Abstract
The recently emerging SARS‐CoV‐2 variant omicron displays an unusual association of 30 mutations, 3 deletions, and 1 insertion. To analyze the impact of this atypic mutational landscape, we constructed a complete structure of the omicron spike protein. Compared with the delta variant, the receptor‐binding domain (RBD) of omicron has an increased electrostatic surface potential, but a decreased affinity for the ACE‐2 receptor. The N‐terminal domain (NTD) has both a decreased surface potential and a lower affinity for lipid rafts. The omicron variant is predicted to be less fusogenic and thus less pathogenic than delta, due to a geometric reorganization of the S1‐S2 cleavage site. Overall, these virological parameters suggest that omicron does not have a significant infectivity advantage over the delta variant. However, in omicron, neutralizing epitopes are greatly affected, suggesting that current vaccines will probably confer little protection against this variant. In conclusion, the puzzling mutational pattern of the omicron variant combines contradictory properties which may either decrease (virological properties) or increase (immunological escape/facilitation) the transmission of this variant in the human population. This Janus‐like phenotype may explain some conflicting reports on the initial assessment of omicron and provide new insights about the molecular mechanisms controlling its dissemination and pathogenesis worldwide.
Keywords: antibody susceptibility, coronavirus, evolution, infection, pathogenesis, SARS coronavirus, virulence, virus classification
HIGHLIGHTS
The spike protein of omicron is analyzed and compared with other SARS‐CoV‐2 variants
Compared with delta, omicron has a lower capacity to fuse with human host cells
Low activity of neutralizing antibodies suggests immunological escape of omicron
1. INTRODUCTION
Omicron (B.1.1.529) is a SARS‐CoV‐2 variant that recently emerged in southern Africa and in several European countries. 1 It was first detected in South Africa at the beginning of November 2021 and as of 16/12/2021, 7277 genomes were available from the GISAID database (https://www.gisaid.org/), mostly from South Africa (n = 1130) and from the United Kingdom (n = 4116). Although in most regions the prevalence of omicron is currently low compared to delta that accounts for 1 387 376 genomes since 01/11/2021, this variant surprised many people as it carries an unusual number of mutations in its spike protein: 30 mutations, 3 deletions, and 1 insertion (Table 1). For comparison, the delta variant B.1.617.2 has only 9 mutations and 1 deletion. Omicron was designated a variant of concern (VOC) on 26/11/2021. 2
Table 1.
Mutational pattern and T‐index of SARS‐CoV‐2 variants delta and omicron
| Variant | Mutations—NTD | Mutations—RBD | Mutations—rod | T‐index | I‐index |
|---|---|---|---|---|---|
| Delta B.1.617.2 | T19R T95I G142D ∆E156 ∆F157 R158G | L452R T478K | D614G P681R D950N | 10.81 | 3.10 |
| Omicron B.1.1.529 | A67V ∆H69 ∆V70 T95I G142D ∆V143 ∆Y144 ∆Y145 ∆N211 L212I +214EPE | G339D S371L S373P S375F K417N N440K G446S S477N T478K E484A Q493R G469S Q498R N501Y Y505H T547K | D614G H655Y N679K P681H N764K D796Y N856K N954K N969K L981F | 3.90 | 5.80 |
Note: Transmissibility index (T‐index) is calculated as follows (details previously published in ref. 3 for alpha, beta, gamma, and delta variants):
T‐index = ΔGmut/ΔGwt [NTD‐ganglioside] × ΔGmut/ΔGwt [RBD‐ACE‐2] × [Surface Potential]NTD × [Surface Potential]RBD
For omicron: T‐index = 0.83 × 0.77 × 1.24 × 4.93 = 3.90
The immune‐escape index (I‐index) is calculated as described in ref. 4
I‐index = 1/2 (∆Gwt/∆Gmut (RBD‐nAb) + ∆Gwt/∆Gmut (NTD‐nAb)). The I‐index of the original original 20B strain is equal to 1.
Abbreviations: NTD, N‐terminal domain; RBD, receptor‐binding domain.
So far, the analysis of omicron has given a series of various and somewhat paradoxical results. First, its mutational pattern does not seem to result from the direct evolution of a known variant, and in particular, it does not derive from the delta variant, which has been dominant worldwide during the last months of 2021. 5 Indeed, the omicron variant lacks the typical L452R mutation which is characteristic of most delta variants. 6 , 7 Second, initial assessments of omicron propagation in South Africa and some European countries (Denmark, UK, and France) indicated that this variant is highly contagious. 8 However, this high transmissibility did not seem to correlate with a clearcut higher affinity of the omicron spike protein for the ACE‐2 receptor: some groups reported a moderate increase of the receptor‐binding domain (RBD) affinity for ACE‐2, 9 , 10 whether, in contrast, others reported a decreased affinity. 11 , 12 To further complexify the problem, a third group concluded that delta and omicron spike proteins display a similar for ACE‐2, due to compensation of mutations that either increase or decrease ACE‐2 binding in the case of omicron. 13 Third, in vitro experiments performed with culture cells also gave mixed results. In some cells, the infectivity and replication of the omicron variant were higher than delta, whereas in other cells opposite results were obtained, with delta being clearly more performant than omicron. 14 , 15 Moreover, several reports suggest that the omicron variant spike confers impaired cell–cell fusion activity, 14 which may correlate with low pathogenicity. 15
In face of such conflicting results, the aim of the present study was to provide a global in silico analysis of the omicron spike protein. To this end, we used a series of molecular modeling approaches to assess the affinity of the RBD for ACE‐2, but also the avidity of the N‐terminal domain (NTD) for lipid raft gangliosides. 3 , 16 We also studied the electrostatic surface potential of both the RBD and the NTD, a critical parameter that controls the kinetic of interaction of the virus with the host cell membrane. 3 Finally, we analyzed the impact of the delta and omicron mutational profiles on the affinity of neutralizing antibodies directed against the RBD and NTD of each variant.
2. METHODS
2.1. Mutational profile extraction
The mutational pattern of SRAS‐CoV‐2 variants was extracted as an excel file from the Los Alamos database (https://cov.lanl.gov/components/sequence/COV/int_sites_tbls.comp).
2.2. Structural model of the omicron spike protein
A complete structure of the spike protein was generated from the original 20B strain (Wuhan + D614G, pdb 7bnm). 17 All gaps in the pdb file were fixed by inserting the missing amino acids with Robetta [https://robetta.bakerlab.org/], a protein structure prediction service. 18 The structure was then submitted to several rounds of energy minimization with the Polak‐Robière algorithm as described previously. 3 This source file model was used to introduce the specific mutational profiles of delta and omicron with the MUTATE tool of Swiss‐PdbViewer. 19
2.3. Electrostatic surface potential
The electrostatic potential was measured by the Molegro Molecular viewer (http://molexus.io/molegro-molecular-viewer). It is expressed as the sum of the Coulomb potentials for each atom of the protein, with a distance‐dependent dielectric constant. Color intensities (negative in red, positive in blue, neutral in white) were quantified with the ImageJ software as described previously. 3 Values >1 are indicative of an electropositive surface.
2.4. Affinity of the spike protein for ACE‐2
The ACE‐2 RBD complex used as reference was obtained from pdb 6M0J (B.1 Wuhan strain). 20 This model was optimized by energy minimization, raising the energy of interaction from ΔG = −229 kJ·mol−1 to ΔG = −343 kJ·mol−1. 3 The affinity of delta and omicron RBDs for ACE‐2 was estimated in comparison with this value after introducing the appropriate mutations in the optimized pdb file 6M0J.
2.5. Avidity of the spike protein for lipid rafts
The structural model of the NTD bound to lipid raft gangliosides (energy of interaction ΔG = −397 kJ·mol−1) was obtained and corrected for gaps as previously described. 3 The avidity of delta and omicron NTDs for lipid raft gangliosides was estimated in comparison with this value after introducing the appropriate mutations in the optimized reference model.
2.6. T‐index analysis
The transmissibility index (T‐index) of a SARS‐CoV‐2 variant combines kinetic and affinity parameters which govern virus binding to host cells. It gives an estimation of the overall interaction of the NTD with lipid raft gangliosides (ΔGmut/ΔGwt [NTD‐ganglioside]), the affinity of the RBD for the ACE‐2 receptor (ΔGmut/ΔGwt [RBD‐ACE‐2]), and the electrostatic surface potential of the NTD and RBD surfaces facing the host cell membrane. 3 It is calculated with the following formula:
ΔGmut/ΔGwt [NTD‐ganglioside] × ΔGmut/ΔGwt [RBD‐ACE‐2] × [Surface Potential]NTD × [Surface Potential]RBD
2.7. I‐index analysis
The immuno‐escape index (I‐index) evaluates the level of resistance of a SARS‐CoV‐2 variant to neutralizing antibodies (nAb) directed against the RBD and the NTD of the Spike protein. 4 It is calculated according to the following formula:
1/2 (ΔGwt/ΔGmut[RBD‐nAb] + ΔGwt/ΔGmut [NTD‐nAb])
2.8. Scanning electron microscopy
The scanning electron microscopy image was obtained from a SARS‐CoV‐2 omicron‐RNA‐positive VeroE6 cell culture supernatant, 21 using a SU5000 microscope (Hitachi High‐Technologies Corporation).
3. RESULTS
3.1. Structural analysis of the spike protein of delta and omicron variants
A structural model of the omicron spike protein was generated and compared to the one of the delta variant. In the case of omicron, mutations heavily affected the NTD, the RBD, as well as other critical regions including the proteolytic cleavage site and the fusion machinery (Figure 1A,B). The geometric reorganization of the protein around the S1‐S2 proteolytic cleavage site of omicron (Figure 1A), due to the H655Y/N679K/P681H triad (vs. the single mutation P681R for delta), suggests that host cell surface proteases such as transmembrane serine protease 2 (TRMPSS2) 22 may not be equally active on both variants.
Figure 1.
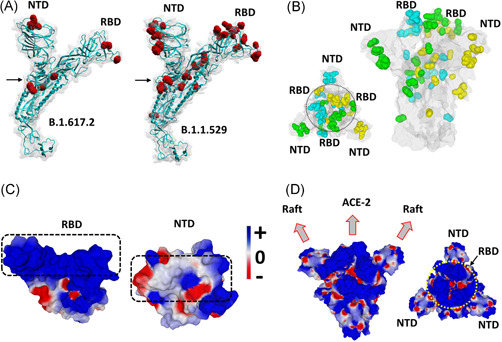
Structural analysis of omicron spike protein. (A) Molecular models of the delta B.1.617.2 and omicron B.1.1.529 spike proteins with mutations highlighted in red and the S1‐S2 cleavage site indicated by an arrow. (B) Spike trimer of the omicron variant as viewed from the host cell surface (left panel, central RBDs indicated within a circle), or perpendicularly to the virus envelope (right panel). Mutations are colored in cyan, green and yellow corresponding, respectively, to spike subunits A, B, and C. The NTD and RBD of each chain is indicated. The protein surface is colored in grey. (C) Electrostatic surface potential of omicron RBD and NTD. The color scale for the electrostatic surface potential (negative in red, positive in blue, neutral in white) is indicated. The regions of the RBD and the NTD, respectively, bound to ACE‐2 and the lipid raft are indicated by dashed rectangles. (D) Two views of the electrostatic surface potential of the trimeric spike of omicron (left panel, lateral view of the trimer perpendicular to viral envelope; right panel, top view of the trimer facing the host cell surface, the central RBDs are indicated by a yellow circle). NTD, N‐terminal domain; RBD, receptor‐binding domain
3.2. T‐index analysis
To learn more about the infectivity and pathogenicity of the omicron variant, we applied to this variant the analysis of the T‐index (transmissibility index) which had allowed us, soon after the appearance of the delta variant in April 2021, to anticipate its expansion globally. 3 This index takes into account the interaction of the NTD domain with the lipid rafts of host cell membranes, 3 , 16 the interaction of the RBD domain with the ACE‐2 receptor, 23 and the electrostatic surface potential of both NTD and RBD which reveals the kinetics of virus binding to target cells. 3 Taken together, these critical parameters control the very first steps of SARS‐CoV‐2 infection, from the initial attraction of the viral particle by the cell surface, to ACE‐2 binding. 3 , 16 Therefore, the T‐index adequately reflects the relative infectivity of a SARS‐CoV‐2 variant compared to any other strain.
In the case of the omicron variant, some of these parameters were increased, but others were decreased when compared with the original 20B strain and to the delta variant. Indeed, the avidity of omicron spike NTD for lipid rafts was decreased by 17% (ΔG = −329 kJ·mol−1 for omicron vs. −397 kJ·mol−1 for the reference NTD). The affinity of the RBD for ACE‐2 was decreased by 23% (ΔG = −264 kJ·mol−1 for omicron vs. −343 kJ·mol−1 for the reference RBD). The electrostatic surface potential of the NTD was decreased by 25% whereas it was increased by 3.8‐fold for the RBD (Figure 1C,D). Based on these parameters, the T‐index of the omicron variant was estimated to be 3.90 (Table 1). This value is high compared to the Wuhan/D614G strain (2.16), 3 but rather low compared to delta variants (10.67 for a variant with E484Q 3 and 10.81 for the B.1.617.2 variant studied here). In fact, the T‐index of the omicron variant is in the same range as that of all other SARS‐CoV‐2 variants tested so far which, with the noticeable exception of the delta variant (T‐index >10), have a T‐index value <4 3 (Figure 2).
Figure 2.
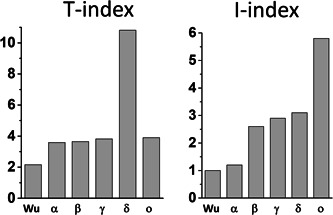
T‐index and I‐index analysis of SARS‐CoV‐2 variants. T‐index values of Wu (Wuhan strain), alpha (α), beta (β), and gamma (γ) were from ref. 3 I‐index values of Wu (Wuhan strain), alpha (α), beta (β), and gamma (γ) were from ref. 4 The T‐index and I‐index of delta (δ) and omicron (ο) were calculated in the present study
A detailed analysis of the omicron spike protein reveals that its pattern of mutations does not obey a clearcut logic. For instance, the omicron RBD surface which faces ACE‐2 is almost uniformly positive (Figure 1C), which does not fit optimally with the surface of ACE‐2 which, although chiefly electronegative, displays several positive spots that will not allow a perfect adjustment with the RBD. 3 This is in line with the lower affinity of the omicron RBD for ACE‐2 (Table 1). Similarly, the surface of the omicron NTD is not positive enough (Figure 1C) to perfectly interact with the electronegative lipid raft surface. 3 Indeed, the global analysis of the electrostatic surface potential of the trimeric spike protein revealed that the central area formed by the three RBDs is highly positive (potential value 4.93), whereas the lateral NTDs, which are far less electropositive (potential value 1.24), displayed a mosaic pattern combining a series of positive, negative and neutral spots (Figure 1D). Thus, the trimeric spike of the omicron variant does not follow the usual concomitant evolution of the NTD and RBD towards higher electropositivity. 3
3.3. I‐index analysis
Taken together, these data suggest that the omicron variant does not have a clearcut transmissibility advantage over the delta variant. However, other parameters may counterbalance these virological properties. In particular, the immune status of the host might be critical. The mutational pattern of the omicron variant greatly affects the spike surface that faces the host cell (Figure 1A,B). Correspondingly, the structural analysis of the omicron spike protein revealed that the main neutralizing epitopes 24 , 25 are greatly affected by omicron mutations (Table 1). Indeed, the immuno‐escape index (I‐index) 4 of omicron is 1.9‐fold higher than the one of delta (5.8 vs. 3.1, respectively, given that the I‐index of the Wuhan strain is 1.0). All other variants have an I‐index <3, showing that as far as immune escape is concerned, this variant is far ahead of all competitors (Figure 2). Hence current vaccines will probably confer little protection against the omicron variant.
3.4. Detection of the omicron variant in viral cultures
In Marseille, we have already deposited 18 genomes in GISAID as of 16/12/2021 (out of 62 for France). The picture (Figure 3) shows a SARS‐CoV‐2 omicron variant isolate from a supernatant of culture on VeroE6 cells. 21 As a matter of fact, this picture is that of the first isolate of an omicron variant obtained from a patient in France. The present structural analysis was conducted promptly, and its results and interpretation were available opportunely when the first cases of the omicron variant were detected in our geographical area and in France. It is therefore part of a necessarily polyphasic approach that allows the broadest characterization of a new variant.
Figure 3.

Scanning electron microscopy of a SARS‐CoV‐2 Omicron‐RNA‐positive culture supernatant. Image was obtained using a SU5000 scanning electron microscope (SEM) (Hitachi High‐Technologies Corporation). A viral particle is indicated by a white arrow
4. DISCUSSION
The main outcome of the present study is that the omicron variant has an atypical mutational pattern combining contradictory properties that may either decrease (intrinsic virological properties) or increase (immunological escape) the transmission of this variant in the human population. To the best of our knowledge, our analysis is the first one that considers all four parameters defined as critical factors of SARS‐CoV‐2 intra‐ or inter‐host transmissibility 3 , 4 , 16 : the electrical potential of both the RBD and the NTD surfaces (kinetic parameter of virus binding to host cell membranes), the avidity of the NTD for gangliosides (interaction with lipid rafts), the affinity of the spike protein for ACE‐2 (receptor recognition) and a statement of neutralizing epitopes (immune escape). Our results agree with previous studies of the omicron spike protein that indicated an increase of the surface potential of the RBD 26 and a decreased affinity of the RBD for ACE‐2. 11 , 12 However, it is fair to mention that other studies showed either increased affinity of omicron RBD for ACE2 9 , 10 or no difference between omicron and delta variants. 13 These discrepancies, which may be related to the methodology used by different authors, may lead to divergent conclusions with respect to the transmissibility of the omicron variant. Among the difficulties, one should be aware that the mutational pattern of the omicron variant is puzzling because it combines some mutations that are part of the backbone of alpha, beta, gamma, delta, or B.1.160 lineages, 27 i.e. a complex patchwork that does not seem to fulfill a biochemical logic.
According to a recent study, it is possible that the spike protein of this variant was subjected to a strong positive selection compatible with host‐jumping. 28 Indeed, the mutations in the omicron RBD significantly overlapped with SARS‐CoV‐2 mutations known to promote adaptation to mouse hosts, particularly through enhanced spike protein binding affinity for mouse ACE‐2. Thus, a tentative scenario could be that the ancestor of the omicron virus jumped from humans to mice, accumulated mutations, then jumped back into humans. 28 According to this model, the mutational pattern of the omicron spike protein should not be interpreted as the result of a gradual improvement of SARS‐CoV‐2 for human host cells, but instead, as a series of compromises required for inter‐species back and forth jumps.
In marked contrast with omicron, the delta variant is characterized by a concomitant and convergent evolution of the NTD and the RBD, leading to a remarkable adaptation for their respective targets (lipid raft and ACE‐2) on human cell membranes. In this respect, it is interesting to mention that two routes of infection have been characterized for SARS‐CoV‐2: cell surface fusion, which requires S1‐S2 cleavage by TMPRSS2 (route 1), and endocytosis, which is TMPRSS2‐independent (route 2). 15 The T‐index adequately reflects the first route, as it takes into account the electrostatic surface potential that controls the binding of the virus to the host cell membrane. 3 As expected, delta (T‐index >10) is fourfold more efficient than omicron (T‐index 3.90) to infect TRMPSS2‐rich Calu‐3 cells through route 1. 15 On the contrary, omicron is 10 fold more efficient than delta to infect HEK cells which only support endosomal entry of SARS‐CoV‐2 (route 2). 15 Overall, these data suggest that delta is optimized for fusion at the cell surface, whereas omicron gains entry through endosomal fusion. This specificity of the omicron variant, which is probably due to the geometric reorganization of the S1‐S2 cleavage site of its spike protein (Figure 1A), is consistent with (i) a lower capacity to fuse infected cells to form syncytia, and (ii) a lower pathogenicity.
The mutations and indels located in the omicron NTD and RBD also dramatically affect key neutralizing epitopes, 24 , 25 suggesting that current vaccines based on the original Wuhan strain will confer very little protection against this variant (Table 1 and Figure 2). In parallel, the facilitating epitope of the NTD 29 , 30 , 31 is almost destroyed by the 3‐amino acid insertion at position 214 (amino acid residues 215‐216‐217), further underscoring the lack of logic in this exacerbated mutational profile. However, even in absence of facilitating antibodies, sub‐neutralizing antibodies may bind to the virus and mediate its entry into host cells by an Fc receptor‐dependent mechanism. 32 Therefore, the neutralizing antibodies elicited upon vaccination may at best not protect against the omicron variant 33 but at worst facilitate its transmission through a classic antibody‐dependent enhancement (ADE) mechanism. This hypothesis is supported by the high prevalence of omicron contaminations in vaccinated people, e.g. in Norway. 34 In Marseille (France), we have already deposited 18 genomes in GISAID as of 16/12/2021 (out of 62 for France). Some of the cases were indeed in vaccinated patients. In addition, some correspond to secondary transmission, which was not observed for the first cases of infection with the Marseille‐1 variant which had emerged in our region in July 2020. 35
5. CONCLUSIONS
This structural analysis of the omicron variant (B.1.1.529) may shed some light on the striking and unusual structural features that control the multiple facets of this newly emerging variant. According to the target cell type, the omicron variant may be either less or more infectious than delta. Due to an inconsistent distribution of its electrostatic potential and to a defect in the S1‐S2 cleavage, omicron is expected to be less pathogenic than delta. Further studies will be necessary to assess whether the ACE‐2 receptor polymorphism 36 , 37 could allow regional breakthroughs of omicron, especially in immunocompromised individuals.
An important question is whether omicron may increase in prevalence and become predominant in case of decreased incidence of delta as was the case for all previous variants to date a few months after their emergence, most probably because of the accumulation of noxious mutations. 38 This may indeed be the case in South Africa, where omicron emerged at the time of a delta disappearance. 39 In contrast, in Denmark, France, and UK, delta was still present when omicron started to spread. 40 In this case, immune escape may facilitate an omicron breakthrough in vaccinated people, 34 which may suggest that the omicron variant has a fitness advantage in presence of neutralizing antibodies generated against the original Wuhan spike protein. Whether these neutralizing antibodies may facilitate the infection by the omicron variant through an ADE mechanism 32 remains to be established.
A limitation of the present analysis is that it compares a newly‐emerging virus to an old one that emerged several months ago. 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 As a matter of fact, despite no clear virological advantage over delta, omicron might increase in prevalence and even become predominant, which has been the fate of all previous variants to date after a few months of circulation due to the accumulation of most often deleterious mutations. Finally, one should be aware that resistance to infection in the upper respiratory tract could be due preferentially to innate mucosal immunity and could not be assessed by measuring circulating antibodies in the blood that do not have access to these mucosal membranes. 50
FUNDING INFORMATION
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée‐Infection 10‐IAHU‐03 and was also supported by Région Provence Alpes Côte d'Azur and European funding FEDER PRIMMI (Fonds Européen de Développement Régional‐Plateformes de Recherche et d'Innovation Mutualisées Méditerranée Infection), FEDER PA 0000320 PRIMMI, and by Hitachi High‐Technologies Corporation, Tokyo, Japan.
CONFLICT OF INTERESTS
Didier Raoult has a conflict of interest being a consultant for Hitachi High‐Technologies Corporation, Tokyo, Japan from 2018 to 2020. All other authors have no conflicts of interest to declare. Funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
AUTHOR CONTRIBUTIONS
Jacques Fantini, Nouara Yahi, and Henri Chahinian: performed the molecular modeling studies. Bernard La Scola: performed the characterization of the omicron variant in cell cultures. Philippe Colson and Nouara Yahi: analyzed the genomic sequences and extracted the mutational patterns of SARS‐CoV‐2 variants. Jacques Fantini and Didier Raoult: designed the study and wrote the manuscript.
Fantini J, Yahi N, Colson P, Chahinian H, La Scola B, Raoult D. The puzzling mutational landscape of the SARS‐2‐variant Omicron. J Med Virol. 2022;94:2019‐2025. 10.1002/jmv.27577
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article.
REFERENCES
- 1. Callaway E. Heavily mutated Omicron variant puts scientists on alert. Nature. 2021;600:21. [DOI] [PubMed] [Google Scholar]
- 2. He X, Hong W, Pan X, Lu G, Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm. 2021;2:838‐845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fantini J, Yahi N, Azzaz F, Chahinian H. Structural dynamics of SARS‐CoV‐2 variants: a health monitoring strategy for anticipating Covid‐19 outbreaks. J Infect. 2021;83:197‐206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaafar R, Boschi C, Aherfi S. High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS‐CoV‐2. Viruses. 2021;13(11):2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tian D, Sun Y, Zhou J, Ye Q. The global epidemic of the SARS‐CoV‐2 delta variant, key spike mutations and immune escape. Front Immunol. 2021;12:751778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nörz D, Grunwald M, Tang HT, et al. Rapid automated screening for SARS‐CoV‐2 B.1.617 lineage variants (Delta/Kappa) through a versatile toolset of qPCR‐Based SNP detection. Diagnostics. 2021;11:1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng X, Garcia‐Knight MA, Khalid M. Transmission, infectivity, and antibody neutralization of an emerging SARS‐CoV‐ 2 variant in California carrying a L452R spike protein mutation. medRxiv preprint (2021). 2021;599:114. 10.1101/2021.03.07.21252647 [DOI] [Google Scholar]
- 8. Espenhain M, Funk T, Overvad M. Epidemiological characterisation of the first 785 SARS‐CoV‐2 Omicron variant cases in Denmark, December 2021. Euro Surveill. 2021;26(50):5537. 10.2807/1560-7917.ES.2021.26.50.2101146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shah M, Woo HG. Omicron: a heavily mutated SARS‐CoV‐2 variant exhibits stronger binding to ACE2 and potently escape approved COVID‐19 therapeutic antibodies. bioRxiv. 2021;5:823. 10.1101/2021.12.04.471200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Golcuk M, Yildiz A, Gur M. The Omicron variant increases the interactions of SARS‐CoV‐2 spike glycoprotein with ACE2. bioRxiv. 2021;11:3696. 10.1101/2021.12.06.471377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Teruel N, Crown M, Bashton M, Najmanovich R. Computational analysis of the effect of SARS‐CoV‐2 variant Omicron Spike protein mutations on dynamics, ACE2 binding and propensity for immune escape. bioRxiv. 2021;1:33. 10.1101/2021.12.14.472622 [DOI] [Google Scholar]
- 12. Jawaid MZ, Baidya A, Mahboubi‐Ardakani R, Davis RL, Cox DL. Simulation of the omicron variant of SARS‐CoV‐2 shows broad antibody escape, weakened ACE2 binding, and modest increase in furin binding. bioRxiv. 2021. 10.1101/2021.12.14.472704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mannar D, Saville JW, Zhu X, et al. SARS‐CoV‐2 Omicron variant: ACE2 binding, Cryo‐EM structure of spike protein‐ACE2 complex and antibody evasion. bioRxiv. 2021. 10.1101/2021.12.19.473380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao H, Lu L, Peng Z. SARS‐CoV‐2 Omicron variant shows less efficient replication and fusion activity when compared with delta variant in TMPRSS2‐expressed cells. Emerg. Microbes & Infect. 2021;83:1‐18. 10.1080/22221751.2021.2023329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Willett BJ, Grove J, MacLean OA, et al. (2021). The hyper‐transmissible SARS‐CoV‐2 Omicron variant exhibits significant antigenic change, vaccine escape and a switch in cell entry mechanism. https://www.gla.ac.uk/media/Media_829360_smxx.pdf
- 16. Fantini J, Chahinian H, Yahi N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID‐19. Biochem Biophys Res Commun. 2021;538:132‐136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Benton DJ, Wrobel AG, Roustan C, et al. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS‐CoV‐2. Proc Natl Acad Sci USA. 2021;118:e2022586118‐e2022586131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526‐W531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guex N, Peitsch MC. SWISS‐MODEL and the Swiss‐PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714‐2723. [DOI] [PubMed] [Google Scholar]
- 20. Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581:215‐220. [DOI] [PubMed] [Google Scholar]
- 21. Wurtz N, Penant G, Jardot P, Duclos N, La Scola B. Culture of SARS‐CoV‐2 in a panel of laboratory cell lines, permissivity, and differences in growth profile. Eur J Clin Microbiol Infect Dis. 2021;40:477‐484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoffmann M, Kleine‐Weber H, Schroeder S, et al. SARS‐CoV‐2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271‐280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS‐CoV‐2. Nature. 2020;581:221‐224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chi X, Yan R, Zhang J, et al. A neutralizing human antibody binds to the N‐terminal domain of the Spike protein of SARS‐CoV‐2. Science. 2020;369:650‐655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones BE, Brown‐Augsburger PL, Corbett KS, et al. The neutralizing antibody, LY‐CoV555, protects against SARS‐CoV‐2 infection in nonhuman primates. Sci Transl Med. 2021;13:eabf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pascarella S, Ciccozzi M, Bianchi, Benvenuto D, Cauda R, Casson A. The electrostatic potential of the Omicron variant spike is higher than in delta and delta‐plus variants: a hint to higher transmissibility? J Med Virol. 2021;121:800. 10.1002/jmv.27528.7 [DOI] [PubMed] [Google Scholar]
- 27. Harvey WT, Carabelli AM, Jackson B, et al. SARS‐CoV‐2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409‐424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wei C, Shan KJ, Wang W, Zhang S, Huan Q, Qian W. Evidence for a mouse origin of the SARS‐CoV‐2 Omicron variant. J Genet Genomics. 2021;S1673‐8527(21):00373‐00378. 10.1016/j.jgg.2021.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yahi N, Chahinian H, Fantini J. Infection‐enhancing anti‐SARS‐CoV‐2 antibodies recognize both the original Wuhan/D614G strain and Delta variants. A potential risk for mass vaccination? J Infect. 2021;83:607‐635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Soh WT, Kishikawa J, et al. An infectivity‐enhancing site on the SARS‐CoV‐2 spike protein targeted by antibodies. Cell. 2021;184:3452‐3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li D, Edwards RJ, Manne K, et al. In vitro and in vivo functions of SARS‐CoV‐2 infection‐enhancing and neutralizing antibodies. Cell. 2021;184:4203‐4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Taylor A, Foo SS, Bruzzone R, Dinh LV, King NJ, Mahalingam S. Fc receptors in antibody‐dependent enhancement of viral infections. Immunol Rev. 2015;268:340‐364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS‐CoV‐2. bioRxiv. 2021. 10.1101/2021.12.14.472719 [DOI] [PubMed] [Google Scholar]
- 34. Brandal LT, MacDonald E, Veneti L, et al. Outbreak caused by the SARS‐CoV‐2 Omicron variant in Norway, November to December 2021. Euro Surveill. 2021;26(50):1560‐7917. 10.2807/1560-7917.ES.2021.26.50.2101147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colson P, Levasseur A, Gautret P, et al. Introduction into the Marseille geographical area of a mild SARS‐CoV‐2 variant originating from sub‐Saharan Africa: an investigational study. Travel Med Infect Dis. 2021;40:101980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Devaux CA, Rolain JM, Raoult D. ACE2 receptor polymorphism: susceptibility to SARS‐CoV‐2, hypertension, multi‐organ failure, and COVID‐19 disease outcome. J Microbiol, Immunol Infect. 2020;53:425‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X. Comparative genetic analysis of the novel coronavirus (2019‐nCoV/SARS‐CoV‐2) receptor ACE2 in different populations. Cell Discov. 2020;6:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. de la Peña M, Elena SF, Moya A. Effect of deleterious mutation‐accumulation on the fitness of RNA bacteriophage MS2. Evolution. 2000;54:686‐691. [DOI] [PubMed] [Google Scholar]
- 39. https://graphics.reuters.com/world-coronavirus-tracker-and-maps/countries-and-territories/south-africa/
- 40. https://graphics.reuters.com/world-coronavirus-tracker-and-maps/
- 41. Miller NL, Clark T, Raman R, asisekharan R. Insights on the mutational landscape of the SARS‐CoV‐2 Omicron variant. bioRxiv: the preprint server for biology2021.12.06.471499. 2021.
- 42. Queen D. Another year another variant: COVID 3.0‐Omicron. Int Wound J. 2022;19:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shanmugaraj B, Malla A, Khorattanakulchai N, Phoolcharoen W. SARS‐CoV‐2 Omicron Variant: could it be another threat? J Med Virol. 2021. 10.1002/jmv.27532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Li X. Omicron: call for updated vaccines. J Med Virol. 2021. 10.1002/jmv.27530 [DOI] [PubMed] [Google Scholar]
- 45. Mohiuddin M, Kasahara K. Investigating the aggressiveness of the COVID‐19 Omicron variant and suggestions for possible treatment options. Respir Med. 2021;191:106716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang X, Wu S, Wu B, et al. SARS‐CoV‐2 Omicron strain exhibits potent capabilities for immune evasion and viral entrance. Signal Transduct Target Ther. 2021;6:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maxmen A. Omicron blindspots: why it's hard to track coronavirus variants. Nature. 2021;600:579. 10.1038/d41586-021-03698-7 [DOI] [PubMed] [Google Scholar]
- 48. Dyer O. Covid‐19: Omicron is causing more infections but fewer hospital admissions than delta, South African data show. BMJ (Clinical research ed.). 2021;375:n3104. [DOI] [PubMed] [Google Scholar]
- 49. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K. Omicron SARS‐CoV‐2 variant: unique features and their impact on pre‐existing antibodies. J. Autoimmunity. 2021;126:102779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sonigo P, Petit C, Arhel NJ (2021). https://www.jle.com/fr/covid19-vacciner-contre-detection-par-PCR-ou-contre-maladie-covid19 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.


