Abstract
Poly(A) tail length is regulated in both the nucleus and cytoplasm. One factor that controls polyadenylation in the cytoplasm is CPEB1, an RNA binding protein that associates with specific mRNA 3′UTR sequences to tether enzymes that add and remove poly(A). Two of these enzymes, the noncanonical poly(A) polymerases GLD2 (TENT2, PAPD4, Wispy) and GLD4 (TENT4B, PAPD5, TRF4, TUT3), interact with CPEB1 to extend poly(A). To identify additional RNA binding proteins that might anchor GLD4 to RNA, we expressed double tagged GLD4 in U87MG cells, which was used for sequential immunoprecipitation and elution followed by mass spectrometry. We identified several RNA binding proteins that coprecipitated with GLD4, among which was FMRP. To assess whether FMRP regulates polyadenylation, we performed TAIL-seq from WT and FMRP-deficient HEK293 cells. Surprisingly, loss of FMRP resulted in an overall increase in poly(A), which was also observed for several specific mRNAs. Conversely, loss of CPEB1 elicited an expected decrease in poly(A), which was examined in cultured neurons. We also examined polyadenylation in wild type (WT) and FMRP-deficient mouse brain cortex by direct RNA nanopore sequencing, which identified RNAs with both increased and decreased poly(A). Our data show that FMRP has a role in mediating poly(A) tail length, which adds to its repertoire of RNA regulation.
Keywords: CPEB1, FMRP, GLD4, polyadenylation, nanopore RNA-seq
INTRODUCTION
Poly(A) tail length exerts a major influence on gene expression by regulating mRNA export, translation, and stability (Jalkanen et al. 2014). Poly(A) is modulated in both the nucleus and cytoplasm by poly(A) polymerases, deadenylating enzymes, and RNA binding proteins (Ivshina et al. 2014; Yu and Kim 2020). One family of RNA binding proteins that determine transcript-specific cytoplasmic polyadenylation are the CPEBs (cytoplasmic polyadenylation element binding proteins). CPEB1, for example, binds the 3′UTR cytoplasmic polyadenylation element (CPE) as well as enzymes that lengthen and shorten poly(A) in oocytes (Barnard et al. 2004; Kim and Richter 2006), neurons (Huang et al. 2002; Udagawa et al. 2012), primary cells (Burns and Richter 2008; Burns et al. 2011), and established cell lines (Novoa et al. 2010). Two CPEB1-associated noncanonical poly(A) polymerases that promote polyadenylation are GLD2 (TENT2, PAPD4, Wispy) and GLD4 (TENT4B, PAPD5, TRF4, TUT3) (Barnard et al. 2004; Burns et al. 2011). Other members of the CPEB family of RNA binding proteins, CPEB2, CPEB3, and CPEB4 also promote polyadenylation (Pavlopoulos et al. 2011; Chen et al. 2018; Parras et al. 2018), although it is not clear whether they tether GLD2 or GLD4 to RNA.
To identify additional RNA binding proteins that might anchor GLD2 or GLD4 to RNA, we double-tagged these enzymes and expressed them in U87MG cells; GLD2 was unstable and not suitable for further analysis. GLD4 levels, however, were sufficient for two rounds of immunoprecipitation and elution followed by mass spectrometry. Compared to mock immunoprecipitation with IgG, we identified 37 RNA binding proteins that coprecipitated with GLD4. One of these proteins is FMRP, the product of the Fragile X Syndrome gene FMR1. FMRP most frequently binds coding regions of mRNAs but 5′ and 3′UTRs as well (Darnell et al. 2011; Maurin et al. 2018; Li et al. 2020a). Through its association with coding sequences, FMRP is thought to inhibit translation by impeding ribosome translocation (Darnell et al. 2011; Udagawa et al. 2013; Shah et al. 2020; Richter and Zhao 2021). The association of FMRP with 3′UTRs has been linked to RNA localization (Goering et al. 2020), and translational repression through complex interactions with miRNAs, AGO2, and the RNA binding protein MOV10 (Kenny et al. 2014). Here, we use TAIL-seq (Chang et al. 2014) from HEK cells to demonstrate that FMRP mostly inhibits polyadenylation. In contrast, direct RNA sequencing of mouse brain cortex indicates that FMRP both inhibits and activates polyadenylation, Moreover, in contrast to FMRP, we show that CPEB1 promotes polyadenylation, as expected. Our results suggest that FMRP regulates poly(A) tail length, which may have implications for the pathophysiology of Fragile X syndrome (Hagerman et al. 2017; Richter and Zhao 2021). These observations may also explain, at least in part, the fact that in mice, CPEB1 ablation “rescues” a number of Fragile X-like phenotypes in Fmr1 knockout (KO) mice (Udagawa et al. 2013). For example, protein synthesis in the Fmr1 KO mouse hippocampus is abnormally high, but in Fmr1/CPEB1 double KO mice, protein synthesis is reduced to normal levels. Going further, one form of synaptic plasticity, metabotropic glutamate receptor-mediated long term depression (mGluR-LTD), is exaggerated in Fmr1 KO hippocampus but is rescued to normal in Fmr1/CPEB1 double KO mice. Although there is a link between the rescue of Fragile X-like phenotypes in Fmr1/CPEB1 double KO mice and ribosome translocation (Udagawa et al. 2013), poly(A) tail length regulation may also be involved. Perhaps these two molecular mechanisms may be linked as well.
RESULTS
GLD4-interacting proteins
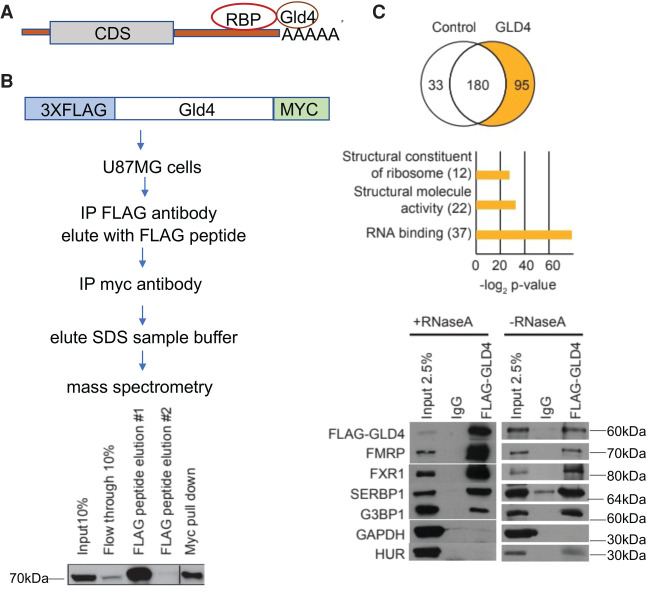
We hypothesized that GLD4 would be tethered to mRNA 3′UTRs by RNA binding proteins, which would regulate polyadenylation, possibly of specific mRNAs (Fig. 1A). To identify GLD4 interacting proteins, we expressed this enzyme, which was tagged with 3XFLAG and MYC epitopes at each end, in U87MG cells, followed by immunoprecipitation with FLAG antibody, elution with FLAG peptide, immunoprecipitation with MYC antibody, elution with SDS-containing sample buffer, and mass spectrometry (Fig. 1B; Supplemental Fig. 1A; Supplemental Table 1). There were 95 coprecipitating proteins in the GLD4-specific eluate, 49 of which have RNA or ribosome associations (Fig. 1C). Thirty-seven of these are related to RNA binding activity, several of which are cytoplasmic RNA binding proteins (Supplemental Fig. 1B). Four of these proteins, FMRP, FXR1, SERBP1, and G3BP1 coprecipitated with GLD4 in the absence or presence of RNase A (Fig. 1C).
FIGURE 1.
Identification of GLD4-interacting proteins. (A) At top is a diagram of RNA binding proteins (RBPs) that might tether GLD4 to 3′UTRs of RNAs. (B) Flow diagram to identify GLD4-associated proteins. U87MG cells were transduced with inducible pTRIPZ lentivirus expressing 3XFLAG-GLD4-myc, and the expression was induced with 1 µg/mL doxycycline for 48 h, followed by sequential FLAG immunoprecipitation, elution, MYC immunoprecipitation, elution, and mass spectrometry. The western at bottom shows amount of of tagged GLD4 present at each step. (C) Relative to an empty vector control (i.e., RFP), there were 95 GLD4-specific coprecipitating proteins, 37 of which were identified as RNA binding proteins by a gene ontology (GO) analysis. The western blots at bottom show coprecipitation of FMRP, FXR1, SERBP1, and G3BP1 with GLD4 in the absence or presence of RNase A. GAPDH and HuR as well as coprecipitation with nonspecific IgG serve as controls.
FMRP control of polyadenylation
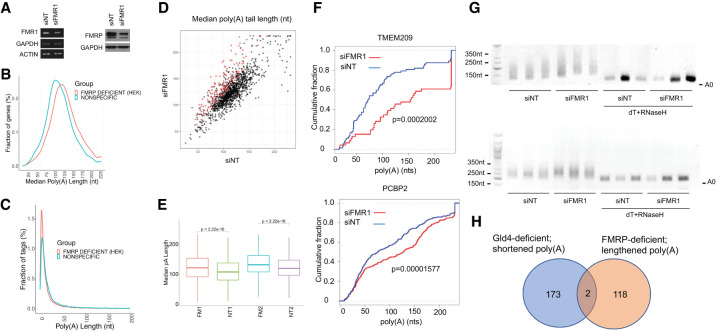
We were particularly intrigued by the coprecipitation of GLD4 with FMRP, which regulates translation at the elongation step by stalling ribosome translocation (Darnell et al. 2011; Udagawa et al. 2013; Shah et al. 2020), at initiation by association with cap-binding factors (Napoli et al. 2008), and through interactions with miRNAs/AGO on mRNA 3′UTRs (Kenny et al. 2014). Based on its interaction of GLD4, we surmised that FMRP would also regulate polyadenylation, possibly as an activator as we hypothesized that it would anchor the enzyme to RNA 3′UTRs much like CPEB1. To assess this, we isolated RNA from HEK293 cells transfected with siRNA against FMR1, which reduced FMRP by 70% relative to a nontargeting (NT) siRNA (Fig. 2A), and generated TAIL-seq libraries as presented in Chang et al. (2014) (Supplemental Table 2). Figure 2B shows the overall distribution of poly(A) tails on RNAs from control (nonspecific siRNA) compared to FMRP-deficient. Control cells had a peak poly(A) tail size of ∼105 nt while FMRP-deficient cells had a peak poly(A) tail size of ∼120 nt, a surprising increase and not the expected decrease. The fraction of sequence tags under each condition was similar (Fig. 2C). A scatter plot of the same TAIL-seq data shows that >100 specific RNAs underwent poly(A) lengthening upon FMRP deficiency (Fig. 2D), but only five underwent significant decreases in poly(A). Figure 2E shows a box plot of the differences in poly(A) between the two replicates—in both cases, FMRP deficiency resulted in an overall increase in mean poly(A) tail length. Figure 2F shows cumulative poly(A) plots of two specific RNAs TMEM209 and PCBP2, upon depletion of FMRP (siFMR1) relative to a nontargeting control (siNT). We validated the poly(A) tail size changes of these specific mRNAs by a PCR-basted RNA ligation poly(A) test (RL-PAT). siRNA for FMR1 induced a size shift in the PCR products of both RNAs (performed in triplicate), but when the poly(A) tails were removed by prior treatment with oligo(dT) and RNase H, there was no size change in the RT-PCR products (Fig. 2G). Therefore, the depletion of FMRP leads to increased poly(A) on specific mRNAs.
FIGURE 2.
FMRP control of polyadenylation in HEK cells. (A) siRNA depletion of FMR1 RNA (left, RT-PCR) and FMRP protein (right, western blot analysis). siNT refers to a nontargeting siRNA. (B) Distribution of poly(A) tail lengths of RNAs versus fraction of genes in FMRP-deficient HEK cells as determined by TAIL-seq. (C) Distribution of poly(A) tail length versus fraction of sequence tags in FMRP-deficient HEK cells. (D) Scatter plot of median poly(A) length of RNAs from FMRP-depleted versus control (nontargeting siRNA). Log2 fold change of the ratio of siFMRP/siNT >1, >0.5, or <0.5 is indicated. (E) Box plot of mean poly(A) tail length in individual replicates of TAIL-seq from FMRP-depleted (FM1 and FM2) control siRNA (NT1 and NT2) (Wilcoxon rank sum test after Bonferroni correction method). (F) Cumulative poly(A) plots of two RNAs (TMEM209 and PCBP2) following FMRP depletion (FM2). P values are from Kolmogorov–Smirnov test. (G) PAT assays for poly(A) tail length for TMEM209 and PCBP2 RNAs. Some samples were treated with oligo (dT) and RNase H to remove poly(A) prior to the PAT assay. (H) Venn diagram for RNAs whose poly(A) tails are regulated by FMRP (this study) or GLD4 (Shin et al. 2017).
We determined whether the RNA substrates, which have elongated poly(A) tails upon FMRP depletion, are the same as those that have shortened poly(A) tails when GLD4 is depleted (Shin et al. 2017). Figure 2H shows that surprisingly, there are only two RNAs whose poly(A) tail length is regulated in opposite directions by these two proteins. However, we note that polyadenylation in GLD4-depleted cells was estimated by binding RNAs to poly(U) agarose beads followed by washing at 50°C and elution at 65°C. This thermal elution procedure, while not suitable for precise poly(A) length determination, is nonetheless adequate for assessing the general changes in tail growth or removal (McFleder et al. 2017). These data indicate that although GLD4 and FMRP associate with one another, their respective activities to regulate poly(A) are not mutually dependent.
To assess whether RNAs whose poly(A) tail lengths are lengthened upon Fmr1 depletion are bound by FMRP, we first determined whether there is overlap between these transcripts and those that were identified by FMRP PAR-CLIP in HEK cells (Ascano et al. 2012). Supplemental Figure 2A shows that there is a small but statistically significant (P < 0.0001) overlap between these two data sets. We also performed FMRP RNA immunoprecipitation (RIP) for the top eight RNAs whose polyadenylation is lengthened by Fmr1 depletion (Supplemental Fig. 2B–D). Only two of these (creatine kinase B, CKB; ATP citrate lyase, ACLY) were coimmunoprecipitated with FMRP relative to a background control (nonspecific IgG). Both of these RNAs, as well as HSP90, which coimmunoprecipitated with FMRP but whose poly(A) tail was unaffected by Fmr1 depletion, are on the FMRP CLIP list (Supplemental Fig. 2C; Darnell et al. 2011). Six other RNAs whose poly(A) tails were regulated by FMRP were not coprecipitated with FMRP above background (Supplemental Fig. 2D). Finally, we performed a HOMER analysis for sequence motifs that might be enriched in the 3′UTRs of mRNAs whose poly(A) tails were lengthened by Fmr1 depletion. Considering the top 100 RNAs in this category, Supplemental Figure 3 shows a number of enriched motifs, some of which resemble previously described FMRP binding motifs (Anderson et al. 2016; Liu et al. 2018). However, a further mutatational analysis is required to determine whether these motifs are FMRP tethering sites for polyadenylation.
Direct RNA sequencing in Fmr1-deficient mouse cortex
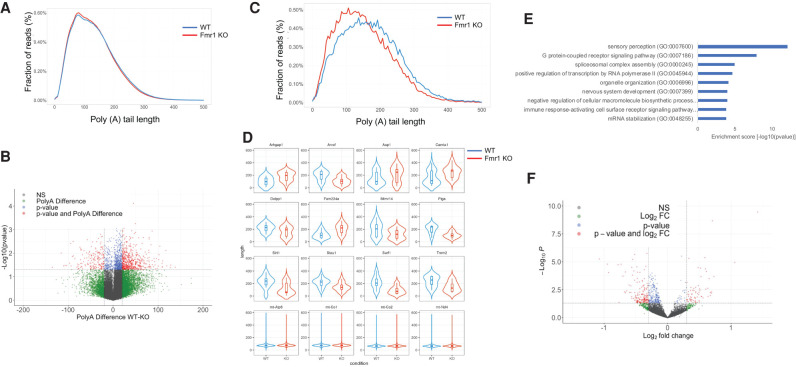
Determination of poly(A) tail size by TAIL-seq relies on an algorithm that accurately reads long polymeric T stretches that is often unreliable with conventional base calling software (Chang et al. 2014). Direct RNA sequencing by nanopore technology (Garalde et al. 2018) reads long poly(A) stretches with reasonably high fidelity (Bilska et al. 2020; Depledge and Wilson 2020; Li et al. 2020b) and does not require PCR amplification, which can be biased to shorter RNAs. Consequently, we performed direct RNA sequencing to assess poly(A) tail length from the brain cortex of WT and Fmr1 knockout (KO) mice (all in biologic triplicates). With an average of 3.3 million reads per brain cortex sample, we detected 30,975 transcripts in common between WT and KO samples originating from 15,126 genes. In addition, 292 genes are expressed exclusively in the Fmr1 KO cortex while 861 genes that are present in the WT cortex are not detected in the KO. Nevertheless, most of these unique transcripts are expressed at very low levels (typically <1 rpm) (Supplemental Table 3). Next, we analyzed all the reads using Nanopolish, an algorithm that estimates tail length based on the low-variance ionic current that is registered as the mRNA tail passes through the nanopore (Workman et al. 2019). Poly(A) tail lengths transcriptome-wide were similar between the two genotypes and ranged from a minimum of 3 to a maximum of ∼1700 adenylate residues. The vast majority of transcripts showed poly(A) tail lengths that ranged from 50–150 adenylate residues and peaked at around 80 residues (Fig. 3A). Using a cutoff of P < 0.05, we detected 1409 isoforms that showed differential genotype-specific poly(A) length changes. Of these, more than half exhibited an average change in tail length that exceeded 20 residues. In contrast, tail lengths of mitochondrial RNAs as well as the vast majority of transcripts were unchanged between the two conditions (Fig. 3D; Supplemental Table 3). Within the set of differentially polyadenylated transcripts, 179 RNAs had longer poly(A) tails in the Fmr1 KO cortex (maximal increase, 135 residues), while 558 RNAs had shorter poly(A) in the Fmr1 KO cortex (maximal decrease averaged 138 residues) (Fig. 3B,C; Supplemental Table 3). Figure 3D shows violin plots for several RNAs demonstrating increased or decreased poly(A) in the Fmr1 KO cortex relative to WT. The poly(A) tails of several mitochondrial RNAs are the same irrespective of genotype. Figure 3E shows a Gene Ontology (GO) analysis of RNAs with altered poly(A), which encode molecules involved in neural function (sensory perception, nervous system), RNA regulation (splicing complex, mRNA stabilization), and other activities. Finally, we also found that the levels of ∼100 RNAs were altered, either up or down, by at least 2.5-fold in Fmr1 KO cortex relative to WT (Fig. 3F; Supplemental Fig. 4). There was no overlap with the RNAs that had altered levels with those that had a change in poly(A) tail length (Supplemental Fig. 4). These data demonstrate that in the mouse cortex, FMRP regulates bidirectional poly(A) tail length.
FIGURE 3.
Determination of poly(A) tail length in Fmr1 KO cortex by direct RNA sequencing using a nanopore platform. (A) Distribution of poly(A) tail lengths of RNAs in WT and Fmr1 KO brain cortex as a fraction of total reads excluding mitochondrial RNAs. (B) Volcano plot showing RNAs that undergo poly(A) tail size changes in WT compared to Fmr1 KO brain cortex. In red, transcripts that show differential poly(A) tail lengths of at least 20 residues (P < 0.05; two tailed t-test). (C) Distribution of poly(A) tail lengths of differentially polyadenylated transcripts (n = 736) versus fraction of total reads in WT and Fmr1 KO cortex. (D) Violin plots with box plot for top differential poly(A) transcripts with P < 0.05 (two tailed t-test) as in B; reads for each transcript were pooled for the two genotypes. (E) Gene Ontology (GO) term analysis of RNAs that undergo poly(A) tail size changes in Fmr1 KO cortext compared to WT. (F) Volcano plot of RNAs that have altered expression levels in Fmr1 KO cortext compared to WT. For all comparisons, 2 WT and 3 Fmr1 KO cortexes were analyzed.
CPEB1 regulation of polyadenylation
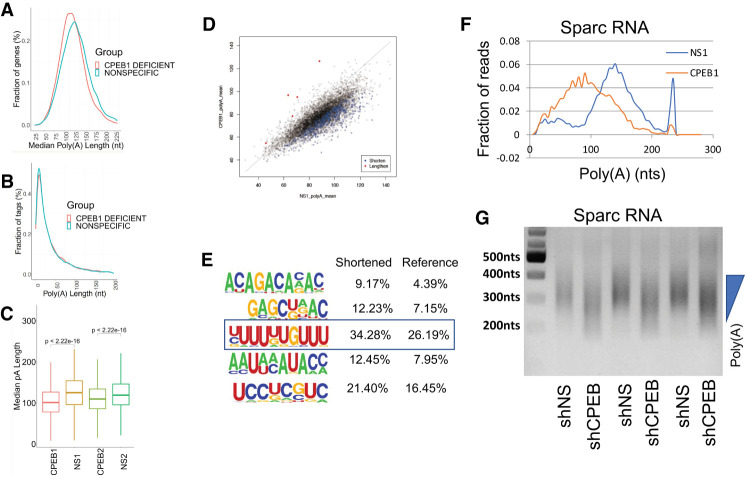
To assess whole transcriptome changes in polyadenylation by another RNA binding protein, we performed TAIL-seq from CPEB1-deficient mouse hippocampal neurons. As expected, CPEB1 deficiency induced an overall shortening of poly(A) (Fig. 4A; Supplemental Table 4) with a similar fraction of sequence tags (Fig. 4B). Box plots of replicate experiments in Figure 4C show that CPEB1 deficiency caused an overall poly(A) reduction of about 25 nt. The scatter plot in Figure 4D demonstrates that 462 RNAs underwent poly(A) shortening in CPEB1-deficient neurons. A motif analysis of the RNAs that underwent poly(A) shortening in CPEB1-deficient neurons shows a few diverse sequences including a U-rich CPE- like element (boxed), which is the binding site for CPEB1. One RNA that displayed prominent poly(A) shortening upon CPEB1 deficiency is SPARC (secreted protein, acidic and rich in cysteine), from a mean length of∼145 nt to a mean length of ∼85 nt (Fig. 4F). Poly(A) shortening of Sparc RNA was validated in an RL-PAT assay performed in biologic triplicate (Fig. 4G). These data demonstrate that in contrast to FMRP, CPEB1 almost exclusively promotes general as well as mRNA-specific poly(A) elongation.
FIGURE 4.
TAIL-seq analysis of poly(A) tail length in cultured neurons depleted of CPEB1. (A) Distribution of poly(A) tail lengths of RNAs versus fraction of genes in CPEB1-deficient rat hippocampal neurons as determined by TAIL-seq. (B) Distribution of poly(A) tail length versus fraction of sequence tags in CPEB1-deficient hippocampal neurons. (C) Box plots of median poly(A) tail distribution in biologic replicates of CPEB1-deficient neurons (CPEB1) compared to a nonspecific siRNA control (NS1 NS2) (Wilcoxon rank sum test after Bonferroni correction method). (D) Scatter plot of poly(A) tail lengths in CPEB1-depleted neurons. (E) Motif analysis of RNAs whose poly(A) tails were shortened following CPEB1 depletion. The boxed motif contains the putative binding site for CPEB1. (F) Distribution of poly(A) of SPARC RNA in CPEB1-deficient neurons. (G) PAT assay of SPARC RNA performed in biologic triplicate from CPEB1-deficient neurons.
DISCUSSION
CPEB1 is the prototypical RNA binding protein that anchors noncanonical poly(A) polymerases to the 3′UTRs to facilitate cytoplasmic polyadenylation. In our quest to identify additional RNA binding proteins with a similar activity to CPEB1, we performed double-tag GLD4 pulldowns and mass spectrometry, which identified 27 RNA binding proteins. Nearly half of these proteins are nucleus-cytoplasm shuttling proteins, including FMRP and its paralog FXR1. These results led us to consider whether these two proteins regulate polyadenylation, particularly FMRP because its absence results in the Fragile X Syndrome, a neurodevelopmental disorder that is the most common inherited form of intellectual impairment. The syndrome is also characterized by speech and developmental delays, perseveration, and autism (Hagerman et al. 2017; Richter and Zhao 2021). FMRP is encoded by FMR1, which in humans, typically harbors ∼50 or fewer CGG triplets in the 5′UTR. However, when these triplets expand to 200 or more, FMR1 is methylated and transcriptionally silenced; thus, loss of FMRP drives the disorder. Although FMRP interacts predominantly with coding regions of mRNAs, it also associates with 3′-UTRs (Darnell et al. 2011; Maurin et al. 2018; Li et al. 2020a), suggesting that it could, like CPEB1, tether noncanonical poly(A) polymerases to regulate polyadenylation.
In HEK293 cells, depletion of FMRP elicits poly(A) elongation for most mRNAs, not an expected shortening that occurs when CPEB1 is depleted (Tay and Richter 2001). This observation suggests that FMRP does not merely tether a constitutively active poly(A) polymerase to RNA 3′UTRs, which is the case for CPEB1. In contrast, FMRP may interfere with the activity of a polymerase directly through protein–protein interactions, or indirectly by altering the activity or expression of other proteins that in turn modulate polyadenylation activity. Because FMRP and GLD4 do not regulate the poly(A) tails of the same RNAs, FMRP likely binds other poly(A) polymerases. In any event, the regulation of poly(A) tail length by FMRP represents a previously little known function of the protein.
In contrast to HEK293 cells, the situation in the mouse brain cortex is more complex in that FMRP inhibits and promotes polyadenylation of specific RNAs. There are several differences in methodologies as well as cell and tissue type that may explain this apparent discrepancy. TAIL-seq, which was used to assess poly(A) in FMRP-depleted HEK293 cells and direct RNA sequencing with a nanopore platform, which was used to determine poly(A) in Fmr1 KO mouse brain cortex, use widely divergent technologies and base calling software. These two approaches would likely account for at least some of the inconsistencies in poly(A) length determination. Probably more important, however, are the different cells/tissues that were used for the analysis. Rapidly dividing transformed HEK293 cells in culture are substantially different than mouse brain cortex containing many cell types that divide slowly if at all. In addition, each cell/tissue type has a different repertoire of factors that may be dysregulated upon FMRP depletion that in turn affects poly(A). For example, the poly(A) binding protein Pab1c is down-regulated in Fmr1 KO brain cortex (Supplemental Fig. 2B) but not in FMRP-depleted HEK cells. Motif analysis identified PABPC1 motifs are significantly enriched on mRNA 3'UTRs with longer poly(A) tails in Fmr1-depleted HEK cells (Supplemental Fig. 3A). This protein might stabilize poly(A) on some mRNAs, and its loss would be expected to allow exonuclease activity to shorten the tails.
Loss of FMRP from the cortex altered the expression of only a small set of mRNAs above or below a 2.5-fold threshold. The most highly affected gene was Transthyretin (TTR), which was also found to be also maximally affected upon human FMR1 overexpression in a transgenic mouse model (Fernández et al. 2012). In the Fmr1 KO cortex, ∼1400 transcripts underwent significant changes in poly(A) tail length and, as expected, most had shorter tails. This shortening did not correlate with mRNA expression because loss of FMRP affected the levels of only a small set of mRNAs (±2.5-fold) (Fig. 3). Interestingly, for transcripts where the poly(A) shortening exceeded 20 residues, steady state tail lengths in WT cortex averaged ∼160–200 residues compared to the global average of ∼80 Å residues. Thus, mRNAs with longer tails at steady state may be targets of FMRP regulation as the average tail length of these transcripts dropped to ∼100 residues in the Fmr1 KO cortex.
Depletion of CPEB1 from Fmr1 KO mice rescues several pathological phenotypes associated with Fragile X Syndrome such as memory impairment, synapse number, and synaptic plasticity (Udagawa et al. 2013). The underlying molecular mechanism(s) of this rescue is not clear, although the transit rates of ribosomes may be involved (Udagawa et al. 2013). The results presented here suggest that poly(A) tail length, at least in principle, could be involved in the rescue. Although poly(A) tail length can regulate the rate of translation initiation, it is unclear to what extent it may influence polypeptide elongation (i.e., rate of ribosome translocation). It is possible these two events are linked, although detailed quantitative measurements would be required to determine whether this is the case. Even so, it is unlikely any one molecular mechanism is responsible for ameliorating Fragile X syndrome in mice especially given that 15 genetic or pharmacologic rescue paradigms have been published (Richter et al. 2015).
Finally, there are 11 terminal nucleotidyl transferase families encoded by the human genome (Liudkovska and Dziembowski 2020; Yu and Kim 2020) and probably at least 1542 RNA binding proteins (Gerstberger et al. 2014). Although not all TENTs catalyze polyadenylation (some promote polyuridylation), their association with different combinations of RNA binding proteins suggests an enormous complexity of polynucleotide addition to mRNA 3′ ends. These post-transcriptional modifications may influence mRNA translation and turnover as well as subcellular localization. Moreover, cell- or tissue-specific signaling events could result in changes in polyadenylation, For example, cell surface receptor signaling induces CPEB1 phosphorylation that in turn promotes polyadenylation (Mendez et al. 2000). FMRP also undergoes receptor signaling-induced phosphorylation, but in this case, causes its destruction (Huang et al. 2015), which would in turn produce alterations in polyadenylation similar to an Fmr1 KO mouse. Therefore, RNA binding proteins in combination with TENTs have the potential to greatly regulate gene expression in time and space.
MATERIALS AND METHODS
Tagged GLD4 immunoprecipitation and mass spectrometry
3XFLAG-GLD4-myc sequence was subcloned from p3xFLAG-myc CMV26 GLD4 (Burns et al. 2011) and inserted into pTRIPZ vector using AgeI and XhoI sites. U87MG cells were transduced with inducible pTRIPZ lentivirus expressing RFP (control) or 3XFLAG-GLD4-myc, and the expression was induced with 1 µg/mL doxycycline for 48 h. Freshly lysed cells were incubated with FLAG antibody conjugated Dynabeads G (Life Technologies), washed and eluted with 3X FLAG peptides (Sigma) followed by a second round of coimmunoprecipitation with myc antibody conjugated Dynabeads (NEB). Affinity purified protein complexes were electrophoresed a short distance into a nondenaturing gel. After in-gel digest, the peptides were run on the QExactive (Thermo Scientific) LC–MS/MS at the proteomics and mass spectrometry facility at the University of Massachusetts Medical School. Data were analyzed with an intensity-based absolute quantification method, and differential protein abundance was calculated as fold change relative to the control using Scaffold software (Proteome Software).
Cell and neuronal culture
U87MG human glioblastoma cells (a gift of Dr. Alonzo Ross, University of Massachusetts Medical School) and HEK293T human embryonic kidney cells (ATCC) were cultured in DMEM supplemented with 10% FBS and 1% antibiotic/antimycotic compounds. Cell transfection was performed with Lipofectamine 2000 (Invitrogen) for U87MG cells and calcium phosphate precipitation for HEK293 cells. The culture of primary hippocampal neurons was performed as described (Huang and Richter 2007) in neurobasal media (Invitrogen) containing B27 supplement (B27 media) and glutamine (1 µg/mL).
siRNA and shRNA depletions
Cells were transfected with siRNAs by Dharmafect 1 and typically incubated 72–96 h. Oligonucleotide siRNA sequences were: siFMR1, 5′-GCACUAAGUUGUCUCUGAU-3′; siFXR1, 5′-CGAGCUGAGUGAUUGGUCA-3′; siAUF1, 5′-GAUUGACGCCAGUAAGAAC-3′. siGLD4 was ON-TARGETplus SMARTpool siRNA from Dharmacon. Lentiviral shRNAs targeting CPEB are described elsewhere (Udagawa et al. 2012). The culture of primary hippocampal neurons was performed as described (Huang and Richter 2007) in neurobasal media (Invitrogen) containing B27 supplement (B27 media) and glutamine (1 µg/mL). After 14 d, hippocampal neurons were infected with lentivirus expressing shRNA against CPEB and nonspecific shRNA as a control. All oligonucleotide seqences are provided in the Supplemental Information except GLD4 and CPEB1, which were ON-TARGET plus SMARTpool siRNAs from Dharmacon.
Coimmunoprecipitation
Cells were cotransfected with cDNAs encoding FLAG-GLD4 and/or GFP-CPEB1. Twenty-four hours later, the cells were lysed with lysis buffer (25 mM Hepes pH 7.4, 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM DTT, 1 mM MgCl2) supplemented with protease (Roche) and phosphatase (Calbiochem) inhibitors. Cell lysates were subjected to coimmunoprecipitation either with or without RNase A (50 µg/mL, Sigma-Aldrich) overnight at 4°C, and the eluate was used for western blot analysis to confirm the interaction.
For other experiments, HEK cells were washed 100 µg/mL cycloheximide (CHX) containing phosphate-buffered saline (PBS) and then lysed in lysis buffer [20 mM Tris-HCl pH 7.5, 5 mM MgCl2, 100 mM KCl, 1 mM DTT, 100 µg/mL CHX, EDTA-free protease inhibitor complex, 1% Triton X-100(v/v)]. The lysates were pipetted and titurated through a 25G needle. The lysates were centrifuged at 4°C for 10 min at 14,000g. The supernatant was precleared with Dynabeads Protein G (Invitrogen) and incubated with FMRP antibody (Abcam, ab17722), and normal rabbit IgG as a negative control. The Dynabeads were added to RNA–protein–antibody complexes and the RNA–protein complexes were eluted with TRIzol. FMRP coimmunoprecipitated RNAs were measured by RT-qPCR with GAPDH as the RNA standard.
Western blotting
Immunoblotting was performed with antibodies against FLAG (Sigma), GAPDH (Cell Signaling), GFP (Abcam), FMRP (Abcam), SYMPLEKIN (BD), eIF4E (BD), AUF1 (a gift of Dr. Gary Brewer, Rutgers University), FXR1 (Bethyl), SERBP1 (Bethyl), G3BP1 (Bethyl), and HuR (Bethyl). Band intensity was quantified using ImageJ densitometry software.
TAIL-seq
The culture of primary hippocampal neurons was performed as described (Huang and Richter 2007) in neurobasal media (Invitrogen) containing B27 supplement (B27 media) and glutamine (1 µg/mL). After 14 d, hippocampal neurons were infected with lentivirus expressing shRNA against CPEB and nonspecific shRNA as a control. After 4 d of infection, cells were harvested and total RNA was extracted. TAIL-seq libraries were prepared, sequenced and analyzed as described (Chang et al. 2014). All TAIL-seq data were assessed in biologic duplicate.
PAT assays
RNA ligation-mediated polyadenylation assays (RL-PAT) were performed as described elsewhere (Yamagishi et al. 2016). In brief, 2–10 µg of total RNA was ligated to 5′ phosphorylated anchor primer by T4 RNA ligase 1 (NEB). The ligated RNAs were used for a reverse transcriptase reaction using anti-anchor primer and Superscript III. A total of 1–2 µL of cDNAs were amplified using a gene-specific forward primer and a universal anchor antisense reverse primer. PCR products were separated in 2% agarose gel to detect poly(A) tail length. All the oligonucleotide sequences used for RL-PAT assay are in Supplemental File 2.
Motif enrichment analysis
The top 100 RNAs whose poly(A) tails were regulated upon Fmr1 depletion as assessed by TAIL-seq were analyzed using HOMER motif analysis (http://homer.ucsd.edu/homer/motif/). The 3′-UTR sequences and coding sequences for these RNAs were extracted and deposited into a fasta format using ENSEMBL BioMart (https://m.ensembl.org/info/data/biomart/index.html). Coding sequences were used as background for the comparison.
Nanopore direct RNA sequencing and sequencing and analysis
RNA was sequenced directly using a Nanopore PromethION instrument. Poly(A) RNA was isolated from mouse brain cortex (WT and Fmr1 KO, both C57BL/6J) using µMACS mRNA Isolation kit (130-075-101). Five hundred nanograms was used for library preparation for direct RNA sequencing with SQK-RNA002 kit (Oxford Nanopore). Sequencing was performed using PromethION with the FLO_PRO002 flow cell (Oxford Nanopore). Base calling of the Fast5 data outputs from PromethION was performed using Guppy (v4.0.11) and then converted into Fastq files. Reads were aligned to the reference mouse transcriptome mm10 (Ensembl) using Minimap2 (v2.17) with the following flags: “-ax map-ont -N 100”, and only primary alignments were kept. Sorting and indexing of reads were performed using Samtools (v1.9). Poly(A) length estimation was performed using Nanopolish (v0.13.2) using the default setting. Transcripts with less than 10 reads per condition were filtered out. t-tests were used on average poly(A) length in each sample to identify statistically significant genotype-dependent poly(A) length changes of at least 20 residues (P < 0.05). Isoform quantification was performed using Salmon (v1.5.2) in alignment-based mode and differential isoform expression was performed using DESEQ2. Nanopore sequencing was performed with biologic triplicates.
DATA DEPOSITION
The Gene Expression Omnibus (GEO) accession number for the data reported in this paper is GSE 188840.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to V. Narry Kim and Hyeshik Chang for TAIL-seq analysis of polyadenylation and Ozkan Aydemir for discussions of bioinformatics. This work was supported by National Institutes of Health (NIH) grants GM046779 and GM135087 (to J.D.R.), Region AURA Pack Ambition international 2020 and ATIP-Avenir grants (to K.P.), 16H06279 PAGS (to Y.S.), and by a PhD fellowship from Fondation Recherche Medicale (to L.C.).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079050.121.
MEET THE FIRST AUTHOR
Lies Chikhaoui.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Lies Chikhaoui is one of the three co-first authors of this paper, “Oppositional poly(A) tail length regulation by FMRP and CPEB1,” along with Jihae Shin and Ki Young Paek. Lies is a PhD student in Kiran Padmanabhan's lab at the Institute for Functional Genomics of Lyon (ENS Lyon). He studies pathways that regulate dynamic mRNA processing using 4th generation direct RNA sequencing.
What are the major results described in your paper and how do they impact this branch of the field?
FMRP (fragile X mental retardation protein) is an RNA binding protein essential for normal cognitive development. My colleagues at UMass Med School found it interacts with the cytoplasmic poly(A) polymerase GLD4. Loss of FMRP in HEK293 cells in their experiments led to overall poly(A) lengthening. Using direct RNA nanopore sequencing to quantify tail length, my analysis showed that in the cortex of FMRP knockout mice, mRNAs had both longer and shorter tails, suggesting that FMRP has an additional previously unappreciated role in poly(A) tail dynamics.
What led you to study RNA or this aspect of RNA science?
While working on circadian transcriptomes in collaboration with the lab of Dr. Yutaka Suzuki, I realized that direct RNA sequencing could fairly accurately determine poly(A) tail length in intact mRNAs without any processing. Therefore, we approached the Richter lab to use the FMRP study as a test bed.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
FMRP knockdown experiments in HEK cells revealed that the protein primarily played a role in poly(A) tail lengthening (using TAIL-seq, an alternative method for tail length determination). Surprisingly, in the mouse cortex, FMRP deletion revealed a regulatory role in tail length determination, with some mRNAs gaining stretches of A's while others ended up with shorter poly(A) tails.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
One of the key events that has had a major impact in my young career was an accidental loss of samples. At the beginning of my PhD, I visited the lab of Dr. Yutaka Suzuki in Japan with the aim to perform ChIP-seq and to learn bioinformatic analyses methods. The samples were damaged during shipping and this stopped my plans dead in their tracks. Instead, a secondary project involving direct RNA nanopore sequencing took off (because the samples survived!), and I have spent the last two years applying this powerful technique to biological problems, including the one on FMRP.
What are your subsequent near- or long-term career plans?
While I was still at university I decided to get some research experience—a year before most of my classmates. At that time, I was absolutely sure that I wanted to do academic research, and I thought it was time to see what the big bad wolf looked like before I started my career, so I went to get some experience in industry. I realized that in fact R&D is just a continuation of what we do in academia, with an incredible amount of knowledge generated by researchers, turned into applications by companies. This experience changed my view of science, and my career plans with it.
From these experiences, I feel I want to follow the path of science down to application, namely to grow and take full advantage of the freedom that a PhD offers, to develop my critical thinking skills in the free and stimulating environment of academia, and then try to have a more direct and concrete impact on people's lives by putting these skills to use in R&D.
I feel that I have taken full advantage of the scientific freedom that an academic environment offers to grow and mature as a scientist. I am now turning to the second stage of my career plan: apply these skills to the benefit of the community. In this regard, I think nanopore tech will serve this purpose greatly as I would like to find game-changing applications.
What were the strongest aspects of your collaboration as co-first authors?
Each one of us brought a different set of skills to interrogate the same scientific problem, and the net result is that we have identified a previously unknown regulatory role for FMRP. This publication for me is the best illustration of how collaboration takes science up to the next level. Each of us had an area of expertise that allowed us to work efficiently on the problem, and we were helped along by experience of the conductors, Dr. Padmanabhan and Dr. Richter.
REFERENCES
- Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. 2016. Identification of consensus binding sites clarifies FMRP binding determinants. Nucl Acids Res 44: 6649–6659. 10.1093/nar/gkw593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascano M, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, et al. 2012. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature 492: 382–386. 10.1038/nature11737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard DC, Ryan K, Manley JL, Richter JD. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651. 10.1016/j.cell.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Bilska A, Kusio-Kobiałka M, Krawczyk PS, Gewartowska O, Tarkowski B, Kobyłecki K, Nowis D, Golab J, Gruchota J, Borsuk E, et al. 2020. Immunoglobulin expression and the humoral immune response is regulated by the non-canonical poly(A) polymerase TENT5C. Nat Commun 11: 2032. 10.1038/s41467-020-15835-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, Richter JD. 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev 22: 3449–3460. 10.1101/gad.1697808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns DM, D'Ambrogio A, Nottrott S, Richter JD. 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473: 105–108. 10.1038/nature09908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Chen HF, Hsu CM, Huang YS. 2018. CPEB2-dependent translation of long 3′-UTR Ucp1 mRNA promotes thermogenesis in brown adipose tissue. EMBO J 37: e99071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. 2011. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 146: 247–261. 10.1016/j.cell.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depledge DP, Wilson AC. 2020. Using direct RNA nanopore sequencing to deconvolute viral transcriptomes. Curr Protoc Microbiol 57: e99. 10.1002/cpmc.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández JJ, Martínez R, Andújar E, Pérez-Alegre M, Costa A, Bonilla-Henao V, Sobrino F, Pintado CÓ, Pintado E. 2012. Gene expression profiles in the cerebellum of transgenic mice over expressing the human FMR1 gene with CGG repeats in the normal range. Genet Mol Res 11: 467–483. 10.4238/2012.March.1.4 [DOI] [PubMed] [Google Scholar]
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, et al. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15: 201–206. 10.1038/nmeth.4577 [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. 2014. A census of human RNA-binding proteins. Nat Rev Genet 15: 829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goering R, Hudish LI, Guzman BB, Raj N, Bassell GJ, Russ HA, Dominguez D, Taliaferro JM. 2020. FMRP promotes RNA localization to neuronal projections through interactions between its RGG domain and G-quadruplex RNA sequences. Elife 9: e52621. 10.7554/eLife.52621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman RJ, Berry-Kravis E, Hazlett HC, Bailey DB, Moine H, Kooy RF, Tassone F, Gantois I, Sonenberg N, Mandel JL, et al. 2017. Fragile X syndrome. Nat Rev Dis Primers 3: 17065. 10.1038/nrdp.2017.65 [DOI] [PubMed] [Google Scholar]
- Huang YS, Richter JD. 2007. Analysis of mRNA translation in cultured hippocampal neurons. Methods Enzymol 431: 143–162. [DOI] [PubMed] [Google Scholar]
- Huang YS, Jung MY, Sarkissian M, Richter JD. 2002. N-methyl-D-aspartate receptor signaling results in Aurora kinase-catalyzed CPEB phosphorylation and αCaMKII mRNA polyadenylation at synapses. EMBO J 21: 2139–2148. 10.1093/emboj/21.9.2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ikeuchi Y, Malumbres M, Bonni A. 2015. A Cdh1-APC/FMRP ubiquitin signaling link drives mGluR-dependent synaptic plasticity in the mammalian brain. Neuron 86: 726–739. 10.1016/j.neuron.2015.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivshina M, Lasko P, Richter JD. 2014. Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30: 393–415. 10.1146/annurev-cellbio-101011-155831 [DOI] [PubMed] [Google Scholar]
- Jalkanen AL, Coleman SJ, Wilusz J. 2014. Determinants and implications of mRNA poly(A) tail size—does this protein make my tail look big? Semin Cell Dev Biol 34: 24–32. 10.1016/j.semcdb.2014.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, Zhou H, Kim M, Skariah G, Khetani RS, Drnevich J, Arcila ML, Kosik KS, Ceman S. 2014. MOV10 and FMRP regulate AGO2 association with microRNA recognition elements. Cell Rep 9: 1729–1741. 10.1016/j.celrep.2014.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183. 10.1016/j.molcel.2006.08.016 [DOI] [PubMed] [Google Scholar]
- Li M, Shin J, Risgaard RD, Parries MJ, Wang J, Chasman D, Liu S, Roy S, Bhattacharyya A, Zhao X. 2020a. Identification of FMR1-regulated molecular networks in human neurodevelopment. Genome Res 30: 361–374. 10.1101/gr.251405.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Ren X, Ding Q, Bi Y, Xie D, Zhao Z. 2020b. Direct full-length RNA sequencing reveals unexpected transcriptome complexity during Caenorhabditis elegans development. Genome Res 30: 287–298. 10.1101/gr.251512.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Li Y, Stackpole EE, Novak A, Gao Y, Zhao Y, Zhao X, Richter JD. 2018. Regulatory discrimination of mRNAs by FMRP controls mouse adult neural stem cell differentiation. Proc Natl Acad Sci 115: E11397–E11405. 10.1073/pnas.1809588115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liudkovska V, Dziembowski A. 2020. Functions and mechanisms of RNA tailing by metazoan terminal nucleotidyltransferases. Wiley Interdiscip Rev RNA 12: e1622. 10.1002/wrna.1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurin T, Lebrigand K, Castagnola S, Paquet A, Jarjat M, Popa A, Grossi M, Rage F, Bardoni B. 2018. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res 46: 6344–6355. 10.1093/nar/gky267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFleder RL, Mansur F, Richter JD. 2017. Dynamic control of dendritic mRNA expression by CNOT7 regulates synaptic efficacy and higher cognitive function. Cell Rep 20: 683–696. 10.1016/j.celrep.2017.06.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. 2000. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404: 302–307. 10.1038/35005126 [DOI] [PubMed] [Google Scholar]
- Napoli I, Mecaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. 2008. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell 134: 1042–1054. 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- Novoa I, Gallego J, Ferreira PG, Mendez R. 2010. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol 12: 447–456. 10.1038/ncb2046 [DOI] [PubMed] [Google Scholar]
- Parras A, Anta H, Santos-Galindo M, Swarup V, Elorza A, Nieto-González JL, Picó S, Hernández IH, Díaz-Hernández JI, Belloc E, et al. 2018. Autism-like phenotype and risk gene mRNA deadenylation by CPEB4 mis-splicing. Nature 560: 441–446. 10.1038/s41586-018-0423-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. 2011. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 147: 1369–1383. 10.1016/j.cell.2011.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Zhao X. 2021. The molecular biology of FMRP: new insights into fragile X syndrome. Nat Rev Neurosci 22: 209–222. 10.1038/s41583-021-00432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Bassell GJ, Klann E. 2015. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci 16: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S, Molinaro G, Liu B, Wang R, Huber KM, Richter JD. 2020. FMRP control of ribosome translocation promotes chromatin modifications and alternative splicing of neuronal genes linked to autism. Cell Rep 30: 4459–4472. 10.1016/j.celrep.2020.02.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Paek KY, Ivshina M, Stackpole EE, Richter JD. 2017. Essential role for non-canonical poly(A) polymerase GLD4 in cytoplasmic polyadenylation and carbohydrate metabolism. Nucleic Acids Res 45: 6793–6804. 10.1093/nar/gkx239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Richter JD. 2001. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell 1: 201–213. 10.1016/S1534-5807(01)00025-9 [DOI] [PubMed] [Google Scholar]
- Udagawa T, Swanger SA, Takeuchi K, Kim JH, Nalavadi V, Shin J, Lorenz LJ, Zukin RS, Bassell GJ, Richter JD. 2012. Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Mol Cell 47: 253–266. 10.1016/j.molcel.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, et al. 2013. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med 19: 1473–1477. 10.1038/nm.3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, et al. 2019. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi R, Tsusaka T, Mitsunaga H, Maehata T, Hoshino S. 2016. The STAR protein QKI-7 recruits PAPD4 to regulate post-transcriptional polyadenylation of target mRNAs. Nucleic Acids Res 44: 2475–2490. 10.1093/nar/gkw118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Kim VN. 2020. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat Rev Mol Cell Biol 21: 542–556. 10.1038/s41580-020-0246-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.