Abstract
We conducted a quantitative structure-activity relationship study using a database of 158 quinolones previously tested against Mycobacterium avium-M. intracellulare complex in order to develop a model capable of predicting the activity of new quinolones against the M. avium-M. intracellulare complex in vitro. Topological indices were used as structural descriptors and were related to anti-M. avium-M. intracellulare complex activity by using the linear discriminant analysis (LDA) statistical technique. The discriminant equation thus obtained correctly classified 137 of the 158 quinolones, including 37 of a test group of 44 randomly chosen compounds. This model was then applied to 24 quinolones, including recently developed fluoroquinolones, whose MICs were subsequently determined in vitro by using the Alamar blue microplate assay; the biological results confirmed the model's predictions. The MICs of these 24 quinolones were then treated by multilinear regression (MLR) to establish a model capable of classifying them according to their in vitro activities. Using this model, a good correlation between measured and predicted MICs was found (r2 = 0.88; r2cv [cross-validation correlation] = 0.82). Moxifloxacin, sparfloxacin, and gatifloxacin were the most potent against the M. avium- M. intracellulare complex, with MICs of 0.2, 0.4, and 0.9 μg/ml, respectively. Finally, virtual modifications of these three drugs were evaluated in LDA and MLR models in order to determine the importance of different substituents in their activity. We conclude that the combination of molecular-topology methods, LDA, and MLR provides an excellent tool for the design of new quinolone structures with enhanced activity.
Mycobacteria belonging to the Mycobacterium avium-M. intracellulare complex are responsible for opportunistic infections in immunocompromised patients, especially those with HIV infection (29), and there is a need for new drugs that can be used for treatment and prophylaxis. Quinolones are good candidates because of their broad antibacterial spectrum, which includes atypical mycobacteria (16, 22, 25, 30), together with their good tissue distribution and intracellular concentration (2). Ciprofloxacin and sparfloxacin are the only quinolones currently used against M. avium-M. intracellulare complex infection, but the incidence of strains resistant to these compounds is increasing, and there is a need for new derivatives.
In addition to in vitro and in vivo tests, which are time-consuming for the M. avium-M. intracellulare complex, powerful methodologies for drug design and drug database screening and selection are now available (7, 19). Equation systems linking structure and activity (QSAR studies) are particularly relevant, and application of the mathematical models thereby obtained to large libraries of computer-generated compounds is known as virtual computational screening (5, 24). An important feature in this method is the use of good structural descriptors that are representative of the molecular features responsible for the relevant biological activity; a very useful technique for describing molecular structure is molecular topology, a two-dimensional QSAR method which takes into account the internal atomic arrangement of compounds. The structure of each molecule is represented by specific subsets of topological indices (TIs). Klopman et al. first developed computer-based predictive models to characterize anti-M. avium-M. intracellulare complex activity (20, 21, 23). The aim of this study was to develop new QSAR models, based on TIs, statistical linear discriminant analysis (LDA), and multilinear regression (MLR) in order to predict the in vitro activity and MICs of quinolones against the M. avium-M. intracellulare complex.
MATERIALS AND METHODS
The study involved a number of steps, which are described in the following paragraphs.
Obtaining structural descriptors by molecular topology.
A database of 158 quinolones with known anti-M. avium-M. intracellulare complex activities has been built up from several articles by Klopman et al. (20, 21, 23). Each quinolone was characterized by a set of 145 TIs specific to each molecule. We used the topological descriptors provided by MOLCONN-Z software, version 3.50 (L. H. Hall, Eastern Nazarene College, Quincy, Mass.), especially the Kier and Hall connectivity indices (up to the 10th order) and the electrotopological indexes (17, 18). We also calculated some descriptors as charge indices (11) using Etopo 11, a computer software developed in our research unit.
Statistical treatment: LDA.
On the basis of the data of Klopman et al. (20, 21, 23), the quinolones studied here were separated into active and inactive compounds according to their MICs around a cutoff of 32 μg/ml. In accordance with the studies of Klopman et al., this cutoff was selected since it allowed a clear differentiation between active and inactive drugs. LDA was then applied to these two groups (except for 44 quinolones reserved as a test group) in order to obtain a mathematical equation linking structural descriptors and activity. LDA is a pattern recognition method providing a classification model based on the combination of variables that best predicts the category or group to which a given compound belongs. The independent variables in this study were the calculated TIs, and the discrimination property was anti-M. avium-M. intracellulare complex activity. The software used for the LDA study was the BMDP 7M package (W. J. Dixon, BMDP Statistical Software, University of California, Berkeley), which randomly chooses the compounds reserved for the test set. The variables used to compute the linear classification functions are chosen in stepwise manner: at each step the variable that makes the larger contribution to the separation of the groups is entered into the discriminant equation (or the variable that makes the smallest contribution is removed). The method used to select the descriptors was based on the F-Snedecor parameter, which allows the assessment of relative importance among the candidate variables (8, 15). The classification criterion was the minimal Mahalanobis distance, which is the distance of each case to the mean of all cases used in the regression equation (15). The quality of the discriminant equation was evaluated using Wilk's U-statistical parameter, a multivariate analysis of variance that tests the equality of group means for the variable(s) in the discriminant equation (15).
PDDs.
Pharmacological distribution diagrams (PDDs) were constructed to determine the intervals of the equation in which the probability of finding active compounds increases (9). PDDs are histograms of calculated values of the mathematical functions in which expectancies appear on the ordinate axis. For an arbitrary interval of values of a given function, we can define an expectancy of activity as Ea = a/(i + 1), where a is the number of active compounds in the interval divided by the total number of active compounds and i is the number of inactive compounds in the interval divided by the total number of inactive compounds. The expectancy of inactivity is defined in a symmetrical way as Ei = i/(a + 1). This representation provides good visualization of the regions of minimum overlap and selects regions in which the probability of finding improved compounds is maximum.
Pharmacological tests.
Twenty-four quinolones that had not been used for the LDA analysis were evaluated with the model in order to establish their potential activities against the M. avium-M. intracellulare complex. These quinolones were selected as having structures representative of the main quinolone structures, including the most recently developed fluoroquinolones (3, 6). In parallel, in vitro studies were performed with the MO-1 strain of M. avium, which was isolated from the blood of a patient with AIDS and which has already been used for in vitro drug susceptibility studies (27). We used the Alamar blue susceptibility test described by Collins and Franzblau (4). This technique uses an oxidation-reduction dye which is an indicator of cell growth and/or viability. The inoculum was prepared from a subculture of M. avium in 10 ml of Middlebrook 7H9 broth containing 0.2% (vol/vol) glycerol, 0.05% (vol/vol) Tween 80, and 10% albumin-dextrose-catalase (ADC) (Difco Laboratories, Detroit, Mich.). The cultures were used after 7 days of incubation at 37°C with 5% CO2. Initial drug dilutions were prepared in the appropriate medium, and subsequent twofold dilutions were made in 0.1 ml of 7H9 broth (plus glycerol and ADC) in 96-well microplates. Control wells consisted of medium alone (0.2 ml) and bacteria alone (0.1 ml of medium plus 0.1 ml of bacteria; final density, 2 × 105 CFU/well). Bacteria (0.1 ml) were added to the wells containing the drug dilutions. Each test was run in duplicate, and control experiments were done with the solvent alone. After 4 days of culture at 37°C with 5% CO2, 20 μl of 10× Alamar blue solution (Interchim, Montluçon, France, or Alamar Biosciences Inc., Sacramento, Calif.) was added to one medium control well and one bacterial control well, and the plates were further incubated for 24 h. When the color changed in the bacterial well (from the blue oxidized form to the pink reduced form), Alamar blue was added to the other wells and plates were incubated for 24 h. Results were read on a spectrophotometer at wavelengths of 540 and 620 nm using an automatic plate reader (Labsystems Multiskan RC, Helsinki, Finland) interfaced to a Macintosh computer. Percent inhibition was calculated as (1 − mean test well optical density [OD]/mean bacterial well OD) × 100. The MIC was the lowest drug concentration yielding at least 90% inhibition. Results obtained in preliminary experiments with anti-M. avium-M. intracellulare complex antibiotics (rifampin, clarithromycin, rifabutin) and quinolones (ofloxacin and sparfloxacin) were in agreement with those obtained by Collins and Franzblau (4).
MLR.
A correlation between the calculated and observed MICs of the 24 quinolones tested was obtained by MLR in order to predict the MICs of new quinolones. The MLR was performed with the 9R module of the BMDP program, which estimates regression equations for best subsets of predictor variables and provides detailed residual analysis. The lower Mallows' Cp was used to identify the best subsets in accordance with the equation Mallows' Cp = RSS/s2 − (n − 2p′), where RSS is the residual sum of squares for the best subset being tested, p′ is the number of independent variables in the subset (including the intercept), n is the number of cases, and s2 is the residual mean square based on the regression using all independent variables (26).
Virtual computational screening.
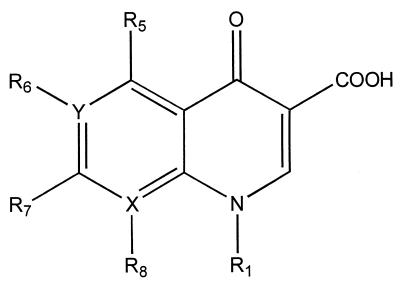
Computational screening was used to determine the influence of quinolone substituents on anti-M. avium-M. intracellulare complex activity. Virtual structures obtained by omission or substitution of radicals R1, R6, R7, or R8 on the three most active quinolones were designed (Fig. 1). Their TIs were calculated, and LDA and MLR equations were applied to predict anti-M. avium-M. intracellulare complex activity or inactivity and MICs.
FIG. 1.
General structure of the quinolones. X and Y can be carbon or nitrogen atoms, and the R1, R5, R6, R7, and R8 groups can be very diverse structures.
RESULTS
LDA.
Using a MIC cutoff of 32 μg/ml, the following equation (M), selected by LDA, classified the compounds as active against M. avium-M. intracellulare complex if M was >0 and inactive if M was <0: M = −2.6 + 20.13χch − 12.94χc + 42.54χcv + 25.66χch − 2.2G3v + 2.4G4v. Statistical parameters were as follows: n = 114, F = 30.79, Wilk's U = 0.37. The indices 4χc and 4χcv represent the quaternary ramifications, 3χch and 6χch reflect the presence of cycles of three and six atoms, respectively, and G3v and G4v furnish information about the transfer of intramolecular charges between atoms separated by distances of 3 and 4, respectively (13). The 3χch index made a marked contribution to the positivity of the equation, reflecting the role of the cyclopropyl substituent at N-1 to anti-M. avium-M. intracellulare complex activity. Sixty-one of 77 quinolones with cyclopropyl substitutions were active in vitro, and all of them had M values that were >0 (except PD135739).
The values of the discriminant function M and the corresponding prediction of activity or inactivity are presented in Table 1 for the training group and in Table 2 for the test group. In the training group (114 compounds), 51 of the 57 active compounds (89.5%) and 49 of the 57 inactive compounds (86.0%) were correctly classified. In the test group, which comprised 44 randomly chosen quinolones, 23 of the 27 active compounds (85.2%) and 14 of the 17 inactive compounds (82.4%) were correctly classified. Overall, the rate of correct classification was 86.7%.
TABLE 1.
Classification of quinolones in the training group by use of the discriminant function obtained by LDAa
| Quinolone | M | Predicted activity | Quinolone | M | Predicted activity | |
|---|---|---|---|---|---|---|
| Active | Inactive | |||||
| AMQ1 | 5.00 | + | PD107572 | −3.55 | − | |
| AMQ2 | 4.05 | + | PD108666 | −9.39 | − | |
| AMQ3 | 4.13 | + | PD108685 | −6.14 | − | |
| AMQ4 | 2.90 | + | PD108758 | −6.58 | − | |
| AMQ5 | 4.88 | + | PD108970 | −7.70 | − | |
| AMQ6 | 4.56 | + | PD109073 | −0.08 | − | |
| PD115311 | −2.45 | − | PD109205 | −6.53 | − | |
| PD117579 | 1.43 | + | PD109631 | 1.18 | + | |
| PD117596 | 1.78 | + | PD109813 | −2.02 | − | |
| PD118013 | 2.08 | + | PD110869 | −3.53 | − | |
| PD119421 | 4.03 | + | PD110920 | −6.13 | − | |
| PD119805 | 3.06 | + | PD111518 | −2.46 | − | |
| PD119977 | 4.48 | + | PD111700 | −5.46 | − | |
| PD120114 | 2.39 | + | PD111754 | −1.39 | − | |
| PD120316 | 3.81 | + | PD111834 | −6.46 | − | |
| PD120683 | 4.24 | + | PD112388 | −3.56 | − | |
| PD121285 | 1.74 | + | PD113721 | −7.14 | − | |
| PD122642 | 0.37 | + | PD114033 | −6.43 | − | |
| PD123982 | 2.91 | + | PD114111 | −1.06 | − | |
| PD125354 | 4.26 | + | PD114507 | −5.32 | − | |
| PD125554 | 7.04 | + | PD114843 | −5.54 | − | |
| PD125999 | 5.35 | + | PD114950 | −7.04 | − | |
| PD126889 | 4.77 | + | PD114980 | −4.86 | − | |
| PD131413 | 1.48 | + | PD115896 | −2.43 | − | |
| PD135042 | 5.49 | + | PD116427 | −5.28 | − | |
| PD135144 | 4.08 | + | PD117186 | −5.69 | − | |
| PD135739 | −0.95 | − | PD118362 | −5.38 | − | |
| PD137156 | 3.59 | + | PD119035 | −4.90 | − | |
| PD137954 | −0.59 | − | PD119244 | −6.90 | − | |
| PD138032 | 4.68 | + | PD119344 | −0.23 | − | |
| PD138926 | 3.39 | + | PD120261 | −1.03 | − | |
| PD139586 | 1.77 | + | PD120262 | −7.13 | − | |
| PD142421 | 8.27 | + | PD120818 | −1.13 | − | |
| PD143170 | 1.55 | + | PD120896 | −7.68 | − | |
| PD144881 | 2.61 | + | PD120978 | −4.46 | − | |
| PD158804 | 3.54 | + | PD121054 | −4.25 | − | |
| PD160788 | 5.85 | + | PD121264 | −4.37 | − | |
| PD160790 | 3.17 | + | PD123972 | 0.78 | + | |
| PD160792 | 3.94 | + | PD125639 | 1.82 | + | |
| PD160793 | 4.57 | + | PD127275 | 1.91 | + | |
| PD160826 | 2.23 | + | PD127367 | 0.90 | + | |
| PD161144 | 5.70 | + | PD130426 | −2.04 | − | |
| PD161314 | 5.18 | + | PD131161 | −2.87 | − | |
| PD161315 | 4.03 | + | PD131199 | −2.91 | − | |
| PD161316 | 6.34 | + | PD135723 | 0.40 | + | |
| PD161649 | 5.64 | + | PD135964 | −5.86 | − | |
| PD161650 | −0.86 | − | PD138362 | −0.75 | − | |
| PD161654 | −0.58 | − | PD139735 | −3.13 | − | |
| PD162279 | 3.03 | + | PD160336 | −0.53 | − | |
| PD162280 | 3.97 | + | PD161147 | −4.18 | − | |
| PD162281 | 4.73 | + | PD161318 | −1.12 | − | |
| PD162282 | 5.22 | + | PD161323 | −2.18 | − | |
| PD163051 | 3.02 | + | PD161546 | −5.00 | − | |
| PD163052 | 3.81 | + | PD162277 | −2.01 | − | |
| PD163054 | 5.16 | + | PD163451 | 0.11 | + | |
| PD163450 | 1.96 | + | PD163655 | −0.08 | − | |
| PD163755 | −2.39 | − | T3761 | 1.06 | + |
TABLE 2.
Classification of quinolones in the test gorup by use of the discriminant function obtained by LDAa
| Quinolone | M | Predicted activity |
|---|---|---|
| Active | ||
| PD116169 | −5.34 | − |
| PD118106 | 3.29 | + |
| PD121960 | 0.39 | + |
| PD122957 | 2.12 | + |
| PD122958 | 0.66 | + |
| PD123767 | 2.57 | + |
| PD125353 | 3.91 | + |
| PD126065 | 7.73 | + |
| PD126592 | 2.24 | + |
| PD127439 | 7.79 | + |
| PD129603 | 4.23 | + |
| PD131575 | 3.74 | + |
| PD135346 | 6.13 | + |
| PD138136 | 0.25 | + |
| PD142198 | 5.61 | + |
| PD143289 | 6.53 | + |
| PD143471 | 2.14 | + |
| PD160338 | −2.69 | − |
| PD160829 | −1.65 | − |
| PD161142 | −1.60 | − |
| PD161148 | 4.43 | + |
| PD161317 | 5.07 | + |
| PD161645 | 5.37 | + |
| PD162287 | 5.86 | + |
| PD163048 | 3.13 | + |
| PD163049 | 1.98 | + |
| PD163050 | 4.29 | + |
| Inactive | ||
| PD109131 | −0.10 | − |
| PD111103 | −4.26 | − |
| PD111752 | −0.41 | − |
| PD114554 | −4.71 | − |
| PD115717 | −7.14 | − |
| PD116507 | −7.75 | − |
| PD116700 | −6.07 | − |
| PD120755 | −5.20 | − |
| PD129626 | 1.82 | + |
| PD134545 | −6.30 | − |
| PD135305 | 4.88 | + |
| PD135522 | −4.39 | − |
| PD136576 | −0.86 | − |
| PD141494 | 2.17 | + |
| PD160352 | −4.79 | − |
| PD161141 | −3.13 | − |
| PD162278 | −1.21 | − |
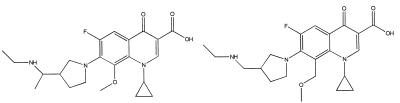
A good example of the discriminating capacity of the model was the result obtained with two quinolones which have the same molecular weight and large structural similarities but different anti-M. avium-M. intracellulare complex activities. The M function was 1.77 for PD139586, which is active in vitro, and −0.75 for PD138362, which is inactive (Fig. 2).
FIG. 2.
Structures of quinolones PD139586 (left) and PD138362 (right).
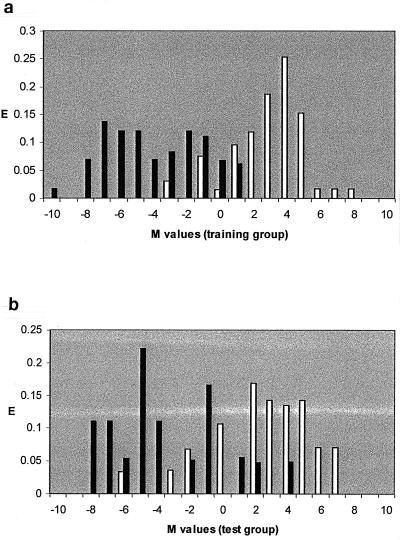
The pharmacological activity distribution diagrams (Fig. 3) show that for M values between −1 and 1.5 the classification of the drugs is uncertain, because of marked overlap of M values of several active and inactive drugs. In contrast, the highest activity expectancy occurred at M values >1.5. As our principal objective was to define a clear-cut difference between active and inactive quinolones, we assumed that quinolones with M values >1.5 were active and those with M values <−1 were inactive. Quinolones with M values between −1 and 1.5 were considered nonclassified.
FIG. 3.
(a) Activity distribution diagram of anti-M. avium activity in the training group (114 compounds), obtained after LDA statistical treatment. (b) Activity distribution diagram of anti-M. avium activity in the test group (44 compounds selected at random), obtained after LDA statistical treatment. E, expectancy of activity or inactivity; white bars, active quinolones; black bars, inactive ones.
When these criteria were applied to the 158 quinolones, only 2 of 50 compounds in the training group (PD125639 and PD127275) and 3 of 23 compounds in the test group (PD129626, PD135305, and PD141494) had M values >1.5 even though they were inactive in vitro. This meant that a cutoff of M = 1.5 yielded overall rates of correct classification of quinolones predicted as active of 96.0% in the training group and 87.0% in the test group.
Predicted and observed activities of 24 commercial quinolones.
The activities of 24 quinolones that had not been used to calculate the mathematical model were estimated, and the results were compared to those subsequently obtained in vitro (Alamar blue tests). Sixteen compounds were classified (Table 3). Assuming that a drug was inactive if it had a MIC of >10 μg/ml, 15 quinolones were correctly classified as active or inactive. The seven compounds that were predicted as active showed in vitro activity at low concentrations (between 0.2 and 5.4 μg/ml). Among the quinolones that were predicted as inactive, only lomefloxacin was misclassified.
TABLE 3.
Comparison of predictions of activity by molecular topology and LDA analysis versus experimental MICs
| Quinolone | LDA analysis result
|
Experimental MIC (μg/ml) | |
|---|---|---|---|
| M value | Predicted activity | ||
| Moxifloxacin | 3.90 | Active | 0.2 |
| Sparfloxacin | 5.06 | Active | 0.4 |
| Gatifloxacin | 4.54 | Active | 0.9 |
| Temafloxacin | −0.21 | N.C.a | 1.0 |
| Levofloxacin | 0.77 | N.C. | 2.1 |
| Ofloxacin | 0.77 | N.C. | 2.5 |
| Trovafloxacin | 1.69 | Active | 2.7 |
| Ciprofloxacin | 7.20 | Active | 2.8 |
| Lomefloxacin | −1.28 | Inactive | 4.5 |
| Clinafloxacin | 2.16 | Active | 5.0 |
| Grepafloxacin | 3.85 | Active | 5.4 |
| Fleroxacin | −0.18 | N.C. | 8.1 |
| Pefloxacin | 1.13 | N.C. | 10.0 |
| Norfloxacin | −0.95 | N.C. | 11.4 |
| Enoxacin | −1.78 | Inactive | 13.7 |
| Acrosoxacin | −0.68 | N.C. | 23.5 |
| Rufloxacin | 0.51 | N.C. | 31.0 |
| Irloxacin | −3.63 | Inactive | 47.2 |
| Pipemidic acid | −1.97 | Inactive | >250.0 |
| Flumequine | −1.96 | Inactive | >250.0 |
| Piromidic acid | −4.43 | Inactive | >250.0 |
| Nalidixic acid | −3.65 | Inactive | >250.0 |
| Cinoxacin | −2.24 | Inactive | >250.0 |
| Oxolinic acid | −3.25 | Inactive | >250.0 |
N.C., not classified.
MLR and virtual computational screening.
Based on the results of in vitro MICs, we used the MLR statistical technique to define a mathematical model able to correlate experimental and calculated MICs. The best correlation was obtained using the following equation: log (1/MIC) = −7.6 + 1.05χp − 2.09χp + 0.30χv. Statistical parameters were as follows: r2 = 0.88 (r2cv [cross-validation correlation] = 0.82), Mallows' Cp = 4.0, standard error = 0.35, P (significance) < 0.0001.
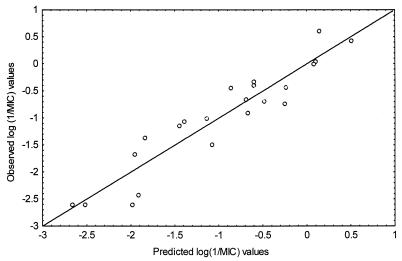
Experimental and calculated MICs of the 24 quinolones are presented, together with their structures, in Table 4, and their correlation is represented graphically in Fig. 4. The most-active quinolones were moxifloxacin, sparfloxacin, and gatifloxacin, with MICs of 0.2, 0.4, and 0.9 μg/ml, respectively, suggesting that the best structural characteristics for anti-M. avium-M. intracellulare complex activity are a quinoline basic nucleus, a fluorine atom at R6, a cyclopropyl group at N-1, and nucleophilic groups (F or OCH3) at R8.
TABLE 4.
Structures of the quinolones tested and MICs
| Quinolone | Basea | Group atb:
|
Anti-MAI activity (μg/ml)c
|
|||||
|---|---|---|---|---|---|---|---|---|
| R1 | R8 | R5 | R6 | R7 | Exper. | Calc. | ||
| Moxifloxacin | A | Cyclopropyl | -OCH3 | H | F | Piperidino-pyrrolidinyl | 0.2 | 0.7 |
| Sparfloxacin | A | Cyclopropyl | F | NH2 | F | 3′,5′-Methylpiperazinyl | 0.4 | 0.3 |
| Gatifloxacin | A | Cyclopropyl | -OCH3 | H | F | 3′-Methylpiperazinyl | 0.9 | 0.8 |
| Temafloxacin | A | 2,4-Difluorophenyl | H | H | F | 3′-Methylpiperazinyl | 1.0 | 0.8 |
| Levofloxacin | A | H | F | 4′-Methylpiperazinyl | 2.1 | 4.0 | ||
| Ofloxacin | A | H | F | 4′-Methylpiperazinyl | 2.5 | 4.0 | ||
| Trovafloxacin | B | 2,4-Difluorophenyl | H | F | Aza-bicyclohexanyl | 2.7 | 1.7 | |
| Ciprofloxacin | A | Cyclopropyl | H | H | F | 1-Piperazinyl | 2.8 | 7.3 |
| Lomefloxacin | A | -CH2CH3 | F | H | F | 3′-Methylpiperazinyl | 4.5 | 4.9 |
| Clinafloxacin | A | Cyclopropyl | Cl | H | F | 3′-Aminopyrrolidinyl | 5.0 | 3.1 |
| Grepafloxacin | A | Cyclopropyl | H | CH3 | F | 3′-Methylpiperazinyl | 5.4 | 1.8 |
| Fleroxacin | A | -CH2CH2F | F | H | F | 4′-Methylpiperazinyl | 8.1 | 4.7 |
| Pefloxacin | A | -CH2CH3 | H | H | F | 4′-Methylpiperazinyl | 10.0 | 13.6 |
| Norfloxacin | A | -CH2CH3 | H | H | F | 1-Piperazinyl | 11.4 | 24.5 |
| Enoxacin | B | -CH2CH3 | H | F | 1-Piperazinyl | 13.7 | 27.7 | |
| Acrosoxacin | A | -CH2CH3 | H | H | H | 4-Pyridinyl | 23.5 | 68.8 |
| Rufloxacin | A | H | F | 1-Piperazinyl | 31.0 | 11.9 | ||
| Irloxacin | A | -CH2CH3 | H | H | F | 1-Pyrrolyl | 47.2 | 89.1 |
| Pipemidic acid | C | -CH2CH3 | H | 1-Piperazinyl | >250.0 | 95.8 | ||
| Flumequine | A | H | F | H | >250.0 | 82.1 | ||
| Piromidic acid | C | -CH2CH3 | H | 1-Pyrrolidinyl | >250.0 | 330.9 | ||
| Nalidixic acid | B | -CH2CH3 | H | H | CH3 | >250.0 | 462.5 | |
| Cinoxacin | D | -CH2CH3 | H | H | >250.0 | 486.3 | ||
| Oxolinic acid | A | -CH2CH3 | H | H | >250.0 | 449.3 | ||
A, B, C, and D, basic nucleus quinoline, 1,8-naphtyridine, pyrido[2,3-d]pyrimidine, and 1,2-cinnoline, respectively.
Levofloxacin R1 to R8, R1-CH(CH3)CH2O-R8; ofloxacin R1 to R8, R1-CH(CH3)CH2O-R8; rufloxacin R1 to R8, R1-CH2CH2S-R8; flumequine R1 to R8, R1-CH(CH3)CH2CH2-R8; cinoxacin R6 to R7, R6-O-CH2-O-R7; oxolinic acid R6 to R7, R6-O-CH2-O-R7.
Exper., experimental; calc, calculated.
FIG. 4.
Correlation between experimental (y-axis) and calculated (x-axis) log (1/MIC) values for 24 quinolones. MICs were determined in vitro by culture and calculated by MLR analysis.
As the LDA and MLR equations were reliably predictive of in vitro activity, they were then applied to virtual structures derived from the three quinolones most active against M. avium-M. intracellulare complex (moxifloxacin, sparfloxacin, and gatifloxacin) by removing or substituting significant radicals. Initial analysis by LDA showed the crucial importance of the N-1 position, as omission of the cyclopropyl radical implied LDA values that were <0 for the three drugs, i.e., anti-M. avium-M. intracellulare complex inactivity. The other changes were less determinant, yielding positive LDA values, and a more accurate analysis was conducted with the MLR technique (Table 5). Replacement of the N-1 cyclopropyl by tert-butyl or 2,4-difluorophenyl radicals implied similar or lower MICs. Deletion of R6 resulted in a three- to fivefold increase in the MIC but not in a complete loss of activity. Piperazine or an equivalent group on R7 was determinant for anti-M. avium-M. intracellulare complex activity, as its replacement by a simple pyrrolidinyl or 3′-amino-pyrrolidinyl radical resulted in predicted MICs 2 to 18 times higher. Similarly, the presence of R8 seemed essential for activity, because its omission implied a very large increase in the MIC in every case.
TABLE 5.
Predicted MICs by computational screening and MLR function on virtual quinolones derived by modification of moxifloxacin, sparfloxacin, and gatifloxacin
| Change | MIC (μg/ml) of:
|
||
|---|---|---|---|
| Moxifloxacin | Sparfloxacin | Gatifloxacin | |
| Unchanged (experimental MIC) | 0.2 | 0.4 | 0.9 |
| R1 changesa | |||
| t-Butyl instead of cyclopropyl | 0.1 | 0.2 | 0.2 |
| 2,4-difluorophenyl instead of cyclopropyl | 0.3 | 0.4 | 0.5 |
| R6 omitted | 1.1 | 1.2 | 2.5 |
| R7 changesa | |||
| Pyrrolidinyl instead of original groups | 3.6 | 4.3 | 3.6 |
| 3′-Amino-pyrrolidinyl instead of original groups | 2.1 | 2.5 | 2.1 |
| R8 omitted | 4.8 | 3.5 | 9.9 |
| R8 changed (CH3 instead OCH3 or F) | 1.1 | 0.4 | 1.9 |
R1 and R7 suppression has no sense, as all the quinolones used in the study had a substituent at these positions.
DISCUSSION
Molecular topology is a very useful tool for describing molecular structures and has been used for efficient analysis of QSAR data (10). This method has proven its utility for the prediction of diverse physical, chemical, and biological properties in different groups of compounds (14) and has also been used with success for the design of new drugs (10, 12, 13). In previous studies Klopman et al. developed a MULTICASE fragment approach that was used to analyze the structure-activity relationships of quinolones against mycobacteria and to discriminate between active and inactive compounds (23). Using LDA, we have confirmed that molecular-topology methods can reliably discriminate between drugs according to their in vitro activities. In addition we show that MICs of active compounds can also be predicted using an MLR model.
LDA on a panel of 158 active and inactive quinolones correctly classified 96 and 87% of the quinolones predicted as active in the training group and test group, respectively, showing the excellent predictive capacity of the model. When LDA was applied to 24 commercial quinolones that had not been used to define the model and whose MICs were subsequently determined in vitro, 16 quinolones were assigned to the active or inactive group and 15 showed the expected activity or inactivity. Eight quinolones were not classified because of intermediate M values from the LDA equation, but it should be noted that these drugs also had intermediate in vitro activities. The most important result was the correct prediction of seven active quinolones which had low in vitro MICs, and specially the correct prediction of three of them (moxifloxacin, sparfloxacin, and gatifloxacin) which had MICs of <1 μg/ml.
In addition to the ability of a model to discriminate between active and inactive compounds, we considered it important that a model be able to predict the effective concentration of quinolones against M. avium. For this purpose, an MLR analysis was developed. It was applied to the in vitro values of the 24 quinolones and defined an equation which accurately correlated experimental and calculated MICs (r2 = 0.88). Furthermore, the very good predictive capacity was confirmed by a cross-validation test (r2cv = 0.82). In conclusion, we consider that a combination of structural description by using TIs and statistical treatment by LDA and MLR can reliably select new quinolones effective against the M. avium-M. intracellulare complex. The obtained prediction models can readily be applied to large databases of quinolones to identify active structures and to estimate MICs.
In addition, the combination of LDA and MLR proved useful to investigate the influence of different quinolone radicals on anti-M. avium-M. intracellulare complex activity. The role of several radicals in quinolone activity has already been demonstrated by Klopman et al. (20, 23), but using MLR allowed estimates of their influence on MICs. Virtual structures were designed from modifications of moxifloxacin, sparfloxacin, and gatifloxacin, and their TIs were calculated. A first study with LDA was sufficiently determinant to show the importance of a cyclopropyl group at the N-1 position, which is present on the three above-mentioned quinolones, as its omission resulted in negative M values, implying total anti-M. avium-M. intracellulare inactivity. Using MLR as a complement to LDA allowed the identification of other structural changes that did not result in negative M values (and thus could not be discriminated by LDA) but that had a significant impact on anti-M. avium-M. intracellulare complex activity, as they induced marked changes in MICs determined with the MLR equation. Specifically, tert-butyl and 2,4-difluorophenyl radicals in the N-1 position produced predicted MICs as good as or better than experimental MICs of moxifloxacin, gatifloxacin, and sparfloxacin, confirming the experiments previously performed by Klopman (20) to test the effectiveness of these groups. A loss of activity was found when the R6 position was not occupied by an F atom, confirming the importance of this radical, which is suspected to be involved in cellular penetration. Nevertheless, its influence could be less than that on other bacteria, since some activity was found against the M. avium-M. intracellulare complex. Anti-M. avium-M. intracellulare complex activity was theoretically decreased by a pyrrolidinyl group or a 3′-amino-pyrrolidinyl group relative to the original groups in R7: a 3′-methyl-piperazinyl group (gatifloxacin), a 3′,5′-dimethyl-piperazinyl group (sparfloxacin), and a pyrrolidinyl group attached to a six-member ring (moxifloxacin). Finally, the determining role of nucleophilic substituents such as F and OCH3 radicals on R8 was demonstrated, as its suppression resulted in a very marked increase in estimated MICs. These results may reflect a unique characteristic of the anti-M. avium-M. intracellulare complex activity of quinolones compared to their activity against other bacteria, in which the influence of the R7 and R8 positions is less marked.
On the basis of these results, these predictive models might prove useful in the design of new quinolones with improved anti-M. avium-M. intracellulare complex activity, providing considerable cost savings relative to in vitro and in vivo experimental models (1).
ACKNOWLEDGMENTS
R. Gozalbes is indebted to the association Ensemble contre le SIDA and the French Ministry of Foreign Affairs for their financial support for this work conducted in the Parasitology-Mycology Laboratory and ITODYS, at University Paris-7, Paris, France. The members of the Molecular Connectivity and Drug Design Research Unit are grateful for the support by Generalitat Valenciana through project GV99-91-1-12.
We thank Bayer Pharma, Esteve, Glaxo Wellcome, Grünenthal, Hoechst-Marion-Roussel, Mediolanum Farmaceutici, Monsanto Searle, Parke-Davis, and Sanofi Winthrop laboratories for supply of compounds and David D. Young for reviewing the manuscript.
REFERENCES
- 1.Brun-Pascaud M, Rajagopalan-Levasseur P, Chau F, Bertrand G, Garry L, Derouin F, Girard P M. Drug evaluation of concurrent Pneumocystis carinii, Toxoplasma gondii, and Mycobacterium avium complex infections in a rat model. Antimicrob Agents Chemother. 1998;42:1068–1072. doi: 10.1128/aac.42.5.1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bryskier A. Fluoroquinolones: mechanisms of action and resistance. Int J Antimicrob Agents. 1993;2:151–184. doi: 10.1016/0924-8579(93)90052-7. [DOI] [PubMed] [Google Scholar]
- 3.Bryskier A, Chantot J F. Classification and structure-activity relationships of fluoroquinolones. Drugs. 1995;49(Suppl. 2):16–28. doi: 10.2165/00003495-199500492-00005. [DOI] [PubMed] [Google Scholar]
- 4.Collins L, Franzblau S. Microplate Alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41:1004–1009. doi: 10.1128/aac.41.5.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Julián-Ortiz J V, Gálvez J, Muñoz-Collado C, García-Domenech R, Gimeno-Cardona C. Virtual Combinatorial Syntheses and Computational Screening of new potential anti-Herpes compounds. J Med Chem. 1999;42:3308–3314. doi: 10.1021/jm981132u. [DOI] [PubMed] [Google Scholar]
- 6.Domagala J M. Structure-activity and structure-side-effect relationships for the quinolone antibacterials. J Antimicrob Chemother. 1994;33:685–706. doi: 10.1093/jac/33.4.685. [DOI] [PubMed] [Google Scholar]
- 7.Doucet J P, Weber J. Computer-aided molecular design: theory and applications. London, United Kingdom: Academic Press; 1996. [Google Scholar]
- 8.Draper N R, Smith H. Applied regression analysis. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1981. pp. 101–102. [Google Scholar]
- 9.Gálvez J, García-Domenech R, de Gregorio Alapont C, de Julián-Ortiz J V, Popa L J. Pharmacological distribution diagrams: a tool for de novo drug design. J Mol Graphics. 1996;14:272–276. doi: 10.1016/s0263-7855(96)00081-1. [DOI] [PubMed] [Google Scholar]
- 10.Gálvez J, García-Domenech R, de Julián-Ortiz J V, Soler R. Topological approach to drug design. J Chem Inf Comput Sci. 1994;32:272–284. doi: 10.1021/ci00024a017. [DOI] [PubMed] [Google Scholar]
- 11.Gálvez J, García R, Salabert M T, Soler R. Charge indexes. New topological descriptors. J Chem Inf Comput Sci. 1994;34:520–525. [Google Scholar]
- 12.Gozalbes R, Gálvez J, García-Domenech R, Derouin F. Molecular search of new active drugs against Toxoplasma gondii. SAR QSAR Environ Res. 1999;10:47–60. doi: 10.1080/10629369908039165. [DOI] [PubMed] [Google Scholar]
- 13.Gozalbes R, Gálvez J, Moreno A, García-Domenech R. Discovery of new antimalarial compounds by use of molecular connectivity techniques. J Pharm Pharmacol. 1999;51:111–117. doi: 10.1211/0022357991772204. [DOI] [PubMed] [Google Scholar]
- 14.Gozalbes R, de Julián-Ortiz J V, Antón-Fos G M, Gálvez J, García-Domenech R. Prediction of chromatographic properties of organophosphorus insecticides by molecular connectivity. Chromatographia. 2000;51:331–337. [Google Scholar]
- 15.Jennrich R, Sampson P. Stepwise discriminant analysis. In: Dixon W J, editor. BMD biomedical computer programs. Berkeley: University of California Press; 1979. pp. 339–357. [Google Scholar]
- 16.Johnson S M, Roberts G D. In vitro activity of ciprofloxacin and ofloxacin against the M. avium-intracellulare complex. Diagn Microbiol Infect Dis. 1987;7:89–91. doi: 10.1016/0732-8893(87)90077-0. [DOI] [PubMed] [Google Scholar]
- 17.Kier L B, Hall L H. Molecular connectivity in structure-activity analysis. Letchworth, England: John Wiley & Sons; 1986. pp. 225–246. [Google Scholar]
- 18.Kier L B, Hall L H. An electrotopological-state index for atoms in molecules. Pharm Res. 1990;7:801–807. doi: 10.1023/a:1015952613760. [DOI] [PubMed] [Google Scholar]
- 19.Kleinberg M L, Wanke L A. New approaches and technologies in drug design and discovery. Am J Health-Syst Pharm. 1995;52:1323–1336. doi: 10.1093/ajhp/52.12.1323. [DOI] [PubMed] [Google Scholar]
- 20.Klopman G, Fercu D, Renau T E, Jacobs M R. N-1-tert-Butyl-substituted quinolones: in vitro anti-Mycobacterium avium activities and structure-activity relationship studies. Antimicrob Agents Chemother. 1996;40:2637–2643. doi: 10.1128/aac.40.11.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klopman G, Li J-Y, Wang S, Pearson A J, Chang K, Jacobs M R, Bajaksouizian S, Ellner J J. In vitro anti-Mycobacterium avium activities of quinolones: predicted active structures and mechanistic considerations. Antimicrob Agents Chemother. 1994;38:1794–1802. doi: 10.1128/aac.38.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klopman G, Wang S, Jacobs M R, Bajaksouizian S, Edmonds K, Ellner J J. Anti-Mycobacterium avium activity of quinolones: in vitro activities. Antimicrob Agents Chemother. 1993;37:1799–1806. doi: 10.1128/aac.37.9.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klopman G, Wang S, Jacobs M R, Ellner J J. Anti-Mycobacterium avium activity of quinolones: structure-activity relationship studies. Antimicrob Agents Chemother. 1993;37:1807–1815. doi: 10.1128/aac.37.9.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahana R. Virtual combinatorial chemistry. Sci Am. 1997;241:56–58. . (In French.) [Google Scholar]
- 25.Leysen D C, Haemers A, Pattyn S R. Mycobacteria and the new quinolones. Antimicrob Agents Chemother. 1989;33:1–5. doi: 10.1128/aac.33.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallows C L. Some comments on Cp. Technometrics. 1973;15:661–675. [Google Scholar]
- 27.Perronne C, Gikas A, Truffot-Pernot C, Grosset J, Vildé J L, Pocidalo J J. Activities of sparfloxacin, azithromycin, temafloxacin, and rifapentine compared with that of clarithromycin against multiplication of Mycobacterium avium complex within human macrophages. Antimicrob Agents Chemother. 1991;35:1356–1359. doi: 10.1128/aac.35.7.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yajko D M, Nassos P S, Hadley W K. Broth microdilution testing of susceptibilities to 30 antimicrobial agents of Mycobacterium avium strains from patients with acquired immune deficiency syndrome. Antimicrob Agents Chemother. 1987;31:1579–1584. doi: 10.1128/aac.31.10.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Young L S, Inderlied C B, Berlin O G W, Gottlied M J. Mycobacterial infections and AIDS patients, with an emphasis on the Mycobacterium avium complex. Rev Infect Dis. 1986;8:1024–1033. doi: 10.1093/clinids/8.6.1024. [DOI] [PubMed] [Google Scholar]
- 30.Young L S, Berlin O G W, Inderlied C B. Activity of ciprofloxacin and other fluorinated quinolones against mycobacteria. Am J Med. 1987;82(Suppl. 4A):23–26. [PubMed] [Google Scholar]