Abstract
The human immunodeficiency virus type 1 (HIV-1) encodes multiple RNA molecules. Transcripts that originate from the proviral 5′ long terminal repeat (LTR) function as messenger RNAs for the expression of 16 different mature viral proteins. In addition, HIV-1 expresses an antisense transcript (Ast) from the 3′LTR, which has both protein-coding and noncoding properties. While the mechanisms that regulate the coding and noncoding activities of Ast remain unknown, post-transcriptional modifications are known to influence RNA stability, interaction with protein partners, and translation capacity. Here, we report the nucleoside modification profile of Ast obtained through liquid chromatography coupled with mass spectrometry (LC-MS) analysis. The epitranscriptome includes a limited set of modified nucleosides but predominantly ribose methylations. A number of these modifications were mapped to specific positions of the sequence through RNA modification mapping procedures. The presence of modifications on Ast is consistent with the RNA-modifying enzymes interacting with Ast. The identification and mapping of Ast post-transcriptional modifications is expected to elucidate the mechanisms through which this versatile molecule can carry out diverse activities in different cell compartments. Manipulation of post-transcriptional modifications on the Ast RNA may have therapeutic implications.
Keywords: HIV-1, antisense RNA, epitranscriptomics, nucleoside analysis, RNA modification mapping
INTRODUCTION
Coding and noncoding RNAs of all organisms are covalently modified with more than 150 biochemical groups ranging from simple methylations to hypermodifications of individual nucleotides (Boccaletto et al. 2018; Boccaletto and Baginski 2021). These post-transcriptional modifications are introduced by stand-alone and RNA-guided cellular enzymes, and they play critical roles in regulating the stability, structure, and function of the target RNA (Decatur and Fournier 2002; Whipple et al. 2011; Sibbritt et al. 2013; Lorenz et al. 2017; Arango et al. 2018; Choe et al. 2018; Price et al. 2020; Roovers et al. 2021). Viral RNAs are no exception. A variety of DNA and RNA viruses express RNA molecules that have been reported to contain methylation of the base in adenosine (m6A: N6-methyladenosine, m1A: 1-methyladenosine), guanosine (m7G: 7-methylguanosine, m1G: 1-methylguanosine), and cytidine (m5C: 5-methylcytosine) residues, ribose methylation of all four residues (Nm), and pseudouridylation (Ψ) (Borchardt et al. 2020; Courtney 2021). Cytidine is also acetylated to form ac4C (N4-acetylcytidine) in a few viral nucleic acids (McIntyre et al. 2018). Modifications in viral RNA are shown to increase the replication of HIV-1 (Lichinchi et al. 2016; Kennedy et al. 2017; Tsai et al. 2020), hepatitis B (Imam et al. 2018), influenza A (Courtney et al. 2017), and SV40 (Tsai et al. 2018). Further, the number of RNA modifications found in viral RNAs are greater than those of cellular RNAs, presumably conferring functional advantage such as regulated gene expression (Tirumuru et al. 2016; Courtney et al. 2019a,b).
Modifications on RNA molecules can be detected and mapped through several approaches (Wiener and Schwartz 2021). Modification mapping by high-throughput methods involves fragmentation of enriched RNA, capture of RNA fragments by modification-specific antibody, subsequent isolation, and deep sequencing (Dominissini et al. 2012). This antibody mapping initially developed for m6A was adapted to identify m1A (Dominissini et al. 2016), m5C (Edelheit et al. 2013), and ac4C (Arango et al. 2018) in RNA. Antibody mapping is inherently noisy and requires additional controls. The mapping strategy results in large footprints of 20–100 nt, where the exact location of modification is not very clear. Therefore, it requires additional methods to validate the proposed sites of modification (Helm and Motorin 2017). Chemical labeling of modified nucleoside that blocks reverse transcription or introduces polymorphism can yield a footprint of 1 nt during biochemical mapping methods. Such methods include bisulfite sequencing to distinguish m5C from C (Schaefer et al. 2009), UV cross-linking of RNA and bound antibody for m6A (Linder et al. 2015), Ψ-seq using N-cyclohexyl-N′-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT) for selective detection of pseudouridines (Carlile et al. 2014; Schwartz et al. 2014), and detection of ribose methylations (Nm) based on their resistance to alkaline treatment (Marchand et al. 2016) or oxidation associated fragmentation (Zhu et al. 2017; Hsu et al. 2019). These methods, however, are technically challenging, because they require very large read-depth, tend to generate artifacts, and are less sensitive to lowly expressed RNA (Courtney 2021).
Liquid-chromatography coupled with mass spectrometry (LC-MS) is a powerful approach that provides an unbiased readout of the residential post-transcriptional modifications (Pomerantz and McCloskey 1990; Jora et al. 2021) of RNA. The analysis involves two steps (Jora et al. 2019). In the first step, analysis of RNA hydrolysate provides a census of post-transcriptional modifications through accurate measurement of chromatographic retention time and mass shift of added chemical group to the ribonucleosides (Jora et al. 2018). In the second step, location of modification group in the sequence is determined by the mass spectrometric sequencing of oligonucleotides, a process referred to as RNA modification mapping (Kowalak et al. 1993). Employment of nucleobase-specific ribonucleases (Thakur et al. 2020) provides predictability of the oligonucleotide digestion products and helps facilitate MS data analysis while identifying the position of the modifying group in the sequence.
Upon integration into the host genome, the human immunodeficiency virus type 1 (HIV-1) proviral genome expresses several types of RNA molecules. Transcripts that originate from the 5′ long terminal repeat (LTR) are called “sense transcripts,” are grouped into three classes depending on their degree of splicing (multiply spliced, singly spliced, and unspliced), and function as messenger RNA (mRNA) for the expression of 16 different mature viral proteins. Unspliced transcripts also function as viral genome and are packaged into newly produced viral particles that will infect new target cells (Ferguson et al. 2002). In addition, the HIV-1 provirus expresses an antisense transcript (Ast) from the 3′LTR (Li et al. 2021). This RNA has a dual role: as mRNA that encodes for the viral protein, ASP (Affram et al. 2019; Gholizadeh et al. 2021), and as long noncoding RNA (lncRNA) that suppresses HIV-1 expression through epigenetic silencing of the 5′LTR (Zapata et al. 2017; Li et al. 2021). The mechanisms that regulate the balance between the two functions of Ast are unknown. Here, we sought to identify and map post-transcriptional modifications of Ast that may have a role in regulating its functions. We report the presence of a defined set of nucleoside modifications in Ast expressed in Jurkat cells.
RESULTS
Ast RNA isolation and sequence confirmation
Post-transcriptional nucleoside modifications in the sense HIV-1 RNA sequence have been shown to influence viral expression and replication (Kong et al. 2019; Ringeard et al. 2019; Tsai et al. 2020, 2021). Prevalence of such modifications in the HIV-1 antisense transcript (Ast) have not been reported. To characterize the ribonucleoside modifications of Ast, we isolated total RNA from a Jurkat cell line stably transduced with a lentiviral vector expressing Ast under the CMV promoter, which we described previously (Zapata et al. 2017). We then used a set of six biotinylated oligonucleotides complementary to Ast (Supplemental Table S1) and Streptavidin magnetic beads to isolate Ast from total RNA. Purified Ast RNA was initially evaluated for its integrity and relative mobility on an agarose gel in contrast to the ribosomal RNA. Figure 1 shows the presence of two discrete bands of RNA that differ in their relative mobility in the purified sample. The slow-migrating band corresponds well with the expected size of full-length Ast RNA (2.6 kb) whose position was intermediate to that of 28S (5 kb) and 18S (1.9 kb) rRNA of total RNA sample. The fast-migrating band could represent a spliced version of Ast RNA consistent with a previous report (Kobayashi-Ishihara et al. 2012). To confirm the identity of the streptavidin-enriched RNA, we performed RT-PCR with primers specific for Ast. As templates, we used total RNA (before and after DNase I treatment), and streptavidin-enriched RNA (before and after RNase T1 treatment). We found that these primers amplified a cDNA product of the expected size (575 bp) from both total and purified RNA preparations (Supplemental Fig. S1). Sequencing confirmed that these PCR products were identical, and that they were Ast amplification products.
FIGURE 1.
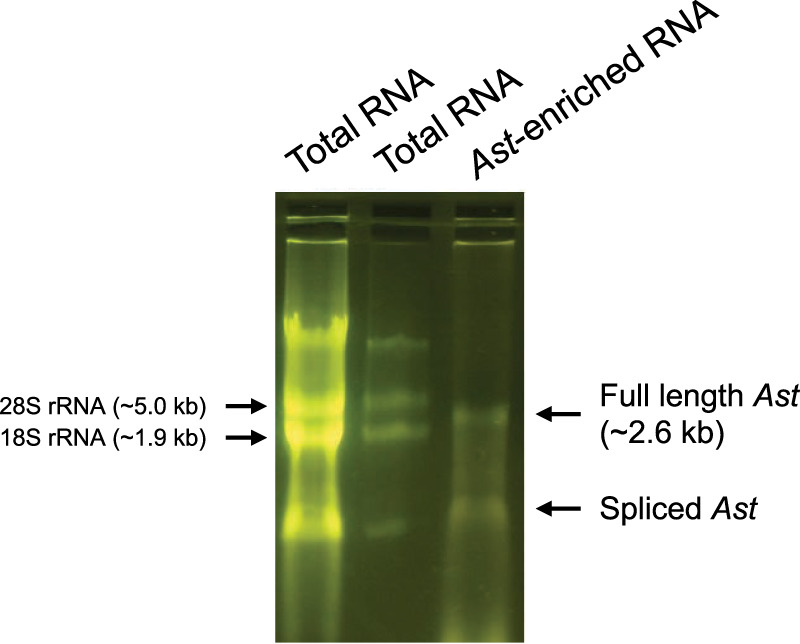
Agarose gel electrophoresis of Ast RNA. Total RNA was extracted from Jurkat-AST cells, and Ast RNA was enriched using streptavidin magnetic beads (lane 3). Total RNA (5 and 1 µg, lanes 1,2) and streptavidin-enriched (1 µg, lane 3) RNA samples were then run on a 1.2% agarose gel. The slow-migrating band in the streptavidin-enriched lane is consistent with the expected size of the full-length Ast RNA. The fast migrating band is likely a splice variant of Ast that has been described previously (Kobayashi-Ishihara et al. 2012).
Identification of post-transcriptional modifications in Ast RNA
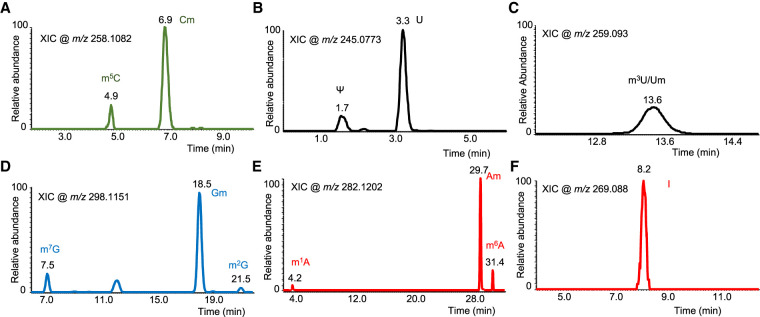
LC-MS analysis of the hydrolysate of the Ast RNA preparation indicated the presence of a small set of post-transcriptional modifications (Table 1). They include nitrogenous base and ribose methylations, inosine (deaminated form of adenosine), and pseudouridylation (Ψ, isomer of uridine). Positional isomers of nitrogenous base methylations are discriminated by the differences in chromatographic retention time (Fig. 2) and high-energy collision induced dissociation (HCD)-based fragmentation of the nucleoside in the MS/MS analysis. Ribose methylations are distinguished by the neutral loss of 146 Da compared to 132 Da for nitrogenous base methylations during tandem mass spectrometry (Supplemental Figs. S2, S3). However, some modifications (indicated by “*” in Table 1) could only be distinguished by the characteristic retention time and mass-to-charge (m/z) values of nucleoside molecular ion (MH+) as they did not trigger MS/MS, presumably due to low signal levels. They include m7G (7-methylguanosine), m3U/Um (3-methyluridine/2′-O-methyuridine, which exhibit identical retention time), I (inosine), and m1A (1-methyladenosine) (Table 1). Methylated uridine, m3U/Um, is the only nucleoside whose signal improved slightly with increased amount of RNA digest injected onto the column (Supplemental Fig. S4). However, its signal abundance still remained below the level that could trigger MS/MS analysis under standard conditions.
TABLE 1.
List of ribonucleoside modifications observed in Ast RNA
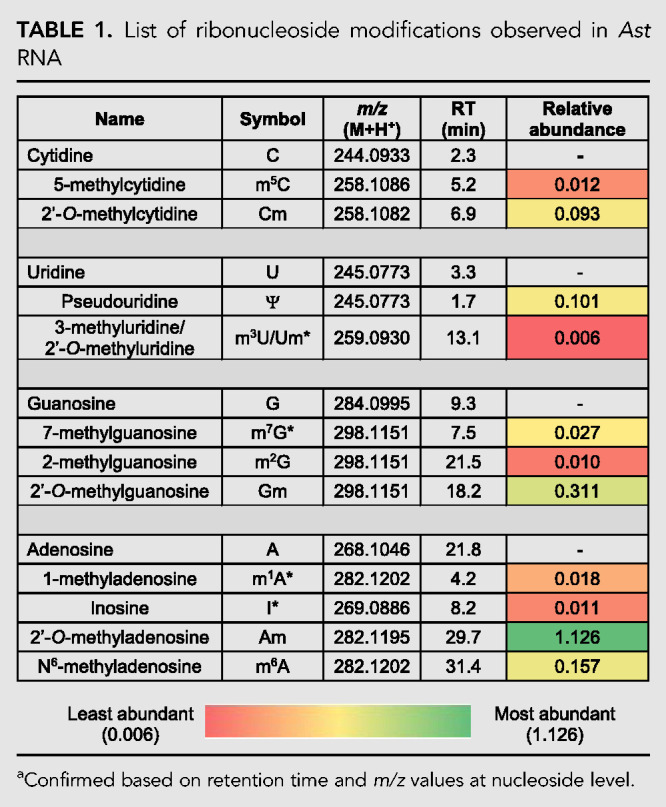
FIGURE 2.
LC-MS based detection of Ast RNA nucleoside modifications. Extracted ion chromatograms (XICs) corresponding to specific mass-to-charge (m/z) values for each class of the modified nucleosides are shown. (A) The cytidine modifications, 5-methylcytide and 2′-O-methylcytidine (m5C and Cm, respectively) share identical m/z values but differ in their retention times. (B,C) Uridine (U) modifications, Ψ (pseudouridine, shorter retention time than uridine, panel B), and 3-methyluridine and 2′-O-methyluridine (m3U and Um, respectively, panel C), which are indistinguishable by retention time and m/z values. (D) Guanosine modifications, 7-methylguanosine, 2′-O-methylguanosine, and 2-methylguanosine (m7G, Gm, and m2G, respectively) differentiated by their retention time and mass spectral behavior. (E,F) Adenosine modifications, 1-methyladenosine, 2′-O-methyladenosine, and N6-methyladenosine (m1A, Am, and m6A, respectively, panel E) and inosine (I, panel F) are distinguished by their retention time and MS spectra.
Although a majority of these modifications are also found in rRNA (except inosine), none of the human rRNA-specific modifications such as N6, N6-dimethyladenosine (m6,6A, Supplemental Fig. S5), 1-methyl-3-(3-amino-3-carboxypropyl)pseudouridine (m1acp3Ψ), or N4-acetylcytidine (ac4C; Taoka et al. 2018) were detected in the purified Ast RNA sample, suggesting that the purified sample is indeed enriched for Ast RNA, and the ribosomal RNA contamination is minimal to the extent that it is not detectable either by gel electrophoresis or ribonucleoside modification analysis.
Mapping of nucleoside modifications in Ast RNA sequence
To further ease the isolation and purification of Ast from cell lysates, we generated a lentiviral vector expressing the Ast transcript fused at the 3′ end to four copies of an artificial RNA aptamer (4 × S1m) that binds streptavidin with very high affinity (Jazurek et al. 2016; Daneshvar et al. 2020). After verification by DNA sequencing, we produced lentiviral particles, which we used to establish a stably transduced Jurkat-derived cell line (Jurkat-AST-4 × S1m). To confirm expression of the AST-4 × S1m fusion transcript, we extracted total RNA from stably transduced cells, and performed RT-qPCR as described (Zapata et al. 2017; Supplemental Fig. S6). The Ast RNA purified from these constructs exhibited identical profile albeit migrating slowly on the gel (Supplemental Fig. S7). The hydrolysate of this Ast RNA isolated via direct streptavidin enrichment from Jurkat-AST-4 × S1m cells also exhibited identical modification census confirming its origin (Supplemental Fig. S8; Supplemental Table S2).
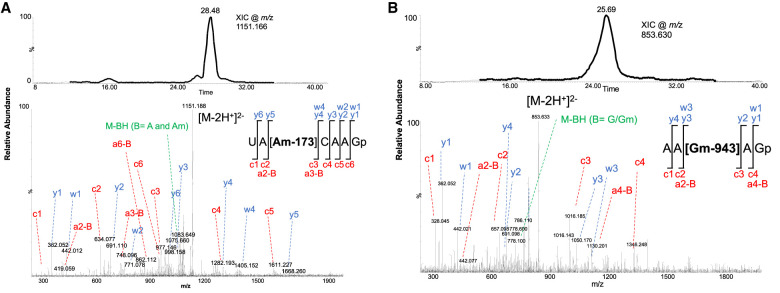
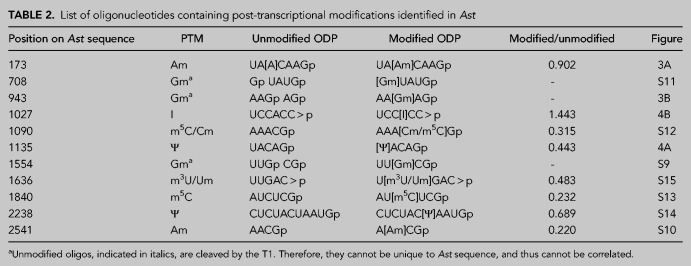
Next, we adapted the LC-MS based RNA modification mapping approach to locate the positions of modified nucleosides. We used G-specific RNase T1 to digest the RNA into oligonucleotides and sequenced them through IP-RP-LC-MS/MS. Initial analysis revealed the presence of four modified oligonucleotides, UA[Am]CAAGp, AA[Gm]AGp (Fig. 3A,B), UU[Gm]CGp (Supplemental Fig. S9) and A[mA]CGp (Supplemental Fig. S10) corresponding to positions 173, 943, 1554, 2541, respectively, in the Ast RNA digest. Tandem spectra of these molecular ions revealed the presence of >90% of the expected fragment ions corresponding to the sequence and assigned the methylation status. Although the nucleobase or ribose methylations lead to identical mass shifts, evaluation of the MS/MS spectra provided clues whether it is base methylation or ribose methylation. Scoring the fragment ions that correspond to the loss of methylated base (mN base) or ribose-methylated nucleoside (Nm) or its phosphate form was considered for assigning the methylation to the base or ribose sugar. However, these data did not represent all the PTMs as fragment ion ambiguity was observed with regard to some pyrimidine modifications at certain locations (see below).
FIGURE 3.
(A) LC-MS based detection of UA[Am]CAAGp from Ast RNA. Extracted ion chromatogram for m/z 1151.166 (top panel). Tandem mass spectrum of oligonucleotide anion (m/z 1151.166) with all the observed sequence informative fragment ions (bottom panel). The fragment ion, y and w series contain the 3′-end of the oligomer while the a-B and c ion series contain 5′ end of oligomer. Fragmentation pattern of the oligomer is depicted. (B) LC-MS based detection of AA[Gm]AGp from Ast RNA. Extracted ion chromatogram for m/z853.63 (top panel). Tandem mass spectrum of oligonucleotide anion (m/z853.63) with all the observed sequence informative fragment ions (bottom panel). Fragmentation pattern of the oligomer is depicted.
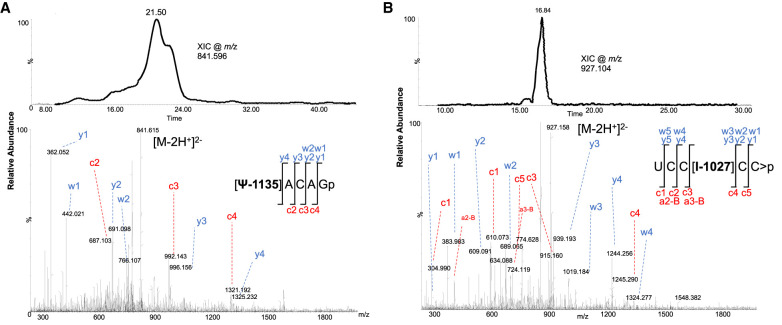
When unmodified oligonucleotides were excluded from triggering MS/MS (Cao and Limbach 2015), additional oligonucleotides with known modifications were evident in the Ast RNA digest. They include, [Gm]UAUGp (Supplemental Fig. S11), AAA[Cm/m5C]Gp (Supplemental Fig. S12), and AU[m5C]UCGp (Supplemental Fig. S13). The MS/MS signals for these oligonucleotides were less abundant in the LC-MS analysis where exclusion strategy was not applied. To detect pseudouridine-containing oligonucleotides, the T1 digest was derivatized with acrylonitrile, and the cyanoethylated (+53 Da) oligonucleotides were detected by LC-MS/MS. This exercise revealed the presence of two oligonucleotides [Ψ]ACAGp (Fig. 4A) and CUCUAC[Ψ]AAUGp (Supplemental Fig. S14) with high quality mass spectra of sequence informative fragment ions.
FIGURE 4.
(A) LC-MS based detection of [Ψ]ACAGp from Ast RNA. Extracted ion chromatogram for m/z 841.596 (top panel). Tandem mass spectrum of oligonucleotide anion (m/z 841.596) with all the observed sequence informative fragment ions (bottom panel). Fragmentation pattern of the oligomer is depicted. (B) LC-MS based detection of UCC[I]CC > p from Ast RNA. Extracted ion chromatogram for m/z 927.104 (top panel). Tandem mass spectrum of oligonucleotide anion (m/z 927.104) with all the observed sequence informative fragment ions (bottom panel). Fragmentation pattern of the oligomer is depicted.
When Ast RNA was digested with RNase cusativin and subjected to LC-MS analysis, two additional oligonucleotides, UCC[I]CC > p (Fig. 4B), U[Um/m3U]GAC > p (Supplemental Fig. S15) were detected. All these oligonucleotides are mapped to unique positions in the Ast RNA sequence (Table 2) that may be considered as signature digestion products for Ast RNA. Further, at least three modifications—namely Am at position 173, Gm at position 708, and mA at position 2541, which were detected in RNase T1 digest—were also recapitulated in cusativin digestion products (Supplemental Table S3).
TABLE 2.
List of oligonucleotides containing post-transcriptional mofdifications identified in Ast
Sequence coverage by modification mapping
When T1 digest of Ast RNA was analyzed without exclusion strategy, 76% of the sequence was recapitulated for both modified and unmodified oligonucleotides of 4-mers or above during our LC-MS analysis. When exclusion strategy was used, an increase in coverage of 9% was observed. LC-MS analysis of cusativin digest resulted in 67% sequence coverage. Lower sequence coverage by cusativin is likely due to longer digestion products, coelution of multiple oligonucleotides causing ion suppression, which lead to poor fragmentation and poor sequence coverage. Inclusion of the coverage by both enzymatic digests and the sequence overlaps indicated >85% sequence coverage (data not shown).
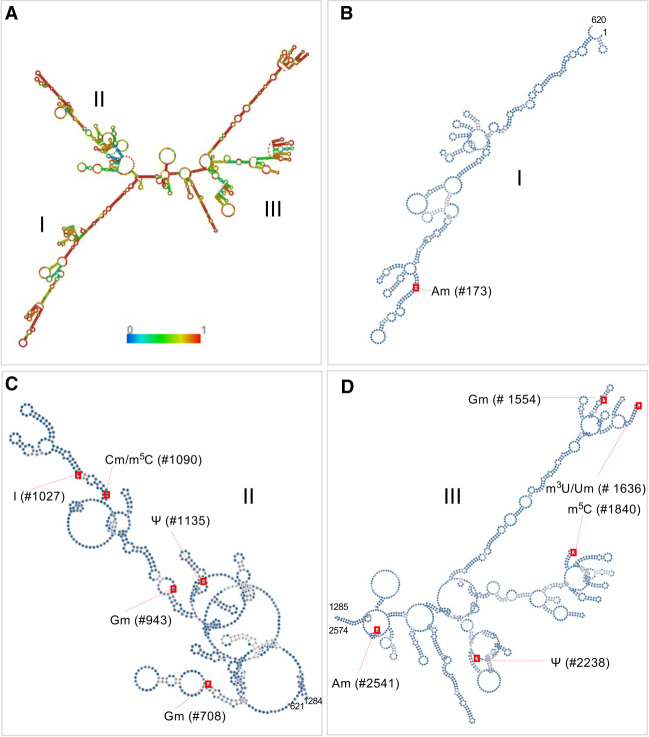
Modification positions in secondary structure
The proposed secondary structure of Ast RNA predicted by RNA Fold WebServer is shown in Figure 5A (Gruber et al. 2008; Lorenz et al. 2011). To better visualize the positions of the modifications, the sequence was subdivided into three regions: Region I (nucleotides 1–620 Fig. 5B), Region II (nucleotides 621–1284, Fig. 5C) and Region III (nucleotides 1285–2574, Fig. 5D), where the modification positions are indicated by red squares. Although modifications are mapped to all three regions of Ast RNA, multiple modifications are concentrated to Region II. The secondary structure of Ast RNA with the aptamer sequence at the 5′ end has also been evaluated for its contribution to the potential changes in secondary structure (Supplemental Fig. S16). No significant differences were observed between the secondary structures of Ast RNA with and without the aptamer.
FIGURE 5.
Putative secondary structure of Ast RNA predicted by using RNA Fold Web Server. (A) Overall secondary structure of Ast RNA, from nucleotides 1 through 2574. The color scale represents the probability of base pairing from 0 to 1. (B) Ast RNA sequence from Region I (nucleotides 1–620), with modification Am at position 173. (C) Ast RNA sequence from Region II (nucleotides 621–1284), with modifications Gm (positions 708 and 943), inosine (position 1027), Cm/m5C (position 1090), and ψ (position 1135). (D) Ast RNA sequence from Region III (nucleotides 1285–2574), with Gm (position 1554), Um/m3U (position 1636), m5C (position 1840), ψ (position 2238), and Am (position 2541).
Interactions of Ast RNA with RNA-modifying enzymes
Finally, we performed RNA immunoprecipitation (RIP) assays using antibodies against the RNA modifying enzymes that catalyze them. We focused on three proteins: the 2′-O-methyltransferases, Fibrillarin (FBL) and FTSJ3, and the pseudouridine synthase 1, dyskerin (DKC1). We prepared whole cell lysates from Jurkat-AST cells under nondenaturing conditions, and then we used antibodies to FBL, FTSJ3, and DKC1 to immunoprecipitate ribonucleoprotein complexes. As control we used preimmune rabbit IgG. After washes and RNA extraction, we used RT-qPCR to assess enrichment of Ast. As a control, we performed RT-qPCR to detect the presence of U1 snRNA. We detected increased levels of Ast in the ribonucleoprotein complexes immunoprecipitated with antibodies to all three enzymes (Fig. 6). The enrichment of Ast was significant both compared to Ast levels detected in ribonucleoprotein complexes that were recovered with preimmune IgG control antibody and compared to the U1 snRNA levels detected in the ribonucleoprotein complexes recovered with the three enzyme-specific antibodies. While a search of the snoRNA database using the Ast RNA sequence did not reveal clear snoRNA candidates that might guide catalysis of the modifications we identified, the interaction of Ast with FBL, FTSJ3, and DKC1 revealed by RIP assay provides further proof to support the presence of the epitranscriptomic modifications reported above.
FIGURE 6.
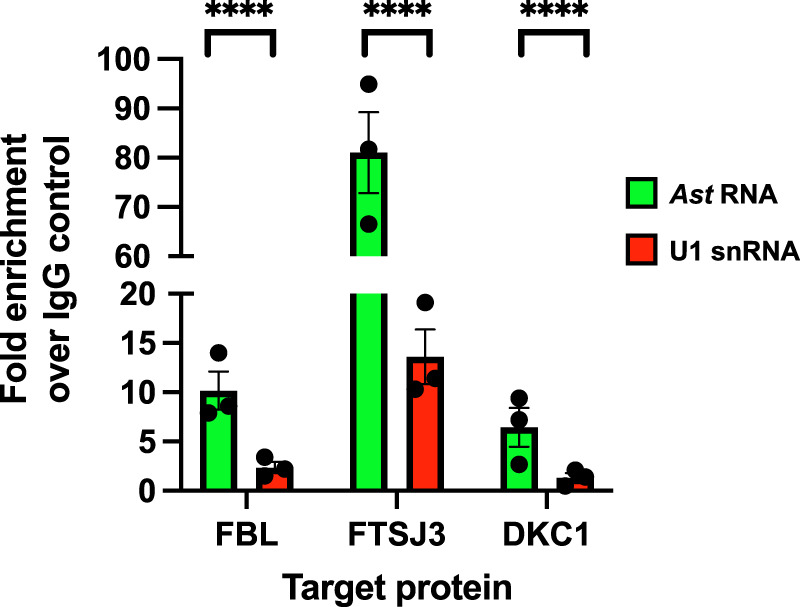
RNA immunoprecipitation (RIP) assay using antibodies directed against the 2′-O-methyltransferases, FBL and FTSJ3, and the pseudouridine synthase 1, DKC1. Whole cell lysates from Jurkat-AST cells were incubated with antibodies to FBL, FTSJ3, DKC1, or preimmune rabbit IgG, and then with Protein-A Dynabeads. Immunocomplexes were washed, and RNA extracted and used in RT-qPCR assays to detect the presence of Ast (green bars) and U1 snRNA (red bars). The data show fold enrichment of Ast or U1 snRNA over the preimmune IgG control levels. Each dot represents an independent immunoprecipitation experiment (three biological replicates for each target protein), and it represents the mean value of three PCR reactions (three technical replicates for each amplification). Bars represent standard error of means (SEM). Statistical analyses were performed by two-way ANOVA (Graph Prism). (****) P < 0.0001.
DISCUSSION
Understanding the role of nucleoside modifications in viral gene expression and infection requires the identification and mapping of the PTMs in the viral nucleic acids. The genomic RNA of HIV-1 has been extensively investigated for the presence of PTMs, including m6A (Lichinchi et al. 2016; Tirumuru et al. 2016; Kennedy et al. 2017; Tsai et al. 2021), m5C (Courtney et al. 2019b), ac4C (Tsai et al. 2020), ribose methylations (Nm, Ringeard et al. 2019), Ψ, m1A, m7G, m1G, and m6,6A (McIntyre et al. 2018). These modifications have been implicated in regulation of viral gene expression, infection, viral RNA stability, avoidance of immune sensing, and viral latency (Courtney 2021). The discovery of an HIV-encoded antisense Ast RNA able to suppress viral replication (Kobayashi-Ishihara et al. 2012) makes it an attractive candidate for epitranscriptomic analysis. Our studies describe the comprehensive profile and mapping of the PTMs associated with highly pure Ast RNA through LC-MS analysis.
The purity of isolated Ast RNA was assessed at multiple stages. Electrophoresis of the purified RNA revealed two species that exhibited migration behavior intermediate to ribosomal RNA components or around 3 kb based on size standards. The preparation also exhibited a second RNA component similar to an RNA product reported previously (Kobayashi-Ishihara et al. 2012). RT-PCR and sequencing analysis of the isolated RNA confirmed its identity as Ast.
Repeated analysis of the hydrolysates of multiple preparations of Ast RNA (without or with streptavidin aptamer) yielded identical modified nucleoside profiles. Modifications were assigned based on the known chromatographic retention time and mass spectral behavior exhibited by known RNA samples, such as E. coli tRNA. Moreover, the HCD fragmentation used in the current study provide spectra that are unique to the specific isomer of methylation (Jora et al. 2018). The Ast RNA exhibited inosine, which is not known to be present in ribosomal RNA, while the rRNA-specific modifications (Taoka et al. 2018) were conspicuously absent suggesting that the preparation is free from rRNAs and the PTMs can be unambiguously assigned to the Ast RNA.
Evaluation of the relative abundance of these modifications indicate predominance of ribose methylations (Nm) compared to base methylations. Ribose methylations are incorporated by either stand-alone methyltransferases (Somme et al. 2014) or the C/D box small nucleolar RNA (Cavaillé et al. 1996; Kiss-László et al. 1996). The Nm modifications are known to stabilize alternative secondary structures through base-pairing interactions to stay in the excited conformation thereby influencing the biological activities (Abou Assi et al. 2020). It would be interesting to know whether these modifications could play similar roles in structural stabilization during suppression of viral replication.
Employment of our optimized modification mapping methodology (Thakur et al. 2020) revealed the positions of modifications in the sequence. Incidentally, all the modified oligonucleotides in Table 2 are unique to the Ast RNA sequence. Methylation of base and ribose yield identical mass, and differentiation of the same at oligonucleotide level is not straightforward. In the current studies, such assignments were made based on the scoring of methylated base losses or loss of ribose methylated nucleoside (Nm) or its phosphate form (Nmp) in the MS/MS spectra. For UA[Am]CAAGp, we did not observe the loss of methylated adenine from molecular ion. Instead, we noticed the presence of molecular ions that correspond to the loss of nucleoside, Am or its phosphate form in the spectra as reported previously (Qiu and McCloskey 1999). Further, observation of abundant signals for a-B (5′-end) and w (3′-end) fragment ions near the ribose methylation sites support this interpretation. Similar behavior was observed for Gm modifications. Moreover, RNase T1 is known to cleave RNA at m2G but not m7G or Gm. Oligonucleotides containing m7G exhibit m7G base loss as the predominant ion in the MS/MS spectra following collision-induced dissociation (Wong et al. 2013). However, we did not observe such behavior in the MS/MS spectra suggesting that this methylation could not be m7G. Since the modification was found to be internal, this cannot be m2G because T1 is known to cleave RNA at this site, leaving the Gm as the most likely modification in these sequences. Higher abundance of ribose methylations in the nucleoside analysis also supports this possibility.
Spectra of ribose methylations for pyrimidines did not exhibit such unequivocal response. For example, in the case of AAA[Cm/m5C]Gp and U[Um/m3U]GAC > p, base losses corresponding to both methylated base and ribose methylations were noticed. Future studies involving RNA-seq technologies may help clarify the exact status (Krogh and Nielsen 2019). Similarly, m1A or m6A modifications cannot be distinguished by the current methodology, and future studies involving pseudo-MS3 methods can help differentiate these possibilities (Nakayama et al. 2019). It should be noted that we applied strict criteria of including oligonucleotides of 4-mers or above for sequence coverage calculations. Relaxing this criterion to 3-mers can fill multiple positions of sequence, which would not only increase the sequence coverage but also the number of modified oligonucleotides that can be reported for Ast RNA (Supplemental Table S4). A comparison of the peak area ratios of the modified and unmodified versions of the oligonucleotides indicated site-specific variations in terms of the degree of modification (Table 2). Future studies may reveal the biochemical or biological significance of these variations including how the cellular environment (e.g., cell type, cell activation/maturation) impacts them, as well as their potential contribution to regulating the alternative protein-coding and noncoding functions of Ast (Zapata et al. 2017; Affram et al. 2019) and the cytoplasmic or nuclear distribution of Ast, where the alternative functions are exerted.
Evidence from multiple studies shows that the HIV-1 antisense transcript Ast is a bifunctional RNA (Gholizadeh et al. 2021). On one hand, Ast functions as a mRNA that encodes the HIV-1 antisense protein, ASP (Clerc et al. 2011; Torresilla et al. 2013; Liu et al. 2019), which we have shown to be expressed on the surface of productively infected cells and on the envelope of viral particles (Affram et al. 2019). On the other hand, studies from our and other groups showed that Ast functions as a ncRNA able to promote HIV-1 latency through epigenetic silencing of the 5′LTR (Kobayashi-Ishihara et al. 2012; Saayman et al. 2014; Zapata et al. 2017). The mechanisms that regulate the balance between the protein-coding and noncoding (regulatory) activities of Ast are not fully understood. However, polyadenylation was shown to impact the nuclear versus cytoplasmic distribution of Ast and, thus, its function (Ma et al. 2021). The characterization of the Ast epitranscriptome may shed new light onto the mechanisms that regulate the subcellular localization and the alternative roles of this transcript.
In summary, we report comprehensive details of the epitranscriptomic profile of the HIV-1 antisense RNA, Ast by an unbiased LC-MS methodology. Ribose methylations were most abundant in this set of modifications while base methylations were at lower abundance level. A complementary LC-MS methodology also located the positions of these modification with high confidence. Multiple modifications were found to be concentrated in one specific region of Ast RNA suggesting a possible role in regulating its activity.
MATERIALS AND METHODS
Reagents
Streptavidin beads and Proteinase K were purchased from New England Biolab (NEB), Direct-zol RNA Kit from Zymo Research. Ribonuclease T1 was procured from Worthington Biochemical Corporation. Cusativin was expressed and purified from E. coli based over an expression system as described before (Addepalli et al. 2017). All other chemicals were procured from Fisher Scientific, unless specified.
Generation and culture of stably transduced Jurkat cell lines
Generation of the Jurkat-derived cell lines stably transduced with a lentiviral vector expressing Ast (J-AST) has been described previously (Zapata et al. 2017). To generate the Jurkat-derived cell line stably transduced with a lentiviral vector expressing Ast fused to streptavidin-binding aptamer sequence, a DNA fragment containing the entire sequence of Ast (2574 nt, Kobayashi-Ishihara et al. 2012) fused in 3′ to four repeats of the streptavidin-binding aptamer, S1m (4 × S1m, Jazurek et al. 2016; Daneshvar et al. 2020) was produced by gene synthesis (GenScript), and cloned into the EcoRI site of the pLVX-Puro lentiviral vector (Takara). Packaging of lentiviral particles was carried out by transfecting the pLVX-AST-4 × S1m vector into Lenti-X 293T cells along with Lenti-X VSV-G Packaging Single Shot (both from Takara). After concentration with PEG-it (System Biosciences), lentiviral particles were used to transduce Jurkat E6-1 cells (ATCC) at a multiplicity of infection (m.o.i) = 20. After 24 h, cells were cultured in the presence of 1 µg/mL puromycin (Takara) to select stably transduced cells, which were then maintained in complete medium containing RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, streptomycin, and 1 µg/mL puromycin. Cultures were passaged every 2–3 d and diluted to 0.5 × 106 cells/mL in fresh complete medium supplemented with puromycin. Stably transduced cells were only maintained for 4–5 wk in culture throughout the experiment before a new low-passage stock was put in culture as described above.
Verification of Ast expression
To confirm expression of Ast, we used the same procedure reported previously (Zapata et al. 2017). Briefly, total RNA was extracted from 5 × 106 stably transduced cells and from the parental Jurkat E6-1 cell line using the Qiagen RNeasy Kit. After further digestion with Turbo DNase, 100–500 ng of total RNA were used for cDNA synthesis using the iScript Select cDNA Synthesis Kit (BioRad), and the AST-tag RT primer with sequence 5′-CTGATCTAGAGGTACCGGATCCAACATGTGGCAGGAAGTAGG-3′ (the tag sequence is underlined). For the qPCR reaction, we utilized the TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer's instructions. The sequence of the forward and reverse primers, and the probe were: 5′-TGATGAACATCTAATTTGTCCACTGA-3′; 5′-CTGATCTAGAGGTACCGGAT-3′; 5′-CCCAATGTATGCCCCTCCCA-3′. For the standard curve, the Ast RNA amplicon carrying the tag sequence was cloned into pUC57. The plasmid was then linearized and utilized at 100–105 copies to generate a PCR standard curve. The number of copies per 3 µL of cDNA was then normalized to the equivalent number of cells as determined by counting by trypan blue exclusion with a hemacytometer.
Ast RNA isolation
For enrichment of Ast RNA expressed in J-AST cells, 200 µg of total RNA isolated as described above was hybridized to 500 pmol mixture of biotinylated oligonucleotides that are reverse complementary to the Ast at positions 171, 571, 971, 1371, 1770, and 2163 (see Supplemental Table S1). The mixture was then incubated with 500 µg of pretreated Streptavidin Magnetic Beads as per the vendor's (NEB) protocol. After incubating the RNA and bead mixture at room temperature for 60 min with gentle shaking, the tube was transferred to a magnet to pull the beads to the magnet side. After discarding the clear liquid with a pipette, the beads were washed six times with wash/binding buffer. After the final wash buffer step, 95 µL of elution buffer (10 mM Tris-HCl pH 7.5; 100 mM NaCl; 1 mM EDTA and 0.5% SDS) and 5 µL of Proteinase K were added, incubated at 50°C for 45 min, 95°C for 10 min, and chilled on ice for 3 min. After magnetic separation of beads, the eluted RNA mixture was transferred to a RNase-free tube. Sample clean-up was performed with Direct-zol RNA Kit (Zymo Research). For enrichment of Ast fused to 4 × S1m aptamer, about 70 µg of total RNA (0.15 µg/μL) in wash/binding buffer was added to 500 µg of pretreated Streptavidin Magnetic Beads, and Ast RNA was isolated as described above.
RT-PCR to confirm the presence of Ast RNA
The sequence of purified Ast RNA was confirmed by reverse transcription and polymerase chain reaction (RT-PCR) using Access Quick RT-PCR Kit (Promega). Purified Ast RNA (100 ng) or total RNA (1 µg) was used as a template during RT reaction. An oligonucleotide, 5′-GGTTTTGCGATTCTAAAATG-3′, reverse complementary to the 3′-end of Ast RNA, was used to prepare cDNA, followed by amplification of the 5′-end of RNA between 3–571 nt using a primer set of 5′-TGGAAAGTCCCCAGCGGAAA-3′ and 5′-GAGCAGTATCTCGAGACCTA-3′. The resulting amplification product was gel purified using QIAquick Gel Extraction Kit (Qiagen), and it was subjected to Sanger sequencing.
Nucleoside analysis by LC-MS
The Ast RNA was hydrolyzed to nucleosides following optimal enzymatic digestion with nuclease P1, phosphodiesterase and alkaline phosphatase (Jora et al. 2018). The resulting hydrolysate was subjected to reversed phase chromatography using a high strength silica column (Acquity UPLC HSS T3, 1.8 µm, 1.0 mm × 50 mm, Waters). The LC method used 5.3 mM ammonium acetate in H2O, pH 5.3, as Mobil Phase A (MPA), and a mixture of acetonitrile/H2O (40:60 v/v) with 5.3 mM ammonium acetate as Mobil Phase B (MPB). The gradient program consisted of 0% B for 6.3 min, 2% B at 13.1 min, 3% B at 16 min, 5% B at 21.4 min, 25% B at 24.6, 50% B at 26.9 min, 75% B at 30.2 min (hold for 0.3 min), 99% B at 33 min (hold for 6 min), then returning to 0% B at 39 min for a re-equilibration of the column for 16 min. A flow rate of 60 µL min−1 was used. The column temperature was set at 30°C. Electrospray conditions and high energy collision dissociation parameters were identical to those described before (Jora et al. 2018, 2021).
RNA modification mapping through LC-MS
The Ast RNA was hydrolyzed to oligonucleotides using nucleobase-specific RNases T1 (Addepalli and Limbach 2016) and cusativin (Addepalli et al. 2017), independently. Digestion mix included 120 mM ammonium acetate, 40 U of T1 or 1 µg of cusativin per 2–3 µg of purified Ast RNA. RNA-enzyme mixture was incubated at 37°C for 120 min for RNase T1, and at 62°C for 90 min for cusativin, respectively. To detect pseudouridines, the T1 digest was derivatized with acrylonitrile in 41% ethanol and 1.1 M TEAA, pH 8.6, at 70°C for 120 min (Addepalli and Limbach 2016). The IP-RP-LC-MS/MS (ion pairing reversed phase liquid chromatography coupled with tandem mass spectrometry) was performed on an Ultimate 3000 (Thermo Scientific) UHPLC system using a Waters peptide BEH C18 column (0.1 × 100 mm, 1.7 µm particle size) at 60°C. MPA consisted of 8 mM TEA and 200 mM HFIP (pH 7.8) in H2O, and MPB consisted of 8 mM TEA and 200 mM HFIP in 1:1 H2O:methanol at a flow rate of 65 µL min−1. After initial hold at 5% B, the gradient was ramped to 55% B at 70 min, 95% B at 75 min with a hold for 5 min before equilibrating the column to initial conditions (5% B) for 30 min. Mass spectrometric analysis was performed in negative ion polarity through an electrospray ionization (ESI) source connected to a Waters Synapt G2-S (quadrupole time-of-flight, Q-TOF) mass spectrometer in sensitivity mode. The ESI source parameters consisted of 2.5 kV source voltage, 30 V sample cone, source and desolvation temperatures at 120°C and 400°C, cone and desolvation gas flow rates at 5 and 800 L h−1, respectively. A scan range of 400 to 2000 m/z (0.5 sec scan time) for MS acquisition and 200 to 2000 m/z (1.0 sec scan time) for tandem mass spectral (MS/MS) acquisition was used. MS/MS spectra were collected under data-dependent acquisition mode using an m/z dependent collision energy profile (Thakur et al. 2020). A second injection that excluded unmodified oligonucleotides from collision-induced dissociation (using a mass exclusion list) during tandem mass spectrometry was also performed. Such a strategy is known to improve the detection of modified oligonucleotides (Cao and Limbach 2015).
RNA immunoprecipitation assay (RIP)
RNA immunoprecipitation was performed according to the method described previously (Rinn et al. 2007). Nuclear isolation buffer was prepared by mixing 1.28 M sucrose; 40 mM Tris-HCl pH 7.5; 20 mM MgCl2; 4% Triton X-100. RIP buffer was prepared by mixing 150 mM KCl, 25 mM Tris pH 7.4, 5 mM EDTA, 0.5 mM DTT, 0.5% IGEPAL, Halt Protease Inhibitor Cocktail (Thermo Scientific), 100 U/mL SUPERASin (Ambion). Cells were grown in RPMI-1640 media. Twenty million cells were harvested by centrifugation at 300g and washed with 1× PBS once, then resuspended in cell lysis buffer (2 mL 1× PBS, 2 mL nuclear isolation buffer, and 6 mL water) and incubated on ice for 20 min with frequent mixing by inverting the tube. The mixture was then centrifuged at 2500g for 10 min to pellet the nuclei. The nuclear pellet was then resuspended in 1 mL RIP buffer and divided into two 500 µL fractions for IP and control IP. Nuclei were disrupted with a Dounce homogenizer with 20 strokes. The nuclear debris was pelleted by centrifugation 13,000 rpm for 10 min. The supernatant was transferred to a fresh Eppendorf tube. Antibodies to FTSJ3 (Bethyl; catalog no. A304-198A), FBL (Proteintech; catalog no. 16021), DKC1 (Bethyl; catalog no. A302-591A), and rabbit IgG (Cell Signaling; catalog no. 2729S) were added in each tube and incubated at 4°C for 2 h with gentle rotation. 40 µL of Protein A-Dynabeads (Invitrogen) were then added to the tube and were incubated at 4°C for another 1 h with gentle rotation. Beads were washed with RIP buffer three times and once with cold PBS using a magnetic separator. Beads were then resuspended in 1 mL triazole for RNA isolation. Fifty nanograms of RNA were used for gene-specific cDNA synthesis using the following primers: 5′-GAACGCAGTCCCCCACTAC-3′ for U1 snRNA and 5′-CTGATCTAGAGGTACCGGATCCAACATGTGGCAGGAAGTAGG-3′ for Ast RNA. The enrichment was quantified by using TaqMan Gene Expression Master Mix (Applied Biosystems) according to the manufacturer's instructions. The sequences of the forward and reverse primers, and the probe for the Ast RNA were: 5′-TGATGAACATCTAATTTGTCCACTGA-3′; 5′-CTGATCTAGAGGTACCGGAT-3′; 5′-CCCAATGTATGCCCCTCCCA-3′. The sequences of the forward and reverse primers, and the probe for the U1 RNA were: 5′-GGCGAGGCTTATCCATTGCA-3′, 5′-GCAGTCGAGTTTCCCACATTTG-3′, and 5′-CCGGATGTGCTGACCC-3′.
Data analysis
In silico digestion of Ast RNA generated 470 oligonucleotide digestion products (ODPs). Removal of duplicate sequences leave 216 ODPs that can be distinguished by tandem mass spectrometry. However, some of them are sequence isomers exhibiting identical mass or m/z values, and their removal leaves 142 ODPs that can be differentiated by unique m/z values. Similarly, cusativin could generate 818 ODPs. Removal of duplicates leaves 197 ODPs that may be distinguished by MS/MS, with 150 of these ODPs that can be differentiated by unique m/z values. Mass spectral data was initially analyzed by RNAModMapper (Yu et al. 2017) followed by manual evaluation of each spectrum.
snoRNA database search
BLAST analysis of Ast RNA against snoRNA database (https://www-snorna.biotoul.fr) was performed to identify H/ACA and C/D box guide snoRNAs that may potentially be involved in pseudouridine and methylation modifications, respectively (Lestrade and Weber 2006).
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to acknowledge the help rendered by Manasses Jora during LC-MS analysis. This research was funded by the National Institute of Allergy and Infectious Diseases (7R01AI144983 to F.R.). The generous support of the University of Cincinnati is also appreciated.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079043.121.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
MEET THE FIRST AUTHORS
Mariana Estevez.

Rui Li.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Mariana Estevez and Rui Li are co-first authors of this paper, “Identification and mapping of post-transcriptional modifications on the HIV-1 antisense transcript Ast in human cells.” Mariana is a PhD candidate in Professor Addepalli's lab at the Department of Chemistry, University of Cincinnati. Her research focuses on the understanding of the biological roles of post-transcriptional modifications and the damaging effects of xenobiotic-induced oxidative stress on cellular and viral RNAs. Rui is a post-doctoral fellow in Dr. Romerio's lab at Johns Hopkins School of Medicine. He is studying the role of the HIV-1 antisense transcript, Ast in viral latency with the goal of exploiting its silencing properties to develop a functional cure for HIV-1 infection.
What are the major results described in your paper and how do they impact this branch of the field?
In this manuscript, we were able to identify and map the post-transcriptional modifications of HIV-1 antisense transcript, Ast RNA, through LC-MS/MS analyses. The incidence of such modifications is supported by evidence of physical interaction between Ast RNA and the RNA modifying enzymes that catalyze these modifications. We hope to harness this knowledge to understand their potential involvement in viral suppression and test their potential as drug targets.
What led you to study RNA or this aspect of RNA science?
ME: RNA has been known to be an intermediate in protein synthesis; however, its potential is realized now with the advent of mRNA vaccines. Post-transcriptional modifications found in RNA modulate the function of different RNAs through structural maintenance, fine-tuning of interactions and control of signal responses. Knowledge of the occurrence of post-transcriptional modifications in viral RNAs is essential to understand their role in viral gene expression regulation. This led me to pursue the field of viral RNA science, so that we can better understand their roles and explore their potential to use them as drug candidates/targets.
RL: I became interested in viral–host interaction, focusing on an RNA virus, since graduate school. After joining Dr. Romerio's lab, I was immediately attracted by HIV-encoded antisense Ast RNA which has been shown to suppress viral replication and regulate latency. Understanding the epitranscriptome of Ast RNA and enzymes that catalyze those modifications would help us to uncover a new layer of viral–host interaction and may also facilitate the development of new therapeutics to control HIV infection and latency.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
ME: Yes. The major difficulty was the optimization of isolation conditions for a good yield of Ast RNA. This required large amounts of total RNA for a series of trial-and-error experiments. An extensive literature search and discussions with my advisor helped me identify suitable conditions for optimal yield and highest purity for subsequent LC-MS analyses. Isolation by two different approaches, without or with an aptamer at either end of the Ast RNA molecule, surprisingly, yielded identical modification information indicating the consistency of the data.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
ME: I always had an innate passion and curiosity about the biochemical workings of the human body. This is one of the reasons why I decided to pursue a PharmD and later continue my studies with a PhD, to become a scientist. Moreover, science has a profound positive impact on people's lives, and I wanted to be part of it. During my PhD, I was intrigued by RNA science, and how much this field is unexplored. I wanted to contribute and expand our understanding of the contributions of RNA in influencing the cells’ mechanisms in normal and disease states.
RL: The first time that I saw virus infected cells under multicolor fluorescence microscopy. It was recorded as a very beautiful image that became the cover of my PhD thesis defense presentation.
If you were able to give one piece of advice to your younger self, what would that be?
ME: Be persistent and bold, and don't give up on your dreams!
RL: I would advise my younger self to be more open-minded and ask for help and suggestions when you suffer.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
ME: My mentors. I had multiple mentors throughout my academic career and they each approached science in a different way. These different exposures allowed me to shape my own philosophy.
RL: My PhD mentor taught me to be persistent, and I learned to be focused on my niche in science from Dr. Romerio.
What are your subsequent near- or long-term career plans?
ME: I would like to continue learning about the RNA world. I am moving toward completion of my PhD studies, and I want to pursue a research and development scientist career at a biotechnology company, where I intend to apply the knowledge that I have acquired thus far and continue contributing to the scientific community.
RL: I am considering employment in a university or a biotech company as an independent scientist. Let's just keep doing what we're doing and see where things go.
What were the strongest aspects of your collaboration as co-first authors?
ME: The strongest aspects of our collaboration were the complementary sets of expertise in each laboratory, and the constant communication between collaborators.
RL: We are equipped with complementary skills and worked as a team to efficiently complete our experiments.
How did you decide to work together as co-first authors?
It was designed by our principal investigators.
REFERENCES
- Abou Assi H, Rangadurai AK, Shi H, Liu B, Clay MC, Erharter K, Kreutz C, Holley CL, Al-Hashimi HM. 2020. 2′-O-methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res 48: 12365–12379. 10.1093/nar/gkaa928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B, Limbach PA. 2016. Pseudouridine in the anticodon of Escherichia coli tRNATyr(QΨA) is catalyzed by the dual specificity enzyme RluF. J Biol Chem 291: 22327–22337. 10.1074/jbc.M116.747865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addepalli B, Venus S, Thakur P, Limbach PA. 2017. Novel ribonuclease activity of cusativin from Cucumis sativus for mapping nucleoside modifications in RNA. Anal Bioanal Chem 409: 5645–5654. 10.1007/s00216-017-0500-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affram Y, Zapata JC, Gholizadeh Z, Tolbert WD, Zhou W, Iglesias-Ussel MD, Pazgier M, Ray K, Latinovic OS, Romerio F. 2019. The HIV-1 antisense protein ASP is a transmembrane protein of the cell surface and an integral protein of the viral envelope. J Virol 93: e00574-19. 10.1128/JVI.00574-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, Hosogane M, Sinclair WR, Nanan KK, Mandler MD, et al. 2018. Acetylation of cytidine in mRNA promotes translation efficiency. Cell 175: 1872–1886.e1824. 10.1016/j.cell.2018.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Baginski B. 2021. MODOMICS: an operational guide to the use of the RNA modification pathways database. Methods Mol Biol 2284: 481–505. 10.1007/978-1-0716-1307-8_26 [DOI] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt EK, Martinez NM, Gilbert WV. 2020. Regulation and function of RNA pseudouridylation in human cells. Annu Rev Genet 54: 309–336. 10.1146/annurev-genet-112618-043830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Limbach PA. 2015. Enhanced detection of post-transcriptional modifications using a mass-exclusion list strategy for RNA modification mapping by LC-MS/MS. Anal Chem 87: 8433–8440. 10.1021/acs.analchem.5b01826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaillé J, Nicoloso M, Bachellerie JP. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383: 732–735. 10.1038/383732a0 [DOI] [PubMed] [Google Scholar]
- Choe J, Lin S, Zhang W, Liu Q, Wang L, Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al. 2018. mRNA circularization by METTL3–eIF3h enhances translation and promotes oncogenesis. Nature 561: 556–560. 10.1038/s41586-018-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerc I, Laverdure S, Torresilla C, Landry S, Borel S, Vargas A, Arpin-Andre C, Gay B, Briant L, Gross A, et al. 2011. Polarized expression of the membrane ASP protein derived from HIV-1 antisense transcription in T cells. Retrovirology 8: 74. 10.1186/1742-4690-8-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG. 2021. Post-transcriptional regulation of viral RNA through epitranscriptional modification. Cells 10: 1129. 10.3390/cells10051129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, Cullen BR. 2017. Epitranscriptomic enhancement of influenza A virus gene expression and replication. Cell Host Microbe 22: 377–386.e375. 10.1016/j.chom.2017.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Chalem A, Bogerd HP, Law BA, Kennedy EM, Holley CL, Cullen BR, Dermody TS, Bieniasz P, Goff S. 2019a. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio 10: e01209. 10.1128/mBio.01209-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, Swanstrom R, Holley CL, Cullen BR. 2019b. Epitranscriptomic addition of m5C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe 26: 217–227.e216. 10.1016/j.chom.2019.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daneshvar K, Ardehali MB, Klein IA, Hsieh FK, Kratkiewicz AJ, Mahpour A, Cancelliere SOL, Zhou C, Cook BM, Li W, et al. 2020. lncRNA DIGIT and BRD3 protein form phase-separated condensates to regulate endoderm differentiation. Nat Cell Biol 22: 1211–1222. 10.1038/s41556-020-0572-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27: 344–351. 10.1016/S0968-0004(02)02109-6 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. 2016. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 530: 441–446. 10.1038/nature16998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. 2013. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 9: e1003602. 10.1371/journal.pgen.1003602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson MR, Rojo DR, von Lindern JJ, O'Brien WA. 2002. HIV-1 replication cycle. Clin Lab Med 22: 611–635. 10.1016/S0272-2712(02)00015-X [DOI] [PubMed] [Google Scholar]
- Gholizadeh Z, Iqbal MS, Li R, Romerio F. 2021. The HIV-1 antisense gene ASP: the new kid on the block. Vaccines (Basel) 9: 513. 10.3390/vaccines9050513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res 36: W70–W74. 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Motorin Y. 2017. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 18: 275–291. 10.1038/nrg.2016.169 [DOI] [PubMed] [Google Scholar]
- Hsu PJ, Fei Q, Dai Q, Shi H, Dominissini D, Ma L, He C. 2019. Single base resolution mapping of 2′-O-methylation sites in human mRNA and in 3′ terminal ends of small RNAs. Methods 156: 85–90. 10.1016/j.ymeth.2018.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, Kim SJ, Mason CE, Horner SM, Siddiqui A. 2018. N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci 115: 8829–8834. 10.1073/pnas.1808319115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazurek M, Ciesiolka A, Starega-Roslan J, Bilinska K, Krzyzosiak WJ. 2016. Identifying proteins that bind to specific RNAs‐‐focus on simple repeat expansion diseases. Nucleic Acids Res 44: 9050–9070. 10.1093/nar/gkw803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jora M, Burns AP, Ross RL, Lobue PA, Zhao R, Palumbo CM, Beal PA, Addepalli B, Limbach PA. 2018. Differentiating positional isomers of nucleoside modifications by higher-energy collisional dissociation mass spectrometry (HCD MS). J Am Soc Mass Spectrom 29: 1745–1756. 10.1007/s13361-018-1999-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jora M, Lobue PA, Ross RL, Williams B, Addepalli B. 2019. Detection of ribonucleoside modifications by liquid chromatography coupled with mass spectrometry. Biochim Biophys Acta Gene Regul Mech 1862: 280–290. 10.1016/j.bbagrm.2018.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jora M, Borland K, Abernathy S, Zhao R, Kelley M, Kellner S, Addepalli B, Limbach PA. 2021. Chemical amination/imination of carbonothiolated nucleosides during RNA hydrolysis. Angew Chem Int Ed Engl 60: 3961–3966. 10.1002/anie.202010793 [DOI] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AVR, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. 2017. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 22: 830. 10.1016/j.chom.2017.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088. 10.1016/S0092-8674(00)81308-2 [DOI] [PubMed] [Google Scholar]
- Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, Nakano K, Yamochi T, Ishida T, Watanabe T. 2012. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 9: 38. 10.1186/1742-4690-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Rivera-Serrano EE, Neidleman JA, Zhu J. 2019. HIV-1 replication benefits from the RNA epitranscriptomic code. J Mol Biol 431: 5032–5038. 10.1016/j.jmb.2019.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalak JA, Pomerantz SC, Crain PF, McCloskey JA. 1993. A novel method for the determination of post-transcriptional modification in RNA by mass spectrometry. Nucleic Acids Res 21: 4577–4585. 10.1093/nar/21.19.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh N, Nielsen H. 2019. Sequencing-based methods for detection and quantitation of ribose methylations in RNA. Methods 156: 5–15. 10.1016/j.ymeth.2018.11.017 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ. 2006. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162. 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Sklutuis R, Groebner JL, Romerio F. 2021. HIV-1 natural antisense transcription and its role in viral persistence. Viruses 13: 795. 10.3390/v13050795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1: 16011. 10.1038/nmicrobiol.2016.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder B, Grozhik AV, Olarerin-George AO, Meydan C, Mason CE, Jaffrey SR. 2015. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods 12: 767–772. 10.1038/nmeth.3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Torresilla C, Xiao Y, Nguyen PT, Cate C, Barbosa K, Rassart E, Cen S, Bourgault S, Barbeau B. 2019. HIV-1 antisense protein of different clades induces autophagy and associates with the autophagy factor p62. J Virol 93: e01757-18. 10.1128/JVI.01757-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol 6: 26. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Lünse CE, Mörl M. 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7: 35. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Yasunaga JI, Shimura K, Takemoto K, Watanabe M, Amano M, Nakata H, Liu B, Zuo X, Matsuoka M. 2021. Human retroviral antisense mRNAs are retained in the nuclei of infected cells for viral persistence. Proc Natl Acad Sci 118: e2014783118. 10.1073/pnas.2014783118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. 2016. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res 44: e135. 10.1093/nar/gkw547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre W, Netzband R, Bonenfant G, Biegel JM, Miller C, Fuchs G, Henderson E, Arra M, Canki M, Fabris D, et al. 2018. Positive-sense RNA viruses reveal the complexity and dynamics of the cellular and viral epitranscriptomes during infection. Nucleic Acids Res 46: 5776–5791. 10.1093/nar/gky029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H, Yamauchi Y, Nobe Y, Sato K, Takahashi N, Shalev-Benami M, Isobe T, Taoka M. 2019. Method for direct mass-spectrometry-based identification of monomethylated RNA nucleoside positional isomers and its application to the analysis of leishmania rRNA. Anal Chem 91: 15634–15643. 10.1021/acs.analchem.9b03735 [DOI] [PubMed] [Google Scholar]
- Pomerantz SC, McCloskey JA. 1990. Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol 193: 796–824. 10.1016/0076-6879(90)93452-Q [DOI] [PubMed] [Google Scholar]
- Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, Mason CE, Horner SM, Wilson AC, Depledge DP, et al. 2020. Direct RNA sequencing reveals m6A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun 11: 6016. 10.1038/s41467-020-19787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F, McCloskey JA. 1999. Selective detection of ribose-methylated nucleotides in RNA by a mass spectrometry-based method. Nucleic Acids Res 27: e20. 10.1093/nar/27.18.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. 2019. FTSJ3 is an RNA 2'-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565: 500–504. 10.1038/s41586-018-0841-4 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Droogmans L, Grosjean H. 2021. Post-transcriptional modifications of conserved nucleotides in the T-loop of tRNA: a tale of functional convergent evolution. Genes (Basel) 12: 140. 10.3390/genes12020140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. 2014. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther 22: 1164–1175. 10.1038/mt.2014.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Lyko F. 2009. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 37: e12. 10.1093/nar/gkn954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibbritt T, Patel HR, Preiss T. 2013. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA 4: 397–422. 10.1002/wrna.1166 [DOI] [PubMed] [Google Scholar]
- Somme J, Van Laer B, Roovers M, Steyaert J, Versées W, Droogmans L. 2014. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA (New York, NY) 20: 1257–1271. 10.1261/rna.044503.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taoka M, Nobe Y, Yamaki Y, Sato K, Ishikawa H, Izumikawa K, Yamauchi Y, Hirota K, Nakayama H, Takahashi N, et al. 2018. Landscape of the complete RNA chemical modifications in the human 80S ribosome. Nucleic Acids Res 46: 9289–9298. 10.1093/nar/gky811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur P, Estevez M, Lobue PA, Limbach PA, Addepalli B. 2020. Improved RNA modification mapping of cellular non-coding RNAs using C- and U-specific RNases. Analyst 145: 816–827. 10.1039/C9AN02111F [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5: e15528. 10.7554/eLife.15528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torresilla C, Larocque E, Landry S, Halin M, Coulombe Y, Masson JY, Mesnard JM, Barbeau B. 2013. Detection of the HIV-1 minus-strand-encoded antisense protein and its association with autophagy. J Virol 87: 5089–5105. 10.1128/JVI.00225-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Courtney DG, Cullen BR. 2018. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog 14: e1006919. 10.1371/journal.ppat.1006919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Jaguva Vasudevan AA, Martinez Campos C, Emery A, Swanstrom R, Cullen BR. 2020. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe 28: 306–312.e306. 10.1016/j.chom.2020.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Bogerd HP, Kennedy EM, Emery A, Swanstrom R, Cullen BR. 2021. Epitranscriptomic addition of m6A regulates HIV-1 RNA stability and alternative splicing. Genes Dev 35: 992–1004. 10.1101/gad.348508.121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. 2011. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 25: 1173–1184. 10.1101/gad.2050711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiener D, Schwartz S. 2021. The epitranscriptome beyond m6A. Nat Rev Genet 22: 119–131. 10.1038/s41576-020-00295-8 [DOI] [PubMed] [Google Scholar]
- Wong SY, Javid B, Addepalli B, Piszczek G, Strader MB, Limbach PA, Barry CE. 2013. Functional role of methylation of G518 of the 16S rRNA 530 loop by GidB in Mycobacterium tuberculosis. Antimicrob Agents Chemother 57: 6311–6318. 10.1128/AAC.00905-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Lobue PA, Cao X, Limbach PA. 2017. RNAModMapper: RNA modification mapping software for analysis of liquid chromatography tandem mass spectrometry data. Anal Chem 89: 10744–10752. 10.1021/acs.analchem.7b01780 [DOI] [PubMed] [Google Scholar]
- Zapata JC, Campilongo F, Barclay RA, DeMarino C, Iglesias-Ussel MD, Kashanchi F, Romerio F. 2017. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 506: 34–44. 10.1016/j.virol.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Pirnie SP, Carmichael GG. 2017. High-throughput and site-specific identification of 2′-O-methylation sites using ribose oxidation sequencing (RibOxi-seq). RNA 23: 1303–1314. 10.1261/rna.061549.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.