Abstract
The apicoplast, a plastid-like organelle of Toxoplasma gondii, is thought to be a unique drug target for quinolones. In this study, we assessed the in vitro activity of quinolones against T. gondii and developed new quantitative structure-activity relationship models able to predict this activity. The anti-Toxoplasma activities of 24 quinolones were examined by means of linear discriminant analysis (LDA) using topological indices as structural descriptors. In parallel, in vitro 50% inhibitory concentrations (IC50s) were determined in tissue culture. A multilinear regression (MLR) analysis was then performed to establish a model capable of classifying quinolones by in vitro activity. LDA and MLR analysis were applied to virtual structures to identify the influence of each atom or substituent of the quinolone ring on anti-Toxoplasma activity. LDA predicted that 20 of the 24 quinolones would be active against T. gondii. This was confirmed in vitro for most of the quinolones. Trovafloxacin, grepafloxacin, gatifloxacin, and moxifloxacin were the quinolones most potent against T. gondii, with IC50s of 0.4, 2.4, 4.1, and 5.1 mg/liter, respectively. Using MLR analysis, a good correlation was found between measured and predicted IC50s (r2 = 0.87, cross-validation r2 = 0.74). MLR analysis showed that the carboxylic group at position C-3 of the quinolone ring was not essential for anti-Toxoplasma activity. In contrast, activity was totally dependent on the presence of a fluorine at position C-6 and was enhanced by the presence of a methyl group at C-5 or an azabicyclohexane at C-7. A nucleophilic substituent at C-8 was essential for the activity of gatifloxacin and moxifloxacin.
The discovery of a novel organelle in apicomplexan parasites and its characterization in Toxoplasma gondii offers new opportunities for pharmacological research on several protozoa of major medical importance (16). This organelle, the apicoplast, is a plastid-like structure which was probably acquired by secondary endosymbiosis from a green alga (15). The function of the apicoplast is still not clear, but the presence of this procaryotic structure within T. gondii presents a unique therapeutic target. Fichera and Roos showed that several antibiotics, such as azithromycin and ciprofloxacin, could inhibit DNA replication within the apicoplast and thus inhibited Toxoplasma growth (6). That study confirmed the previously well-known effect of macrolides on T. gondii but also revealed fluoroquinolones as candidate anti-Toxoplasma drugs. However, other studies performed in vitro and in vivo failed to confirm the activity of ciprofloxacin against T. gondii and showed that, among the fluoroquinolones, only trovafloxacin and some of its derivatives inhibited Toxoplasma growth at micromolar concentrations (9, 10). Better knowledge of the structure-activity relationships of quinolones against T. gondii is thus needed.
The aims of this work were (i) to assess quinolone activity against T. gondii by using a previously described model of virtual prediction (8) and by testing the inhibitory effects of 24 quinolones and fluoroquinolones in vitro, (ii) to establish quantitative structure-activity relationship (QSAR) models based on molecular topology and multilinear regression (MLR) analysis in order to predict the 50% inhibitory concentrations (IC50s) of quinolones for T. gondii, and (iii) to identify the basic chemical structures responsible for the anti-T. gondii activity of quinolones by using atom level topological indices and by testing computer-generated virtual structures of quinolones (2, 13).
MATERIALS AND METHODS
The 24 quinolones studied were cinoxacin, enoxacin, flumequin, nalidixic acid, norfloxacin, oxolinic acid, pipemidic acid, piromidic acid, sparfloxacin, temafloxacin, trovafloxacin (Sigma Aldrich, Paris, France), ciprofloxacin, moxifloxacin (Bayer Pharma), irloxacin (Laboratorios Dr. Esteve), grepafloxacin (Glaxo Wellcome), gatifloxacin (Grünenthal), levofloxacin, ofloxacin (Hoechst Marion Roussel), rufloxacin (Mediolanum Farmaceutici), lomefloxacin (Monsanto Searle), clinafloxacin (Parke-Davis), fleroxacin (Roche), pefloxacin (Roger Bellon), and acroxacin (Sanofi Winthrop).
Assessment of quinolone anti-Toxoplasma activity by LDA.
We used a mathematical model previously described for virtual identification of anti-T. gondii drugs (8). Briefly, linear discriminant analysis (LDA) is a pattern recognition method which provides a classification model based on the combination of variables that best predicts the category (active or inactive) to which a given compound belongs. The independent variables in this study were topological indices (TIs) that were calculated for each drug, and the discrimination property was in vitro anti-T. gondii activity. Two LDA equations (T1 and T2) were obtained. Equation T1 discriminates drugs that are active against T. gondii (T1 > 0) from any other drug with no antiprotozoal activity (T1 < 0). Equation T2 separates anti-Toxoplasma drugs (T2 > 0) from antiprotozoals with no anti-Toxoplasma activity (T2 < 0). Both equations were reliably predictive of in vitro activity, as more than 90% of the drugs included in the test groups have been correctly classified by their anti-Toxoplasma activity (8).
In vitro assessment of quinolone anti-Toxoplasma activity.
Stock solutions of each drug were prepared at 2 mg/ml in dimethyl sulfoxide, and serial dilutions were then prepared in distilled water.
In vitro studies were performed with the virulent RH strain of T. gondii, which was maintained in mice by intraperitoneal passage every 2 days. For each experiment, tachyzoites were collected from the peritoneal cavity and then resuspended in physiological saline. Tissue culture and drug tests were carried out using MRC5 fibroblasts as previously described (3), with minor modifications. Briefly, confluent monolayers prepared in 96-well tissue culture plates were inoculated with 2,000 fresh tachyzoites. After 4 h, drugs at various concentrations were added to the culture medium and the plates were incubated for a further 72 h. Each drug was tested at 10 concentrations ranging from 0.01 to 200 mg/liter (final concentration in the culture). Each concentration was tested in eight replicate wells and in two replicate culture plates. Each culture plate comprised eight negative control wells (without T. gondii) and eight positive control wells (without a drug). After incubation, the plates were examined microscopically for cytopathic effects and then fixed with cold methanol for 5 min. Toxoplasma growth was assessed by enzyme-linked immunoassay directly on fixed cultures by using a peroxidase-labeled monoclonal antibody directed against the T. gondii SAG-1 surface protein. After addition of the substrate, spectrophotometric readings were performed at a wavelength of 405 nm with blanking on the negative control wells. For each well, the results were expressed as optical density values. The optical density values were plotted as a function of the logarithm of the concentration, and a linear regression model was used to summarize the concentration-effect relationship and to determine the IC50 (3).
MLR.
The 24 quinolones were characterized by using a set of 145 TIs specific for each molecule (11). We used topological descriptors provided by the MOLCONN-Z software, version 3.50 (L. H. Hall, Eastern Nazarene College, Quincy, Mass.), and especially the Kier & Hall connectivity indexes (up to 10th order). We also calculated some descriptors as charge indexes (7) using the Etopo 11 software developed in our research unit.
The calculated TIs were related by MLR to the observed IC50s of the 24 quinolones to predict the IC50s of new quinolones. MLR was performed with the 9R module of the BMDP program (W. J. Dixon, BMDP Statistical Software, University of California, Berkeley), which estimates regression equations for best subsets of predictor variables and provides detailed residual analysis. The lower Mallows Cp was used to identify the best subsets. Mallows' Cp = RSS/s2 − (n − 2p′), where RSS is the residual sum of squares for the best subset being tested, p′ is the number of independent variables in the subset (including the intercept), n is the number of cases, and s2 is the residual mean square based on the regression using all independent variables (14).
Topological superposition with atomic E-state indices to identify basic pharmacophore structures.
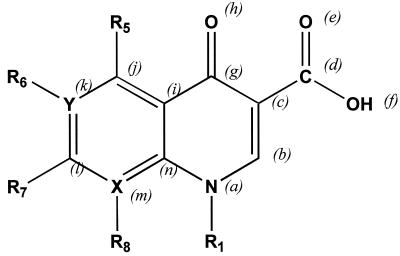
Usual structural descriptions based on topological descriptors are based mainly on the whole molecule, and the QSAR models thereby obtained therefore offer little insight into drug pharmacophores. We therefore used of a new kind of topological descriptor—E-state indices—specific for each atom (12). Briefly, the E-state index of a given atom reflects its electronic and topological features, taking into account the interaction with the rest of the molecule, particularly the relationship between valence and sigma electrons. These topological indices were related to the anti-T. gondii activities of the 24 quinolones. We focused on atoms which are always present in the basic structure of quinolones. The E-state indices were calculated for each atom in the set (numbered as shown in Fig. 1) and were related by MLR to the IC50s of the 24 quinolones.
FIG. 1.
General quinolone structure. X and Y can be carbon or nitrogen atoms, and the R1, R5, R6, R7, and R8 groups can be very diverse structures. The basic structure selected to study the pharmacophoric structure by using E-state indices is indicated by italic letters, representing the atoms studied.
Virtual computational screening.
Computational screening was used to determine the influence of quinolone substituents on anti-Toxoplasma activity and to help select new quinolones with improved efficacy. Virtual structures were designed by omission or substitution of radical R1, R5, R6, R7, or R8 on the most active quinolones tested (trovafloxacin, grepafloxacin, gatifloxacin, and moxifloxacin). Figure 1 shows the general quinolone structure and radical numbers. The TIs were calculated, and LDA and MLR equations were used to determine their activity or inactivity and IC50s, respectively.
RESULTS
Anti-Toxoplasma activities of quinolones determined by LDA and in vitro tests.
The T1 and T2 equations obtained by LDA were applied to the 24 quinolone structures (Table 1). Among the 24 quinolones tested, 20 had positive T1 values, indicating theoretical activity against T. gondii. T2 values were negative for 21 of the 24 compounds, indicating that the anti-Toxoplasma effects of these drugs cannot be distinguished from general antiprotozoan activity. Most of the quinolones tested had growth-inhibitory activity in vitro, although some were effective only at high concentrations. IC50s ranged from 0.4 mg/liter for trovafloxacin to >100 mg/liter for cinoxacin and levofloxacin (Table 2).
TABLE 1.
LDA values of several quinolones submitted to equations T1 and T2
| Quinolone | T1 | T2 |
|---|---|---|
| Acrosoxacin | −1.1 | −10.2 |
| Cinoxacin | 0.5 | −2.5 |
| Clinafloxacin | −2.8 | −12.8 |
| Ciprofloxacin | 0.4 | −11.1 |
| Enoxacin | 4.4 | −0.9 |
| Fleroxacin | 2.3 | −3.6 |
| Flumequin | 1.2 | −5.7 |
| Gatifloxacin | 4.1 | −10.4 |
| Grepafloxacin | 2.3 | 5.8 |
| Irloxacin | 3.3 | −10.6 |
| Levofloxacin | 7.3 | −5.1 |
| Lomefloxacin | 2.5 | −3.8 |
| Moxifloxacin | 2.5 | −14.6 |
| Nalidixic acid | −2.3 | 3.0 |
| Norfloxacin | 3.5 | −6.2 |
| Ofloxacin | 7.3 | −5.1 |
| Oxolinic acid | 0.4 | −5.6 |
| Pefloxacin | 4.0 | −3.4 |
| Pipemidic acid | 4.6 | 0.1 |
| Piromidic acid | 3.8 | −0.3 |
| Rufloxacin | 6.8 | −9.9 |
| Sparfloxacin | −1.7 | −11.5 |
| Temafloxacin | 2.3 | −19.0 |
| Trovafloxacin | 5.1 | −17.0 |
TABLE 2.
Structures of the 24 quinolones studied, in order of decreasing experimental and calculated IC50s against T. gondii
| Quinolone | Basea | R1 | R8 | R5 | R6 | R7 | IC50 (mg/liter)
|
|
|---|---|---|---|---|---|---|---|---|
| Exptl | Calculated | |||||||
| Trovafloxacin | B | 2,4-Difluorophenyl | H | F | Azabicyclohexane | 0.4 | 0.5 | |
| Grepafloxacin | A | Cyclopropyl | H | CH3 | F | 3′-Methylpiperazine | 2.4 | 4.4 |
| Gatifloxacin | A | Cyclopropyl | −OCH3 | H | F | 3′-Methylpiperazine | 4.1 | 9.0 |
| Moxifloxacin | A | Cyclopropyl | −OCH3 | H | F | Piperidinopyrrolidine | 5.1 | 2.5 |
| Temafloxacin | A | 2,4-Difluorophenyl | H | H | F | 3′-Methylpiperazine | 11.5 | 17.3 |
| Clinafloxacin | A | Cyclopropyl | Cl | H | F | 3′-Aminopyrrolidine | 15.0 | 15.6 |
| Acrosoxacin | A | −CH2CH3 | H | H | H | 4′-Pyridine | 20.3 | 27.9 |
| Enoxacin | B | −CH2CH3 | H | F | Piperazine | 20.3 | 26.1 | |
| Lomefloxacin | A | −CH2CH3 | F | H | F | 3′-Methylpiperazine | 21.2 | 21.2 |
| Rufloxacin | A | —b | —b | H | F | Piperazine | 22.3 | 20.6 |
| Irloxacin | A | −CH2CH3 | H | H | F | Pyrrole | 22.4 | 24.2 |
| Piromidic acid | C | −CH2CH3 | H | Pyrrolidine | 26.2 | 40.7 | ||
| Sparfloxacin | A | Cyclopropyl | F | NH2 | F | 3′,5′-Methylpiperazine | 39.5 | 20.1 |
| Flumequin | A | —c | —c | H | F | H | 40.6 | 43.8 |
| Fleroxacin | A | −CH2CH2F | F | H | F | 4′-Methylpiperazine | 46.8 | 44.6 |
| Oxolinic acid | A | −CH2CH3 | H | H | —d | —d | 47.2 | 46.5 |
| Norfloxacin | A | −CH2CH3 | H | H | F | Piperazine | 48.3 | 47.7 |
| Ofloxacin | A | —e | —e | H | F | 4′-Methylpiperazine | 53.6 | NCf |
| Nalidixic acid | B | −CH2CH3 | H | H | CH3 | 73.6 | 68.7 | |
| Ciprofloxacin | A | Cyclopropyl | H | H | F | Piperazine | 79.4 | 27.9 |
| Pefloxacin | A | −CH2CH3 | H | H | F | 4′-Methylpiperazine | 77.7 | 141.7 |
| Pipemidic acid | C | −CH2CH3 | H | Piperazine | 116.4 | 39.3 | ||
| Levofloxacin | A | —e | —e | H | F | 4′-Methylpiperazine | 159.6 | NC |
| Cinoxacin | D | −CH2CH3 | H | H | —d | —d | 200.0 | NC |
| K1g | B | 2,4-Difluorophenyl | H | F | 2-Methyl-6-amino-3aza-bicyclo[3.1.0.]hexyl | 0.2 | 0.2 | |
| K2 | B | 2,4-Difluorophenyl | H | F | 6-Aminomethyl-3aza-bicyclo[3.1.0.]hexyl | 0.2 | 0.6 | |
| K3 | B | 2,4-Difluorophenyl | CH3 | F | 6-Amino-3-azabicyclo[3.1.0.]hexyl | 0.3 | 0.1 | |
| K4 | B | Cyclopropyl | H | F | 6-Amino-3-azabicyclo[3.1.0.]hexyl | 0.6 | 0.8 | |
| K5 | B | 2,4-Difluorophenyl | H | F | 6-Amino-3-azabicyclo[3.1.0.]hexyl | 0.9 | 0.4 | |
| K6 | A | 2,4-Difluorophenyl | H | H | F | 6-Amino-3-azabicyclo[3.1.0.]hexyl | 1.1 | 0.7 |
| K7 | B | 2,4-Difluorophenyl | H | F | 2-Methyl-6-aminomethyl-3aza-bicyclo[3.1.0.]hexyl | 1.5 | 0.2 | |
| K8 | B | 2,4-Difluorophenyl | H | F | 5-Amino-3-azabicyclo[3.1.0.]hexyl | 3.0 | 5.2 | |
| K9 | B | Cyclopropyl | H | F | 6-Methylamino-3aza-bicyclo[3.1.0.]hexyl | 4.3 | 1.0 | |
| K10 | B | Cyclopropyl | H | F | 5-Methylamino-3-azabicyclo[3.1.0.]hexyl | 4.5 | 12.1 | |
| K11 | B | 2,4-Difluorophenyl | H | F | 6-Methylamino-3aza-bicyclo[3.1.0.]hexyl | 4.3 | 0.5 | |
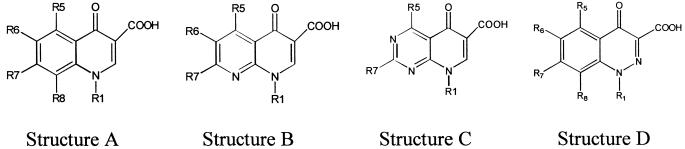
Structures A, B, C, and D represent the basic quinoline nucleus, 1,8-naphthyridine, pyrido[2,3-d]pyrimidine, and 1,2-cinnoline, respectively.
R1-CH2CH2S-R8.
R1-CH(CH3)CH2CH2-R8.
R6-O-CH2-O-R7.
R1-CH(CH3)CH2-O-R8.
NC, not calculated.
Quinolone structures K1 to K11 and IC50s were described by Khan et al. (10).
MLR analysis.
From the results of in vitro testing of the 24 quinolones, the MLR technique was used to establish a QSAR model able to correlate the chemical structures with in vitro activity. In this analysis, levofloxacin and ofloxacin were not considered because they have the same plane formula, implying the same TIs. Preliminary analysis also led us to remove cinoxacin, as its particular structure and experimental IC50 (200 mg/liter) resulted in an incorrect model.
The best equation obtained with the remaining 21 quinolones was log (1/IC50) = −6.1 + 0.3G2 − 0.6G3 − 9.3J4 + 18.1J4v + 0.3PR1. The statistical parameters were as follows: r2 = 0.87 (cross-validation r2 [r2cv] = 0.74), Mallows' Cp = 6.0, standard error = 0.24, and P < 0.0001.
The principal selected descriptors were charge indexes (G2, G3, J4, and J4v), which take into account the distribution of intramolecular charges at different topological distances (7). Their presence is logical because of the multiple points of union to the basic quinolone structure and the presence of heteroatoms such as N and F. The other selected index was PR1, which reflects the degree of ramification of the structure (PR1 increases with ramification, and the IC50 falls as a result).
The experimental and calculated IC50s of the 24 quinolones are presented, together with their structures, in Table 2. The correlation is represented graphically in Fig. 2.
FIG. 2.
Comparison between experimental (y axis) and calculated (x axis) log (1/IC50) values for 24 quinolones. IC50s were determined in vitro by culture and calculated by MLR analysis.
From both the experiments and the mathematical model, four fluoroquinolones emerged as more active than the other compounds, as their IC50s were below 10 mg/liter. Trovafloxacin was the most active drug, with experimental and calculated IC50s below 0.5 mg/liter, followed by grepafloxacin, gatifloxacin, and moxifloxacin.
To further validate the predictive model, the MLR equation was also applied to 11 trovafloxacin analogs whose structures and IC50s were recently published (10). Although the technique used to determine the IC50 was slightly different from that used in our study, very good agreement was obtained for 10 of 11 compounds between our predicted IC50s and those obtained experimentally by Khan et al. (10).
Identification of pharmacophoric structure.
To identify the basic quinolone atoms which contribute the most to anti-Toxoplasma activity, fourteen E-state indices representing all of the atoms in the basic structure (designated as shown in Fig. 1) were related to the IC50s of the 24 quinolones. The best equation was IC50 = 871.5 + 55.4 S (b) − 72.5 S (h) + 4.3 S (k) − 24.7 S (l). The statistical parameters were n = 22, r2 = 0.74, and r2cv = 0.42.
The statistical parameters indicate that the atomic position only partially correlates with the IC50. The mean values (and ranges) of the indices were 1.39 (0.90 to 3.85) for S (b), 12.37 (11.91 to 12.78) for S (h), 0.13 (−1.02 to 4.24) for S (k), and 0.40 (−0.28 to 1.42) for S (l). This equation reveals the atomic positions which most contribute to lowering of the calculated IC50. The most contributory structures were the carbonyl group—represented by (h)—and position 2, both of which are basic quinolone structures, and, to a lesser extent, positions 6 and 7, which usually bear a fluorine atom and a substitutive radical, respectively. The carboxylic group at position 3 did not appear in the equation.
Virtual computational screening of some analogs of the most active quinolones was used to identify the influence of radicals on anti-Toxoplasma activity. As the LDA and MLR equations were reliably predictive of in vitro activity, they were then applied to virtual structures derived from the four quinolones most active against T. gondii, i.e. trovafloxacin, grepafloxacin, gatifloxacin, and moxifloxacin. Several major substituents (R1, R5, R6, R7, and R8) were removed or replaced, and the in vitro anti-Toxoplasma activities of these virtual compounds were then estimated by LDA and MLR analysis.
LDA resulted in positive T1 values and negative T2 values for almost all of the virtual quinolones tested, showing that the basic quinolone structure accounts for anti-Toxoplasma and antiprotozoan activity.
MLR analysis yielded estimated IC50s of the virtual compounds (Table 3). The results revealed the importance of the substituents at R5, R6, and R7. The presence of a fluorine at R6 was crucial, as its omission from the four most active quinolones resulted in an increase in the IC50 to >100 mg/liter. When the R5 methyl group was removed from grepafloxacin, there was a 15-fold increase in the IC50; when it was added to the structure of trovafloxacin, gatifloxacin or moxifloxacin, the IC50 fell 5- to 7-fold. At R7, we replaced the original groups with several substituents that are present in other quinolones. With trovafloxacin, all of the changes resulted in significant loss of activity. In contrast, with grepafloxacin, gatifloxacin, and moxifloxacin, replacement of the original R7 substituent with an azabicyclohexane group resulted in a 10- to 12-fold increase in activity, showing the importance of this radical in anti-Toxoplasma activity. Similarly, we found that the presence of a nucleophilic substituent at R8 was important for the activity of gatifloxacin and moxifloxacin, as its omission resulted in a three- to ninefold increase in the IC50. The presence of a cyclopropyl or 2,4-difluorophenyl at R1 was associated with better activity than was that of a methyl, ethyl, or t-butyl radical.
TABLE 3.
Computational screening of MLR function applied to virtual quinolones derived from trovafloxacin, grepafloxacin, gatifloxacin, and moxifloxacin
| Changea | IC50 (mg/liter) by MLR analysis
|
|||
|---|---|---|---|---|
| TVXb | GPXc | GTXd | MOXe | |
| Exptl IC50 | 0.4 | 2.4 | 4.1 | 5.1 |
| N1 changes | ||||
| 2,4-Difluorophenyl instead of cyclopropyl | 2.5 | 4.9 | 2.3 | |
| Cyclopropyl instead of 2,4-difluorophenyl | 0.8 | |||
| Methyl instead of original groups | 3.6 | 6.9 | 11.9 | 6.4 |
| Ethyl instead of original groups | 2.9 | 7.5 | 13.0 | 7.0 |
| t-butyl instead of original groups | 1.8 | 7.8 | 15.5 | 8.2 |
| C-5 changes | ||||
| H instead of methyl group | 37.9 | |||
| Methyl group instead of H | 0.06 | 0.79 | 0.44 | |
| C-6 omitted | >100.0 | >100.0 | >100.0 | 100.0 |
| C-7 changes | ||||
| 3′-Amino-pyrrolidinyl instead of original groups | 10.2 | 5.2 | 9.5 | 9.5 |
| Pyrrolidinyl instead of original groups | 12.6 | 3.2 | 5.9 | 5.9 |
| Piperazine instead of original groups | 13.3 | 3.6 | 6.7 | 6.7 |
| Methyl instead of original groups | 16.1 | 2.7 | 7.4 | 7.4 |
| Aza-bicyclohexane instead of original groups | 0.21 | 0.39 | 0.39 | |
| C-8 omitted | 36.3 | 17.5 | ||
R1 and R7 suppression is irrelevant, as all of the quinolones studied had a substituent at these positions.
TVX, trovafloxacin.
GPX, grepafloxacin.
GTX, gatifloxacin.
MOX, moxifloxacin.
DISCUSSION
The results of this study confirm that quinolones are active against T. gondii (6, 9, 10) and show that this activity can be predicted using molecular topology methods. The LDA model, which we have previously used to identify antiprotozoan and anti-Toxoplasma drugs (8), showed that 20 of the 24 quinolones or fluoroquinolones studied were predicted to be active against T. gondii. When chemical structures defined by TIs were entered into an equation which distinguished anti-Toxoplasma drugs from other antiprotozoan drugs, the values were negative for 21 of the 24 compounds, indicating that the anti-Toxoplasma efficacy of these drugs could not be distinguished from general antiprotozoan activity. Despite the fact that four quinolones were misclassified with T1 and three were misclassified with T2 (probably due to the heterogeneity of the database used to build the model), these results suggest that, beside their anti-Toxoplasma activity, quinolones are also effective against other protozoa. This supports the hypothesis that several protozoa, and more specifically those belonging to the phylum Apicomplexa, have a quinolone target in common. It is also in keeping with several reports on the in vitro and in vivo activities of some fluoroquinolones against Plasmodium falciparum (4).
The predicted activity of quinolones against T. gondii was confirmed in vitro. An inhibitory effect was found with most of the quinolones tested, although sometimes only at high concentrations. Twenty of the 24 quinolones had IC50s above 10 mg/liter; this may explain why Khan et al. (9), who used concentrations below 10 mg/liter, found that ciprofloxacin, fleroxacin, ofloxacin, temafloxacin, and tosufloxacin were not active. However, like those authors, we found that trovafloxacin was highly active, with an IC50 of 0.4 mg/liter. We also found that another three fluoroquinolones (grepafloxacin, gatifloxacin, and moxifloxacin) potently inhibited Toxoplasma growth, with IC50s below 5 mg/liter. These results indicate that only a few quinolones are candidates for the treatment of toxoplasmosis and that more effective compounds need to be developed.
To identify more active quinolones, the experimental IC50s were related to a large number of TIs by using the MLR method. The equation thus obtained accurately matched experimental and calculated IC50s (r2 = 0.87), and the very good predictive capacity of the model was confirmed by the cross-validation test (r2cv = 0.74). Furthermore, when we examined 11 trovafloxacin analogs whose IC50s had not been determined in our laboratory, very good agreement was observed between our predicted IC50s of 10 compounds and those determined experimentally by Khan et al. (10).
The LDA and MLR models were then used to identify the pharmacophoric structures responsible for the anti-Toxoplasma activity of quinolones. Two complementary approaches were used to examine the respective roles of each atom in the quinolone ring and that of the substituent radicals. We first used new atomic E-state indices which provide information on the electronic and topological structure at the atomic level (12). The regression equation revealed that the C-14 position of the quinolone ring markedly contributed to lowering of the calculated IC50. This reflects the importance of the carbonyl position in anti-Toxoplasma activity. Surprisingly, the carboxyl group, which is essential for gyrase binding in bacteria (1, 5), did not appears in the MLR equation, suggesting that this group is not so crucial for anti-Toxoplasma activity.
Next, virtual modifications of trovafloxacin, grepafloxacin, gatifloxacin, and moxifloxacin were submitted to LDA and MLR analysis to investigate the influence of different radicals on anti-Toxoplasma activity. The presence of a fluorine at R6 was fundamental for anti-Toxoplasma activity, as its omission resulted in a total lack of activity. The presence of a cyclopropyl or a 2,4-difluorophenyl group at R1 appeared to be related to the anti-Toxoplasma activity of the four quinolones tested, as other substituents resulted in an increase in the IC50. The presence of an R8 substituent was important for the activity of grepafloxacin and moxifloxacin. All changes in the R7 radical resulted in lower activity. It is of note that changing the 3′-aminopyrrolidinyl substituent on trovafloxacin resulted in a marked loss of activity; in fact, this structure is that of tosufloxacin, which has been reported to be inactive in vitro (10), thus further validating the predictive value of our QSAR models. Finally, we showed the importance of a methyl group at C-5 and an aza-bicyclohexane at R7, as their presence or addition markedly enhanced anti-Toxoplasma activity.
In conclusion, the combination of LDA using topological descriptors and MLR statistical analysis can contribute to the design of new quinolones with improved anti-Toxoplasma activity and possibly identify drugs with a broader spectrum of antiprotozoan activity. Computational screening of thousands of virtual molecules using this method in a search for optimal substitutions is readily feasible and is far less costly than combinatory chemistry and in vitro screening.
ACKNOWLEDGMENTS
R. Gozalbes is indebted to the association Ensemble contre le SIDA and the French Ministry of Foreign Affairs for their financial support of this work, which was conducted in the Parasitology-Mycology Laboratory and ITODYS, University of Paris VII, Paris, France. The members of the Molecular Connectivity & Drug Design Research Unit are grateful for the support given by Generalitat Valenciana through the project GV99-91-1-12.
We thank Bayer Pharma, Esteve, Glaxo Wellcome, Grünenthal, Hoechst Marion Roussel, Mediolanum Farmaceutici, Monsanto Searle, Parke-Davis, Roche, Roger Bellon, and Sanofi Winthrop Laboratories for supplying compounds. We thank David Young for reviewing the manuscript.
REFERENCES
- 1.Bryskier A, Chantot J F. Classification and structure-activity relationships of fluoroquinolones. Drugs. 1995;49(Suppl. 2):16–28. doi: 10.2165/00003495-199500492-00005. [DOI] [PubMed] [Google Scholar]
- 2.de Julián-Ortiz J V, Gálvez J, Muñoz-Collado C, García-Domenech R, Gimeno-Cardona C. Virtual combinatorial syntheses and computational screening of new potential anti-herpes compounds. J Med Chem. 1999;42:3308–3314. doi: 10.1021/jm981132u. [DOI] [PubMed] [Google Scholar]
- 3.Derouin F, Chastang C. Enzyme immunoassay to assess effect of antimicrobial agents on Toxoplasma gondii. Antimicrob Agents Chemother. 1988;32:303–307. doi: 10.1128/aac.32.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Divo A A, Sartorelli A C, Patton C L, Bia F J. Activity of fluoroquinolone antibiotics against Plasmodium falciparum in vitro. Antimicrob Agents Chemother. 1988;32:1182–1186. doi: 10.1128/aac.32.8.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Domagala J M. Structure-activity and structure-side-effect relationships for the quinolone antibacterials. J Antimicrob Chemother. 1994;33:685–706. doi: 10.1093/jac/33.4.685. [DOI] [PubMed] [Google Scholar]
- 6.Fichera M E, Roos D S. A plastid organelle as a drug target in apicomplexan parasites. Nature. 1997;390:407–409. doi: 10.1038/37132. [DOI] [PubMed] [Google Scholar]
- 7.Gálvez J, García R, Salabert M T, Soler R. Charge indexes. New topological descriptors. J Chem Inf Comput Sci. 1994;34:520–525. [Google Scholar]
- 8.Gozalbes R, Gálvez J, García-Domenech R, Derouin F. Molecular search of new active drugs against Toxoplasma gondii. Struct-Act Relat Quant Struct-Act Relat Environ Res. 1999;10:47–60. doi: 10.1080/10629369908039165. [DOI] [PubMed] [Google Scholar]
- 9.Khan A A, Slifer T, Araujo F G, Remington J S. Trovafloxacin is active against Toxoplasma gondii. Antimicrob Agents Chemother. 1996;40:1855–1859. doi: 10.1128/aac.40.8.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan A A, Araujo F G, Brighty K E, Gootz T D, Remington J S. Anti-Toxoplasma gondii activities and structure-activity relationships of novel fluoroquinolones related to trovafloxacin. Antimicrob Agents Chemother. 1999;43:1783–1787. doi: 10.1128/aac.43.7.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kier L B, Hall L H. Molecular connectivity in structure-activity analysis. Letchworth, England: Research studies press. John Wiley & Sons; 1986. pp. 225–246. [Google Scholar]
- 12.Kier L B, Hall L H. An electrotopological-state index for atoms in molecules. Pharm Res. 1990;7:801–807. doi: 10.1023/a:1015952613760. [DOI] [PubMed] [Google Scholar]
- 13.Lahana R. Virtual combinatorial chemistry. Sci Am. 1997;241:56–58. . (French edition.) [Google Scholar]
- 14.Mallows C L. Some comments on Cp. Technometrics. 1973;15:661–675. [Google Scholar]
- 15.Roos D S, Crawford M J, Donald R G K, Kissinger J C, Klimczak L J, Striepen B. Origin, targeting, and function of the apicomplexan plastid. Curr Opin Microbiol. 1999;2:426–432. doi: 10.1016/S1369-5274(99)80075-7. [DOI] [PubMed] [Google Scholar]
- 16.Soldati D. The apicoplast as a potential therapeutic target in Toxoplasma and other apicomplexan parasites. Parasitol Today. 1999;15:5–7. doi: 10.1016/s0169-4758(98)01363-5. [DOI] [PubMed] [Google Scholar]