Abstract
The 5′UTR part of coronavirus genomes plays key roles in the viral replication cycle and translation of viral mRNAs. The first 75–80 nt, also called the leader sequence, are identical for genomic mRNA and subgenomic mRNAs. Recently, it was shown that cooperative actions of a 5′UTR segment and the nonstructural protein NSP1 are essential for both the inhibition of host mRNAs and for specific translation of viral mRNAs. Here, sequence analyses of both the 5′UTR RNA segment and the NSP1 protein have been done for several coronaviruses, with special attention to the betacoronaviruses. The conclusions are: (i) precise specific molecular signatures can be found in both the RNA and the NSP1 protein; (ii) both types of signatures correlate between each other. Indeed, definite sequence motifs in the RNA correlate with sequence motifs in the protein, indicating a coevolution between the 5′UTR and NSP1 in betacoronaviruses. Experimental mutational data on 5′UTR and NSP1 from SARS-CoV-2 using cell-free translation extracts support these conclusions and show that some conserved key residues in the amino-terminal half of the NSP1 protein are essential for evasion to the inhibitory effect of NSP1 on translation.
Keywords: SARS-CoV-2, NSP1, SL1, 5′UTR, translation, ribosome
INTRODUCTION
The coronaviruses belong to the Coronaviridae family (order Nidovirales, kingdom Orthornavirae), which is subdivided into four Genera: alpha, beta, gamma, and deltacoronavirus. Here we analyze in some detail the betacoronaviruses. The betacoronaviruses are subdivided into four Subgenera: the Embecovirus, the Hibecovirus, the Merbecovirus, and the Sarbecovirus (Lefkowitz et al. 2018; Gorbalenya et al. 2020; Gulyaeva and Gorbalenya 2021). Among those, we will focus on the Sarbecovirus, the subgenus to which the SARS-CoV-1 and the SARS-CoV-2 belong. In Sarbecovirus, the genomic material is a positive single-stranded RNA molecule, 26.2–31.7 kb. The RNA strand is 5′-capped and polyadenylated at the 3′ end (Yogo et al. 1977; Lai and Stohlman 1981). The genome encodes two long open reading frames (ORF1a and ORF1b) at the 5′ side and several ORFs that are expressed in the late phase of infection from subgenomic RNAs (sgRNAs) (Brian and Baric 2005). Translation of ORF1a and ORF1b from the whole ss(+)RNA are the first events of the infectious process. The polyproteins synthesized from ORF1a and ORF1ab are processed into 16 nonstructural proteins (NSP1–NSP16). Besides the positive single strand genomic RNA, nine subgenomic RNAs (S, 3a, E, M, 6, 7a, 7b, 8, and N) are produced during the late phase of the infection by SARS-CoV-2 (Kim et al. 2020) and they all contain the common so-called 5′ leader sequence (nucleotides 1 to 75) (Kim et al. 2020).
The secondary structure of the 5′UTR of SARS-CoV-2 has been studied theoretically (Rangan et al. 2020) and in solution (Miao et al. 2021). The first 80 nt common to all viral transcripts fold in a series of three hairpins: SL1, SL2, and SL3. Recent structural studies have shown that NSP1 binds tightly to the small ribosome subunit and blocks the entry of the mRNA channel, thereby inhibiting cellular translation (Schubert et al. 2020; Thoms et al. 2020; Yuan et al. 2020; Lapointe et al. 2021). Experiments with reporter systems have further shown that with the sole presence of SL1 in the 5′UTR, the translation inhibition induced by NSP1 is relieved, thereby allowing translation (Tidu et al. 2021). SL1 promotes NSP1 evasion by acting on the NSP1 carboxy-terminal domain enabling viral RNA accommodation in the ribosome for translation. The interaction between the SL1 RNA hairpin and the NSP1 carboxy-terminal domain occurs while NSP1 remains bound on the ribosome. It was therefore concluded that NSP1 acts as a ribosome gatekeeper to impair cellular translation and specifically promote viral translation (Tidu et al. 2021). For the 5′ leader to overcome the translational inhibition imposed by NSP1, specific interactions between SL1 and NSP1 are thus expected. Here, we show that the analysis of sequence comparisons points to a coevolution of the sequences of SL1 and NSP1 in coronaviruses. In addition, in vitro translation experiments are supportive of the idea that the amino-terminal region of NSP1 contains conserved residues required to evade the translation inhibition imposed by NSP1.
The available sequence comparisons focus on the viral proteins and rarely consider the noncoding segments of the RNA genome. It was thus also interesting to compare the deduced relationships and phylogenies between strains. Comparisons between the 5′UTR of betacoronaviruses allowed us to establish a classification of SL1 into four classes in sarbecoviruses, SARS-CoV-1 harbors a type I SL1 whereas SARS-CoV-2 has a type III SL1. Concomitantly, we analysed the NSP1 proteins of betacoronavirus. NSP1 from SARS-CoV-1 and -2 present few but significant differences in the three domains of the proteins. By swapping key residues from SARS-CoV-1 into both SL1 and NSP1 from SARS-CoV-2, we bring experimental evidence that NSP1 and SL1 from the 5′UTR coevolved as suggested by sequence alignments in the Sarbecovirus family.
RESULTS
Sequence alignments for the 5′UTR RNA
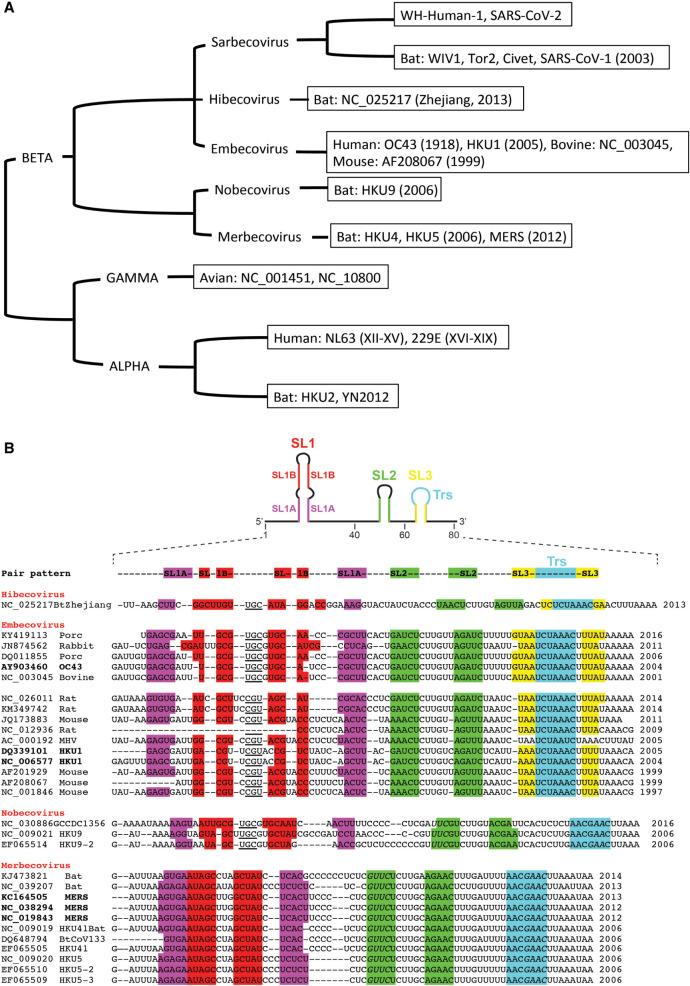
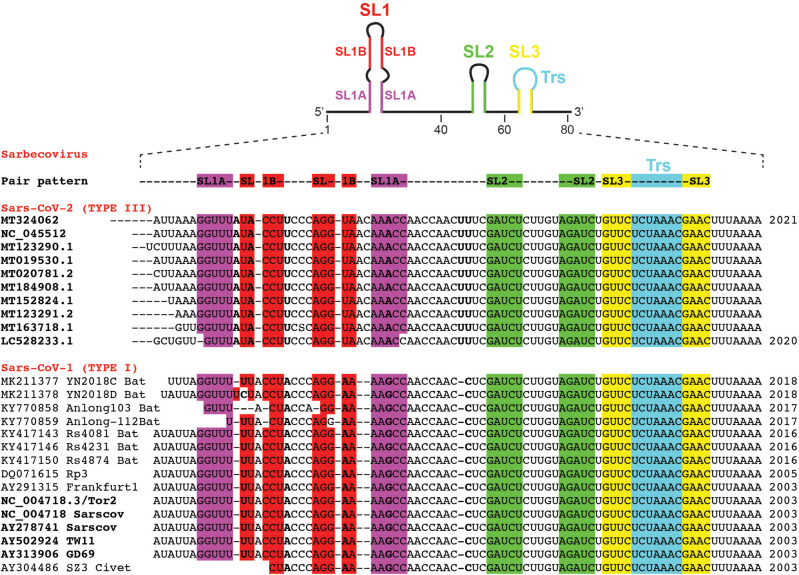
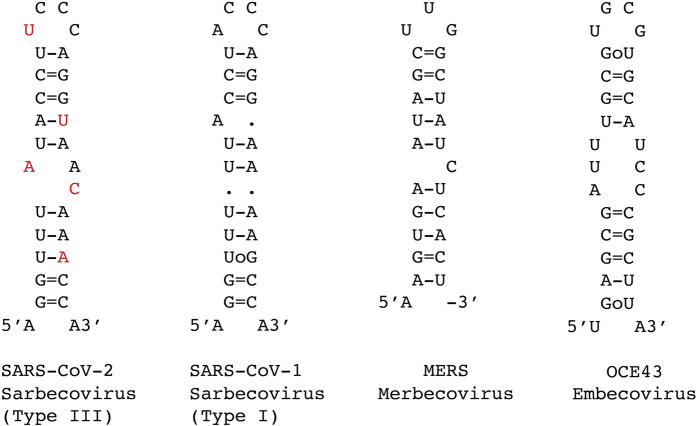
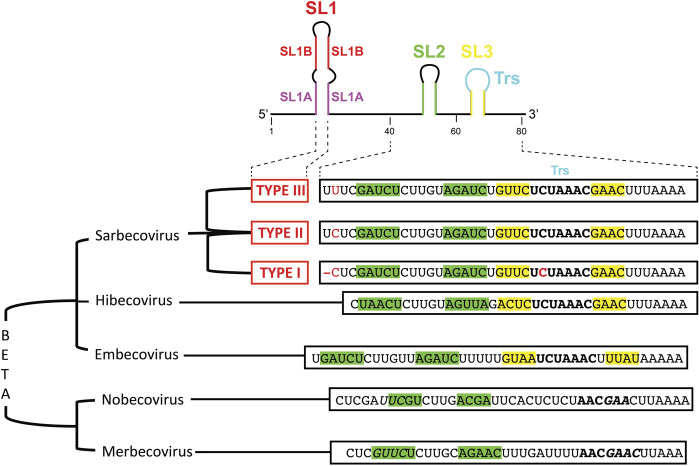
To analyze the genome variability of the 5′UTRs of coronaviruses, we have aligned the 5′ proximal 75–80 nt from the corresponding genomic RNAs. Figure 1 displays sequence alignments for chosen subsets of betacoronaviruses infecting various species (a rough tree of the genus and subgenus considered here with some typical ID for the sequenced strains is shown in Fig. 1A), and Figure 2 shows the sequence alignments of the sarbecovirus subfamily that contains SARS-CoV-1 and SARS-CoV-2. The general organization of the 5′UTR into three hairpins SL1–SL2–SL3 is highly conserved in coronaviruses. However, the highest variability is observed in SL1. This allows the establishment of a classification of the coronaviruses according to their type of SL1. For simplicity, for the sarbecovirus subfamily, we refer to type I to type IV, with type I gathering the SARS-CoV-1 sequences, type III, the SARS-Cov-2, and type II the sequences displaying intermediate situations (see below), and type IV other distant sequences. The resulting consensus secondary structures of examples of the main SL1 structures are shown in Figure 3. Unfortunately, many sequences present in databases do not contain these most 5′ ends of the genomes. This is especially disturbing when attempting to delineate a temporal evolution of the viruses. However, as will be shown later, the very high conservation of the NSP1 sequences allows determining with a high probability the corresponding sequence of the 5′UTR.
FIGURE 1.
(A) A rough phylogenetic tree of the coronavirus family. Some strain names are indicated along the branches. Sequences can be found in the alignments shown in Figures 1B and 2. Based in part on Smith et al. (2014). (B) Selected sets of aligned sequences of the first 80 nt of betacoronavirus genomes, except the Sarbecovirus, which are shown in Figure 2 (for a rough tree of coronaviruses, see Supplemental Figure S1). The color code is such that base paired segments are colored identically (purple, SL1A, red, SL1B, green, SL2, and yellow, SL3, with the TRS sequence in cyan). Sequences in bold were isolated from infected humans. The dates are those of database deposition.
FIGURE 2.
Selected sets of aligned sequences of the first 80 nt of SARS-CoV-1 and SARS-CoV-2 genomes from the betacoronavirus Sarbecovirus. Same annotations as in Figure 1. The nucleotides in bold vary between types I and III.
FIGURE 3.
Consensus secondary structures derived from the sequence alignments for the SL1 hairpin present in SARS-CoV-2 (also called type III), SARS-CoV-1 (also called type I), Merbecovirus, and Embecovirus. The nucleotides in red show the five point-mutations between the type I and type III SL1 hairpins.
The alphacoronaviruses are the oldest known coronaviruses infecting humans (Smith et al. 2014). Here, we will discuss neither the alphacoronaviruses, nor the gammacoronaviruses (Fig. 1A). The Embecovirus OC43 strain emerged between the end of the nineteenth and the beginning of the twentieth centuries (Vijgen et al. 2005). The HKU1, the origin of which is unknown, belongs also to this subgenus (Woo et al. 2005a,b). SARS-CoV (also called SARS-CoV-1) (Peiris et al. 2003), a Sarbecovirus, was identified in 2003 and MERS-CoV (Zaki et al. 2012), a Merbecovirus, in 2012. It is difficult to discuss the Hibecovirus subgenus because the sequence is unique (Wu et al. 2016). However, one can note that the hairpin loop of SL1 has similarities (UGC) with that of the Nobecoviruses, the hairpin loop of SL2 is identical to that of Nobecoviruses, and the TRS segment AACGAAC is present in both with an absence of the strong base pairs that form SL3. These two subgenera have been observed only in bats.
The Embecovirus subgenus displays a larger variation in sequence types. This may reflect the fact that the first sequence dates to 1997. One can distinguish two subgroups depending on the apical loop of SL1, either UGC (like in the Nobe- and Hibecovirus) or CGU (Fig. 1B). In some sequences, these residues could be part of the stable tetraloop of the type UNCG, but not in all; thus, some alternative or dynamic conformations may exist. The SL3 stem forms three to four base pairs. Correlated variations appear between the SL2 stem and the single strand between SL2 and SL3. The sequences of the Merbecovirus subgenus display a much larger conservation with a 3-nt loop, often -CUA (or five, since we cannot be sure that the framing nucleotides form a usual Watson–Crick pair). Again, SL3 does not appear to form a stable helix and the TRS is most probably single-stranded. The SL1 stem is stronger than in the Embecovirus. The TRS sequences are identical with those of the Nobe- and Merbecovirus but different from those of the Hibe- and Embecovirus.
Figure 2 shows sequences related to the Sarbecovirus subgenus. Two subgroups can be clearly distinguished, the SARS-CoV-1 and SARS-CoV-2. The sequence conservation is high and for SARS-CoV-1 it extends over 15 years. The similarities between the two subgroups are extensive: SL2 and SL3 are identical and the stem of SL1 presents only a change from a UoG to a U–A pair. The main differences are in SL1B, the first nucleotide of the SL1 hairpin loop, two positions 3′ of SL2; following the 3′ end of SL1, after the conserved stretch AACCAAC, there is a conserved UU in SARS-CoV-2 that is replaced by a single C in SARS-CoV-1. Figure 3 presents the resulting consensus secondary structures for the SL1 hairpin in the SARS-CoV-2, SARS-CoV-1, Merbecovirus, and Embecovirus. Some variations departing from the main types of sequences can be found. The number of sequences, from SARS-CoV-2 isolated from infected humans, with variations is small compared to the huge number of available sequences (search done using BLAST against the Coronavirus data base of Genbank). The variations occur mainly in SL1 (additional base pair in SL1B) (Supplemental Fig. S1). The variations of SARS-CoV-1 sequences occur either in the segment preceding SL2 (addition of C or changes between C and U) or in the loop of SL3 (C to U change). All those sequences were from viruses isolated from bats (Supplemental Fig. S1).
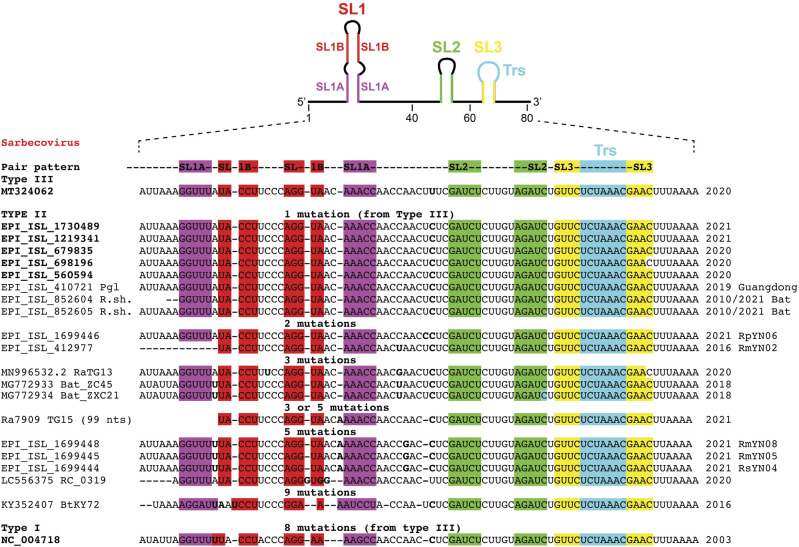
However, some sequences appear to be in-between SARS-CoV-1 and SARS-CoV-2 (Fig. 4). We named them type II sarbecoviruses. The names of the ID strains are given in Figure 4 since these are often discussed regarding the origins of the SARS-CoV-2 virus (see discussion below). The sequences deviate from the SARS-CoV-2 type between one and nine mutations. Five recent sequences come from infected humans, and all the other sequences are from bats with one from a pangolin (Guangdong). The sequences isolated from infected humans deviate by one mutation (a U to C mutation in the upstream sequence of SL2) like the Pangolin from Guandong (Liu et al. 2020) and, interestingly, two recent sequences from viruses isolated from R. shameli bats (Hul et al. 2021). The famous RaTG13 (Zhou et al. 2020b) sequence deviates by three mutations (like ZC45 and ZXC21 [Hu et al. 2018]), while RpYN06 and RmYN02 (Zhou et al. 2020a) deviate by two mutations only. The recently published sequence TG15 (Guo et al. 2021) deviates at least by three and maybe five mutations like RsYN04, RmYN05, or RmYN08 (Zhou et al. 2021). The sequences from the Guangxi pangolins (Wacharapluesadee et al. 2021) depart still further (these are called type IV for convenience, see Supplemental Fig. S2). For ease of comparison, the consensus secondary structures for types I to IV SL1 hairpins are shown in Supplemental Figure S3.
FIGURE 4.
Aligned sequences of the first 80 nt of SARS-related genomes, called type II, isolated from various sources. They display variations with respect to both SARS-CoV-1 (type I) and SARS-CoV-2 (type III) sequences. The variations occur either in SL1 or in the upstream segment before SL2. Same annotations as in Figure 1.
Sequence alignments for the NSP1 protein
The NSP1 sequences for the Sarbecovirus subgenus align very well. On the other hand, the sequences of the Sarbecovirus do not align well with those of either the Merbecovirus or the Embecovirus (a Clustal-W multiple alignment is shown in Supplemental Fig. S4). The HKU1 sequence (Embecovirus) has a long additional carboxy-terminal end that is not shown here. The alignment yields 16 aligned identical residues (highlighted in cyan in Supplemental Fig. S4). The 16 aligned identical residues were not observed to vary in the alignments of Sarbecovirus types I, II, and III. Among those residues known experimentally to be functionally important, residues Y/F157 and R/K175 (indicated by #) align meaningfully.
Residues R124, K125, Y154, F157, K164, H165, R171, R175 are conserved throughout SARS-CoV-1 and SARS-CoV-2. Mutations K164A and H165A of SARS-CoV-1 NSP1 abolish binding to the 40S subunit (Kamitani et al. 2009). This is also true for SARS-CoV-2 NSP1; mutations of the key residues within the region 153–178 result in the loss of NSP1 binding to the ribosome (Supplemental Fig. S5; Schubert et al. 2020; Thoms et al. 2020). Likewise, the mutations R124A and K125A inhibit SARS-CoV-1 NSP1 ability to promote translation inhibition (Lokugamage et al. 2012). The NSP1 and SL1 sequences are therefore coupled so that a NSP1 sequence is correlated with a specific type of SL1 hairpin in Sarbecovirus. In the Sarbecovirus, most variations in the 5′UTR occur between type I (like SARS-CoV-1) and types II or III (like SARS-CoV-2) (Supplemental Fig. S3). Clearly, as shown by the NSP1 alignments, type II (Supplemental Figs. S6, S7) is closer to type III than to type I NSP1 (9 NSP1 mutations between II and III with up to 22 NSP1 mutations between I and II). This is also the case for type II and type III SL1 (Supplemental Fig. S3). However, there are conservations between type I and type II NSP1 sequences that do not occur in type III NSP1 sequences. Figure 5A illustrates the conservations of the NSP1 proteins on the alignments and Figure 5B on the available three-dimensional structures from SARS-CoV-1 and SARS-CoV-2. Further variants are discussed in the Supplemental Material section. Further descriptions and discussions about sequence variants in the 5′UTR and NSP1 protein can be found in the Supplemental Section.
FIGURE 5.
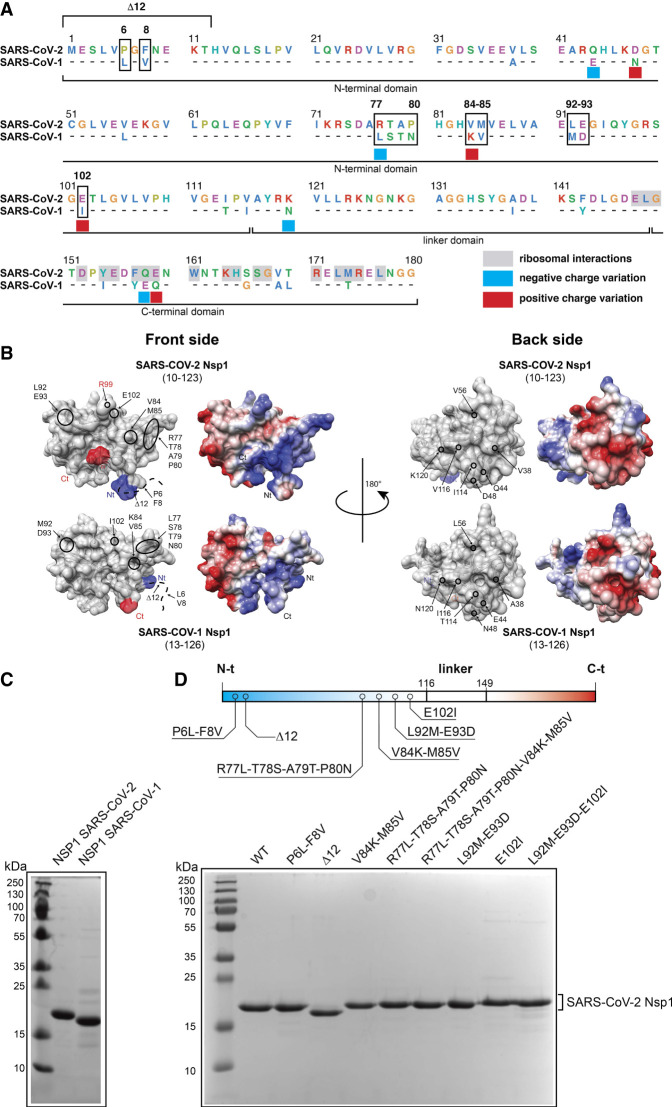
NSP1 proteins from SARS-CoV-1 and SARS-CoV-2. (A) Protein sequence alignment of SARS-CoV-2 and SARS-CoV-1 NSP1 proteins. For SARS-CoV-1, only the divergent amino acids are shown. The amino acids are shown according to the following color code: negatively charged amino acids in pink, hydrophobic amino acids in blue, positively charged amino acids in green, aromatic amino acids in cyan, glycines and prolines in orange. Residues involved in interactions with ribosomal components are boxed in gray in SARS-CoV-2 (Schubert et al. 2020; Thoms et al. 2020; Yuan et al. 2020). Negative charge variations from SARS-CoV-2 to SARS-CoV-1 are indicated by blue squares, and positive charge variations are indicated by red squares. Residues that have been mutated in this study are boxed in black. The NSP1 proteins are subdivided in three domains, the amino-terminal domain, the central linker domain, and a carboxy-terminal domain. (B) Surface representation of crystal structure of SARS-CoV-2 NSP1 from residues E10 to L123 (PDB: 7K7P) (Clark et al. 2021) and NMR structure of SARS-CoV-1 NSP1 from residues H13 to G126 (PDB: 2HSX) (Almeida et al. 2007) from the front side (left panels) and from the back side (right panels). Both structures have been aligned and represented with the amino-terminal end in blue and the carboxy-terminal end in red. Divergent residues from SARS-CoV-1 that have been inserted in SARS-CoV-2 are circled in black. Next to each structure, the electrostatic surfaces of the protein with negative and positive charges are colored in red and blue, respectively. The position of residue R99 in SARS-CoV-2 NSP1 is indicated in red. (C) SDS PAGE analysis of purified recombinant wild-type SARS-CoV-1 and SARS-CoV-2 NSP1 proteins. (D) Linear representation of the three domains of NSP1, the Nt-domain in blue, the linker domain and the Ct-domain in red. The SARS-CoV-2 residues that have been mutated are marked. SDS PAGE analysis of purified recombinant wild-type and mutant SARS-CoV-2 NSP1 proteins.
Amino-acid swapping between SARS-CoV-1 and SARS-CoV-2 NSP1
Previous mutations have shown that conserved amino acids in the linker (R124A/K125A) and carboxy-terminal (K164A/H165A) regions prevent NSP1 to inhibit mRNA translation in both SARS-CoV-1 (Kamitani et al. 2009; Lokugamage et al. 2012) and SARS-CoV-2 (see Supplemental Fig. S4 of Tidu et al. 2021) and, thus, following the structural data (Schubert et al. 2020; Thoms et al. 2020; Yuan et al. 2020) should inhibit the binding of NSP1 to the mRNA channel of ribosomes.
NSP1 inhibits translation by blocking the mRNA channel on the ribosome. However, translation evasion is mediated by SL1 in the 5′UTR of viral mRNA transcripts (Tidu et al. 2021). We first purified recombinant NSP1 from SARS-CoV-1 and SARS-CoV-2 (Fig. 5B,C) and, using a canonical β-globin reporter mRNA, we found that the level of translation inhibition of SARS-CoV-1 NSP1 is significantly less than that with SARS-CoV-2 NSP1 (Supplemental Fig. S8A). Similarly, the translation evasion of cognate viral mRNA constructs was also less efficient with SARS-CoV-1 NSP1 than with SARS-CoV-2 NSP1 (Supplemental Fig. S8B,C). Considering these differences and the overall folding differences present in the available structural data, we undertook residues swapping experiments between NSP1 proteins and SL1 from both viruses to assess the functional links between them. We decided to keep the SARS-CoV-2 NSP1 background, because it is the most efficient both in translation inhibition and in viral translation evasion, and residues from SARS-CoV-1 NSP1 were inserted into the SARS-CoV-2 NSP1 background. In total, eight SARS-CoV-2 recombinant NSP1 proteins with residues from SARS-CoV-1 were expressed and purified.
The key positions were chosen in the following way. We looked at two sides of NSP1 by structural comparisons between the available PDB models of SARS-CoV-1 and SARS-CoV-2 NSP1 proteins (Fig. 5B): the front side (left panels of Fig. 5B) and the back side (right panels of Fig. 5B). The previously studied R99A mutation (Mendez et al. 2021) demonstrated that this mutation abrogates viral translation evasion while reducing the translation inhibition activity of NSP1 against cellular mRNA. Since R99 is located on the front side of the NSP1, this suggests that SL1 might interact by contacts with residues located on the same front side. Our hypothesis is that the SARS-CoV-2 NSP1 surface coevolved with its 5′UTR mRNA by adjusting this front side for better SL1 recognition. In agreement with this hypothesis, there are more positive charges (susceptible to interact with negatively charged RNA) present on the front side of the protein than on the back side (see electrostatic surfaces of the right panels of Fig. 5B) for both SARS-CoV-1 and SARS-CoV-2 NSP1. Therefore, all residues swapped from SARS-CoV-1 into a SARS-CoV-2 background are located on the front side of NSP1. The remaining amino acid differences between SARS-CoV-1 and SARS-CoV-2 NSP1 proteins are located on back side of the proteins (right panels of Fig. 5B) and for that reason were not considered in this study.
Previous experiments had shown that the first twelve amino-terminal amino acids are critical for the cellular functions of NSP1 during viral replication (Yuan et al. 2020). Therefore, we also generated a truncated version of SARS-CoV-2 NSP1 called Δ12. Similarly, we introduced SARS-CoV-1 residues into the SARS-CoV-2 background at positions 6 and 8, both localized within the deleted part of Δ12. Figure 5D shows the purified eight recombinant NSP1 mutants.
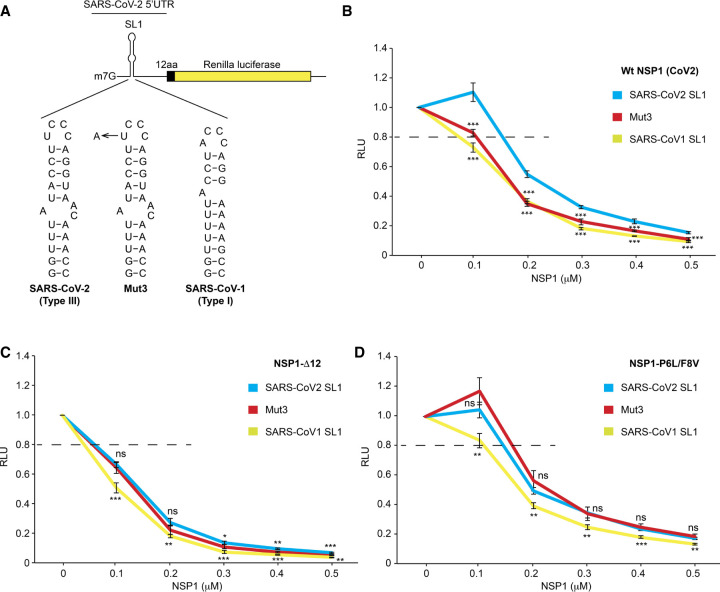
Next, we tested their ability to promote translation in cell-free translation assays of a reporter containing the Renilla sequence with the WT 5′UTR SL1 of SARS-CoV-1, SARS-CoV-2, and a one point-mutation in the SARS-CoV-2 sequence of the apical loop to mimic the SARS-CoV-1 apical loop (Mut3, see Fig. 6A). First, we noticed a slight stimulation with 0.1 µM WT SARS-CoV-2 NSP1 with mRNA containing the WT SL1. Although we cannot offer a direct explanation of this reproducible phenomenon, we speculate that the slight boost in translation results from the increased pool of ribosomes released from endogeneous cellular mRNAs that were specifically inhibited by NSP1. With WT SARS-CoV-2 NSP1, the SL1 mutant Mut3 is significantly less efficient for translation evasion to NSP1 and behaves like the SARS-CoV-1 SL1, as could be expected from its similarity with the SARS-CoV-1 hairpin loop (Fig. 6B; Supplemental Fig. S9). Previous experiments had shown that the first twelve amino-terminal amino acids are critical for the cellular functions of NSP1 during viral replication (Yuan et al. 2020). In agreement with these studies, the removal of the first 12 amino-terminal amino acids of the SARS-CoV-2 NSP1 protein (Fig. 6C; Supplemental Fig. S9) leads to a marked decrease in the efficiency of the translational evasion to inhibition with a less pronounced differential effect on the three tested SL1 hairpins. Thus, the deletion of the amino-terminal region does not prevent ribosomal binding, but it does prevent the evasion from the inhibition mediated by SL1 hairpin. In the presence of the NSP1 mutant P6L/F8V, where the amino acid reversals follow the conservation of the NSP1 sequence observed in SARS-CoV-1, the evasion to inhibition is improved especially for Mut3 and the SARS-CoV-1 hairpins (Fig. 6D; Supplemental Fig. S9). These latter experiments show that the amino-terminal region is necessary for evasion of the inhibitory effect and that the two residues at 6 and 8 either specifically directly interact with or contribute to interactions between NSP1 and the SL1 hairpin. We also tested the mutant P6L/F8V with the whole 5′UTR of SARS-CoV-1 and observed that the viral translation evasion is as low as with both SARS-CoV-1 and SARS-CoV-2 NSP1. This confirms that the SARS-CoV-1 SL1 in its 5′UTR context does not promote efficient viral evasion (Supplemental Fig. S10).
FIGURE 6.
Evasion to NSP1-mediated translation inhibition with NSP1 variants containing mutations in the amino-terminal domain of NSP1. (A) Cartoon depicting the reporter mRNAs used for translation in RRL, they contain the SARS-CoV-2 5′UTR and the first 12 amino-terminal codons of NSP1 fused to the Renilla luciferase coding sequence. The reporter mRNA contains the wild-type SL1 hairpin or mut3 which has a U to A substitution in the loop or the SARS-CoV-1 SL1. (B) Curves representing the average relative activities measured after translation in RRL in the absence or presence of 0.1, 0.2, 0.3, 0.4, or 0.5 µM of wild-type NSP1, (C) Δ12, and (D) P6L/F8V. The averages are obtained from four independent experiments. Standard deviations or translational activity for each transcript are shown and calculated from four independent experiments. ns: nonsignificant; (*) 0.05 > P-value >0.01; (**) 0.01 > P-value >0.001; (***) P-value <0.001; based on Student's t-test. To facilitate comparisons between NSP1 proteins, the results have also been presented according to the hairpin SL1 tested (Supplemental Fig. S9).
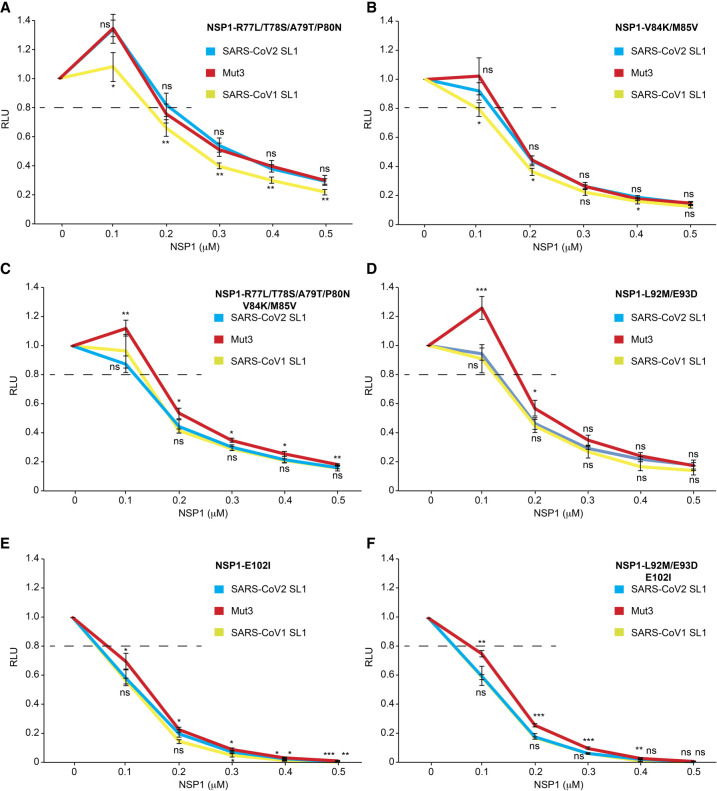
The NSP1 mutant V84K/M85V displays stronger effects than the NSP1 mutant R77L/T78S/A79T/P80N since their combination gives similar values than the double mutant V84K/M85V (compare Fig. 7A–C; Supplemental Fig. S11). Interestingly, the NSP1 mutant R77L/T78S/A79T/P80N is the only mutant with an apparent reduction of the blockage of the mRNA channel (Fig. 7A; Supplemental Fig. S11).
FIGURE 7.
Evasion to NSP1-mediated translation inhibition with NSP1 variants containing mutations in the middle of NSP1. (A) Curves showing relative light unit (RLU) measured after translation in RRL in the absence or presence of 0.1, 0.2, 0.3, 0.4, or 0.5 µM of R77L/T78S/A79T/P80N, (B) V84K/M85V, (C) R77L/T78S/A79T/P80N/V84K/M85V, (D) L92M/E93D, (E) E102I, and (F) L92M/E93D/E102I. The averages are obtained from four independent experiments. Standard deviations or translational activity for each transcript are shown and calculated from four independent experiments. ns: nonsignificant; (*) 0.05 > P-value >0.01; (**) 0.01 > P-value >0.001; (***) P-value <0.001; based on Student's t-test. To facilitate comparisons between NSP1 proteins, the results have also been presented according to the hairpin SL1 tested (Supplemental Fig. S11).
We next introduced other amino acids typical of SARS-CoV-1 into the SARS-CoV-2 NSP1 protein in the region necessary for the conformational change that facilitates evasion of the blocking of the mRNA channel. Thus, the mutation E102I (Fig. 7E; Supplemental Fig. S11), at the end of the amino-terminal segment before the linker region (Fig. 5B,D), strongly prevents evasion from the inhibitory effect of NSP1, meaning again that the NSP1–E102I mutant binds tightly to the initiation complex and is not displaced by the SL1 5′UTR region. The latter mutant has a stronger dominant effect than the double NSP1 mutant L92M/E93D, since the combination of the three point-mutations, L92M/E93D and E102I, has effects like those observed with the single mutant E102I (compare Fig. 7D–F). Interestingly, E102 is very close to R99 (Fig. 5B) and our result is in good agreement with the previously described R99A mutation, which has a lower propensity to promote viral translation evasion (Mendez et al. 2021). Altogether, Mut3 is promoting better evasion to NSP1 inhibition when specific residues from SARS-CoV-1 are introduced into SARS-CoV-2 NSP1 thereby confirming the functional link between these specific residues and the first nucleotide of the apical loop of SL1.
DISCUSSION
Here, sequence analysis has been performed on a short noncoding RNA segment at the 5′UTR segment of the beta-coronaviruses, and especially the Sarbecovirus family responsible for a devastating pandemic that started at the end of 2019 in China. Although such sequence elements are not known to contribute to either infectivity or cell penetration, they are essential for the translation of the viral genome and for its replicative proliferation. The RNA analysis is complemented by the sequence analysis of the first nonstructural protein translated by the virus, NSP1, that has been shown to be involved, together with the 5′UTR RNA, in the control of ribosomal translation initiation (Schubert et al. 2020; Thoms et al. 2020; Mendez et al. 2021; Tidu et al. 2021). We contribute further data showing that both the 5′UTR and the NSP1 protein are linked functionally and therefore are forced to coevolve to interact cooperatively with highly conserved elements of the ribosomal machinery and initiation cofactor proteins. The selection pressure on both the RNA and the protein is patent in the analyzed sequences, as revealed by the patterns of conservation and variation in the NSP1 sequences for each type of 5′UTR and by the experimental mutational data on both the NSP1 protein and the SL1 hairpin of the 5′UTR that are presented. The experimental mutational data confirm that the carboxy-terminal region is central to NSP1 binding to the mRNA channel, while the amino-terminal domain and the linker region contributes both to the conformational rearrangement and to binding to the SL1 hairpin of the 5′UTR that promotes evasion from the NSP1 inhibition. In short, the data concur with a model in which the Nt-domain interacts directly or indirectly with SL1 and thereby promotes remodeling of the Ct-domain out of the mRNA channel of the ribosome. The data support also the view that the linker domain mediates the communication between these two domains during viral evasion.
The question of the origins of the coronavirus that initiated the present pandemic is much debated. Although the data presented here are limited, they do set strong molecular constraints on the viral sequences for successful translation and replication in specific living cells. The various types of the 5′UTR first 80 nt reproduce the main branches of the various phylogenetic trees deduced from whole genome comparisons or functional genomic segments. Figure 8 shows an overall summary of key 5′UTR consensus sequences following the accepted phylogenetic tree.
FIGURE 8.
A rough phylogenetic tree of the Genus beta of the Coronaviridae with the main subgenus species (for more details, see Supplemental Fig. S1A). The consensus sequences derived here for the first 80 nt of the 5′ leader are shown on each branch, with the SL1 nomenclature indicated on the right. The color code is the same as in the alignments (green, SL2; yellow and cyan, SL3). TRS sequences are in bold. In italics are shown potential alternative pairings. In red are shown nucleotides that vary between types I, II, and III in the 5′ segment upstream of SL2.
Finally, one may remark the following. In the 5′UTR, there are seven mutations between type I and II: six mutations in SL1 and a 1-nt U insertion in the 3′ segment preceding the SL2 hairpin. Between type II and III sequences derived from infected humans, there is a single mutation of a C into a U in the same 3′-end part of SL2 (but there are more mutations from bat derived sequences, see Fig. 4). The differences may appear minimal, but their maintenance within each subgroup indicate that they are meaningful. These structures must fold, interact with proteins, and unfold with an appropriate dynamic that depends on the free energies of the hairpins, which themselves depend on the cellular environment (ions, temperature, …) that vary between species. Besides, and importantly, the formation of alternative structures in dynamic equilibrium, some of which are also key for functional initiation, cannot be excluded and should be investigated.
The experimental data show that the coevolved complex between NSP1 and the 5′UTR has virus specificity and that mutations in one component may affect their mutual interactions. In the absence of precise structural information, many unanswered questions remain on the complex interactions involving the 5′UTR leader sequence, the NSP1 protein and the ribosomal machinery, like (i) why did the 5′UTR and NSP1 sequences mutate from the SARS-CoV-1 in the SARS-CoV-2 genomes; (ii) how to explain the weak variations in the SARS-CoV-2 5′UTR genomes? For SARS-CoV-1, similar sequences of the 5′UTR and NSP1 proteins were observed from viruses isolated from bats or infected humans and, thus, both are adapted to ribosomal translation cofactors in both species. However, the SARS-CoV-2 5′UTR and NSP1 protein are only observed in infected humans (with later some other mammals like minks, probably infected by humans) and, up to now, they were not observed in bats (see Supplemental Material). In this work, we did not study all the steps in which the first 80 nt of the 5′UTR are involved, and some of the observed conserved elements may play a role in processes not experimentally probed yet.
MATERIALS AND METHODS
Sequences
Viral sequences were retrieved form NCBI Genbank (http://www.ncbi.nlm.nih.gov), the Gisaid repository (http://www.GISAID.org) (Elbe and Buckland-Merrett 2017), or the National Genomics Data Center, China National Center for Bioinformation (Beijing Institute of Genomics, Chinese Academy of Sciences) (https://bigd.big.ac.cn/) and aligned manually. For the RNA, the alignments were done on the first 80 nt of the genome (when available) and the complete sequence was considered for the protein. The NSP1 protein sequences were derived using the EMBOSS program (https://www.ebi.ac.uk/Tools/st/emboss_transeq/) based on the published genomes. The correspondence between the accession numbers and the usual or common ID strain is given in the figures and tables.
In vitro transcription
The three reporter constructs were transcribed by run-off in vitro transcription with T7 RNA polymerase from DNA templates amplified by PCR. A DNA fragment containing the 900 first nucleotides of SARS-COV-2 (accession number: MN908947.3) was used as a template for PCR amplifications. The transcription was performed in the presence of m7GpppG cap analog (Jena Bioscience) to generate capped mRNAs as previously described (Tidu et al. 2021).
In vitro translation
In vitro translation with cell-free translation extracts were performed using self-made rabbit reticulocyte lysates (RRL) as previously described (Tidu et al. 2021). Briefly, reactions were incubated at 30°C for 60 min and included 200 nM of each transcript. Aliquots of translation reactions were analyzed for Renilla luciferase activity on a luminometer.
NSP1 overexpression and purification
NSP1 and derivatives (P6L/F8V-Δ12-R77L/T78S/A79T-P80N-V84K/M85V-L92M/E93D) were produced as previously described (Tidu et al. 2021) with slight modifications. Briefly, the plasmid pET-His-GST-TEV-LIC-(2GT) was purchased from Addgene. The fusion proteins were expressed in E. coli BL21 Rosetta (DE3) pLysS cells. Cells were grown at 37°C to a cell density of OD600 = 0.6 and then induced with 0.2 mM IPTG during 3 h at 37°C. After induction, cells were harvested and the cell pellets were frozen at −80°C. Frozen pellets were resuspended in EQ/W buffer (40 mM Na phosphate pH 7.2, 500 mM NaCl, 30 mM imidazole) supplemented with 0.1% Triton X-100, cOmplete Protease Inhibitor Cocktail (Merck) and incubated on ice for 30 min with 1 mg/mL lysozyme. After lysis by sonication, the cell lysate was centrifuged at 3200g at 4°C for 10 min to remove the large debris, and the supernatant was further centrifuged at 150,000g at 4°C for 30 min. The supernatant was applied to Ni-NTA Superflow resin (Qiagen) equilibrated in buffer EQ/W. After column washing, 6xHis-GST-TEV-NSP1 proteins were eluted from the resin by buffer EQ/W with gradient increase of imidazole from 30 to 500 mM. Fractions containing 6xHis-GST-TEV-NSP1 were pooled and dialyzed against buffer EQ/W without imidazole overnight. Next, the 6xHis-GST-TEV-NSP1 fraction was loaded on Glutathione HiCap resin (Qiagen) equilibrated with the dialysis buffer, and proteins were eluted by the same buffer with a gradient of glutathione from 0 to 50 mM. Fractions containing 6xHis-GST-TEV-NSP1 fusion proteins were pooled and subjected to TEV protease cleavage overnight at 4°C (50/1 fusion/TEV molar ratio). NSP1 proteins were separated from the 6xHis-GST domains using a last purification step on Ni-NTA resin that retained His-tagged GST and TEV proteins. The pure NSP1 proteins were concentrated and stored in buffer that contains 50% glycerol at −20°C.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work is funded by Agence Nationale pour la Recherche (ANR-17-CE12-0025-01, ANR-17-CE11-0024, ANR-20-COVI-0078), Fondation pour la Recherche Médicale (project CoronaIRES), Fondation Bettencourt Schueller, University of Strasbourg and the Centre National de la Recherche Scientifique.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078972.121.
Freely available online through the RNA Open Access option.
REFERENCES
- Almeida MS, Johnson MA, Herrmann T, Geralt M, Wüthrich K. 2007. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J Virol 81: 3151–3161. 10.1128/JVI.01939-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brian DA, Baric RS. 2005. Coronavirus genome structure and replication. Curr Top Microbiol Immunol 287: 1–30. 10.1007/3-540-26765-4_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LK, Green TJ, Petit CM. 2021. Structure of nonstructural protein 1 from SARS-CoV-2. J Virol 95: e02019-20. 10.1128/JVI.02019-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S, Buckland-Merrett G. 2017. Data, disease and diplomacy: GISAID's innovative contribution to global health. Glob Challenges 1: 33–46. 10.1002/gch2.1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, et al. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5: 536–544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyaeva AA, Gorbalenya AE. 2021. A nidovirus perspective on SARS-CoV-2. Biochem Biophys Res Commun 538: 24–34. 10.1016/j.bbrc.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Hu B, Si H-R, Zhu Y, Zhang W, Li B, Li A, Geng R, Lin H-F, Yang X-L, et al. 2021. Identification of a novel lineage bat SARS-related coronaviruses that use bat ACE2 receptor. Emerg Microbes Infect 10: 1507–1514. 10.1080/22221751.2021.1956373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, Yang L, Ding C, Zhu X, Lv R, et al. 2018. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect 7: 154. 10.1038/s41426-018-0155-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hul V, Delaune D, Karlsson EA, Hassanin A, Tey PO, Baidaliuk A, Gámbaro F, Tu VT, Keatts L, Mazet J, et al. 2021. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. bioRxiv 10.1101/2021.01.26.428212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16: 1134–1140. 10.1038/nsmb.1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181: 914–921.e10. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MM, Stohlman SA. 1981. Comparative analysis of RNA genomes of mouse hepatitis viruses. J Virol 38: 661–670. 10.1128/jvi.38.2.661-670.1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapointe CP, Grosely R, Johnson AG, Wang J, Fernández IS, Puglisi JD. 2021. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc Natl Acad Sci 118: e2017715118. 10.1073/pnas.2017715118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. 2018. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46: D708–D717. 10.1093/nar/gkx932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jiang JZ, Wan XF, Hua Y, Li L, Zhou J, Wang X, Hou F, Chen J, Zou J, et al. 2020. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog 16: e1008421. 10.1371/journal.ppat.1008421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage KG, Narayanan K, Huang C, Makino S. 2012. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol 86: 13598–13608. 10.1128/JVI.01958-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez AS, Ly M, González-Sánchez AM, Hartenian E, Ingolia NT, Cate JH, Glaunsinger BA. 2021. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep 37: 109841. 10.1016/j.celrep.2021.109841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Tidu A, Eriani G, Martin F. 2021. Secondary structure of the SARS-CoV-2 5′-UTR. RNA Biol 18: 447–456. 10.1080/15476286.2020.1814556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361: 1319–1325. 10.1016/S0140-6736(03)13077-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan R, Zheludev IN, Hagey RJ, Pham EA, Wayment-Steele HK, Glenn JS, Das R. 2020. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: a first look. RNA 26: 937–959. 10.1261/rna.076141.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Mühlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27: 959–966. 10.1038/s41594-020-0511-8 [DOI] [PubMed] [Google Scholar]
- Smith EC, Sexton NR, Denison MR. 2014. Thinking outside the triangle: replication fidelity of the largest RNA viruses. Annu Rev Virol 1: 111–132. 10.1146/annurev-virology-031413-085507 [DOI] [PubMed] [Google Scholar]
- Thoms M, Buschauer R, Ameismeier M, Koepke L, Denk T, Hirschenberger M, Kratzat H, Hayn M, MacKens-Kiani T, Cheng J, et al. 2020. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 369: 1249–1256. 10.1126/science.abc8665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidu A, Janvier A, Schaeffer L, Sosnowski P, Kuhn L, Hammann P, Westhof E, Eriani G, Martin F. 2021. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 27: 253–264. 10.1261/rna.078121.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moës E, Thoelen I, Wollants E, Lemey P, Vandamme A-M, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79: 1595–1604. 10.1128/JVI.79.3.1595-1604.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, et al. 2021. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun 12: 972. 10.1038/s41467-021-21240-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Huang Y, Lau SKP, Tsoi HW, Yuen KY. 2005a. In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol Immunol 49: 899–908. 10.1111/j.1348-0421.2005.tb03681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PCY, Lau SKP, Huang Y, Tsoi HW, Chan KH, Yuen KY. 2005b. Phylogenetic and recombination analysis of coronavirus HKU1, a novel coronavirus from patients with pneumonia. Arch Virol 150: 2299–2311. 10.1007/s00705-005-0573-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Yang L, Ren X, He G, Zhang J, Yang J, Qian Z, Dong J, Sun L, Zhu Y, et al. 2016. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J 10: 609–620. 10.1038/ismej.2015.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogo Y, Hirano N, Hino S, Shibuta H, Matumoto M. 1977. Polyadenylate in the virion RNA of mouse hepatitis virus. J Biochem 82: 1103–1108. 10.1093/oxfordjournals.jbchem.a131782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Peng L, Park JJ, Hu Y, Devarkar SC, Dong MB, Shen Q, Wu S, Chen S, Lomakin IB, et al. 2020. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol Cell 80: 1055–1066.e6. 10.1016/j.molcel.2020.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367: 1814–1820. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- Zhou H, Chen X, Hu T, Li J, Song H, Liu Y, Wang P, Liu D, Yang J, Holmes EC, et al. 2020a. A novel bat coronavirus closely related to SARS-CoV-2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Curr Biol 30: 2196–2203.e3. 10.1016/j.cub.2020.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Lou YX, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. 2020b. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, Hu T, Song H, Zhao R, Chen Y, et al. 2021. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 184: 4380–4391.e14. 10.1016/j.cell.2021.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.