Abstract
PIWI-interacting RNAs (piRNAs), which are germ cell-specific small RNAs, are essential for spermatogenesis. In fetal mouse germ cells, piRNAs are synthesized from sense and antisense RNAs of transposable element sequences for retrotransposon silencing. In a previous study, we reported that transgenic mice expressing antisense-Dnmt3L under the control of the Miwi2 promoter (Tg-Miwi2P-asDnmt3L) exhibited piRNA-mediated DNMT3L down-regulation. In this study, two transgene integration loci (B3 and E1) were identified on chromosome 18 of the Tg-Miwi2P-asDnmt3L mice; these loci were weak piRNA clusters. Crossbreeding was performed to obtain mice with the transgene cassette inserted into a single locus. DNMT3L was silenced and spermatogenesis was severely impaired in mice with the transgene cassette inserted at the B3 locus (Tg-B mice). In contrast, spermatogenesis in mice bearing the transgene at the E1 locus (Tg-E mice) was normal. The number of piRNAs for Dnmt3L in Tg-B mice was eightfold higher than that in Tg-E mice. Therefore, both gene silencing and impaired spermatogenesis depended on the transgene copy number rather than on the insertion loci. Additionally, the endogenous Dnmt3L promoter was not methylated in Tg mice, suggesting that Dnmt3L silencing was caused by post-transcriptional gene silencing. Based on these data, we discuss a piRNA-dependent gene silencing mechanism against novel gene insertions.
Keywords: piRNA, PIWI, transcriptional gene silencing (TGS), post-transcriptional gene silencing (PTGS), germ cell
INTRODUCTION
PIWI-interacting RNAs (piRNAs), a class of germ cell-specific small RNAs, regulate spermatogenesis and maintain genome integrity by repressing transposons (Castaneda et al. 2011; Chuma and Nakano 2013; Toth et al. 2016). Approximately half of the human and mouse genomes comprise transposons and transposon fragments as part of their repetitive sequences (Mouse Genome Sequencing et al. 2002). The transcription of transposons is repressed through piRNA-dependent DNA methylation and repressive histone modifications that play crucial roles in mouse spermatogenesis (Aravin et al. 2007, 2008; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008; De Fazio et al. 2011; Reuter et al. 2011; Seisenberger et al. 2012; Zheng and Wang 2012; Di Giacomo et al. 2013; Pezic et al. 2014). In addition to their role in epigenetic gene silencing, piRNAs mediate the RNA degradation of target transposons. piRNA-PIWI family protein complexes mediate transcriptional gene silencing (TGS) and post-transcriptional gene silencing (PTGS) (Senti et al. 2015; Czech and Hannon 2016).
There are three members of the mouse PIWI family of proteins, namely, mouse Piwi (MIWI), MIWI-like (MILI), and MIWI2, all of which are required for spermatogenesis (Kuramochi-Miyagawa et al. 2001, 2004; Deng and Lin 2002; Carmell et al. 2007). In fetal male germ cells, the vast majority of piRNAs are derived from retrotransposon genes, and two PIWI family proteins, MILI and MIWI2, are involved in this process. Fetal piRNAs are mainly generated from transcripts of piRNA-producing regions called piRNA clusters, although the molecular mechanisms underlying piRNA clusters are currently unknown. The piRNA clusters expressed in the fetal testis are genomic regions that accumulate loss-of-function transposon fragments or full-length transposons (Girard et al. 2006; Aravin et al. 2007; Brennecke et al. 2007; Zanni et al. 2013). Primary piRNAs, which exhibit a sequence bias for uridine at the first position of the 5′ end (1U bias), were produced and incorporated into MILI. Next, piRNA-bound MILI, which functions as an endonuclease (slicer activity), cleaves the complementary target RNAs annealed to the primary piRNAs between nucleotides 10 and 11 (Aravin et al. 2007, 2008). The secondary piRNA produced from MILI cleavage exhibits a sequence bias for adenine at the tenth position (10A bias) and is further incorporated into MILI or MIWI2 (Aravin et al. 2008; De Fazio et al. 2011). piRNA-bound MILI represses transposon expression in the cytoplasm through the cleavage of transposon RNAs. In contrast, piRNA-bound MIWI2 migrates to the nucleus, where piRNAs guide binding to nascent RNAs and induce DNA methylation of retrotransposon genes, such as LINE and IAP (Carmell et al. 2007; Molaro et al. 2014; Manakov et al. 2015).
DNA methylation is one of the gene expression control mechanisms that regulates cell differentiation and development. Germ cells undergo global epigenetic reprogramming, including DNA methylation. In mice, DNA methylation is globally erased by embryonic day 12.5 (E12.5) during the development of germ cells; consequently, parental epigenetic imprinting is erased (Hajkova et al. 2002; Sasaki and Matsui 2008). Subsequently, global de novo DNA methylation occurs in ∼76% of the genome of mouse gonocytes, the male germ cells, around E16.5 (Molaro et al. 2014). DNMT3L and DNMT3A, which are involved in de novo DNA methylation, target transposable elements and paternally imprinted genes (H19, Dlk1, and Rasgrf1) (Li et al. 2004; Kato et al. 2007; La Salle et al. 2007; Watanabe et al. 2011). Notably, as DNA methylation of retrotransposon genes is dependent on piRNAs, a similar phenotype is observed in MILI-deficient mice, in which almost no piRNAs are produced.
Previously, we demonstrated that the expression of sense and antisense RNAs in fetal male germ cells is sufficient for piRNA production and subsequent de novo DNA methylation (Itou et al. 2015). We previously generated transgenic (Tg) mice harboring the antisense-EGFP gene under the control of the Miwi2 promoter (Tg-Miwi2P-asEGFP). Miwi2 is expressed only in the male fetal germ cells. Crossing Tg-Miwi2P-asEGFP mice with EGFP-expressing Tg mice resulted in EGFP silencing in the progeny. Next, Tg mice expressing antisense-Dnmt3L under the control of the Miwi2 promoter (Tg-Miwi2P-asDnmt3L) were generated. Tg-Miwi2P-asDnmt3L mice exhibited down-regulated endogenous Dnmt3L RNA expression and a phenotype similar to that of DNMT3L-deficient mice. In these Tg mice, piRNAs against EGFP and Dnmt3L were produced, and DNA methylation occurred in their corresponding genomic loci. However, the piRNAs of these Tg mice were mapped not only to the target genes EGFP and Dnmt3L, but also to Miwi2 promoter regions. These data indicated that the transcription of the piRNA precursors against asEGFP and asDnmt3L was not only regulated by the Miwi2 promoter but also by some unknown promoters outside of the inserted Tg cassette. It is reasonable to assume that these transgenes have been incorporated into piRNA clusters (Shoji and Katsuma 2015; Kuramochi-Miyagawa and Nakano 2015).
In this study, whole-genome sequencing of Tg-Miwi2P-asDnmt3L mice led to the identification of two insertion sites: the B3 and E1 loci on chromosome 18 (ch18). The analysis of the two loci revealed that each of them had different roles in piRNA production and, consequently, in mouse phenotypes. The E1 locus, which is presumably a weak piRNA cluster, produced some piRNAs. However, the mice harboring the transgene at the E1 locus were fertile. In contrast, the mice harboring the transgene at the B3 locus (a very weak piRNA cluster) exhibited infertility due to the post-transcriptional silencing of Dnmt3L. Further analyses revealed that piRNA-mediated Dnmt3L silencing was dependent on the transgene copy number rather than on the transgene insertion locus.
RESULTS
Miwi2P-asDnmt3L transgene insertion locus in the mouse genome
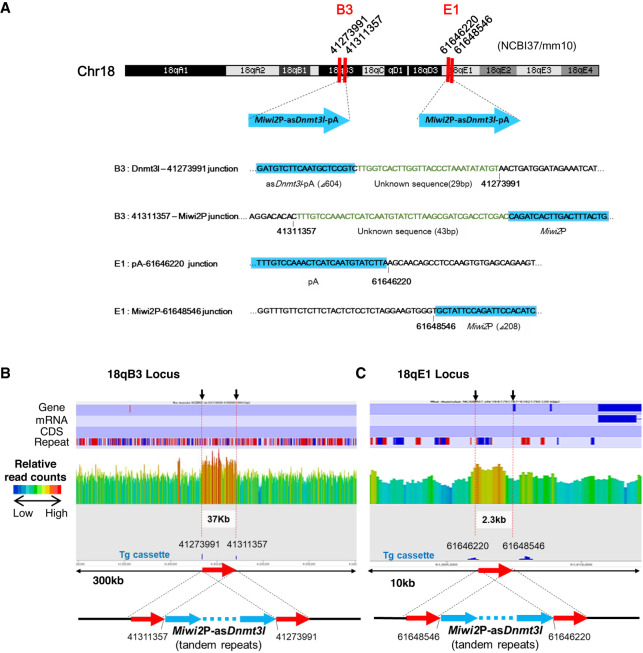
Whole-genome DNA sequencing of Tg-Miwi2P-asDnmt3L mice was performed to examine whether the Tg cassette insertion loci corresponded to piRNA clusters. Four junction sites between the transgene and chromosome (two at the B3 locus and two at the E1 locus on ch18) were identified (Fig. 1A). Dnmt3L-polyA (Δ604)—41273991 (NCBI/mm10) and Miwi2P—41311357 (NCBI/mm10) were detected at the B3 locus, while the polyA—61646220 (NCBI/mm10) and Miwi2P—61648546 (NCBI/mm10) junctions were detected at the E1 locus (Fig. 1A).
FIGURE 1.
Transgene insertion sites on chromosome 18 (ch18) of the mouse genome. (A) The four transgene insertion sites found on ch18 and the sequences of the junctions are shown. The numbers represent the location on the genome (NCBI/mm10 assembly). Genomic sequences and unknown sequences are shown in black and green letters, respectively. Transgene sequences are highlighted in blue. (B,C) Read counts around the insertion loci were determined using whole-genome sequencing. The overall lengths of B and C are 300 kb for Ch18qB3 and 10 kb for Ch18qE1, respectively. The read counts show a 100-bp window. The regions flanked by the insertion sites of the transgenic cassette in both B3 and E1 regions have a high genomic read count (red arrow). This indicates a model in which the transgene is sandwiched between duplicated genomic regions.
Transgene insertions did not reduce the mapping reads by half in any of the two genomic areas. The DNA sequencing read counts around the junction sites of the transgene increased by 1.5-fold in both loci compared to other regions of the same chromosome (Fig. 1B,C). These findings indicate that the 37 and 2.3 kb regions of the 18qB3 and 18qE1 loci, respectively, underwent duplication. Polymerase chain reaction (PCR) was performed using primers designed to flank the 2.3 kb region (Supplemental Fig. S1A). The amplified sequences confirmed duplication (Supplemental Fig. S1B). The duplication of the B3 locus could not be confirmed due to the length of the duplicated region. Therefore, similar to E1, B3 was assumed to have undergone duplication.
piRNAs of the flanking regions of transgene insertion sites
To determine whether these two loci were part of any piRNA cluster, previously reported data on MILI-associated and MIWI2-associated piRNAs of E16.5 wild-type (WT) and MILIDAH (MILI endonuclease-impaired mutant) mice in the piRNA database (piRBase: http://regulatoryrna.org/database/piRNA/index.html) were analyzed (De Fazio et al. 2011; Zhang et al. 2014). The expression level of primary piRNAs in MILIDAH mice was higher than that in WT mice because mutant mice cannot produce secondary piRNAs. Some MILI- and MIWI2-bound piRNAs were detected around the E1 locus in both WT and MILIDAH mice (Supplemental Fig. S1C). In contrast, only a small number of piRNAs were annotated around the B3 locus. The number of piRNAs or coverage, and the average values of each category in the 50 kb regions surrounding the E1 and B3 loci are shown in Supplemental Figure S1C.
Next, these two regions were analyzed using piRNAQuest: http://bicresources.jcbose.ac.in/zhumur/pirnaquest/index.html, which annotates piRNA clusters (Sarkar et al. 2014). Each of the two regions was included in a piRNA cluster, although both cluster scores were low (Supplemental Table S1). These data suggest that the E1 and B3 loci are weak piRNA clusters.
Some sense and antisense piRNAs were detected in both 50 kb regions surrounding the insertion sites in WT mice (Supplemental Fig. S1C,D). Therefore, the piRNAs (23–30 nt) in the 2.3 kb duplicated region of the E1 locus and the 2.3 kb regions adjacent to the insertion sites of the B3 locus (B3–5′ and B3–3′) were analyzed (Supplemental Fig. S1D). The number of each piRNA was normalized based on the length and number of total small RNAs per million after excluding miRNAs and tRNAs. The 2.5 kb of the Miwi2 promoter used for the transgene was not a piRNA cluster region, considering that very few piRNAs were mapped to this region; therefore, it was used as the negative control. In contrast, the number of antisense piRNAs was high in the E1 and B3–5′ regions, whereas that of sense-strand piRNAs was low. These findings indicate that the insertion sites have the potential to produce antisense piRNAs, and that antisense piRNAs are transcribed in this region. Thus, both regions were considered weak piRNA clusters.
Phenotypes of Tg mice with single transgene insertions
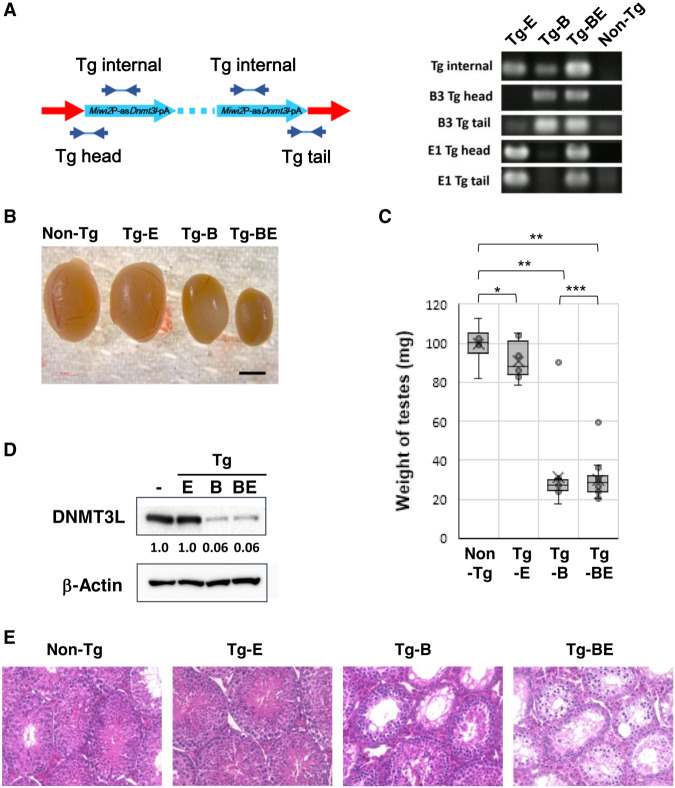
To determine which locus was critical for silencing Dnmt3L, crossbreeding was performed to segregate each insertion into different mice of the progeny. Segregation was confirmed by PCR (Fig. 2A). Mice with the transgene cassette inserted at the B3 locus (Tg-B) exhibited lower testis size and weight relative to body weight than the control mice. In contrast, mice with the cassette at the E1 locus (Tg-E) showed a normal phenotype (Fig. 2B,C).
FIGURE 2.
Transgenic mice (Tg) phenotypes. (A) Schematic illustration of polymerase chain reaction (PCR) primer annealing sites to confirm Tg cassette insertion sites. “Tg internal” legends indicate the junctions between Miwi2 promoter and asDnmt3L. “Tg head” shows the junction between genomic DNA and the Miwi2 promoter at Ch18qB3 and E1 sites, and “Tg tail” shows the junction between the tail of the Tg cassette and genomic DNA at each insertion site. The primer sequences are shown in Supplemental Table S4. The PCR products were subjected to agarose gel electrophoresis with Tris-acetate EDTA buffer (right panel). (B) Morphology of testes of adult mice with transgenes inserted at the E1 locus (Tg-E), B3 locus (Tg-B), and B3 and E1 loci (Tg-BE). Black bar indicates a 2 mm scale. (C) Weights of the testes of Tg-E, Tg-B, and Tg-BE mice. Each dot represents the weight of each testis. The range of the whisker lines represents the minimum and maximum values; outliers were excluded. The box ranges indicate the interquartile range (IQR), the horizontal bars in the box indicate the median, and the “X” marks indicate the mean weights. (98.4, 90.8, 32.0, and 29.8 mg in non-Tg, Tg-E, Tg-B, and Tg-BE mice, respectively; t-test; [*] P = 0.15; [**] P < 0.01; [***] P = 0.72). (D) DNMT3L expression in mice testes at E16.5 was analyzed using western blotting. Upper panel shows DNMT3L, and lower panel shows β-actin. Each lane was loaded with protein extracts from one-half of a testis. The band intensity of DNMT3L was normalized to that of β-actin, as well as to that of DNMT3L in non-Tg mice. Analysis was performed using a GelAnalyzer 2010. (E) Hematoxylin and eosin-stained histological sections showing developmental differences between non-Tg, Tg-E, Tg-B, and Tg-BE mouse testes.
Western blot analysis revealed that the expression of endogenous DNMT3L was down-regulated in the testes of Tg-B mice at E16.5, and in Tg mice with transgenes inserted at both E1 and B3 loci (Tg-BE; Tg-Miwi2P-asDnmt3L). However, the expression level of DNMT3L in Tg-E mice was similar to that in the control mice (Fig. 2D). Hematoxylin and eosin (HE) staining revealed severe spermatogenesis defects in the testes of Tg-B and Tg-BE mice. In contrast, the testes of Tg-E mice did not exhibit any histological defects (Fig. 2E). These findings suggest that the phenotype of Tg-Miwi2P-asDnmt3L mice is only due to the insertion in the B3 locus. We then examined the meiotic chromosome spreads and stained them with antibodies against SCP1 and SCP3 to evaluate meiotic defects in DNMT3L mutants. We found that most meiotic cells in Tg-B mice were at the zygotene or zygotene-like stages, similar to the phenotype of DNMT3L-deficient mice.
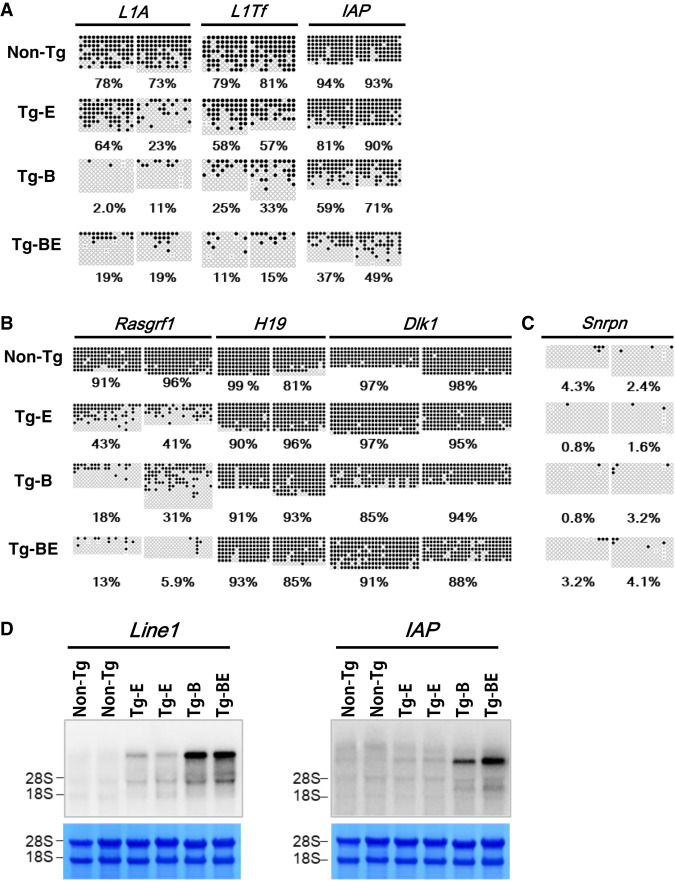
Next, bisulfite sequencing of the target genes of DNMT3L, such as retrotransposons and imprinted genes, was performed. Germ cells were sorted based on the epithelial adhesion molecule Ep-CAM, a spermatogonial stem cell marker, at postnatal day 10 (P10) (Kanatsu-Shinohara et al. 2011). We found that retrotransposon genes (LINE1 type A, LINE1 Tf family, and LTR element IAP1Δ1) and paternally imprinted genes (H19, Dlk1, and Rasgrf1) were methylated in the germ cells of non-Tg mice (Fig. 3A,B). However, consistent with our previous study, retrotransposon genes and Rasgrf1 in the germ cells of Tg-BE mice exhibited decreased methylation levels. Similarly, germ cells of Tg-B mice exhibited decreased levels of DNA methylation. Although Tg-E mice did not exhibit any histological defects at the different stages of spermatogenesis, their germ cells exhibited partial methylation (Fig. 3A,B).
FIGURE 3.
Bisulfite sequencing and northern blot of DNMT3L de novo DNA methylation targets. (A) Methylation levels of retrotransposon genes (L1A, LINE1 type A; L1Tf, LINE1 Tf; and IAP, IAP1Δ1) in germ cells of non-Tg and mice with transgenes inserted at the E1 locus (Tg-E), B3 locus (Tg-B), and B3 and E1 loci (Tg-BE). (B) Methylation levels of paternally imprinted genes (Rasgrf1, H19, and Dlk1). (C) Methylation levels of Snrpn, a maternally imprinted gene, representing sorted cell purity. Black and white circles indicate methylated and unmethylated CpGs, respectively. (D) Northern blotting of LINE1 and IAP in testes of 3 wk in the non-Tg and Tg mice. To compare the amount of RNA loaded, the filters were stained with methylene blue after transfer to show the amount of 28S and 18S.
We performed northern blotting to evaluate the transcription levels of the transposable elements LINE1 and IAP and their correlation with the bisulfite sequencing data. As shown in Figure 3D, both LINE1 and IAP were highly expressed in Tg-B and Tg-BE cells. In contrast, Tg-E showed a lower expression. This indicates that decreased DNA methylation and increased transcription levels of DNMT3L target genes in Tg-Miwi2P-asDnmt3L (Tg-BE) mice were mainly caused by the insertion in B3. However, the phenotypic differences between Tg-B and Tg-E mice could not be explained by the insertion site of the transgene, given that both insertion loci were weak piRNA clusters. Therefore, transgene copy numbers were examined.
Copy number of the antisense-Dnmt3L transgene and its correlation with phenotype
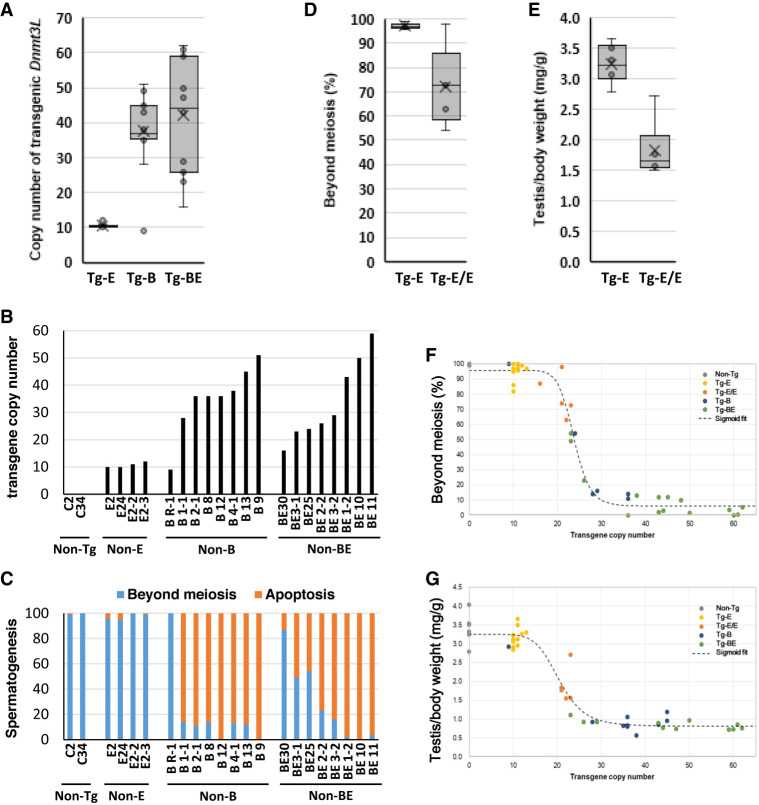
Genomic quantitative PCR (qPCR) analysis revealed that Tg-E mice harbored approximately 10 copies of the transgene cassette. In contrast, 11 of the 12 Tg-B mice harbored 28–51 copies, and 1 mouse harbored only 9 copies. The copy number of the transgene cassette in Tg-BE mice varied from 16 to 61 (Fig. 4A). The reasons for these differences are unclear. One possible explanation is the occurrence of recombination events among tandemly arrayed transgenes, similar to the recombination observed in SINE and LINE transposons.
FIGURE 4.
Copy numbers of asDnmt3L transgenes and their correlation with phenotypes. (A) asDnmt3L transgene copy numbers in mice with transgenes inserted at the E1 locus (Tg-E), B3 locus (Tg-B), and B3 and E1 loci (Tg-BE) were confirmed using quantitative polymerase chain reaction. Each dot indicates the copy number. The range of the whisker lines indicates the minimum and maximum values, excluding outliers. The box range indicates the interquartile range (IQR), the horizontal bars in the box indicate the median, and the “X” marks indicate the mean value. (10, 38, and 42 copies in Tg-E, Tg-B, and Tg-BE mice, respectively). (B) Transgene copy numbers represented through a bar graph. (C) Percentage of cells in seminiferous tubules that have undergone meiosis. The blue and red bars represent the cells that have completed meiosis and those that are arrested at meiosis, respectively. (D) Progression of spermatogenesis in mice with single or double alleles of the transgene cassette inserted at the E1 locus (Tg-E and Tg-E/E mice, respectively). Each dot represents the percentage of cells in the tubules of a mouse that have completed meiosis. The range of the whisker lines indicates the minimum and maximum values excluding outliers. The box range indicates the interquartile range (IQR), the horizontal bars in the box indicate the median, and the “X” marks indicate the mean value (97% and 72% in cells with single and double transgenic alleles, respectively). Student's t-test; (*) P < 0.01. (E) Testicular weight relative to the body weight of mice with single or double transgenic alleles. Each dot represents testis weight relative to body weight (mg/g) and horizontal bars represent averages (3.2 and 1.8 mg/g for mice with single and double transgenic alleles, respectively). Student's t-test; (*) P < 0.01. (F,G) Correlation between Dnmt3L copy number and the progression of spermatogenesis (F) or testicular weight relative to body weight (G). Black, yellow, orange, blue, and green dots indicate cells derived from non-Tg mice, Tg-E mice, mice with double transgenic alleles (Tg-E/E), Tg-B mice, and Tg-BE mice, respectively. The fitting curves were adjusted to a sigmoidal model (Min = 6.03, Max = 95.81, half-maximal effective concentration [EC50] = 23.72, R2 = 0.98 for F; and Min = 0.82, Max = 3.25, EC50 = 20.5, R2 = 0.94 for G).
To determine the correlation between copy number and phenotype, HE staining was performed, and the proportion of cells arrested at meiosis and those that had completed it in the seminiferous tubules was examined (Fig. 4B–C; Supplemental Fig. S2A,B). Tg-E mice exhibited normal spermatogenic progression. In contrast, most Tg-B mice exhibited severe spermatogenic defects; the exception was the mouse with a low copy number (that with nine copies), which exhibited normal spermatogenesis. In Tg-BE mice, the severity of spermatogenic defects was almost directly proportional to the transgene copy number. These findings demonstrate that high transgene copy numbers are correlated with the degree of spermatogenic defects.
Next, the phenotypes of Tg-B mice with low transgene copy numbers and Tg-E mice with high transgene copy numbers were examined. Crossing female Tg-B mice with WT male mice resulted in mice harboring various copies of the transgene. However, Tg-B mice with transgene copy numbers below 40 were not obtained. Crossing male and female Tg-E mice resulted in mice with twofold the number of transgenes on both alleles of the E locus (Tg-E/E mice). In Tg-E/E mice, 30%–50% of the cells in the seminiferous tubules were arrested during meiosis (Fig. 4D; Supplemental Fig. S2C). Additionally, testis size and weight relative to body weight were significantly decreased in Tg-E/E mice (Fig. 4E; Supplemental Fig. S2C). The correlation between transgene copy number and the progression of spermatogenesis or testis weight is shown in Figure 4F,G; Supplemental Figure S2D. It was observed that 10 copies of the transgene did not affect spermatogenesis, whereas more than 25 completely inhibited it. Interestingly, Tg mice harboring approximately 20 copies of the transgene exhibited partial inhibition of spermatogenesis. These data suggest that the phenotype is dependent on the copy number of the transgene rather than on the transgene insertion locus.
piRNA expression in the fetal testes of Tg-BE, Tg-B, and Tg-E mice
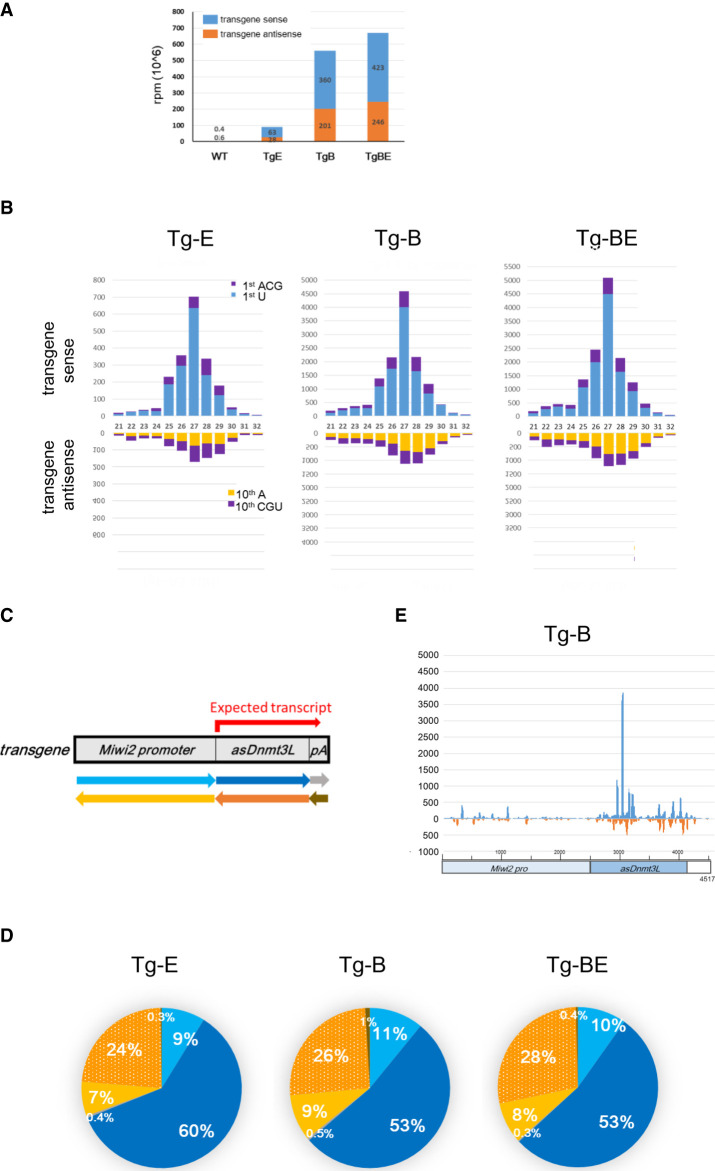
Next, the expression of piRNAs against Dnmt3L was examined. Small RNAs isolated from the testes at E16.5, were subjected to deep sequencing analysis. Sequences with sizes ranging from 15 to 45 nt were selected. After removing both miRNAs and tRNAs, transgene-annotated small RNAs with up to two mismatches were selected (Supplemental Fig. S3A). Tg-BE and Tg-B mice exhibited similar numbers of transgene-annotated piRNAs. However, the number of piRNAs in Tg-E mice was approximately eightfold lower than that in Tg-B mice (Fig. 5A; Supplemental Table S2). These data suggest that piRNA copy number determines the silencing of DNMT3L and, consequently, the phenotype.
FIGURE 5.
Small RNAs from Miwi2P-asDnmt3L in fetal testes. (A) Numbers of small RNAs annotated as from the Miwi2P-asDnmt3L transgene among total reads in the testes of nontransgenic (Tg) mice and mice with transgene cassettes inserted at the E1 locus (Tg-E), B3 locus (Tg-B), and B3 and E1 loci (Tg-BE) at E16.5. Blue bars show the small RNAs from the sense Miwi2 promoter and antisense Dnmt3L (transgene sense strand), while the orange bars show the small RNAs from the antisense Miwi2 promoter and sense Dnmt3L (transgene antisense strand). (B) Size distribution of transgene-annotated small RNAs. The upper side represents the transgene sense strand where blue and purple indicate small RNAs with and without uracil as their first nucleotide, respectively. The lower side represents the transgene antisense strand where orange and purple indicate small RNAs with and without adenine at the tenth position, respectively. (C) Schematic diagram showing the directions of small RNAs. Red arrow indicates the expected transcript of asDnmt3L driven by the Miwi2 promoter. Light blue, blue, and gray arrows indicate transgene sense strands, while yellow, orange, and brown arrows indicate transgene antisense strands. (D) Ratio of transgene-annotated small RNAs in the fetal testis of Tg mice. The colors of the pie charts correspond to the colors of the arrows in C. The shaded orange color contains piRNAs from the transgene and the endogenous Dnmt3L. (E) Mapping of piRNAs of the Tg-B mice onto the transgene cassette sequence. The upper and lower sides represent the sense strand in blue and the antisense strand in orange, respectively.
Analysis of the first and tenth nucleotides of the transgene-annotated small RNAs from Tg mice revealed that the 1U bias in the transgene sense strand was more pronounced than that in the antisense strand. The antisense strand exhibited a 10A bias (Supplemental Fig. S3B). The length distribution and proportion of 1U in the sense strand and of 10A in the antisense strand were similar in both Tg-B and Tg-E mice (Fig. 5B). The analysis of small RNAs with 1U bias revealed a prominent peak of 27 nt in the sense strand but not in the antisense strand.
Figure 5C,D shows the proportion of piRNAs derived from the sense transcripts of the Miwi2 promoter (light blue) and asDnmt3L (blue) and from the antisense transcripts of the Miwi2 promoter (yellow) and sense Dnmt3L (orange). piRNAs corresponding to the first three types of transcripts derived from the transgene, given that the expression of these piRNAs was extremely low in control mice testes. However, it was impossible to distinguish between piRNAs from the sense transcript of the endogenous Dnmt3L gene and those from the antisense transcript of the asDnmt3L transgene. Histograms showing the piRNA levels of the transgene sequence are shown in Figure 5E (Tg-B) and Supplemental Figure S4F (piRNA data from Tg-E, Tg-BE, and previous publications). The number of piRNAs for antisense Dnmt3L was higher than that for the Miwi2 promoter. Hence, most piRNAs of antisense Dnmt3L should derive from transcripts driven by the Miwi2 promoter.
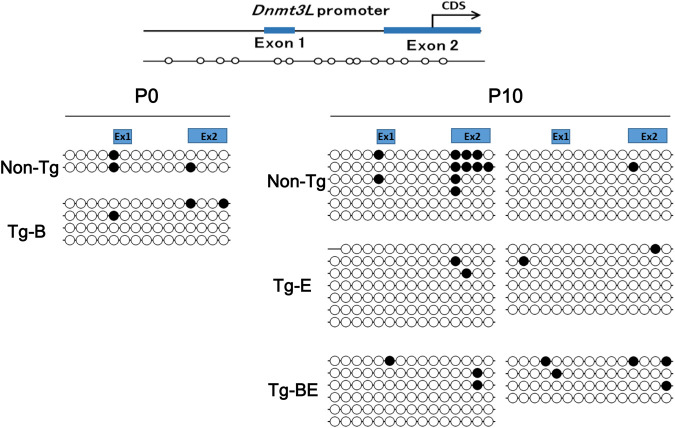
Previously, we reported that endogenous Dnmt3L silencing can be attributed to piRNA-mediated DNA methylation. Hence, DNA methylation of the promoter region of Dnmt3L was examined using bisulfite sequencing. Methylation was not observed in any of the Tg-E, Tg-B, or Tg-BE lines (Fig. 6). However, as shown in Supplemental Figure S3C, the nuclear localization of MIWI2 was the same in the control and Tg-B mice. These data showed that MIWI2-mediated silencing was not affected in Tg-B mice.
FIGURE 6.
Bisulfite sequencing of Dnmt3L promoter. The top panel shows a diagram of the Dnmt3L promoter and its methylation positions (white circles). Methylation levels of the Dnmt3L promoter in germ cells of 0-d-old (P0) nontransgenic (Non-Tg) mice and of mice with a transgene cassette inserted at the B3 locus (Tg-B) as well as in those of 10-d-old (P10) non-Tg mice and mice with transgene cassettes inserted at the E1 locus (Tg-E) and at both the E1 and B3 loci (Tg-BE). Black and white circles indicate methylated and unmethylated CpGs, respectively.
DISCUSSION
Previously, we reported that the expression of antisense transcripts from a Miwi2P-asDnmt3L transgene promoted piRNA-mediated endogenous gene silencing in fetal male germ cells (Itou et al. 2015). However, it was pointed out that the Tg cassette was incorporated into piRNA clusters, and its expression was affected by the insertion loci (Shoji and Katsuma 2015). In this study, whole-genome sequencing and piRNA next-generation sequencing analyses revealed that, although the transgenes were incorporated into weak piRNA clusters, the copy numbers of the inserted transgenes were more important for silencing than the insertion loci.
Transgene insertion sites, copy numbers, and silencing
We defined that the insertion sites of the transgene were weak piRNA clusters because piRNAs derived from the neighboring region of the insertion sites were relatively abundant in the wild-type mouse (Supplemental Fig. S1C,D). Moreover, the whole genome DNA sequencing data indicated that the majority of the transgenes were inserted in a tandem orientation, and only 1% showed head-to-head or tail-to-tail, namely, inverted orientation. PCR analysis failed to detect inverted orientation insertion in our hands, showing that there should be few inverted junctions. Inverted orientation insertions can induce the expression of antisense transcripts, but tandem orientation insertions do not originate from molecular mechanisms. Therefore, we hypothesized that the antisense strand of the transgene was transcribed as a piRNA precursor, albeit in a small amount, by an unknown promoter outside of the inserted Tg. This supports the idea that the insertion regions are weak clusters. However, phenotypic differences between Tg-B and Tg-E mice could not be explained by the insertion site of the transgene, but rather depended on the copy number. Specifically, the copy number of the transgene determines the amount of piRNAs produced and the subsequent piRNA-mediated silencing.
The transgene was inserted into two loci on ch18 of Tg-Miwi2P-asDnmt3L mice. Therefore, crossbreeding was performed to obtain mice with a transgene inserted in only one of the two loci. We found that different mice exhibited different transgene copy numbers. However, no transgenic insertion sites containing more than 60 copies were observed. Therefore, the number of transgenes in these sites either had some limitations or tended to decrease. Moreover, although the copy number of Tg-E mice was stable, those of Tg-B and Tg-BE mice showed some tendency to change. These results suggest that a high copy number of the transgene was not appropriately maintained. After several passages, the copy number was set to approximately 40, maybe due to structural stability issues.
Further analyses revealed that the silencing of Dnmt3L was dependent on the copy number of the transgene and not on the insertion site (Figs. 4F,G; Supplemental Fig. S2D). The high copy number of the transgenes may increase the transcription of asDnmt3L under the control of the Miwi2 promoter, leading to enhanced expression of piRNAs with an asDnmt3L sequence and, consequently, to the silencing of the endogenous Dnmt3L. The findings of this study indicate that a transgene copy number below 10 is not sufficient to silence DNMT3L expression and arrest spermatogenesis. The silencing of Dnmt3L is determined by the balance between the expression levels of endogenous genes and the number of piRNAs.
In addition to the possibility that the transgene was inserted into a piRNA cluster, pre-existing genome defense mechanisms (either from the piRNA cluster or from other genomic locations) may have been activated in response to the insertion. High-copy number tandem repeats of the transgenes may have been recognized as novel piRNA clusters. Nevertheless, the high copy number of the inserted transgene of the antisense strand is important for gene silencing in the fetal testis.
Timing of DNMT3L suppression
Dnmt3L expression was silenced, and the expression of DNMT3L protein was down-regulated at E16.5, in Tg-B and Tg-BE mice (Fig. 2D). Consequently, the methylation levels of DNMT3L targets, such as retrotransposons and Rasgrf1, were diminished (Fig. 3). However, the DNA methylation levels of H19 and Dlk1, which are also DNMT3L targets, were unaffected. piRNA-dependent DNA methylation is slightly delayed compared to piRNA-independent DNA methylation, such as that of H19 and Dlk1 (Molaro et al. 2014). Therefore, piRNA-dependent silencing of Dnmt3L in Tg-Miwi2P-asDnmt3L mice may occur after the DNA methylation of H19 and Dlk1. Meanwhile, DNA methylation of LINE1 is delayed when compared with that of IAP (Kuramochi-Miyagawa et al. 2008; Molaro et al. 2014). In addition, we observed that the reduction in DNA methylation of IAP was not as severe as that of the LINE1 elements L1A and L1Tf (Fig. 3A), which could be explained by the DNA methylation timing differences between LINE1 and IAP.
DNA methylation of DNMT3L target genes in Tg-E mice, which did not exhibit any histological defects, was partially impaired at P10. The endogenous Dnmt3L in the testis at E16.5, was partially silenced because the number of piRNAs for antisense Dnmt3L was low. However, threshold DNA methylation levels of DNMT3L target genes may be required to impair spermatogenesis.
Interpretation of the discrepancies between studies
The methylation level of the endogenous Dnmt3L promoter and the piRNA next-generation sequencing data reported in this study were not consistent with those of a previous study (Supplemental Table S3; Itou et al. 2015). In that study, the endogenous Dnmt3L promoter was methylated, whereas methylation was not observed here. However, gene silencing was observed in both studies. This could be attributed to DNA methylation-mediated silencing in the previous study and only to PTGS in this study.
In this study, the percentage of antisense Dnmt3L piRNAs with 1U bias was high (80%), and the peak length of the piRNAs was 27 nt, which suggested that the antisense Dnmt3L piRNAs were primary piRNAs bound to MILI (Fig. 5B). The endogenous Dnmt3L RNAs may have been targeted by MILI with complementary piRNA sequences and undergone MILI-mediated degradation in the cytoplasm, which resulted in PTGS. In the previous study (Itou et al. 2015), the percentage of antisense Dnmt3L piRNAs with 10A bias was high (49%), and the peak length of the piRNA was 28 nt, which suggested that antisense Dnmt3L piRNAs were secondary piRNAs bound to MIWI2 (Supplemental Fig. S3B–E). MIWI2-bound piRNAs were translocated to the nucleus where they bound Dnmt3L nascent RNA, resulting in DNA methylation of Dnmt3L. Considering the lack of slicer activity of MIWI2, it is likely that silencing at the post-transcriptional level is determined by whether the piRNA against antisense Dnmt3L binds to MILI. Regarding silencing at the transcriptional level, the binding of piRNA against antisense Dnmt3L to MIWI2 as a secondary piRNA is thought to be important. Taken together, although the data are circumstantial, we consider that DNA methylation was not present in the current Tg mice due to the low levels of MIWI2-binding antisense Dnmt3L piRNAs for transcriptional silencing.
The mice examined in this study exhibited only PTGS. In our previous study, we speculated that both PTGS and TGS had occurred in Tg mice because the methylation of the endogenous Dnmt3L promoter was partial (Itou et al. 2015). Additionally, almost two-thirds (61%) of all antisense Dnmt3L piRNAs exhibited a 1U bias with a peak length of 27 nt (Supplemental Fig. S3B,C), suggesting that these piRNAs were primary piRNAs bound to MILI. Thus, both TGS by DNA methylation and PTGS by target RNA degradation play an important role in piRNA-mediated silencing in the fetal testis, where transcripts of both sense and antisense strands of retrotransposons are abundant. However, PTGS may be sufficient for silencing endogenous genes by piRNAs on the antisense strand, as suggested in the current study.
MATERIALS AND METHODS
Mice breeding
All animal experiments were performed in accordance with the general guidelines of The Institute of Experimental Animal Science, Osaka University Medical School. Tg-Miwi2P-asDnmt3L mice were generated by injecting the Miwi2 promoter-antisense Dnmt3L-polyA transgene cassette (Miwi2P-asDnmt3L) into fertilized mouse eggs (Itou et al. 2015). Transgene-positive female mice were crossed with WT B6 male mice. Mice generated by crossing with WT B6 mice more than five times were used in this study. Primer sequences used for genotyping are listed in Supplemental Table S4.
Whole-genome sequencing
The genomic DNA of Tg-Miwi2P-asDnmt3L mice was subjected to whole-genome sequencing. Paired-end sequencing of approximately 150 bases was performed using an Illumina HiSeq X system (Hokkaido System Science). Sequencing data were analyzed using the Bowtie 2. Paired sequences consisting of one element that mapped against the Miwi2P-asDnmt3L transgene and one that mapped against the genome were extracted, whereas those pairs whose elements mapped against the transgene only were excluded. The extracted sequences were mapped against the mouse genome. Most of these sequences were mapped to the genomic regions of Miwi2 and Dnmt3L. The four regions considered to be insertion sites were detected on ch18. The precise insertion sites were confirmed by PCR sequencing with primers targeting the vicinity of the insertion site and inside the transgene. The BioSample accession number of DDBJ is SAMD00324390.
Western blotting
Fetal testes were lysed with 30 µL of radioimmunoprecipitation buffer (50 mM Tris-HCl pH 8.0, 0.1% sodium dodecyl sulfate, 150 mM NaCl, 0.5% DOC, and 1% NP-40) and 10 µL of 4× sample buffer (250 mM Tris-HCl pH 6.8, 20% β-mercaptoethanol, 8% sucrose, and 0.01% bromophenol blue). The lysates were boiled at 98°C for 3 min and incubated with Benzonase nuclease (Novagen) at 22°C for 5 min. Testes samples (a half of a testis each) were boiled at 98°C for 3 min and loaded onto a 10% polyacrylamide gel. The resolved proteins were transferred to PVDF Immobilon-P membranes (0.45 µm) (Millipore). The membrane was then probed with either anti-DNMT3L (E1Y7Q CST#13451; Cell Signaling Technology) (1:1000) or anti-β-actin (Clone AC-15, A5441; Sigma-Aldrich) (1:2500) antibodies at 4°C overnight. Next, the membranes were washed with Tris-buffered saline containing 0.1% Tween-20 (TBS-T) and incubated with the secondary antibody (ECL-anti-rabbit-IgG-HRP, NA9340V; or ECL-anti-mouse-IgG-HRP, NA9310V; GE Healthcare) (1:2000) for 1 h. The membranes were washed, and the immunoreactive signals were detected using the Bio-Rad Clarity Western ECL Substrate #1705060 (Bio-Rad) and a ChemiDoc MP Imaging System (Bio-Rad).
HE staining of paraffin-embedded sections and spermatid counting
The testes were fixed with Bouin's solution overnight at 4°C. Next, the samples were dehydrated with EtOH, equalized with xylene, and embedded in paraffin. The samples were sectioned and subjected to hematoxylin and eosin staining. The images of the sections were captured using a BZ-X710 microscope (KEYENCE). The number of cells in the seminiferous tubules was counted using the ImageJ Plugins Analyze-Cell Counter. The presence of even a few spermatids in the seminiferous tubule was scored as “beyond meiosis” even if there were many cells arrested at meiosis during spermatogenesis. Partial spermatogenesis was scored in more than 200 seminiferous tubules.
Meiotic chromosomal spread analysis
The testes were freshly dissected from adult mice (7 wk) and decapsulated. The testes were ground in PBS, and the cells were collected and centrifuged. The cell pellets were washed several times in PBS and suspended in hypotonic buffer (30 mM Tris at pH 7.5, 17 mM Tris sodium citrate, 5 mM EDTA, and 50 mM sucrose). After keeping for 5 min, the cells were centrifuged and the supernatant discarded, followed by making suspension in 100 mM sucrose. The cell suspension was spread onto slides dipped in formaldehyde solution (1% formaldehyde, 0.15% Triton X-100 in water adjusted with sodium borate to pH 9.2). Slides were then placed in a humidified chamber overnight and air-dried at room temperature. For immunostaining, slides were rinsed for 5 min in PBS, and then incubated for 30 min in blocking buffer (3% BSA, 10% normal goat serum in PBS). The slides were incubated with primary antibodies for Scp1 (NB300-229, Novus) and Scp3 (sc-74569, Santa Cruz Biotechnology) in blocking buffer overnight at 4°C, followed by incubation with goat anti-mouse IgG (H + L) Alexa Fluor 568 (A11004) and goat anti-rabbit IgG (H + L) Alexa Fluor 488 (A11008) (Invitrogen) secondary antibodies and DAPI for 1 h at room temperature. The slides were then washed with PBS and mounted.
Immunohistochemical staining of embryonic testes
E16.5 testes were fixed by incubation with 4% paraformaldehyde (PFA) in PBS for 30 min at 4°C. Cryosections were permeabilized with 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 30 min. Sections were traversed with HistoVT One (Nacalai) at 70°C for 20 min. Slides were treated with blocking buffer (3% BSA and 10% normal goat serum in PBS) for 30 min at room temperature. Following treatment with primary antibodies for MILI (clone17.8, Active motif) or MIWI2 (25D11) (Kojima-Kita et al. 2016) overnight at 4°C, the sections were incubated with secondary antibodies for 1 h at room temperature.
Germ cell sorting and bisulfite sequencing
The resected testes were treated with collagenase B (Roche) and DNase I, followed by treatment with trypsin and DNase I. The samples were washed with phosphate-buffered saline, passed through a cell strainer (FALCON #352235), and sorted using a FACSAria II flow cytometer (BD Bioscience). The germ cells from 10-d-old mouse testes were purified by sorting with phycoerythrin (PE)-conjugated anti-mouse CD326 (Ep-CAM) antibody (BioLegend). Alternatively, germ cells from the testes of 0-d-old mice crossed with Tg-OctEGFP mice were purified by sorting with an anti-EGFP antibody. Genomic DNA was purified and treated with bisulfite using the EpiTect Fast DNA Bisulfite Kit (Qiagen). To test the sorting quality, the methylation level of Snrpn, a maternally imprinted gene that is hypomethylated in male germ cells, was examined. The methylation level of Snrpn was close to 0% in all samples, which indicated high germ cell purity. The primer sequences used for amplification of the bisulfite target sites are listed in Supplemental Table S7.
Northern blotting
Total RNA was prepared from 3-wk and adult testes using Isogen (Nippon Gene). Northern blotting analysis was performed as previously described in a previous study (Kuramochi-Miyagawa et al. 2008). Five micrograms of total RNA per lane were run through the gel. The probes of IAP and Line-1 were as follows: the 3′ noncoding region and part of the 3′LTR region of IAP (nucleotides 6486-6793 in GenBank accession no. M17551) and the common region of type A and Tf of Line-1 (nucleotides 2211–2625 in D84391). To compare the amount of RNA loaded, the filters were stained with 0.04% methylene blue/0.5 M sodium acetate for 5–10′ at room temperature, gently shaking after transfer and rinsed with H2O to show the amount of 28S and 18S.
Measurement of transgene copy number using qPCR
To quantify the transgene copy number, qPCR was performed using a primer set targeting Dnmt3L that can detect both the endogenous Dnmt3L and the antisense Dnmt3L Tg cassette. qPCR was performed using a THUNDERBIRD SYBR qPCR Mix (Toyobo) and a CFX384 Touch Real-Time PCR analysis system (Bio-Rad). The expression level of Dnmt3L was normalized to that of mTert (murine telomerase), which does not exhibit any sequence similarity with WT genomic DNA. The copy number of Dnmt3L in non-Tg mice is two. Thus, the copy number in the Tg mice was subtracted by two to obtain the copy number of the transgene cassette. Primer sequences are listed in Supplemental Table S6.
Small RNAs in fetal testes of Tg-Miwi2P-asDnmt3L
Total RNA was isolated from the testes of each Tg mouse at E16.5 using ISOGEN (Nippon Gene). Small RNA libraries were generated using a TruSeq small RNA Library Prep Kit and analyzed using an Illumina HiSeq System (Macrogen). The raw sequence data in all samples were processed using CLC Genomics Workbench software (Filgen) to trim miRNAs (miRbase) and tRNAs (GtRNAdb), allowing up to two mismatches after removing adapter sequences, low-quality reads, and reads with lengths <15 nt and >45 nt. The small RNA reads of length 15–45 nt were mapped to the Miwi2P-asDnmt3L transgene sequence, allowing up to two mismatches (Supplemental Table S2). This strategy is summarized in Supplemental Figure S3A.
Small RNAs derived from the genomic region around the transgene insertion sites were extracted from the fetal testis library of WT and non-Tg mice (Fig. S1D). The small RNA reads of length 15–45 nt were mapped against a 2.3-kb genomic region before and after the 18qB3 transgene insertion site (B3–3′ and B3–5′) and an overlapping 2.3 kb genomic region (E1) on both sides of the 18qE1 transgene.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Ms. N. Asada for technical assistance and Ms. S. Tuda for secretarial work. This work was supported in part by a Grant-in-Aid for Scientific Research (B) (#19H03421) and Innovative Areas (#26112511) from the Japan Society for the Promotion of Science (JSPS) and AMED-CREST (#120701467) from the Japan Agency for Medical Research and Development.
Author contributions: L.S.P. and S.K.M. performed the experiments and analyzed the data. L. S. P., S.K.M., and T.N. designed the experiments and wrote the manuscript. I.N. supported the whole-genome sequencing analysis.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078905.121.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
SePil Lee.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. SePil Lee is the first author of this paper, “Effects of transgene insertion loci and copy number on Dnmt3L gene silencing through antisense transgene-derived PIWI-interacting RNAs.” SePil received his Ph.D. at Osaka University, and is interested in developmental biology and epigenetics.
What are the major results described in your paper and how do they impact this branch of the field?
We previously investigated antisense transgene-derived artificial piRNA-mediated gene-silencing systems. Based on the findings, we concluded that in the antisense transgene-mediated gene silencing system in mice, integration on a strong piRNA cluster is not the only way to gene silencing, but a high copy number of antisense transgene is sufficient.
What led you to study RNA or this aspect of RNA science?
Despite that we proposed the artificial piRNA system, some points were unclear. Is a simple expressed antisense transgene sufficient to generate artificial piRNA for transcriptional gene silencing? Our research focused on the relationship between piRNA generation and the phenotype aspect of locus effects.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
When phenotypes were found to be opposite to that expected based on a piRNA cluster database analysis, I was surprised and excited, because it meant that some unexpected factor caused the results.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
When pluripotent stem cells were discovered, I imagined their infinite possibilities, which led me to my path as a scientist. Then I was interested in aging, and I thought germ cells, retrotransposon, and stem cells could be the key.
If you were able to give one piece of advice to your younger self, what would that be?
You're doing better than you thought. Just trust yourself and be on your side. And, there is no meaningless failure. What you learn from failure can be as meaningful as success. Even if you feel that life is not going your way, keep struggling.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
From Toru Nakano I learned the fundamentals of all scientific approaches in a broad sense, and I now see how useful they are to me. Satomi Kuramochi-Miyagawa directly instructed and coached me, and taught me the fundamentals of becoming a scientist. Shinpei Yamaguchi had sharp logic often pointing out something that no one had thought of, and I think of him as a role model. I hope to be like these people someday.
What are your subsequent near- or long-term career plans?
Not yet clear; however, I would like to research aging and rejuvenation based on epigenetics.
REFERENCES
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747. 10.1126/science.1142612 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc'his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799. 10.1016/j.molcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Castaneda J, Genzor P, Bortvin A. 2011. piRNAs, transposon silencing, and germline genome integrity. Mutat Res 714: 95–104. 10.1016/j.mrfmmm.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Chuma S, Nakano T. 2013. piRNA and spermatogenesis in mice. Philos Trans R Soc Lond B Biol Sci 368: 20110338. 10.1098/rstb.2011.0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. 2016. One loop to rule them all: the ping-pong cycle and piRNA-guided silencing. Trends Biochem Sci 41: 324–337. 10.1016/j.tibs.2015.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O'Carroll D. 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263. 10.1038/nature10547 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. . miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Saini H, De Fazio S, Carrieri C, Morgan M, Vasiliauskaite L, Benes V, Enright AJ, O'Carroll D. 2013. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol Cell 50: 601–608. 10.1016/j.molcel.2013.04.026 [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. 10.1038/nature04917 [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA. 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23. 10.1016/S0925-4773(02)00181-8 [DOI] [PubMed] [Google Scholar]
- Itou D, Shiromoto Y, Yukiho SY, Ishii C, Nishimura T, Ogonuki N, Ogura A, Hasuwa H, Fujihara Y, Kuramochi-Miyagawa S, et al. 2015. Induction of DNA methylation by artificial piRNA production in male germ cells. Curr Biol 25: 901–906. 10.1016/j.cub.2015.01.060 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Takashima S, Ishii K, Shinohara T. 2011. Dynamic changes in EPCAM expression during spermatogonial stem cell differentiation in the mouse testis. PLoS One 6: e23663. 10.1371/journal.pone.0023663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kaneda M, Hata K, Kumaki K, Hisano M, Kohara Y, Okano M, Li E, Nozaki M, Sasaki H. 2007. Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse. Hum Mol Genet 16: 2272–2280. 10.1093/hmg/ddm179 [DOI] [PubMed] [Google Scholar]
- Kojima-Kita K, Kuramochi-Miyagawa S, Nagamori I, Ogonuki N, Ogura A, Hasuwa H, Akazawa T, Inoue N, Nakano T. 2016. MIWI2 as an effector of DNA methylation and gene silencing in embryonic male germ cells. Cell Rep 16: 2819–2828. 10.1016/j.celrep.2016.08.027 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Nakano T. 2015. Reply to shoji and katsuma. Curr Biol 25: R710. 10.1016/j.cub.2015.07.020 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. 2001. Two mouse piwi-related genes: miwi and mili. Mech Dev 108: 121–133. 10.1016/S0925-4773(01)00499-3 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. 10.1242/dev.00973 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917. 10.1101/gad.1640708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Oakes CC, Neaga OR, Bourc'his D, Bestor TH, Trasler JM. 2007. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol 7: 104. 10.1186/1471-213X-7-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. 2004. Timing of establishment of paternal methylation imprints in the mouse. Genomics 84: 952–960. 10.1016/j.ygeno.2004.08.012 [DOI] [PubMed] [Google Scholar]
- Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. 2015. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep 12: 1234–1243. 10.1016/j.celrep.2015.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. 2014. Two waves of de novo methylation during mouse germ cell development. Genes Dev 28: 1544–1549. 10.1101/gad.244350.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium, Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Pezic D, Manakov SA, Sachidanandam R, Aravin AA. 2014. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev 28: 1410–1428. 10.1101/gad.240895.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai RS. 2011. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480: 264–267. 10.1038/nature10672 [DOI] [PubMed] [Google Scholar]
- Sarkar A, Maji RK, Saha S, Ghosh Z. 2014. piRNAQuest: searching the piRNAome for silencers. BMC Genomics 15: 555. 10.1186/1471-2164-15-555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y. 2008. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 9: 129–140. 10.1038/nrg2295 [DOI] [PubMed] [Google Scholar]
- Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. 2012. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell 48: 849–862. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Jurczak D, Sachidanandam R, Brennecke J. 2015. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev 29: 1747–1762. 10.1101/gad.267252.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji K, Katsuma S. 2015. Is the expression of sense and antisense transgenes really sufficient for artificial piRNA production? Curr Biol 25: R708–R710. 10.1016/j.cub.2015.07.001 [DOI] [PubMed] [Google Scholar]
- Toth KF, Pezic D, Stuwe E, Webster A. 2016. The piRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol 886: 51–77. 10.1007/978-94-017-7417-8_4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. 2011. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852. 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanni V, Eymery A, Coiffet M, Zytnicki M, Luyten I, Quesneville H, Vaury C, Jensen S. 2013. Distribution, evolution, and diversity of retrotransposons at the flamenco locus reflect the regulatory properties of piRNA clusters. Proc Natl Acad Sci 110: 19842–19847. 10.1073/pnas.1313677110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Si X, Skogerbo G, Wang J, Cui D, Li Y, Sun X, Liu L, Sun B, Chen R, et al. 2014. piRBase: a web resource assisting piRNA functional study. Database (Oxford) 2014: bau110. 10.1093/database/bau110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Wang PJ. 2012. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8: e1003038. 10.1371/journal.pgen.1003038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.