Abstract
As severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) evolves, it accumulates mutations, which are changes in the genetic code. Because this virus has built‐in RNA repair mechanisms, it generates mutations more slowly than some other RNA viruses. Thousands of mutations have emerged since the beginning of the pandemic throughout all virus genomes sequenced to date, resulting in thousands of distinct variants. However, some variants have recently been discovered that appear to increase transmissibility and may affect illness pervasiveness. In this study, we investigated SARS‐CoV‐2 variants and how countries intervene with them. We also depicted the top 25 countries where the Omicron variant is prevalent, with the UK, US, Denmark, France, and Australia having the top five places as of January 13, 2022. The perception of SARS‐CoV‐2 variants was investigated in those five countries, and the propagation rate of the Omicron variant was determined to be 51%, 29%, 26%, 15%, and 44%, respectively, indicating that the Omicron variant is the most prevalent among the others. Then, a study of SARS‐CoV‐2 infection test rate based on tests conducted per one million populations with a number of sequences in those five countries reveals that 25%, 73%, 1.6%, 4.8%, and 1.5%, respectively, it suggests that viral testing should be increased in all five countries since it will help to determine the precise distribution of variants and aid governments in making policy decisions for public safety. We anticipated the production of new variants strains. This study implies that limiting disease transmissions, such as acquiring a coronavirus disease 2019 vaccine and booster doses for those aged 18 and older, as well as wearing the mask in public places, is the best strategy to prevent the emergence of new variants.
Keywords: perception of variants, Omicron dispersal, propagation of variants, SARS‐CoV‐2 variants, tests rate
Research Highlights
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants, as well as how countries around the world respond to them, were explored.
The perception of SARS‐CoV‐2 variants was discovered in the top five countries where the Omicron variant is prevalent as of January 13, 2022, including the UK, the United States, Denmark, France, and Australia.
The propagation rate of the Omicron variant was revealed in those five countries.
Based on tests conducted per one million populations with the number of sequences, an analysis of the SARS‐CoV‐2 test rate in those five countries was presented.
1. INTRODUCTION
Viruses such as severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) evolve over time, with mutations in the underlying gene occurring during genome replication. A lineage would be a collection of influenza variants with clades by means of major hereditary ties. A variation is distinguished from other SARS‐CoV‐2 viruses via one additional mutation. Scientists evaluate genomic variations across viruses to find variants and how they are related to one another to improve community epidemic investigations and comprehend national patterns. 1 , 2 Since the commencement of the coronavirus disease 2019 (COVID‐19) pandemic, SARS‐CoV‐2 genetic lineages had emerged and transmitted over the globe. 3 , 4 The US Department of Health and Human Services (HHS) established the SARS‐CoV‐2 Interagency Group (SIG) to focus on the rapid characterization of emerging variants and actively monitor their potential impact on critical SARS‐CoV‐2 countermeasures, such as immunizations, pharmaceuticals, and testing. SARS‐CoV‐2 variations are divided into four categories under the SIG Variant classification scheme: variants being monitored (VBM), variants of interest (VOI), variants of concern (VOC), and variants of high consequence (VOHC). 5
Variants classified as VBM are those for which data implies prospective or concise implications on endorsed or authorized medical preventive actions or those that are linked to more acute infection or rising propagation and are no longer detectable; such variants need not constitute a serious as well as consequential danger to public health. The World Health Organization (WHO) has classified the following Pango lineage variants as VBM since about September 21, 2021: Alpha (B.1.1.7 and Q lineages), Beta (B.1.351), Gamma (P.1 and descendent lineages), Epsilon (B.1.427 & B.1.429), Eta (B.1.525), Iota (B.1.526), Kappa (B.1.617.1), and Mu (P. (B.1.621, B.1.621.1). A VOI is a genetic variant with specific biological indicators that have been linked to changes in antibody binding, decreased neutralization besides monoclonal antibodies formed against early illnesses or mammograms, possibly lower lethality, potential diagnostic effect, or clinical manifestations. 6 , 7 The VOI may necessitate one or more adequate prevention efforts, such as improved timeline tracking, improved experimental analysis, or epidemiologic studies interrogations to determine how seamlessly the virus infects all others, the extent of symptoms, antitumor activity, or if medically endorsed or permitted antiviral drugs to provide prevention. No SARS‐CoV‐2 variants have been recognized as VOI as of yet. VOC would be a variant since there is a high prevalence of infectivity, greater acute illnesses and a considerable decline in the eradication of antibody levels developed from preceding disease or inoculation, diminished effectiveness of therapies or vaccinations. Concerning variants may necessitate one or more public health responses, such as formal notice to WHO under the International Health Regulations, submitting to the CDC, specific geographical attempts to control dispersion, enhanced testing, or investigations to assess the efficacy of inoculations and therapies against the variant. Considerations would involve the production of novel tests or the change of vaccinations or therapies depending on the variant's characteristics.
Delta and Omicron are two current variants of concern. In late 2020, India became the first country to identify the Delta variant. On May 31, 2021, the Delta variant was found, and by November 22, 2021, it had spread to 179 countries. In June 2021, the World Health Organization (WHO) stated that the Delta variant would become the most common stress worldwide. It seems to have mutations T478K, P681R, and L452R in the gene encoding the SARS‐CoV‐2 spike protein, which become known to alter the virus's infectiousness and how it can be neutralized with antigens to recently circulating COVID‐19 virus variants. It is believed as one of the very highly contagious serious infections ever discovered. 8 , 9 , 10 , 11 , 12 Public Health England (PHE) reported a 10.8% supplementary attack rate in domestic encounters of non‐travel and unidentified incidents for Delta in August 2021, compared with 10.2% for the Alpha variant; the rate of incidence in those 386 835 Delta individuals is 0.3%, with 46% of incidence and 6% of mortalities being unvaccinated and under the age of 50. 13 , 14 The variant is suspected of playing a role in the devastating second wave of the epidemic that began in February 2021 in India. 15 Fiji, the UK, and South Africa all suffered the third wave as a result of it. It has also contributed to an upsurge in daily attacks in Asia, the United States, Australia, and New Zealand by late July. 16 , 17 , 18 , 19 , 20 , 21 , 22
Omicron, another COVID variant, came next. It would be the most recent variant, as of December 2021. On November 24, 2021, South Africa was the first to report it to the World Health Organization (WHO). The WHO called it “Omicron” on November 26, 2021, and classified it as a variant of concern. The Omicron variant features an absolutely enormous genetic variation, several of which are unique, as well as a substantial proportion from which impair the spike protein attacked by most COVID‐19 vaccines during the period of its discovery. 23 , 24 , 25 Omicron is presumed to be quite infectious than previous variants of concern, spreading 70 times faster in the alveoli than certain preceding variants, and yet this is less capable of infiltrating profound bronchial membrane, which may help us understand why there is significant prevention of serious illnesses hospitalization. 26 , 27 The variation had been confirmed in over 80 countries as of December 16, 2021. Omicron, according to the World Health Organization, was likely with numerous nations as mid‐December, whether or not it had been found. 28 , 29
2. MATERIALS AND METHODS
In this study, we used two data sets for our exploratory analysis of SARS‐CoV‐2 variants: (i) Global Initiative on Sharing Avian Influenza Data (GISAID) and (ii) Worldometer. The data is acquired as of January 15, 2022.
The GISAID manages the world's biggest open‐access archive of SARS‐CoV‐2 sequences. GISAID's SARS‐CoV‐2 database has over 7 360 219 contributions from 170 distinct nations as of mid‐January 2022, after originally providing the first comprehensive genome sequences of SARS‐CoV‐2 on January 10, 2020. We obtained several variants data from this database to explore the propagation of SARS‐CoV‐2 variants. 30
Worldometer is a public and open‐access initiative that serves worldwide COVID‐19 statistics to a large number of concerned citizens all around the world; from this, we gathered data on SARS‐CoV‐2 infection tests conducted around the world for our research. 31
We used numerous graphs and visualizations to analyze our data to provide deep knowledge of SARS‐CoV‐2 variants, particularly the most recent variant, Omicron, around the world; this allows people to grasp the epidemiological essence of SARS‐CoV‐2 variants. The purpose of this article on SARS‐CoV‐2 variants is to synthesize existing research, collect pertinent data, and enable readers to make sense of the published data and early research on the recent spread of SARS‐CoV‐2 variants.
To determine the distribution of Omicron sequences across countries, we used the number of Omicron variant sequences from each country and location and produced a graph between them. To examine the global perception of SARS‐CoV‐2 variants, we considered the inception and last identified (sampling date) period of each variant and accompanying sequence from each nation and created a visual for them. We considered the number of days accumulated for each variant between their inceptions and last identified (sampling date) period, as well as the relating number of sequences from each country, to determine the propagation of SARS‐CoV‐2 variants in each country across the globe, allowing us to interpret variant propagation country by country. To study the SARS‐CoV‐2 test completion in countries, we examined tests per 1 million populations and the total number of sequences of variants in each nation, which determined the test %, with these able to construct the graph to present the test rate versus countries. The experimental works were developed on a Windows PC with the Python programming language.
3. RESULTS AND DISCUSSION
The distribution of Omicron variant sequences unfolded in 170 nations worldwide, with Figure 1 depicting the dispersion of the top 25 countries worldwide, which includes 21 K and 21 L kinds of Omicron variants. As a result, the UK takes top position with 124 532 sequences (46%), followed by the United States with 84 431 sequences (31%), for a total of 18 253 sequences (7%) Denmark is ranked third, followed by France in fourth place with 3863 sequences (1%) and Australia in the fifth position with 3628 sequences (1%) as of January 13, 2022.
Figure 1.
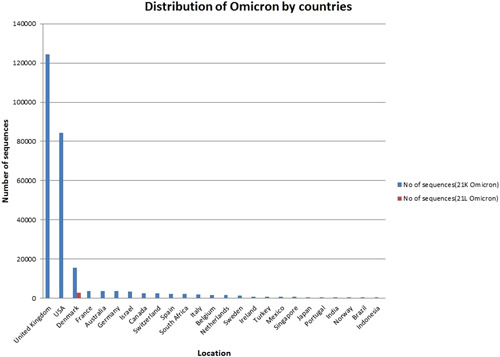
Based on data set 1, Global Initiative on Sharing Avian Influenza Data (GISAID), display the top 25 countries distribution of Omicron variants (21 K and 21 L) around the globe from 170 countries between May 11, 2020 and January 13, 2022
Figure 2 depicts how the top five countries with the highest occurrence of the Omicron variant perceived other variants includes 21A (Delta), 21I (Delta), 21J (Delta), 21K (Omicron), 21L (Omicron), 20H (Beta, V2), 20J (Gamma, V3), 20I (Alpha, V1), 21B (Kappa), 21D (Eta), 21H (Mu), 21F (Iota), 21C (Epsilon), 21G (Lambda), 20E (EU1), 20A.EU2, S:677H.Robin1, S:677 P.Pelican, 20B/S:732A, 20A/S:98F, 20A/S:126A, 20A/S:439K, 20B/S:626S, 20C/S:80Y, and 20B/S:1122L between March 30, 2020 and January 13, 2022. Here, we'll look at the spread of variants in five countries: the UK, the US, Denmark, France, and Australia, where the recent variant Omicron distribution is substantial.
Figure 2.

Shows global perceptions of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants in the top five countries with the highest prevalence of the Omicron between May 11, 2020 and January 13, 2022, based on data set 1, Global Initiative on Sharing Avian Influenza Data (GISAID)
Out of 1 570 879 sequences in the UK, 21J Delta accounts for 68%, 20I Alpha for 17%, 21K Omicron for 8%, 20E EU1 for 5%, and 21I Delta for 1%. 21J Delta accounts for 65% of 1 876 940 sequences in the United States, 20I Alpha 13%, 21J Delta 6%, 21K Omicron 4%, 21C Epsilon 4%, 21A Delta 3%, 20J Gamma 2%, 21F Iota 2%, and 20B/S:732A 1%. In Denmark, 21J Delta accounts for 53% of 316 889 sequences, 20I Alpha for 22%, 20E EU1 for 10%, 21K Omicron 5%, 21I Delta for 2%, 20E EU2 for 2%, 20A/S:439K for 2%, 20A/S:98F, and 21K Omicron for 1%. In France, 21J Delta accounts for 60% of the 153 357 sequences, 20I Alpha for 21%, 20H Beta for 4%, 20A EU2 for 2%, 21I Delta for 3%, 21K Omicron for 3%, 21A Delta for 2%, 20E EU1 for 2%, and 20J Gamma for 1%. Out of 34 360 sequences in Australia, 21J Delta accounts for 85%, 21K Omicron 11%, and 20I Alpha 2%.
Following that, the propagation coefficient is computed for those five nations to assess the spread intensity of the variants using the number of sequences and days determined by the variant identified period and sample period (Tables 1 and 2).
Table 1.
Displays the number of sequences of each SARS‐CoV‐2 variant that occurred in the UK and the United States between the identified period (first_seq) and the sampling period (last_seq) based on data set 1, GISAID
| Variant | UK | USA | ||||
|---|---|---|---|---|---|---|
| Number of sequences | first_seq | last_seq | Number of sequences | first_seq | last_seq | |
| 20I (Alpha, V1) | 26 2782 | 20‐09‐2020 | 26‐11‐2021 | 242 177 | 21‐10‐2020 | 18‐12‐2021 |
| 20H (Beta, V2) | 940 | 10‐12‐2020 | 26‐11‐2021 | 3133 | 01‐01‐2021 | 09‐12‐2021 |
| 20J (Gamma, V3) | 225 | 04‐02‐2021 | 30‐11‐2021 | 29 367 | 09‐01‐2021 | 09‐12‐2021 |
| 21A (Delta) | 4338 | 23‐03‐2021 | 26‐12‐2021 | 51 745 | 09‐02‐2021 | 01‐01‐2022 |
| 21I (Delta) | 20 803 | 27‐03‐2021 | 01‐01‐2022 | 110 074 | 16‐03‐2021 | 05‐01‐2022 |
| 21J (Delta) | 1 065 977 | 13‐11‐2020 | 05‐01‐2022 | 1 213 604 | 05‐11‐2020 | 07‐01‐2022 |
| 21K (Omicron) | 124 499 | 23‐10‐2021 | 09‐01‐2022 | 84 425 | 21‐11‐2021 | 07‐01‐2022 |
| 21L (Omicron) | 33 | 06‐12‐2021 | 04‐01‐2022 | 6 | 14‐12‐2021 | 03‐01‐2022 |
| 21B (Kappa) | 452 | 22‐02‐2021 | 14‐09‐2021 | 333 | 25‐02‐2021 | 09‐12‐2021 |
| 21D (Eta) | 484 | 15‐12‐2020 | 26‐07‐2021 | 1359 | 30‐12‐2020 | 18‐07‐2021 |
| 21F (Iota) | 21 | 21‐01‐2021 | 01‐05‐2021 | 41 870 | 18‐11‐2020 | 18‐12‐2021 |
| 21G (Lambda) | 9 | 23‐02‐2021 | 28‐10‐2021 | 1240 | 20‐01‐2021 | 09‐12‐2021 |
| 21H (Mu) | 113 | 23‐05‐2021 | 13‐12‐2021 | 6185 | 11‐03‐2021 | 24‐12‐2021 |
| 20B/S:732A | 20 | 12‐12‐2020 | 17‐04‐2021 | 13 244 | 05‐11‐2020 | 29‐10‐2021 |
| 20A/S:126A | 22 | 01‐03‐2021 | 02‐08‐2021 | 26 | 25‐03‐2021 | 19‐06‐2021 |
| 20E (EU1) | 82 158 | 18‐07‐2020 | 22‐11‐2021 | 287 | 18‐08‐2020 | 29‐09‐2021 |
| 21C (Epsilon) | 24 | 10‐12‐2020 | 10‐05‐2021 | 66 347 | 11‐08‐2020 | 09‐12‐2021 |
| 20A/S:439K | 3783 | 03‐07‐2020 | 26‐06‐2021 | 87 | 19‐09‐2020 | 25‐10‐2021 |
| S:677H.Robin1 | – | – | – | 6477 | 13‐08‐2020 | 24‐08‐2021 |
| S:677P.Pelican | 1 | 18‐04‐2021 | 18‐04‐2021 | 4846 | 02‐08‐2020 | 14‐09‐2021 |
| 20A.EU2 | 2889 | 04‐08‐2020 | 02‐01‐2022 | 88 | 31‐08‐2020 | 29‐12‐2021 |
| 20A/S:98F | 818 | 03‐08‐2020 | 23‐05‐2021 | 20 | 22‐10‐2020 | 20‐05‐2021 |
| 20C/S:80Y | 284 | 22‐07‐2020 | 13‐09‐2021 | – | – | – |
| 20B/S:626S | 183 | 30‐07‐2020 | 29‐01‐2021 | – | – | – |
| 20B/S:1122L | 21 | 16‐07‐2020 | 15‐11‐2020 | – | – | – |
Abbreviations: GISAID, Global Initiative on Sharing Avian Influenza Data; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Table 2.
Display the number of sequences of each SARS‐CoV‐2 variant that occurred in the Denmark, France, and Australia between the identified period (first_seq) and the sampling period (last_seq) based on data set 1, GISAID
| Variant | Denmark | France | Australia | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of sequences | first_seq | last_seq | Number of sequences | first_seq | last_seq | Number of sequences | first_seq | last_seq | |
| 20I (Alpha, V1) | 63 799 | 09‐11‐2020 | 13‐09‐2021 | 32 650 | 18‐10‐2020 | 12‐12‐2021 | 613 | 30‐11‐2020 | 23‐08‐2021 |
| 20H (Beta, V2) | 130 | 04‐01‐2021 | 19‐07‐2021 | 6178 | 22‐12‐2020 | 14‐12‐2021 | 96 | 10‐12‐2020 | 30‐06‐2021 |
| 20J (Gamma, V3) | 67 | 12‐01‐2021 | 30‐07‐2021 | 1095 | 22‐01‐2021 | 27‐09‐2021 | 8 | 01‐03‐2021 | 25‐04‐2021 |
| 21A (Delta) | 1189 | 29‐03‐2021 | 18‐12‐2021 | 2307 | 06‐04‐2021 | 29‐11‐2021 | 145 | 18‐03‐2021 | 05‐12‐2021 |
| 21I (Delta) | 6328 | 05‐04‐2021 | 02‐01‐2022 | 5197 | 20‐04‐2021 | 29‐12‐2021 | 167 | 04‐04‐2021 | 22‐12‐2021 |
| 21J (Delta) | 151 753 | 05‐04‐2021 | 05‐01‐2022 | 91 861 | 12‐01‐2021 | 03‐01‐2022 | 29 360 | 03‐04‐2021 | 04‐01‐2022 |
| 21K (Omicron) | 15 467 | 22‐11‐2021 | 05‐01‐2022 | 3861 | 12‐11‐2021 | 04‐01‐2022 | 3624 | 26‐11‐2021 | 05‐01‐2022 |
| 21L (Omicron) | 2786 | 05‐12‐2021 | 05‐01‐2022 | 2 | 21‐12‐2021 | 27‐12‐2021 | 4 | 01‐12‐2021 | 04‐01‐2022 |
| 21B (Kappa) | 28 | 08‐03‐2021 | 31‐05‐2021 | 15 | 21‐02‐2021 | 01‐06‐2021 | 156 | 16‐03‐2021 | 26‐06‐2021 |
| 21D (Eta) | 613 | 11‐01‐2021 | 24‐05‐2021 | 738 | 06‐01‐2021 | 22‐10‐2021 | 17 | 06‐01‐2021 | 31‐05‐2021 |
| 21F (Iota) | 8 | 22‐03‐2021 | 26‐04‐2021 | 8 | 16‐03‐2021 | 12‐07‐2021 | 5 | 24‐02‐2021 | 21‐05‐2021 |
| 21G (Lambda) | 9 | 12‐04‐2021 | 11‐08‐2021 | 67 | 06‐04‐2021 | 12‐10‐2021 | 1 | 03‐04‐2021 | 03‐04‐2021 |
| 21H (Mu) | 12 | 05‐04‐2021 | 05‐08‐2021 | 25 | 28‐04‐2021 | 31‐08‐2021 | 49 | 13‐05‐2021 | 05‐06‐2021 |
| 20B/S:732A | 187 | 28‐12‐2020 | 24‐05‐2021 | 44 | 09‐01‐2021 | 20‐05‐2021 | 6 | 06‐04‐2021 | 01‐05‐2021 |
| 20A/S:126A | 0 | 136 | 25‐02‐2021 | 29‐07‐2021 | 0 | ||||
| 20E (EU1) | 27 380 | 03‐08‐2020 | 31‐05‐2021 | 2322 | 30‐04‐2020 | 12‐10‐2021 | 41 | 20‐08‐2020 | 07‐03‐2021 |
| 21C (Epsilon) | 37 | 04‐01‐2021 | 22‐03‐2021 | 9 | 26‐01‐2021 | 22‐07‐2021 | 22 | 08‐12‐2020 | 12‐04‐2021 |
| 20A/S:439K | 5459 | 10‐08‐2020 | 22‐09‐2021 | 307 | 09‐08‐2020 | 14‐04‐2021 | 15 | 08‐09‐2020 | 20‐03‐2021 |
| S:677H.Robin1 | – | – | – | – | – | – | 2 | 15‐12‐2020 | 21‐12‐2020 |
| S:677P.Pelican | 1 | 23‐11‐2020 | 23‐11‐2020 | – | – | – | 2 | 11‐12‐2020 | 11‐12‐2020 |
| 20A.EU2 | 5299 | 27‐07‐2020 | 11‐05‐2021 | 6055 | 09‐07‐2020 | 23‐12‐2021 | 20 | 09‐09‐2020 | 29‐03‐2021 |
| 20A/S:98F | 2958 | 10‐08‐2020 | 22‐03‐2021 | 246 | 18‐08‐2020 | 22‐06‐2021 | 7 | 19‐09‐2020 | 05‐12‐2020 |
| 20C/S:80Y | 63 | 03‐08‐2020 | 28‐09‐2020 | 233 | 24‐07‐2020 | 01‐09‐2021 | – | – | – |
| 20B/S:626S | 545 | 27‐07‐2020 | 22‐02‐2021 | 1 | 11‐08‐2020 | 11‐08‐2020 | – | – | – |
| 20B/S:1122L | 121 | 17‐08‐2020 | 18‐01‐2021 | – | – | – | – | – | – |
Abbreviations: GISAID, Global Initiative on Sharing Avian Influenza Data; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
The propagation rate of variants in the UK can be observed from Figure 3A, where the 21J Delta variant was first detected on November 13, 2020, with 1 065 977 sequences found until January 5, 2022. It takes 2550 of propagation coefficient for 418 days, suggesting a propagation rate of 51%. The 21 K Omicron variant was discovered on October 23, 2021, with 124 499 sequences discovered until January 9, 2022. It has a 1596 propagation coefficient and perhaps 32% to propagate for 78 days. The 21I Alpha variant was discovered on September 20, 2020, with 262 782 sequences discovered until November 26, 2021. It takes 608 of propagation coefficient for 432 days, suggesting a propagation rate of 12%. The 20E EU1 variant was found on July 18, 2020, with 82,158 sequences discovered till November 22, 2021. It takes 167 of the propagation coefficient, and possibly 3%, to propagate over 492 days.
Figure 3.

Shows the propagation rate of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) variants in various countries based on Tables 1 and 2 (data set 1, Global Initiative on Sharing Avian Influenza Data [GISAID]), (A) UK, (B) USA, (C) Denmark, (D) France, and (E) Australia
The propagation rate of variants in the United States can be shown in Figure 3B, where the 21J Delta variant was first detected on November 5, 2020, with 1 213 604 sequences discovered until January 7, 2022. It takes a 2836 propagation coefficient for 428 days, suggesting a propagation rate of 46%. The 21K Omicron variant was discovered for the first time on November 21, 2021, with 84 425 sequences discovered until January 7, 2022. To propagate over 47 days, it has a 1596 propagation coefficient and perhaps 29%. The 21I Alpha variant was discovered for the first time on October 21, 2020, with 242 177 sequences discovered through December 18, 2021. It takes 573 of propagation coefficient for 423 days, suggesting a propagation rate of 9%. The 21I Delta variant was identified on March 16, 2021, with 110 074 sequences discovered till January 5, 2022. It takes 373 propagation coefficient and perhaps 6% to propagate for 295 days. The 21A Delta variant was found on February 9, 2021, with 51 745 sequences revealed until January 1, 2022. It takes 159 propagation coefficients, and possibly 3%, to propagate over 326 days.
The propagation rate of variants in Denmark can be understood from Figure 3C; the 21 J Delta variant was first detected on April 5, 2021, with 151 753 sequences discovered till January 5, 2022. It takes 552 propagation coefficient for 275 days, suggesting a propagation rate of 40%. The 21K Omicron variant was discovered on November 22, 2021, with a total of 15 467 sequences discovered through January 5, 2022. To propagate over 44 days, 352 of the propagation coefficient is bound and possibly 26%. The 21I Alpha variant was found on November 9, 2020, with 63 799 sequences discovered till September 13, 2021. It takes a 207 propagation coefficient for 308 days, suggesting a propagation rate of 15%. The 20E EU1 variation was observed on August 3, 2020, with 27 380 sequences discovered through May 31, 2021. It requires a 301 propagation coefficient, and maybe 7%, to propagate over 301 days. The 21L Omicron difference was found on December 5, 2021, with 2786 sequences discovered till January 5, 2022. It requires 90 percent of the propagation coefficient, and maybe 7%, to propagate over 31 days.
The propagation rate of variants in France can be observed from Figure 3D; the 21J Delta variant was first detected on January 12, 2021, with 91 861 sequences discovered till January 3, 2022. It takes 258 of propagation coefficient for 356 days, suggesting a propagation rate of 53%. The 21K Omicron variant was detected for the first time on November 12, 2021, with 3861 sequences discovered until January 4, 2022. It takes 73 of the propagation coefficient, and maybe 15%, to propagate over 53 days. The 21I Alpha variant was revealed on October 18, 2020, and 32 650 sequences were discovered till December 12, 2021. It takes 78 of the propagation coefficient for 420 days, suggesting a propagation rate of 16%. The 21I Delta variant was discovered for the first time on April 20, 2021, with 5,197 sequences discovered through December 29, 2021. It takes 21 of the propagation coefficient, and maybe 4%, to propagate over 253 days. The 20H Beta variant was discovered for the first time on December 22, 2020, with 6178 sequences discovered through December 14, 2021. It took 17 of the propagation coefficient, and maybe 4%, to propagate over 357 days.
The propagation rate of variants in Australia could be shown in Figure 3E, where the 21J Delta variant was first detected on April 3, 2021, with 29 360 sequences discovered till January 4, 2022. It takes 106 of propagation coefficient for 276 days, suggesting a propagation rate of 52%. The 21K Omicron variant was revealed on November 26, 2021, with 3624 sequences discovered till January 5, 2022. It took 91 of the propagation coefficient, and perhaps 44%, to propagate during 40 days.
Following that, an analysis was undertaken on tests completed for SARS‐CoV‐2 infection in those five nations. Many different types of tests are utilized to identify SARS‐CoV‐2, and its attributes differ. Certain tests yield findings quickly (within minutes), while others take hours to process. Some tests are extremely delicate; others are very distinctive. Influenza tests, particularly NAATs and antigen tests, are employed as diagnostic tests to confirm SARS‐CoV‐2 infection and to assist a person's healthcare services. Viral tests could potentially be employed as diagnostic testing to prevent the spread of SARS‐CoV‐2 by detecting sick people may require to be isolated among others.
The evaluation of SARS‐CoV‐2 tests should be dependent on the environment in which they are administered, such as the prevalence of SARS‐CoV‐2 in the community becoming tested and the status (evidence, complaints, and connections) of the individual being tested. Table 3 shows the number of tests per million inhabitants in those five nations, as well as the number of sequences. Figure 4 depicts the results of the tests based on the percentages calculated in Table 3. As of January 13, 2022, the rate of tests despite the number of sequences with tests per 1 million people in the UK, USA, Denmark, France, and Australia was 25%, 73%, 1.6%, 4.8%, and 1.5%, respectively. This finding suggests that viral testing should be expanded in all five nations. This will aid in determining the accurate propagation of variants and will assist governments in making policy decisions.
Table 3.
Shows the number of SARS‐CoV‐2 infection tests performed per million population and the number of sequences found in the UK, USA, Denmark, France, and Australia, based on data sets 1 (GISAID) and 2 (Worldometer)
| Name of the country | Tests/1 million population | Number of sequences |
|---|---|---|
| UK | 6 286 888 | 1 570 879 |
| USA | 2 564 277 | 1 876 940 |
| Denmark | 19400 378 | 316 889 |
| France | 3 229 555 | 153 357 |
| Australia | 2255 277 | 34 360 |
Abbreviations: GISAID, Global Initiative on Sharing Avian Influenza Data; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 4.
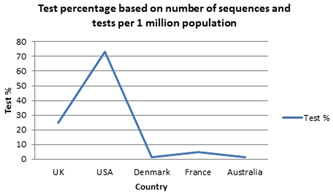
Shows the percentage of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) infection tests in the UK, United States, Denmark, France, and Australia based on Table 3
4. CONCLUSION
SARS‐CoV‐2 variations have been detected since May 2020, and different variants have been identified till this day, with the WHO labeling the majority of them. According to this study, as of mid‐January 2022, the Omicron variant was spread across 170 countries in the world, with the UK moving ahead with 46%, the United States coming in second with 31%, Denmark falling in third with 7%, France goes in fourth with 1%, and Australia coming in fifth with 1%. The propagation rate of Omicron variants in the UK, United States, Denmark, France, and Australia is 51%, 29%, 26%, 15%, and 44%, respectively, findings indicating that Omicron may soon dominate others. The SARS‐CoV‐2 infection test rate based on tests per one million people with the number of sequences in those five countries is 25%, 73%, 1.6%, 4.8%, and 1.5%, respectively. These results suggest that tests should be increased quickly to determine the precise distribution of variants and aid governments in making policy decisions for public safety. As a result, we urge that individuals 18 and older acquire a COVID‐19 vaccination and booster doses, as well as wear mask to prevent the spread of new variants. This study also aids researchers in determining the propagation of SARS‐CoV‐2 variants in other countries, and also future mutations around the world.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Both authors contributed equally.
Rathinasamy M, Kandhasamy S. An exploratory study on the propagation of SARS‐CoV‐2 variants: omicron is the most predominant variant. J Med Virol. 2022;94:2414‐2421. 10.1002/jmv.27634
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in GISAID database at https://covariants.org/.
REFERENCES
- 1. Burki T. Understanding variants of SARS‐CoV‐2. The Lancet. 2021;397(10273), 462. 10.1016/S0140-6736(21)00298-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: Evidence that D614G increases infectivity of the COVID‐19 Virus. Cell. 2020;182(4):812‐827. e19. 10.1016/2Fj.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou B, Thao TTN, Hoffmann D, et al. SARS‐CoV‐2 spike D614G change enhances replication and transmission. Nature. 2021;592:122‐127. 10.1038/s41586-021-03361-1 [DOI] [PubMed] [Google Scholar]
- 4. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(1):64‐75. 10.1016/j.cell.2020.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walensky RP, Walke HT, Fauci AS. SARS‐CoV‐2 Variants of concern in the United States – challenges and opportunities. JAMA. 2021;325(11):1037‐1038. 10.1001/jama.2021.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Epidemiological Update : Occurrence of variants of SARS‐CoV‐2 in the Americas − 26 January 2021. Pan American Health Organization. 2021; https://www.paho.org/en/documents/epidemiological-update-occurrence-variants-sars-cov-2-americas-26-january-2021
- 7. PANGO Lineages . SARS‐CoV‐2 lineages. 2021. Accessed: December 18, 2021. https://cov-lineages.org/global_report.html
- 8. Liu Y, Rocklöv J. The reproductive number of the Delta variant of SARS‐CoV‐2 is far higher compared to the ancestral SARS‐CoV‐2 virus. J Travel Med. 2021;28(7):taab124. 10.1093/jtm/taab124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Callaway Ewen. Delta coronavirus variant: scientists brace for impact. Nature. 2021;595(7865):17‐18. 10.1038/d41586-021-01696-3 [DOI] [PubMed] [Google Scholar]
- 10. Rambaut A, Holmes EC, O'toole Á, et al. A dynamic nomenclature proposal for SARS‐CoV‐2 lineages to assist genomic epidemiology. Nat Microbiol. 2020;5:1403‐1407. 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci U S A. 2020;117(21):11727‐11734. 10.1073/pnas.2003138117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Starr Tyler N, Greaney Allison J, Dingens Adam S, Bloom Jesse D. Complete map of SARS‐CoV‐2 RBD mutations that escape the monoclonal antibody LY‐CoV555 and its cocktail with LY‐CoV016. Cell Reports Medicine. 2021;2(4):100255. 10.1101/2021.02.17.431683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Technical briefing 20 SARS‐CoV‐2 variants of concern and variants under investigation in England. Public Health England. 6 August 2021. Accessed: December 15, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1009243/Technical_Briefing_20.pdf
- 14. SARS‐CoV‐2 variants of concern and variants under investigation in England 2021 Public Health England. Technical briefing, August 20, 2021. Last accessed December 15, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1012644/Technical_Briefing_21.pdf
- 15. Thangaraj JWV, Yadav P, Kumar CG, et al. Predominance of delta variant among the COVID‐19 vaccinated and unvaccinated individuals, India. J Infect. 2022;84(1):94‐118. 10.1016/j.jinf.2021.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mlcochova P, Kemp SA, Dhar MS, et al. SARS‐CoV‐2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599(7883):114‐119. 10.1038/s41586-021-03944-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. He X, He C, Hong W, Zhang K, Wei X. The challenges of COVID‐19 Delta variant: prevention and vaccine development. MedComm(2020). 2021;2(4):846‐854. 10.1002/mco2.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shiehzadegan S, Alaghemand N, Fox M, Venketaraman V. Analysis of the Delta variant B.1.617.2 COVID‐19. ClinPract. 2021;11(4):778‐784. 10.3390/clinpract11040093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid‐19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585‐594. 10.1056/NEJMoa2108891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kenyon G. Australia's struggle with the delta variant. Lancet Infect Dis. 2021;21(10):1358. 10.1016/S1473-3099(21)00579-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Eichler N, Thornley C, Swadi T, et al. Transmission of severe acute respiratory syndrome coronavirus 2 during border quarantine and Air Travel, New Zealand (Aotearoa). Emerg Infect Dis. 2021;27(5):1274‐1278. 10.3201/eid2705.210514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mishra S, Mindermann S, Sharma M, et al. Changing composition of SARS‐CoV‐2 lineages and rise of Delta variant in England. E Clinical Medicine. 2021;39:101064. 10.1016/j.eclinm.2021.101064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang L, Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J Med Virol. 2021. 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 24. Torjesen Ingrid . Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. 10.1136/bmj.n2943 [DOI] [PubMed] [Google Scholar]
- 25. Thakur V, Ratho RK. OMICRON (B.1.1.529): a new SARS‐CoV‐2 variant of concern mounting worldwide fear. J Med Virol. 2021. 10.1002/jmv.27541 [DOI] [PubMed] [Google Scholar]
- 26. Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS‐CoV‐2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol. 2021. 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 27. Kumar S, Thambiraja TS, Karuppanan K, Subramaniam G. Omicron and Delta variant of SARS‐CoV‐2: A comparative computational study of spike protein. J Med Virol. 2021. 10.1002/jmv.27526 [DOI] [PubMed] [Google Scholar]
- 28. Poudel S, Ishak A, Perez‐Fernandez J, et al. Highly mutated SARS‐CoV‐2 Omicron variant sparks significant concern among global experts ‐ What is known so far? Travel Med Infect Dis. 2021;45:102234. 10.1016/j.tmaid.2021.102234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He X, Hong W, Pan X, Lu G, Wei X. SARS‐CoV‐2 Omicron variant: characteristics and prevention. MedComm (2020). 2021;2(4):838‐845. 10.1002/mco2.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hodcroft EB CoVariants: SARS‐CoV‐2 mutations and variants of interest. 2021. Accessed: January 15, 2022. URL: https://covariants.org/
- 31. Worldometer . COVID‐19 Coronavirus Pandemic: Reported Cases and Deaths by Country or Territory. 2022. Accessed: January 15, 2022. https://www.worldometers.info/coronavirus/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GISAID database at https://covariants.org/.


