995;8:557–584. doi: 10.1128/cmr.8.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mackenzie F M, Gould I M. Extended spectrum β-lactamases. J Infect. 1998;36:255–258. doi: 10.1016/s0163-4453(98)94027-0. [DOI] [PubMed] [Google Scholar]
- 20.Massova I, Mobashery S. Molecular bases for interactions between β-lactam antibiotics and β-lactamases. Accounts Chem Res. 1997;30:162–168. [Google Scholar]
- 21.Matagne A, Lamotte-Brasseur J, Frere J M. Catalytic properties of class A β-lactamases: efficiency and diversity. Biochem J. 1998;330:581–598. doi: 10.1042/bj3300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maveyraud L, Saves I, Burletschiltz O, Swaren P, Masson J M, Delaire M, Mourey L, Prome J C, Samama J P. Structural basis of extended spectrum TEM β-lactamases: crystallographic, kinetic, and mass spectrometric investigations of enzyme mutants. J Biol Chem. 1996;271:10482–10489. doi: 10.1074/jbc.271.18.10482. [DOI] [PubMed] [Google Scholar]
- 23.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2-A6. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; ninth informational supplement. M100-S9. 19, no. 1. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1999. [Google Scholar]
- 25.Nicolas M-H, Jarlier V, Honore N, Philippon A, Cole S T. Molecular characterization of the gene encoding SHV-3 β-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1989;33:2096–2100. doi: 10.1128/aac.33.12.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nüesch-Inderbinen M T, Hächler H, Kayser F H. New system based on site-directed mutagenesis for highly accurate comparison of resistance levels conferred by SHV β-lactamases. Antimicrob Agents Chemother. 1995;39:1726–1730. doi: 10.1128/aac.39.8.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peduzzi J, Barthelemy M, Tiwari K, Mattioni D, Labia R. Structural features related to hydrolytic activity against ceftazidime of plasmid-mediated SHV-type CAZ-5 β-lactamase. Antimicrob Agents Chemother. 1989;33:2160–2163. doi: 10.1128/aac.33.12.2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrosino J F, Palzkill T. Systematic mutagenesis of the active site omega loop of TEM-1 β-lactamase. J Bacteriol. 1996;178:1821–1828. doi: 10.1128/jb.178.7.1821-1828.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philippon A, Labia R, Jacoby G. Extended-spectrum β-lactamases. Antimicrob Agents Chemother. 1989;33:1131–1136. doi: 10.1128/aac.33.8.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podbielski A, Schönling J, Melzer B, Haase G. Different promoters of SHV-2 and SHV-2a β-lactamase lead to diverse levels of cefotaxime resistance in their bacterial producers. J Gen Microbiol. 1991;137:1667–1675. doi: 10.1099/00221287-137-7-1667. [DOI] [PubMed] [Google Scholar]
- 31.Rasheed J K, Jay C, Metchock B, Berkowitz F, Weigel L, Crellin J, Steward C, Hill B, Medeiros A A, Tenover F C. Evolution of extended-spectrum β-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/aac.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirot D. Extended-spectrum plasmid-mediated β-lactamases. J Antimicrob Chemother. 1995;36(Suppl. A):19–34. doi: 10.1093/jac/36.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 35.Thomson K S, Sanders C C. A simple and reliable method to screen isolates of Escherichia coli and Klebsiella pneumoniae for the production of TEM- and SHV-derived extended-spectrum β-lactamases. Clin Microbiol Infect. 1997;3:549–554. doi: 10.1111/j.1469-0691.1997.tb00306.x. [DOI] [PubMed] [Google Scholar]
- 36.Venkatachalam K V, Huang W Z, Larocco M, Palzkill T. Characterization of TEM-1 β-lactamase mutants from positions 238 to 241 with increased catalytic efficiency for ceftazidime. J Biol Chem. 1994;269:23444–23450. [PubMed] [Google Scholar]
- 37.Yuan M F, Hall L, Savelkoul P, Vandenbroucke G C, Livermore D M. SHV-13, a novel extended-spectrum β-lactamase, in Klebsiella pneumoniae isolates from patients in an intensive care unit in Amsterdam. Antimicrob Agents Chemother. 2000;44:1081–1084. doi: 10.1128/aac.44.4.1081-1084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]