Bacterial microcompartments are organelle-like structures that enhance a variety of metabolic functions in diverse bacteria. Composed entirely of proteins, thousands of homologous hexameric shell proteins tesselate to form facets while pentameric proteins form the vertices of a polyhedral shell that encapsulates various enzymes, substrates and cofactors. Recent structural data have highlighted nuanced variations in the sequence and topology of microcompartment shell proteins, emphasizing how variation and specialization enable the construction of complex molecular machines. Recent studies engineering synthetic miniaturized microcompartment shells provide additional frameworks for dissecting principals of microcompartment structure and assembly. This review updates our current understanding of bacterial microcompartment shell proteins, providing new insights and highlighting outstanding questions.
INTRODUCTION
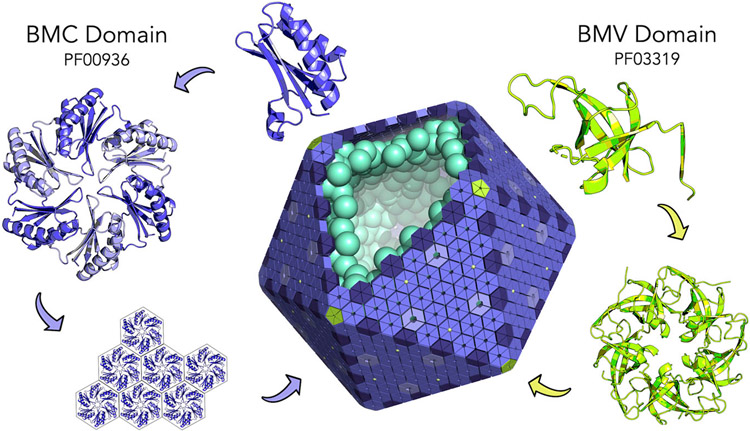
Bacterial Microcompartments (MCPs or alternatively BMCs) are a class of supramolecular structures found in approximately 20% of bacteria [1]. Ranging in size from roughly 100 - 400 nm in diameter, MCPs encapsulate and optimize a myriad of metabolic pathways by concentrating together enzymes and substrates to accelerate catalysis and to prevent the escape of toxic or volatile intermediates [2-8]. Unlike membrane-bound eukaryotic organelles, MCPs are composed entirely of proteins. So-called BMC proteins (based on Pfam PF00936) form hexameric building blocks shaped like hexagonal disks (Figure 1). These tessellate side by side to form flat extended facets [9-13] with lateral associations driven by highly conserved perimeter residues [12,14-16]. In addition, and distinct from BMC proteins, pentameric BMV proteins (Pfam PF03319) form the vertices of the polyhedral shell, which is in some cases roughly icosahedral (Figure 1) [17-19]. The heterogenous shells of MCPs are formed from two to seven BMC paralogs that are often expressed within a single operon [4,7,8]. Narrow pores located at the center of BMC proteins provide channels for the diffusion of specific substrates and cofactors. The external protein shell remains the hallmark of all MCPs.
Figure 1:
Assembly principles of bacterial microcompartments (MCPs). The BMC protein domain (purple) oligomerizes to form hexameric disks. Hundreds of hexamers tesselate side by side to form the nearly flat facets of the outer microcompartment shell (center). The BMV domain (chartreuse) is a distinctly different protein component, which oligomerizes to form pentamers at the polyhedral vertices of the microcompartment shell. Encapsulated enzymes are diagrammed in cyan. This idealized microcompartment shell is shown as a regular icosahedron. Most types of MCPs are polymorphic and less regular in shape.
Despite their structural similarity, MCPs are functionally diverse, carrying out various multi-step metabolic reactions in different bacteria. MCPs can be broadly classified into two major categories: carboxysomes and metabolosomes. The founding MCP type, carboxysomes, utilize bicarbonate (HCO3) and ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) in order to enhance CO2 fixation [20-22]. By contrast, metabolosomes metabolize a variety of substrates. Two types of B12-dependent metabolosomes have been extensively studied and shown to degrade 1,2-propanediol (Pdu MCP) and ethanolamine (EuT MCP) [4,8,21]. Another recently discovered class of MCPs, glycyl radical enzyme metabolosomes (GRMs), perform glycyl radical chemistry using distinct signature enzymes that define an assortment of subclasses, GRMs 1-5 [1,23-26]. Some GRMs metabolize choline (Cut MCPs, Type I and Type II), some consume 1,2-propanediol in a B12-independent manner (Grp MCP) and others process fucose and rhamnose [23,27-30*]. Additionally, recent discoveries have begun to characterize MCPs that use S-1-amino-2-propanol-kinase to process aminoacetone (AAUMs or formerly RMMs), MCPs that degrade ethanol (Etu MCPs) and a new class of MCPs that are predicted to degrade xanthine [1,25,31-33*].
This review focuses on current structural data, highlighting and updating our understanding of the roles of bacterial microcompartment shell proteins. We draw attention to the unusual variations in sequence and topology in BMC shell proteins and emphasize structural polymorphisms in certain subsets, which are likely to relate to functional specialization. Finally, we investigate the implications of recently characterized mini MCP shells, addressing strengths in identifying MCP assembly principals and overall shell topology.
STRUCTURAL FEATURES AND VARIATIONS IN BMC SHELL PROTEINS
To date, the structures of some 110 MCP shell proteins have been deposited in the protein data bank (PDB) [34]. A specialized database focusing on MCPs has recently been established to facilitate the analysis and study of their structures [35**]. Here we summarize the structural features of BMC shell proteins and update the tertiary topological variations discovered so far.
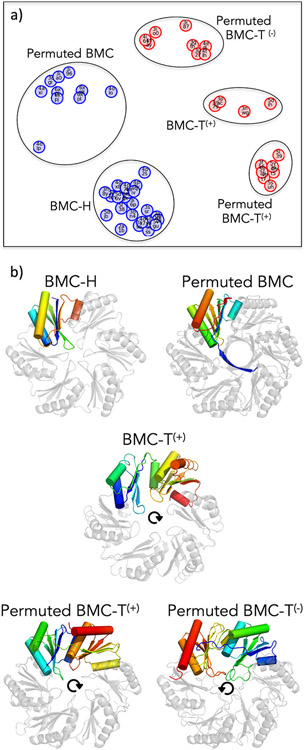
Though structurally similar, BMC shell proteins exhibit topological differences of various types (Figure 2a). The canonical BMC protein is comprised of a roughly 100 amino acid domain (Pf00936), and is referred to as BMC-H for its hexameric assembly (Figure 2b). The hexagonal disks formed by BMC proteins have distinctly shaped top and bottom faces, with one relatively flat face and the other bearing a central depression that creates a concave surface. Looking down the flat face and following the sequence from the N to C-terminus, the secondary structure elements of BMC-H proteins are arranged in a roughly clockwise fashion (Figure 2b). A unique subset of hexameric BMC domain-containing shell proteins, Permuted BMCs, have been discovered to have cyclically permuted sequences and structures. While they possess a similar overall tertiary structure, the circular permutation results in differently poised N and C-termini relative to their BMC-H counterparts [36-38*]. In cases that have been structurally characterized (including Permuted BMCs from Eut, Pdu and Cut MCP types), a novel extension at the N-terminus forms a right-handed 6-stranded beta-barrel (with one strand from each subunit) protruding from the otherwise flat face (Figure 2b). Another unique variation of BMC shell proteins has arisen from gene-duplication events, thereby producing tandem domain structures. So-called BMC-T proteins, comprising two BMC domains, oligomerize to form trimeric pseudohexamers (Figure 2b) whose overall architectures closely resemble a canonical hexameric BMC disk [13,39-41].
Figure 2:
Different BMC shell proteins exhibit varied tertiary structures. (a) Based on comparisons of three-dimensional similarity with sequential ordering enforced, BMC shell proteins of known structure (PDB codes shown) cluster into five distinct populations, representing subtypes within the two major families: BMC-H (blue) which assemble as hexamers from a single BMC domain and BMC-T (red) which assemble as trimers from two BMC domains. In the case of BMC-Ts, superscript notations denote whether the sequential domains are arranged clockwise (+) or counterclockwise (−) in a disk, as shown in panel b. The graph layout is a stochastic optimization placing protein structures at separation distances specified by their coordinate overlap deviations (including penalties for non-overlapping residues) as nearly as possible in two-dimensional space; the overall orientation is arbitrary. (b) Rainbow-colored chains (N-terminus blue, C-terminus red) and arrows highlight the domain organizations in different BMC subtypes. The five BMC shell proteins shown are derived from the Carboxysome (BMC-H; PDB 2EWH), Choline Utilization Type II MCP (Permuted BMC; PDB 6XPI), the Propanediol Utilization MCP (BMC-T(+); PDB 3N79), the Aminoacetone Utilization MCP (BMC-T(−); PDB 5L39) and the Ethanolamine Utilization MCP (Permuted BMC-T(−); PDB 3I82).
Three-dimensional all-against-all comparisons between the known BMC structures reveal further types of variation, particularly among the BMC-T proteins (Figure 2). These relate to surprising differences in the way the sequential tandem domains are arranged, and whether the individual domains are permuted. Remarkably, different BMC-T proteins present sequentially connected BMC domains arranged in either a clockwise or counterclockwise fashion in the context of the trimeric (pseudohexameric) disk. These varied forms can be accommodated with a more finely articulated naming convention; BMC-T(+), Permuted BMC-T(+) and Permuted BMC-T(−). Here the superscript conveys the clockwise (+) or counterclockwise (−) ordering of domains when viewed from the flat face (Figure 2b). The greatest number of BMC-T structures deposited in the PDB are of the Permuted BMC-T(−) type, with a non-exhaustive list including PDBs 3GFH, 3I82, 3MPV, 4FAY, 4FDZ and 6ARD [37,40,42-45]. Despite sequence variations, all known BMC-Ts retain the overall BMC architecture, with pseudohexameric shapes compatible for exchange with BMC-H hexamers within a complete MCP [46]. The specific advantages conferred by distinct variations of the BMC protein in a shell are still not fully understood, but important functional distinctions are believed to relate to overall assembly, interactions with interior enzymes (and likely other cellular proteins), and molecular transport across the shell.
The central pores of BMC shell proteins provide routes for the diffusion of molecules across the MCP shell. Mutagenesis experiments suggest that the narrow pores in BMC-H hexamers are the primary routes of substrate influx [47-49*]. Different MCP types operate on and thus transport different substrates, suggesting that sequence and structural variations in the pores of BMC proteins are likely important for diverse metabolic functions. The pores of BMC-H hexamers are typically narrow (roughly 4 and 7 Å in diameter). Absent structural evidence for large conformational transitions, the narrow pores in BMC-H hexamers are presumed to be relatively static. Electrostatic properties of BMC pores have been analyzed, with particular implications for their roles in MCPs with charged substrates (e.g. the carboxysome [bicarbonate], Eut [ethanolamine], and Aaum [aminoacetone]) [9,10,19,50]. Several recent molecular dynamics (MD) and flux modeling studies have begun to examine the atomic details and mathematical aspects of pore transport. Important questions concern the degree to which pores in BMC-H proteins are selective for their cognate metabolic substrate. Optimal metabolic function would presumably occur with a combination of facile substrate influx and restricted metabolic intermediate efflux. MD studies on the PduA (BMC-H) protein suggested a modest level of selection in this regard, with a preference for its propanediol substrate that is 3 to 10 times greater than its propionaldehyde intermediate [51]. Similar MD studies on carboxysome shell proteins have also reported a range of selectivity for its substrate, with values in one case as high as 1000 times greater than its corresponding intermediate [49*]. Interestingly, transport and metabolic flux modeling on both the carboxysome and the Pdu MCP have emphasized that high selectivity might not be critical for function if internal consumption of the intermediate is sufficiently rapid [52,53]. Nonetheless, the pores of BMC-H proteins present useful targets for modulating MCP function, including by mutagenesis to occlude pores or to insert non-native 4Fe-4S clusters [47,54]. Additional structural studies on more remote BMC homologs could shed further light on transport mechanisms. Based on sequence alignment, several BMC shell proteins from a recently-proposed xanthine MCP [33*] appear to have three to four residue insertions near the loop region, opening the possibility for identifying novel pore features and functionalities.
BMC-T proteins present intriguing features and additional puzzles related to transport. A general theme is that the evolution of trimeric BMC assemblies appears to have allowed for greater versatility at the pore because of lower symmetry; only three instead of six equivalent residues need to be accommodated near the center. Indeed, examples of both Permuted BMC-T(+) and Permuted BMC-T(−) proteins have been shown to form trimers in which the central pore can apparently convert between open and closed forms, with important implications for the transport of larger substrates or cofactors [37,40-42,50,55,56]. The potential presence of larger pores (between 12 – 14 Å [42,55]) in the shell presents a dilemma, as retaining key metabolic intermediates is essential for proper MCP function. Different ideas have been put forward on the subject. In some cases it appears that the large BMC-T pores are regulated and could be occluded by allosteric binding events, e.g. by substrates when the MCP is active [42,50,57,58]. As described below, alternative mechanisms of opening and closing have been proposed in other cases [37,40-42,50,55,56,58]. Interestingly, examples of the BMC-T(+) type include cases where the central pore presents three symmetry-related cysteine residues (one from each protein chain) for coordinating an 4Fe-4S cluster [54,57,59].
POLYMORPHISMS IN QUATERNARY STRUCTURE
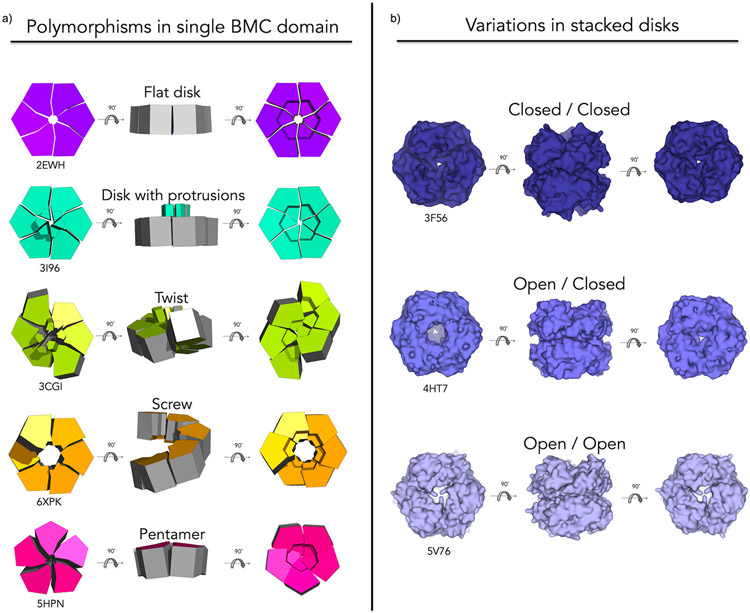
MCP shell proteins display a surprising degree of flexibility (Figure 3). This is particularly true of Permuted BMC proteins. An early study found that the EutS shell protein crystallized in two forms: 1) a canonical flat disk and 2) a hexamer with a twisting or bending deformation down the two-fold axis of symmetry (Fig. 3a) [37]. Recent structural characterization of another Permuted BMC homolog, CutR from a Choline Utilization Type II MCP, showed other forms of flexibility including the formation of flat disks and screw-type helical assemblies of varying pitch (Figure 3a) [38*]. In another instance, a synthetic Permuted BMC – a version of PduA (BMC-H) that was engineered to introduce an artificial circular permutation – rearranged to form a cyclic homopentamer, despite retaining the BMC fold (Figure 3a) [60]. Interestingly, such structural polymorphism does not appear unique to Permuted BMCs. BMV shell proteins are understood to serve as the pentameric component required for (Gaussian) shell curvature and closure [17,18,61], yet the EutN protein (from the BMV family, PF03319) was found in two separate crystallographic experiments to be capable of forming cyclic homohexamers (PDB ID 2HD3 and 2Z9H) [62].
Figure 3:
BMC shell proteins exhibit quaternary structure variations and several modes of flexibility. (a) Geometric renderings highlight variations that have been observed especially within the Permuted BMC protein type including: flat disks (purple), flat disks with beta-barrel protrusions (turquoise), twisted structures (chartreuse), six-fold screws (orange) and rearrangement to pentamers (magenta). (b) Proteins of the Permuted BMC-T(+) type are often observed as stacked disks with variations in pore openness/occlusion. Stacked disks have been reported with two closed pores (deep blue), one open and one closed pore (slate) or two open pores (light blue).
BMC-Ts also appear capable of alternative quaternary conformations (Figure 3b). Specifically, all structurally characterized members of the Permuted BMC-T(+) subset have been observed to assemble as stacked disks (a dimer of trimers), creating a large central cavity accessible by pores on opposite ends [41,50,55,56]. While their biological relevance has yet to be confirmed, their recurrence in multiple studies suggests their potential importance. Stacked disks are observed in crystal structures of recombinantly expressed and purified Permuted BMC-T(+) type proteins from alpha-carboxysomes (CsoS1D), beta-carboxysomes (CcmP), aminoacetone utilization MCPs and other MCPs of unknown function [41,50,55,58]. Double disks have also been observed by crystallography and cryo-EM in recombinantly purified mini MCP shells [46**,56], further described below. One intriguing hypothesis is that they could serve as a gated airlock system for transport [41]. Variability in the open and closed states of the pores of stacked disks have been noted in numerous studies. Some stacked disks have two open pores (PDB ID 4HT7), some have one open and one closed pore (PDB ID 3F56, 3FCH, 4HT5, 5LSR and 5V75) and some have two closed pores (PDB ID 3NWG, 5L39, 5LT5, 5V76 and 5SUH) [41,50,55,56]. The presence of stacked disks with two open pores would counter an airlock mechanism, but crystallographic observations call for cautious interpretation on this issue, owing both to conditions of the crystalline state and challenges in adequately capturing important dynamic behavior.
LARGE RECOMBINANT ASSEMBLY FORMS
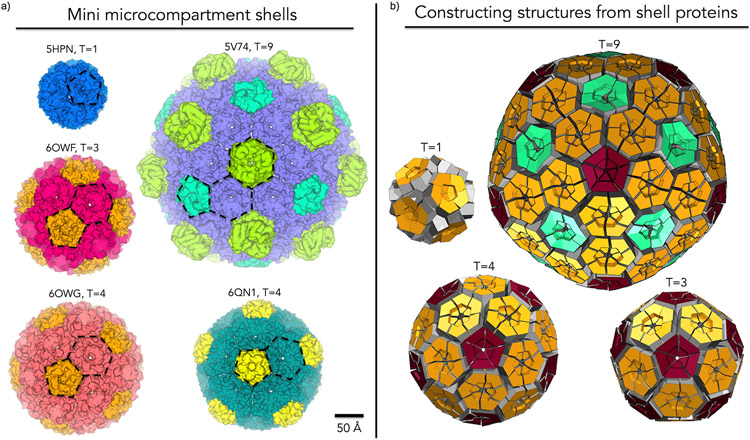
Attempts to assemble larger protein species, including work to mix different shell proteins, date to the first BMC protein structural studies [9]. Subsequent efforts to develop experimental procedures and suitable combinations of shell proteins have led to remarkable successes in purifying and characterizing what can be described as miniaturized synthetic shells (Figure 4a). Several examples have been obtained, built from either a single component or multiple shell components and ranging in size from 130 Å to 400 Å in diameter [24**,46**,56,60,63**]. These miniaturized synthetic MCP shells have helped to support and more finely articulate assembly principles that were formulated from studies on individual shell components. Of particular note, these synthetic miniaturized MCPs have confirmed the roles of BMV proteins as polyhedral vertices and the role of lateral associations between hexameric units through conserved interactions at their perimeters.
Figure 4:
Gallery of miniaturized MCP shells highlighting diversity in shape, size and number of components. (a) First columns (top to bottom): PDB: 5HPN – Shell from an engineered circularly permuted BMC shell protein, PduA, which formed a pentamer; PDB: 6OWF – Shell constructed from one BMV and one BMC-H from a beta-carboxysome, T=3; PDB: 6OWG – Shell constructed from one BMV and one BMC-H from a beta-carboxysome, T=4. Second column (top to bottom): PDB: 5V74 – 6.5 MDa shell constructed from one BMV, one BMC-H and three types of presently indistinguishable BMC-T proteins from a Haliangium ochraceum MCP. PDB: 6QN1 – mini GRM2 shell constructed from one BMV and three presently indistinguishable BMC-Hs. (b) Geometric models representing different icosahedral triangulation patterns observed. BMV (red), BMC-H (orange), and BMC-T(aquamarine). In the T=1 case an engineered BMC-H protein rearranged to form pentameric units.
These miniaturized structures have also led to surprises. The first observed mini shell was obtained serendipitously from an engineering experiment wherein a synthetically permuted version of an otherwise ordinary BMC-H protein, PduA, formed a 130 Å dodecahedron from twelve cyclic homopentamers [60]. More deliberate assembly studies based on mixtures of various BMC and BMV proteins have produced a range of structures. A 6.5 MDa mini shell was constructed from one BMC-H, one BMV and three BMC-T proteins from an MCP of unknown function. This 400 Å diameter, icosahedral shell (triangulation number T=9) highlighted the dynamic ability of different BMC-domain containing proteins to occupy different positions in the icosahedral shell [46**,56]. Studies on constructing mini shells from beta-carboxysome shell proteins resulted in a variety of structures including 210 Å T=3, 245 Å T=4 and a 310 Å prolate T=4, Q=6 icosahedral mini shells. A broad diversity in shape, size and morphology was observed despite using only one BMC-H and one BMV protein [63**]. The most recent study led to the structural characterization of a 250 Å T=4 shell constructed from GRM2 proteins. This work utilized three BMC-Hs, one BMV and numerous enzymatic proteins in varying combinations. In addition to the T=4 shell that was characterized in detail, this work revealed diversity in shape and size [24**]. While they were not necessary for the formation of closed structures, different enzymes appeared to be hierarchically involved in the formation of larger particles, though their structures could not be resolved by cryo-EM [24**].
The experimental studies on miniaturized shells have emphasized the importance of identifying a suitable composition of BMC (and BMV) paralogs for assembly. In some cases, the resolution of the structural studies has not fully distinguished the identities of similar paralogs in shells that contain complex mixtures of BMC proteins [24**,46**,56]. Thus, some uncertainties remain in modeling the key atomic interactions between components; models where distinctions between multiple BMC paralogs are ambiguous exhibit surface complementarity between BMC and BMV components with values that are somewhat lower than seen in well-resolved BMC-BMC interfaces (for example, 0.4 vs 0.6 or higher).
An important and unexpected observation from the structures of miniaturized synthetic MCPs concerns the orientation of the protein layer forming the shell. To date, all the cases on characterized mini shells show that BMC proteins are oriented with their concave faces oriented outward towards the cytosol. Early structural studies on BMC and BMV proteins offered both orientations (concave in or concave out) as possibilities [17], but biochemical and mutational studies provided evidence to suggest that concave faces interact with internal enzymes [64,65], which would require that flat faces to be oriented outward towards the cytosol. Additional biochemical data could be vital for clarifying this outstanding issue. In order to elucidate the exact arrangements of shell proteins, higher resolution structural data on intact native MCPs produced in-situ and containing interior enzymes will be essential. Encapsulated enzymes are particularly critical, given the important roles that interior enzymes have been shown to play in organizing the external shell in some systems [24**,66-68*].
Several Cryo-electron tomography (cryo-ET) studies have begun to pursue in-situ MCP structural elucidation. Multiple cryo-ET studies on carboxysomes, which are the most geometrically regular of the MCP types, have confirmed their roughly icosahedral shape revealing nearly flat facets and identifiable edges, though detailed structural features of the individual shell proteins, including shell protein orientation, have not been resolved [69-71]. Moreover, some degree of order has been seen for the encapsulated RuBisCO molecules [69,70,72-74]. The metabolosome MCPs present even greater challenges owing to their more irregular, polymorphic shapes [75]. Even the degree to which their polyhedral architectures might be described within the broader scheme of irregular icosahedra remains unclear. Recent studies highlight the possible need to bring new kinds of analysis to this problem [68*,76].
CONCLUSION AND OUTLOOK
Bacterial microcompartments are extraordinary examples of how complex protein assemblies have evolved to provide subcellular organization and compartmentalization in bacterial cells. Their ability to form robust supramolecular architectures from a complex mixture of homologous shell proteins rivals similar phenomena seen in large viruses. A great deal of structural data has revealed nuances in topological variations, conformational flexibility and quaternary polymorphisms in MCP shell proteins, highlighting the role that duplication has played in supporting functional diversification. Studies on miniaturized MCPs have affirmed models for larger scale shell architecture, though the absence of internal enzymes presents an important gap that will ultimately need to be bridged in order to understand native MCPs. In particular, the shells of metabolosome MCPs are considerably more irregular compared to the icosahedral and nearly icosahedral assembly models presently available. Cryo-ET studies pushed to higher resolution limits may be essential for achieving a fuller understanding of MCPs. In parallel, computational simulations could be informative regarding biophysical parameters that might govern assembly architecture. New large-scale assembly simulation methods are beginning to provide insights along this line [77*,78].
Acknowledgements
This work was supported by NIH/NIAID Grant R01AI081146. JMO is an HHMI Gilliam Fellow.
Footnotes
The authors declare no competing interests.
REFERNECES
- 1.Jorda J, Lopez D, Wheatley NM, Yeates TO: Using comparative genomics to uncover new kinds of protein-based metabolic organelles in bacteria. Protein Science 2013, 22:179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shively JM, Ball F, Brown DH, Saunders RE: Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 1973, 182:584–586. [DOI] [PubMed] [Google Scholar]
- 3.English RS, Lorbach SC, Qin X, Shively JM: Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Molecular Microbiology 1994, 12:647–654. [DOI] [PubMed] [Google Scholar]
- 4.Bobik TA, Lehman BP, Yeates TO: Bacterial microcompartments: widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Molecular Microbiology 2015, 98:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bobik TA, Havemann GD, Busch RJ, Williams DS, Aldrich HC: The Propanediol Utilization (pdu) Operon of Salmonella enterica Serovar Typhimurium LT2 Includes Genes Necessary for Formation of Polyhedral Organelles Involved in Coenzyme B12-Dependent 1,2-Propanediol Degradation. Journal of Bacteriology 1999, 181:5967–5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havemann GD, Sampson EM, Bobik TA: PduA Is a Shell Protein of Polyhedral Organelles Involved in Coenzyme B12-Dependent Degradation of 1,2-Propanediol in Salmonella enterica Serovar Typhimurium LT2. Journal of Bacteriology 2002, 184:1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M: Bacterial microcompartments. Nature Reviews Microbiology 2018, doi: 10.1038/nrmicro.2018.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stewart KL, Stewart AM, Bobik TA: Prokaryotic Organelles: Bacterial Microcompartments in E. coli and Salmonella. EcoSal Plus 2020, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M, Yeates TO: Protein Structures Forming the Shell of Primitive Bacterial Organelles. Science 2005, 309:936–938. [DOI] [PubMed] [Google Scholar]
- 10.Tsai Y, Sawaya MR, Cannon GC, Cai F, Williams EB, Heinhorst S, Kerfeld CA, Yeates TO: Structural Analysis of CsoS1A and the Protein Shell of the Halothiobacillus neapolitanus Carboxysome. PLoS Biol 2007, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dryden KA, Crowley CS, Tanaka S, Yeates TO, Yeager M: Two-dimensional crystals of carboxysome shell proteins recapitulate the hexagonal packing of three-dimensional crystals. Protein Sci 2009, 18:2629–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sutter M, Faulkner M, Aussignargues C, Paasch BC, Barrett S, Kerfeld CA, Liu L-N: Visualization of Bacterial Microcompartment Facet Assembly Using High-Speed Atomic Force Microscopy. Nano Lett 2016, 16:1590–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yeates TO, Jorda J, Bobik TA: The Shells of BMC-Type Microcompartment Organelles in Bacteria. Journal of Molecular Microbiology and Biotechnology 2013, 23:290–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinha S, Cheng S, Sung YW, McNamara DE, Sawaya MR, Yeates TO, Bobik TA: Alanine Scanning Mutagenesis Identifies an Asparagine–Arginine–Lysine Triad Essential to Assembly of the Shell of the Pdu Microcompartment. Journal of Molecular Biology 2014, 426:2328–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang A, Frank S, Brown I, Warren MJ, Pickersgill RW: Structural Insights into Higher Order Assembly and Function of the Bacterial Microcompartment Protein PduA. J Biol Chem 2014, 289:22377–22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uddin I, Frank S, Warren MJ, Pickersgill RW: A Generic Self-Assembly Process in Microcompartments and Synthetic Protein Nanotubes. Small 2018, 14:1704020. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka S, Kerfeld CA, Sawaya MR, Cai F, Heinhorst S, Cannon GC, Yeates TO: Atomic-Level Models of the Bacterial Carboxysome Shell. Science 2008, 319:1083–1086. [DOI] [PubMed] [Google Scholar]
- 18.Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO: Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci 2013, 22:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sutter M, Wilson SC, Deutsch S, Kerfeld CA: Two new high-resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth Res 2013, 118:9–16. [DOI] [PubMed] [Google Scholar]
- 20.Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM: Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nature Reviews Microbiology 2008, 6:681–691. [DOI] [PubMed] [Google Scholar]
- 21.Kerfeld CA, Heinhorst S, Cannon GC: Bacterial microcompartments. Annu Rev Microbiol 2010, 64:391–408. [DOI] [PubMed] [Google Scholar]
- 22.Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM: Microcompartments in Prokaryotes: Carboxysomes and Related Polyhedra. Appl Environ Microbiol 2001, 67:5351–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herring TI, Harris TN, Chowdhury C, Mohanty SK, Bobik TA: A Bacterial Microcompartment Is Used for Choline Fermentation by Escherichia coli 536. J Bacteriol 2018, 200:e00764–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalnins G, Cesle E-E, Jansons J, Liepins J, Filimonenko A, Tars K: Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nature Communications 2020, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axen SD, Erbilgin O, Kerfeld CA: A Taxonomy of Bacterial Microcompartment Loci Constructed by a Novel Scoring Method. PLOS Computational Biology 2014, 10:e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferlez B, Sutter M, Kerfeld CA: Glycyl Radical Enzyme-Associated Microcompartments: Redox-Replete Bacterial Organelles. mBio 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petit E, LaTouf WG, Coppi MV, Warnick TA, Currie D, Romashko I, Deshpande S, Haas K, Alvelo-Maurosa JG, Wardman C, et al. : Involvement of a Bacterial Microcompartment in the Metabolism of Fucose and Rhamnose by Clostridium phytofermentans. PLOS ONE 2013, 8:e54337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson MC, Wheatley NM, Jorda J, Sawaya MR, Gidaniyan SD, Ahmed H, Yang Z, McCarty KN, Whitelegge JP, Yeates TO: Identification of a Unique Fe-S Cluster Binding Site in a Glycyl-Radical Type Microcompartment Shell Protein. J Mol Biol 2014, 426:3287–3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zarzycki J, Sutter M, Cortina NS, Erb TJ, Kerfeld CA: In Vitro Characterization and Concerted Function of Three Core Enzymes of a Glycyl Radical Enzyme - Associated Bacterial Microcompartment. Sci Rep 2017, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lundin AP, Stewart KL, Stewart AM, Herring TI, Chowdhury C, Bobik TA: Genetic Characterization of a Glycyl Radical Microcompartment Used for 1,2-Propanediol Fermentation by Uropathogenic Escherichia coli CFT073. Journal of Bacteriology 2020, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallette E, Kimber MS: Structure and Kinetics of the S-(+)-1-Amino-2-propanol Dehydrogenase from the RMM Microcompartment of Mycobacterium smegmatis. Biochemistry 2018, 57:3780–3789. [DOI] [PubMed] [Google Scholar]
- 32.Mallette E, Kimber MS: Structural and kinetic characterization of (S)-1-amino-2-propanol kinase from the aminoacetone utilization microcompartment of Mycobacterium smegmatis. J Biol Chem 2018, doi: 10.1074/jbc.RA118.005485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravcheev DA, Moussu L, Smajic S, Thiele I: Comparative Genomic Analysis Reveals Novel Microcompartment-Associated Metabolic Pathways in the Human Gut Microbiome. Front Genet 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burley SK, Berman HM, Kleywegt GJ, Markley JL, Nakamura H, Velankar S: Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive. In Protein Crystallography: Methods and Protocols. Edited by Wlodawer A, Dauter Z, Jaskolski M. Springer; 2017:627–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochoa JM, Bair K, Holton T, Bobik TA, Yeates TO: MCPdb: The Bacterial Microcompartment Database. bioRxiv 2021, doi: 10.1101/2021.01.09.426059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crowley CS, Sawaya MR, Bobik TA, Yeates TO: Structure of the PduU Shell Protein from the Pdu Microcompartment of Salmonella. Structure 2008, 16:1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka S, Sawaya MR, Yeates TO: Structure and Mechanisms of a Protein-Based Organelle in Escherichia coli. Science 2010, 327:81–84. [DOI] [PubMed] [Google Scholar]
- 38.Ochoa JM, Nguyen VN, Nie M, Sawaya MR, Bobik TA, Yeates TO: Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization. Protein Science 2020, 29:2201–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tanaka S, Sawaya MR, Phillips M, Yeates TO: Insights from multiple structures of the shell proteins from the β-carboxysome. Protein Science 2009, 18:108–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sagermann M, Ohtaki A, Nikolakakis K: Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A 2009, 106:8883–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klein MG, Zwart P, Bagby SC, Cai F, Chisholm SW, Heinhorst S, Cannon GC, Kerfeld CA: Identification and Structural Analysis of a Novel Carboxysome Shell Protein with Implications for Metabolite Transport. Journal of Molecular Biology 2009, 392:319–333. [DOI] [PubMed] [Google Scholar]
- 42.Thompson MC, Cascio D, Leibly DJ, Yeates TO: An allosteric model for control of pore opening by substrate binding in the EutL microcompartment shell protein. Protein Science 2015, 24:956–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takenoya M, Nikolakakis K, Sagermann M: Crystallographic Insights into the Pore Structures and Mechanisms of the EutL and EutM Shell Proteins of the Ethanolamine-Utilizing Microcompartment of Escherichia coli. J Bacteriol 2010, 192:6056–6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pang A, Liang M, Prentice MB, Pickersgill RW: Substrate channels revealed in the trimeric Lactobacillus reuteri bacterial microcompartment shell protein PduB. Acta Crystallogr D Biol Crystallogr 2012, 68:1642–1652. [DOI] [PubMed] [Google Scholar]
- 45.Thompson MC, Cascio D, Yeates TO: Microfocus diffraction from different regions of a protein crystal: structural variations and unit-cell polymorphism. Acta Crystallogr D Struct Biol 2018, 74:411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Greber BJ, Sutter M, Kerfeld CA: The Plasticity of Molecular Interactions Governs Bacterial Microcompartment Shell Assembly. Structure 2019, 27:749–763.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdhury C, Chun S, Pang A, Sawaya MR, Sinha S, Yeates TO, Bobik TA: Selective molecular transport through the protein shell of a bacterial microcompartment organelle. PNAS 2015, 112:2990–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slininger Lee MF, Jakobson CM, Tullman-Ercek D: Evidence for Improved Encapsulated Pathway Behavior in a Bacterial Microcompartment through Shell Protein Engineering. ACS Synth Biol 2017, 6:1880–1891. [DOI] [PubMed] [Google Scholar]
- 49.Faulkner M, Szabó I, Weetman SL, Sicard F, Huber RG, Bond PJ, Rosta E, Liu L-N: Molecular simulations unravel the molecular principles that mediate selective permeability of carboxysome shell protein. Scientific Reports 2020, 10:17501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallette E, Kimber MS: A Complete Structural Inventory of the Mycobacterial Microcompartment Shell Proteins Constrains Models of Global Architecture and Transport. Journal of Biological Chemistry 2017, 292:1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park J, Chun S, Bobik TA, Houk KN, Yeates TO: Molecular Dynamics Simulations of Selective Metabolite Transport across the Propanediol Bacterial Microcompartment Shell. J Phys Chem B 2017, 121:8149–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mangan NM, Flamholz A, Hood RD, Milo R, Savage DF: pH determines the energetic efficiency of the cyanobacterial CO2 concentrating mechanism. PNAS 2016, 113:E5354–E5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jakobson CM, Tullman-Ercek D, Slininger MF, Mangan NM: A systems-level model reveals that 1,2-Propanediol utilization microcompartments enhance pathway flux through intermediate sequestration. PLOS Computational Biology 2017, 13:e1005525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aussignargues C, Pandelia M-E, Sutter M, Plegaria JS, Zarzycki J, Turmo A, Huang J, Ducat DC, Hegg EL, Gibney BR, et al. : Structure and Function of a Bacterial Microcompartment Shell Protein Engineered to Bind a [4Fe-4S] Cluster. J Am Chem Soc 2016, 138:5262–5270. [DOI] [PubMed] [Google Scholar]
- 55.Cai F, Sutter M, Cameron JC, Stanley DN, Kinney JN, Kerfeld CA: The Structure of CcmP, a Tandem Bacterial Microcompartment Domain Protein from the β-Carboxysome, Forms a Subcompartment Within a Microcompartment. J Biol Chem 2013, 288:16055–16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sutter M, Greber B, Aussignargues C, Kerfeld CA: Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 2017, 356:1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pang A, Warren MJ, Pickersgill RW: Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe–4S cluster-binding site. Acta Cryst D, Acta Cryst Sect D, Acta Crystallogr D, Acta Crystallogr Sect D, Acta Crystallogr D Biol Crystallogr, Acta Crystallogr Sect D Biol Crystallogr 2011, 67:91–96. [DOI] [PubMed] [Google Scholar]
- 58.Larsson AM, Hasse D, Valegård K, Andersson I: Crystal structures of β-carboxysome shell protein CcmP: ligand binding correlates with the closed or open central pore. Journal of Experimental Botany 2017, 68:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO: Structural Insight into the Mechanisms of Transport across the Salmonella enterica Pdu Microcompartment Shell. J Biol Chem 2010, 285:37838–37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jorda J, Leibly DJ, Thompson MC, Yeates TO: Structure of a novel 13 nm dodecahedral nanocage assembled from a redesigned bacterial microcompartment shell protein. Chem Commun 2016, 52:5041–5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai F, Menon BB, Cannon GC, Curry KJ, Shively JM, Heinhorst S: The Pentameric Vertex Proteins Are Necessary for the Icosahedral Carboxysome Shell to Function as a CO 2 Leakage Barrier. PLOS ONE 2009, 4:e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forouhar F, Kuzin A, Seetharaman J, Lee I, Zhou W, Abashidze M, Chen Y, Yong W, Janjua H, Fang Y, et al. : Functional insights from structural genomics. Journal of structural and functional genomics 2007, 8:37–44. [DOI] [PubMed] [Google Scholar]
- 63.Sutter M, Laughlin TG, Sloan NB, Serwas D, Davies KM, Kerfeld CA: Structure of a Synthetic β-Carboxysome Shell. Plant Physiology 2019, 181:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fan C, Cheng S, Liu Y, Escobar CM, Crowley CS, Jefferson RE, Yeates TO, Bobik TA: Short N-terminal sequences package proteins into bacterial microcompartments. PNAS 2010, 107:7509–7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson MC, Crowley CS, Kopstein J, Bobik TA, Yeates TO: Structure of a bacterial microcompartment shell protein bound to a cobalamin cofactor. Acta Crystallographica Section F Structural Biology Communications 2014, 70:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen AH, Afonso B, Silver PA, Savage DF: Spatial and Temporal Organization of Chromosome Duplication and Segregation in the Cyanobacterium Synechococcus elongatus PCC 7942. PLOS ONE 2012, 7:e47837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA: Biogenesis of a Bacterial Organelle: The Carboxysome Assembly Pathway. Cell 2013, 155:1131–1140. [DOI] [PubMed] [Google Scholar]
- 68.Juodeikis R, Lee MJ, Mayer M, Mantell J, Brown IR, Verkade P, Woolfson DN, Prentice MB, Frank S, Warren MJ: Effect of metabolosome encapsulation peptides on enzyme activity, coaggregation, incorporation, and bacterial microcompartment formation. MicrobiologyOpen 2020, 9:e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM: Structure of Halothiobacillus neapolitanus Carboxysomes by Cryo-electron Tomography. Journal of Molecular Biology 2006, 364:526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ: Organization, Structure, and Assembly of α-Carboxysomes Determined by Electron Cryotomography of Intact Cells. Journal of Molecular Biology 2010, 396:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, et al. : Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 2013, 502:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dai W, Chen M, Myers C, Ludtke SJ, Pettitt BM, King JA, Schmid MF, Chiu W: Visualizing Individual RuBisCO and Its Assembly into Carboxysomes in Marine Cyanobacteria by Cryo-Electron Tomography. Journal of Molecular Biology 2018, 430:4156–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ: The Structure of Isolated Synechococcus Strain WH8102 Carboxysomes as Revealed by Electron Cryotomography. Journal of Molecular Biology 2007, 372:764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, et al. : Visualizing Virus Assembly Intermediates Inside Marine Cyanobacteria. Nature 2013, 502:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kennedy NW, Hershewe JM, Nichols TM, Roth EW, Wilke CD, Mills CE, Jewett MC, Tullman-Ercek D: Apparent size and morphology of bacterial microcompartments varies with technique. PLOS ONE 2020, 15:e0226395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bag S, Prentice MB, Liang M, Warren MJ, Roy Choudhury K: Classification of polyhedral shapes from individual anisotropically resolved cryo-electron tomography reconstructions. BMC Bioinformatics 2016, 17:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mohajerani F, Hagan MF: The role of the encapsulated cargo in microcompartment assembly. PLOS Computational Biology 2018, 14:e1006351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mohajerani F, Sayer E, Neil C, Inlow K, Hagan MF: Mechanisms of scaffold-mediated microcompartment assembly and size-control. bioRxiv 2020, doi: 10.1101/2020.10.14.338509. [DOI] [PMC free article] [PubMed] [Google Scholar]