Abstract
Penicillin tolerance is an incompletely understood phenomenon that allows bacteria to resist drug-induced killing. Tolerance was studied with independent Streptococcus gordonii mutants generated by cyclic exposure to 500 times the MIC of penicillin. Parent cultures lost 4 to 5 log10 CFU/ml of viable counts/24 h. In contrast, each of four independent mutant cultures lost ≤2 log10 CFU/ml/24 h. The mutants had unchanged penicillin-binding proteins but contained increased amounts of two proteins with respective masses of ca. 50 and 45 kDa. One mutant (Tol1) was further characterized. The two proteins showing increased levels were homologous to the arginine deiminase and ornithine carbamoyl transferase of other gram-positive bacteria and were encoded by an operon that was >80% similar to the arginine-deiminase (arc) operon of these organisms. Partial nucleotide sequencing and insertion inactivation of the S. gordonii arc locus indicated that tolerance was not a direct consequence of arc alteration. On the other hand, genetic transformation of tolerance by Tol1 DNA always conferred arc deregulation. In nontolerant recipients, arc was repressed during exponential growth and up-regulated during postexponential growth. In tolerant transformants, arc was constitutively expressed. Tol1 DNA transformed tolerance at the same rate as transformation of a point mutation (10−2 to 10−3). The tolerance mutation mapped on a specific chromosomal fragment but was physically distant from arc. Importantly, arc deregulation was observed in most (6 of 10) of additional independent penicillin-tolerant mutants. Thus, although not exclusive, the association between arc deregulation and tolerance was not fortuitous. Since penicillin selection mimicked the antibiotic pressure operating in the clinical environment, arc deregulation might be an important correlate of naturally occurring tolerance and help in understanding the mechanism(s) underlying this clinically problematic phenotype.
Bacteria have developed at least two mechanisms to escape the bacteriostatic and/or bactericidal effect of penicillin and related drugs: (i) antibiotic resistance and (ii) antibiotic tolerance. Resistant bacteria have the ability to grow in the presence of antibiotic concentrations that are much greater than the minimal concentration of the drug required to inhibit the growth of susceptible organisms, the so-called minimal inhibitory concentration. However, when resistant bacteria are exposed to penicillin concentrations greater than their new, increased MIC, they usually remain susceptible to antibiotic-induced killing (16, 22, 38). Tolerant bacteria, on the other hand, have unchanged MICs but have a drastically decreased susceptibility to drug-induced killing (16, 27, 38).
Resistant bacteria are of great clinical concern, because infections due to such organisms often result in a failure of antimicrobial therapy. The mechanisms of beta-lactam resistance have been extensively studied. They include beta-lactamase production and/or alteration of the membrane-bound penicillin-binding proteins in gram-positive bacteria (8, 12) and alterations in the outer membrane permeability in gram-negative organisms (15, 28). Tolerant bacteria are also of clinical relevance and were linked to treatment failures in both experimental and clinical investigations (9, 20, 31, 34). However, in contrast to resistance, the mechanism by which tolerant bacteria escape penicillin-induced killing is not completely understood.
Previous investigations of penicillin-induced killing and penicillin tolerance suggested that the tolerance phenotype was due to alterations in the bacterial autolytic system. Many kinds of bacteria respond to penicillin treatment by massive bacterial lysis due to antibiotic-induced deregulation of intrinsic cell wall hydrolases, or autolysins. Since bacterial lysis is accompanied by cell death, it was presumed that penicillin-induced autolysis was the very mechanism of drug-induced killing. Accordingly, mutants defective in autolysis were expected simultaneously to resist both drug-induced lysis and killing. However, several studies indicated that autolysis is not the sole mechanism of penicillin-induced killing. Certain bacteria, such as Streptococcus pyogenes and other gram-positive cocci, are killed extensively by penicillin in spite of the fact that they are not lysed by the antibiotic (6, 14, 25). Moreover, specific blockage of penicillin-induced lysis in Streptococcus pneumoniae and Staphylococcus aureus decreased drug-induced killing only marginally, thus suggesting that other, autolysis-independent pathways were involved in penicillin-induced lethality (11, 13, 26, 31, 36, 37).
Recently, a breakthrough study indicated that vancomycin- and penicillin-tolerant pneumococci carried alterations in the two-component regulatory system VncS-VncR, which was not directly related to autolysis (31). However, additional alterations responsible for antibiotic tolerance were also described, thus indicating that there is more than one mechanism for bacterial survival of antibiotics (26, 29, 30). In the present study we explored the existence of such a putative mechanism(s) by using a penicillin-susceptible but lysis-defective strain of Streptococcus gordonii and a series of its penicillin-tolerant derivatives. Physiological, biochemical, and genetic properties of the parent and mutant cells and of their backcross transformants were compared, and an operon indirectly involved in tolerance is described.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The S. gordonii strains used in this study are described in Table 1. Other bacteria included a clinical isolate of S. pyogenes (strain UAA7) and Escherichia coli XL-1 blue. Streptococci were grown at 37°C either in brain heart infusion broth (Difco Laboratories, Detroit, Mich.) without aeration or on Columbia agar (Becton Dickinson Microbiology Systems, Cockeysville, Md.) supplemented with 3% blood. E. coli was grown in Luria- Bertani broth or on Luria-Bertani agar. Growth of the cultures was followed both by measurement of optical density at a wavelength of 620 nm (OD620) with a spectrophotometer (Sequoia-Turner, Montainville, Calif.) and by colony counts. When appropriate, antibiotics were added to the medium at the following concentrations: streptomycin, 100 mg/liter; rifampin, 4 mg/liter; erythromycin, 5 mg/liter for streptococci and 500 mg/liter for E. coli; and ampicillin, 50 mg/liter. Bacterial stocks were stored at −70°C in broth supplemented with 10% (vol/vol) glycerol.
TABLE 1.
S. gordonii strains used in the study
| Designation | Relevant genotype | Phenotype and/or utilization | Source or reference |
|---|---|---|---|
| Challis | Wild type | Reference strain | 33 |
| SG1 | Smr mutant of Challis | Used for tolerance selection; recipient in genetic backcrosses | This work |
| SG2 | Rfr mutant of Challis | Control (Rfr) DNA donor | This work |
| SG101 | SG1 arcA::pJDC9a | arcA-negative; Emr Smr | This work |
| SG102 | SG1 arcB::pJDC9a | arcB-negative; Emr Smr | This work |
| SG103 | SG1 arcB::luc-ermb | Luc+ Emr Smr; used to study arc regulation | This work |
| Tol1 to Tol4 | SG1 tolerant mutants | Selected by penicillin cycling | This work |
| Tol101 | Tol1 arcA::pJDC9a | arcA-negative; Emr Smr | This work |
| Tol102 | Tol1 arcB::pJDC9a | arcB-negative; Emr Smr | This work |
| Tol103 | Tol1 arcB::luc-ermb | Luc+ Emr Smr; used to study arc regulation | This work |
arcA and arcB were inactivated by insertion duplication with the suicide vector pJCD9 (4).
To construct the arcB::luc-erm transcriptional fusion, a copy of the promoterless luc gene containing its ribosome-binding site was targeted into arcB by using the pJDC9 vector, as described in Materials and Methods.
Antibiotics and chemicals.
Penicillin G was purchased from Hoechst-Pharma (Zurich, Switzerland); rifampin was purchased from Novartis (Basel, Switzerland); restriction enzymes (Boehringer Mannheim, Mannheim, Germany), Taq DNA polymerase, and T4 DNA ligase (Gibco BRL, Gaithersburg, Md.) were used according to the manufacturer's recommendations; all other chemicals were reagent-grade, commercially available products.
Antibiotic susceptibility and time-kill curves.
The MICs of the test antibiotics were determined by a standard macrodilution method (40). Time-kill curves were determined by adding appropriate concentrations of antibiotics to bacterial cultures in the exponential phase of growth (OD620 = 0.2) (9). Antibiotic carryover on the agar plates was avoided as described previously (9, 10). Colonies were counted after 48 h of incubation at 37°C.
Selection of penicillin-tolerant derivatives of S. gordonii.
The selection method has been previously described (9, 26, 27). In brief, nonmutagenized 10-ml cultures of the Smr parent strain SG1 were grown to an OD620 of 0.2 and treated with a final concentration of 2 mg of penicillin/liter (i.e., 500 times the MIC) for 18 to 20 h at 37°C. This concentration is readily achieved in the serum of humans during standard penicillin therapy (1). Streptomycin was kept in the medium to avoid contamination with other bacteria during penicillin cycling. The surviving bacteria were washed by centrifugation and resuspension and regrown in drug-free medium. After eight such cycles, single colonies were isolated and characterized for tolerance, stability, and biochemical and genetic properties.
Determination of bacterial proteins by 2D gel electrophoresis and N-terminal amino acid microsequencing.
Two-dimensional (2D) gel electrophoresis was run by slight modifications of a published method (21). One-hundred-milliliter cultures grown to the exponential phase were harvested by centrifugation, washed twice in ice-cold NaCl, and incubated for 45 min at 37°C in lysis buffer (50 mM phosphate buffer [pH 7.5], 10 mM MgCl2, 10 μg of DNase/ml, 10 μg of RNase/ml, 0.5 μg of lysozyme/ml, and 0.1 μg of mutanolysine/ml). Lysis (≥90%) was assessed by phase-contrast microscopy. Cell debris was removed by low-speed centrifugation, and proteins in the supernatant were precipitated with 0.015% (wt/vol) deoxycholate and 7.2% (wt/vol) trichloroacetic acid. For isoelectrofocusing, the protein pellets were dissolved to a final concentration of 400 to 600 mg/liter in 100-μl volumes of sample buffer (8 M urea, 65 mM dithiothreitol, 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, 40 mM Tris, trace bromophenol blue), and applied to a nonlinear pH gradient pH 3-10 strip (Pharmacia Biotech, Uppsala, Sweden). Migration started with a linear voltage increase from 300 to 3,500 V for 3 h, followed by a constant voltage of 3,500 V for 27 h. After isoelectrofocusing, the strips were soaked for 10 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer (50 mM Tris-HCl [pH 6.8], 6 M urea, 5% 2-mercaptoethanol, 30% glycerol, 2% sodium dodecyl sulfate, trace bromophenol blue), placed on the top of a vertical piperazine diacrylyl-acrylamide gradient gel (from 9 to 16%), and left to migrate at 40 mA/gel for 3 to 4 h. Proteins were detected by silver staining. The cross-linker PDA and sodium thiosulfate (final concentration, 0.025%) were added to the gel to optimize resolution.
For N-terminal sequencing, the proteins were blotted onto a polyvinylidene difluoride membrane (Immobilon PSQ; Millipore Inc., Bedford, Mass.), stained with 0.1% amido black, cut out of the membranes, and subjected to automated Edman degradation on an ABI (Applied Biosystems, Foster City, Calif.) 494 Procise sequencer. The derived sequences were compared with the SwissProt protein database by a BLAST search using the GCG software package (Program Manual for the Wisconsin Package, Version 8; Genetics Computer Group, Madison, Wis.).
DNA transformation and genetic analysis.
DNA and competent cells were prepared by previously described methods (19, 33, 35). Plasmids were purified with a plasmid purification kit (Qiagen GmbH, Hilden, Germany). They included the pGemT-Easy vector (Promega Corporation, Madison, Wis.), used for DNA amplification and sequencing, and the suicide vector pJDC9 (4), carrying an erythromycin resistance (Emr) marker and used for insertion duplication mutagenesis in S. gordonii. Insertion mutagenesis was achieved by subcloning internal fragments of the target genes into the polycloning site of pJDC9, followed by transformation into competent cells. Transformants carrying an insert were selected for Emr, and disappearance of the appropriate gene product was assessed by 2D gel electrophoresis. PCR amplification was performed with a Perkin Elmer Cetus (Foster City, Calif.) thermocycler. Oligonucleotide primers were obtained from Life Technologies AG (Basel, Switzerland) or Microsynth GmbH (Balgach, Switzerland). Nucleotide sequencing was performed by Microsynth GmbH. In certain experiments the technique of inverse PCR was used. Partial Sau3A digests (between 500 and 3,000 bp) of the S. gordonii chromosome were self-ligated and amplified. Primers are indicated in Fig. 4. Thermocycling conditions were 94°C for 1 min, 50°C for 1 min, and 72°C for 3 min, with a final extension of 10 min at 72°C. PCR fragments were controlled by more internal primers. Bands were subcloned in the pGemT-Easy vector for nucleotide sequencing.
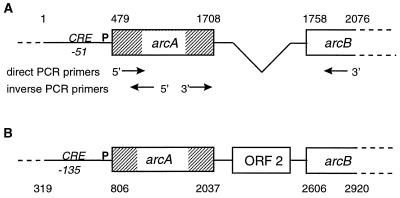
FIG. 4.
Partial map of the S. gordonii arc operon (A) and its S. pyogenes homologue (B). The sequence encompasses the 479 bp preceding the ATG of the arcA ORF, the whole arcA ORF, and the first 318 bp of the arcB ORF. The first bp of the S. gordonii sequence is aligned with bp 319 of the S. pyogenes sequence (accession number X55659). The boxes represent the genes present in the operon. Base pair numbering is indicated at the top and bottom of the figure. Numbers indicate the beginning and the end of the alignment, the start and the stop codons of the arcA genes, and the start codon of the arcB genes. Note that the S. gordonii operon is missing a 582-bp stretch comprising a 440-bp ORF (ORF 2) of unknown function present in S. pyogenes. In S. gordonii, the arcA and arcB genes are separated by a short 49-bp stretch (bp 1708 to 1758) with a putative promoter sequence. P indicates the putative operon promoters. CRE indicates the location of a putative catabolic repression element, present in both S. gordonii and S. pyogenes, at bp −51 and −135 of the start codon, respectively. The arrows indicate the positions of the primers utilized for direct and inverse PCR used to determine the S. gordonii sequence. For direct PCR, the forward primer (5′-TCCACAGACCCGGTAAAG-3′) started at bp 537, and the reverse primer (5′-CAATCCCATCAAAC-3′) started at bp 2076. Inverse PCR utilized two divergent primers, the first (5′-TCATCAAAGAGAAGACGTTC-3′) starting at bp 603 and the second (5′-TGGTAGAATCAGACTATCC-3′) starting at bp 912.
Construction of a luciferase-based transcriptional-fusion system.
An internal fragment of arcB (bp 1813 to 2076; see Fig. 4) was inserted into the multicloning site of pJDC9. The promoterless firefly luciferase (luc) gene containing its ribosome-binding site (kindly provided by A. Podbielsky, University of Ulm, Ulm, Germany) was then subcloned downstream of this fragment, and the construct was integrated into the arcB gene by insertion duplication. For light emission, 100-μl portions of bacterial cultures were added to tubes containing 250 μl of Na-citrate (pH 5.5), (0.1 M). Then, 50 μl of 1 mM beetle luciferine (Promega Corporation) diluted in 25 mM glycylglycine and 15 mM MgSO4 buffer was added to the mixture by the autoinjector of the luminometer (EG & G Berchtold, Bad Wildbad, Germany). After 1 s of equilibration, light was measured for 10 s.
Pulsed-field gel electrophoresis (PFGE).
Plugs of genomic DNA were prepared as described previously (2, 24) in 1.6% low-melting-point agarose (Boehringer Mannheim) and digested with SmaI. Electrophoresis was run on a contour-clamped homogeneous electric field DR III apparatus (Bio-Rad, Richmond, Calif.) for a total of 24 h. The run was divided into three steps with a voltage of 6 V/cm and an angle of 120°C. The first step began with a linear switch time of 1 to 10 s during 10 h, the second step continued with a switch-time of 10 to 15 s during 7.5 h, and the third step continued with a switch time of 15 to 17 s during 6.5 h. DNA fragments of interest were extracted from the gel by digesting the agarose with β-agarose (New England Biolabs Inc., Beverly, Mass.), followed by precipitation with standard techniques.
Nucleotide sequence accession number.
The partial nucleotide sequence of the S. gordonii arc operon, encompassing the promoter region, the gene encoding Adi (arcA), and the first 317 bp of the gene encoding Oct (arcB), was submitted to the DDBJ/EMBL/GenBank databases (accession number AF172730).
RESULTS
Selection and characterization of penicillin-tolerant derivatives of S. gordonii.
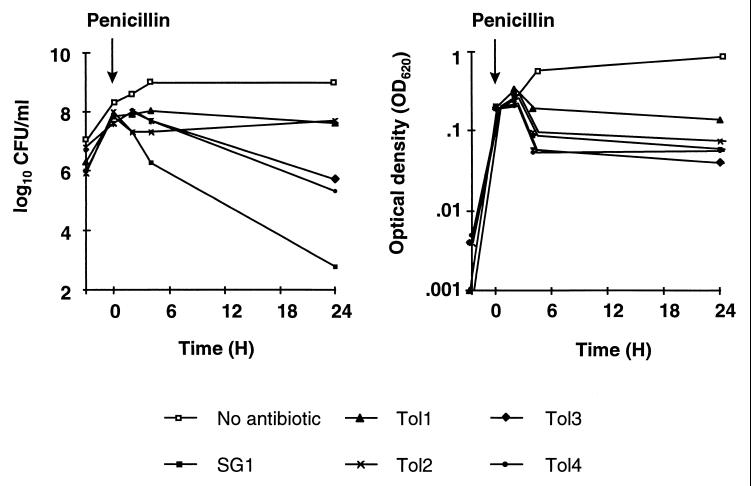
Cyclic exposure of the parent SG1 strain to penicillin selected for tolerance. The left panel of Fig. 1 depicts the loss of viability of SG1 and of four independently selected tolerant mutants exposed to 500 times the MIC of penicillin for 24 h. SG1 lost 4 to 5 log CFU/ml/24 h. In contrast, two tolerant derivatives (Tol1 and Tol2) lost <1 log CFU/ml, and two of them (Tol3 and Tol4) lost only 2 log CFU/ml during the same time. All four mutants were stable after serial passage for up to 35 generation times in drug-free medium. Moreover, the differential tolerance patterns between Tol1 and Tol2 and between Tol3 and Tol4 were also preserved after such passages.
FIG. 1.
Penicillin-induced killing (left panel) and lysis (right panel) of S. gordonii exposed to 500 times the MIC of the drug. Both cultures of the SG1 parent cells and of four independent penicillin-tolerant derivatives (Tol1 to Tol4) were grown to the exponential growth phase, and penicillin was added (arrow) at an OD620 of 0.2. The cultures were monitored for a loss of viable cells and a loss in OD620 as described in Materials and Methods.
One mutant (Tol1) was further characterized. In spite of the mutant's penicillin tolerance, MICs of several beta-lactam and non-beta-lactam antibiotics for Tol1 were unchanged (Table 2). Yet it was cross-tolerant to all beta-lactams and to most non-beta-lactam molecules tested. Exceptions were gentamicin, which was bactericidal against both the SG1 parent strain and the Tol1 tolerant mutant, and vancomycin, which lacked any bactericidal effect. Similar results were obtained with mutants Tol2 to Tol4 (data not presented).
TABLE 2.
MICs of several beta-lactams and unrelated drugs for the parent strain and the Tol1 mutant and loss of viability after 24 h of drug treatment
| Antibiotic | MIC (mg/liter)a for:
|
Loss of viability (log10 CFU/ml)b
|
||
|---|---|---|---|---|
| Parent SG1 | Tol1 | Parent SG1 | Tol1 | |
| Penicillin G | 0.004 | 0.008 | 4–5 | <1 |
| Amoxicillin | 0.008 | 0.008 | 3 | <1 |
| Imipenem | <0.004 | <0.004 | 5–6 | 1 |
| Cefuroxime | 0.008 | 0.016 | 1–2 | <1 |
| Ceftriaxone | 0.008 | 0.008 | 2 | 1 |
| Ceftazidime | 0.064 | 0.125 | 2 | <1 |
| Cefepime | 0.016 | 0.032 | 1.5 | <1 |
| Ciprofloxacin | 1 | 1 | 5–6 | 2–3 |
| Sparfloxacin | 0.25 | 0.25 | 5 | 3 |
| Vancomycin | 1 | 1 | 0 | 0 |
| Rifampin | 0.125 | 0.125 | 1–2 | <1 |
| Streptomycin | >1,000 | >1,000 | NDc | ND |
| Gentamicin | 8 | 8 | ≥6 | ≥6 |
As with Tol1, for the additional mutants Tol2 through Tol4 the MICs of these test drugs were unchanged.
Loss of viability in log10 CFU/ml after 24 h of treatment with of 20 times the MIC of gentamicin, rifampin, and ciprofloxacin, 16 times the MIC of sparfloxacin, or 500 times the MICs of penicillin, amoxicillin, imipenem, ceftriaxone, cefuroxime, ceftazidime, cefepime, and vancomycin.
ND, not determined.
Importantly, the 3 to 4 log10 difference in penicillin-induced killing between the SG1 parent strain and Tol1 was not accompanied by a sizable difference in bacterial lysis. This is apparent in Fig. 1 (right panel) and was confirmed in 16 additional experiments. The percent decrease in optical density during 24 h of penicillin treatment was similar between the two organisms, i.e., 66% ± 24% (mean ± standard error [SE] of 16 determinations) in cultures of SG1, compared to 62% ± 20% in cultures of Tol1. This contrasted with a dramatic difference in loss of viability, which was 4.48 ± 1.39 (mean ± SE log10 CFU/ml/24 h) in cultures of SG1 and only 0.6 ± 0.1 in cultures of Tol1. Thus, autolysis was not the major cause of killing, and other, lysis-independent bactericidal mechanisms were likely to be involved.
Genetic transformation of tolerance.
To test whether penicillin tolerance was transformable, competent SG1 cells were exposed either to Tol1 DNA or to control DNA from the Rfr mutant SG2 (Table 1). The rate of transformation to Rfr was between 10−2 and 10−3. The transformed cultures were subjected to penicillin cycling (Fig. 2) and monitored for conversion to tolerance (26). If tolerance was transformable, Tol1-treated cultures should become enriched in tolerant transformants that would be selected during penicillin cycling. In contrast, control cultures should not become enriched in tolerant transformants. In more than 20 independent experiments, competent cells treated with Tol1 DNA converted to tolerance at the second or third penicillin cycle. In contrast, the cultures exposed to control DNA required at least 8 cycles to show a decrease in penicillin-induced killing.
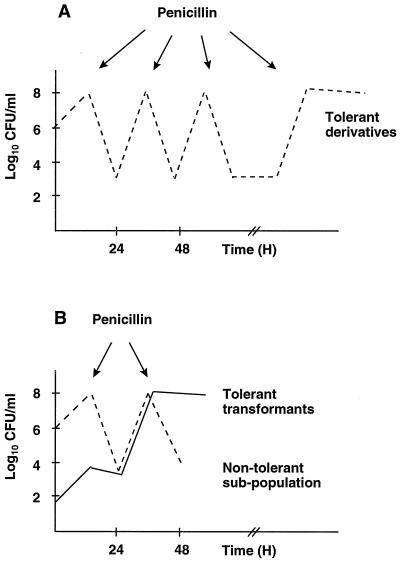
FIG. 2.
Selection of penicillin-tolerant derivatives of S. gordonii by cyclic exposure to penicillin. Panel A illustrates the situation with the SG1 parent cells. Individual broth cultures were grown to an OD620 of 0.2 and treated for 20 h with 500 times the MIC of penicillin. Surviving bacteria were then washed free of the drug and allowed to regrow in antibiotic-free medium before a new cycle of penicillin treatment was started. Repeating such enrichment cycles eventually selected for derivatives that were tolerant to the bactericidal effect of the drug. Panel B illustrates the situation after transformation of the kill-susceptible SG1 parent strain with chromosomal DNA prepared from the tolerant mutant Tol1. The transformed culture contains tolerant transformants (continuous line). Exposure to penicillin resulted in the killing of the nontolerant population and the selection of the penicillin-tolerant transformants (see the text for details).
To evaluate the rate of tolerance transformation, cultures of the SG1 parent strain were mixed with serial 10-fold dilutions of Tol1 and then cycled with penicillin. At the first cycle, the culture containing 10−1 Tol1 cells already demonstrated tolerance (loss of ≤2 log10 CFU/ml/24 h), whereas the other dilutions lost ca. 4 log10 CFU/ml in the same period of time. After the second cycle, the tube containing 10−2 Tol1 cells converted to tolerance. The tube containing 10−3 Tol1 cells converted to tolerance at the third cycle. Compared to DNA transformation, these dilution experiments suggested that transformation of tolerance took place at a rate of 10−2 and 10−3, i.e., equivalent to that of the Rfr point mutation.
Alterations in the protein content of tolerant S. gordonii mutants.
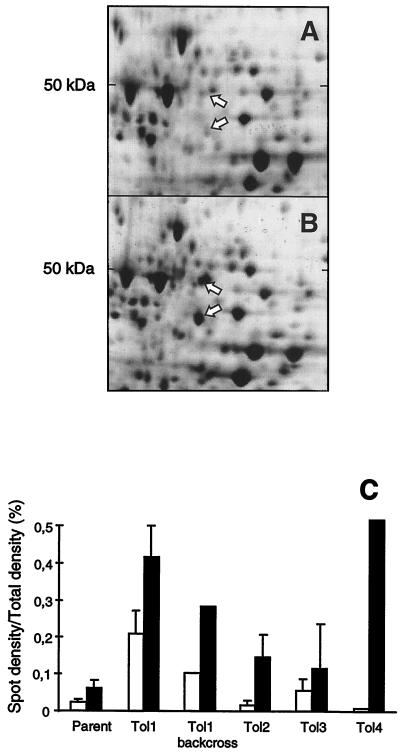
Neither of the four Tol1 to Tol4 mutants had altered penicillin-binding proteins (data not shown). Therefore, whole bacterial proteins were analyzed by 2D-gel electrophoresis. Figures 3A and B present the results for the SG1 parent and the Tol1 mutant analyzed by this technique. Compared to SG1, Tol1 contained two protein spots of increased intensity, of 50 and 45 kDa. Figure 3C indicates that some increase for at least one of these two spots was also observed in the three other mutants (Tol2 to Tol4) and in a backcross transformant of Tol1 (lower panel of Fig. 1).
FIG. 3.
2D-gel electrophoresis (A and B) and quantification of specific protein spots (C) in various S. gordonii isolates. Total protein extracts of the parent strain SG1 (A) and the Tol1 mutant (B) were extracted from cultures in the exponential growth phase (OD620 = 0.2). Electrophoresis conditions and staining were as described in Materials and Methods. Arrows indicate the two protein spots demonstrating an increased intensity in the tolerant mutants. The upper, ca. 50-kDa spot was identified as an arginine deiminase homologue, and the lower, ca. 47-kDa spot was identified as an ornithine carbamoyl transferase homologue. Panel C presents the intensity of the 50-kDa spot (open columns) and the 45-kDa spot (black columns) of the parent SG1, the tolerant mutants Tol1 to Tol4, and a backcross transformant of Tol1. The densities of the spots were determined by scanning densitometry using the Image software (National Institutes of Health, Bethesda, Md.) and were expressed as a percentage of the total gel density. Error bars indicate the mean ± SE of three independent experiments.
The sequence of the 22 N-terminal amino acids of the 50-kDa species was PIEVFSEIGKLKKVMLHRPGKE. This was 93% similar to the streptococcal acid glycoprotein of S. pyogenes (SwissProt accession number P16962) (19) and more than 80% similar to its arginine deiminase (Adi) homologue (7) in Lactobacillus sake (41), Clostridium perfringens (32), and Bacillus licheniformis (23). The sequence of the 28 N-terminal amino acids of the 45-kDa species was VFQGRSFLAEKDFTRAELYLGLSAHLD. This was 95% similar to the ornithine carbamoyl transferase (Oct) of both S. pyogenes (SwissProt accession number P16964) (18) and the other organisms mentioned above (23, 32, 41). In these bacteria, the two genes cluster in the arc operon and the proteins are encoded with a polarity of Adi first and Oct second. If a similar organization also existed in S. gordonii, the concurrent increase of these two gene products in Tol1 would be a logical consequence of deregulation of the putative arc operon in this organism. Therefore, this locus was characterized.
Structure of the S. gordonii arc operon.
Figure 4 presents an alignment of the S. gordonii arc locus and its S. pyogenes homologue (accession number X55659). Compared to S. pyogenes, the S. gordonii arc locus lacked an open reading frame (ORF) between the arcA and arcB genes. In S. pyogenes, these determinants are separated by 569 bp comprising a 440-bp ORF (ORF2) of unknown function (19). In S. gordonii the stop codon of arcA and the ATG of arcB were separated by 49 bp that had no putative promoter sequence. Thus, the two genes could be controlled by a common promoter located upstream of the arcA gene.
Importantly, there was no dissimilarity between the partial sequence of the arc operons of the SG1 parent strain and the Tol1 mutant. This suggested that the tolerance mutation in Tol1 was located either in the 5′-regulatory sequence of the locus or elsewhere on the chromosome. Two types of experiments were performed to clarify this issue. First, the arcA and arcB genes were disrupted to determine whether they were directly or indirectly implicated in tolerance. Second, a luciferase-based transcriptional fusion system was constructed to investigate the regulation of the arc locus in both SG1 and Tol1.
Phenotype of S. gordonii mutants inactivated in the arcA and arcB genes.
Insertion inactivation was achieved by subcloning internal fragments of either arcA (bp 584 to 1003) or arcB (bp 1813 to 2076) into pJDC9, followed by transformation into competent Tol1 cells. Tol1 was used because it overexpressed the two gene products, thereby allowing detection of their decrease in inactivated mutants. Recombinants were tested both for the loss of these products (the 50-kDa and 45-kDa proteins) on 2D-gels and for tolerance.
Interruption of the arcB gene resulted in the disappearance of its 45-kDa product from the gel. Interruption of the arcA gene resulted in the simultaneous disappearance of both its own 50-kDa product and the arcB product (I. Caldelari and P. Moreillon, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. A-72, 1999). This confirmed the polarity of the determinants. However, while successful gene inactivation was achieved, the mutants were altered neither in bacterial growth nor in the tolerance phenotype, whether SG1 parent cells or Tol1 cells were used as the recipient (data not presented). Thus, the arcA and arcB genes were not directly implicated in tolerance. On the other hand, the fact that they were overexpressed in several independent tolerant mutants suggested that they might represent a marker of tolerance, which could help unravel more crucial mutation(s). This hypothesis was investigated next.
Expression of the arc locus in SG1 and Tol1.
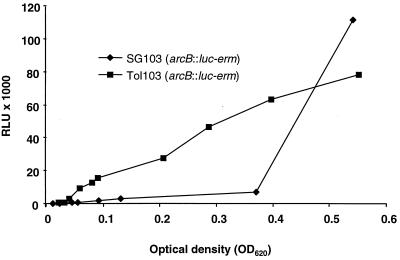
A transcriptional fusion between the arcB ORF and a promoterless luc gene containing its ribosome-binding site was constructed as described in Materials and Methods. Emission of light by the bacteria was measured along the entire range of growth. For the SG1 parent strain, light emission was low during exponential growth and increased in postexponential growth (Fig. 5), indicating that the arc locus was regulated. In Tol1, in contrast, light emission was proportional to bacterial density during the whole growth cycle, indicating that the operon was deregulated and constitutively expressed. The results were very reproducible and were consistent with the increased production of the arcA and arcB products detected by 2D-gel electrophoresis (Fig. 3). We next tested whether arc deregulation correlated with tolerance transformation in genetic backcross experiments.
FIG. 5.
Promoter expression of the arc operon determined by light emission by the SG1 parent and the Tol1 mutant containing a luciferase transcriptional fusion (Table 1, strains SG103 and Tol103, respectively). Broth cultures were monitored along the whole growth phase. Light emission was expressed in relative light units (RLU) and plotted against the cultures' optical densities (OD620).
Functional association and genetic linkage between arc deregulation and tolerance.
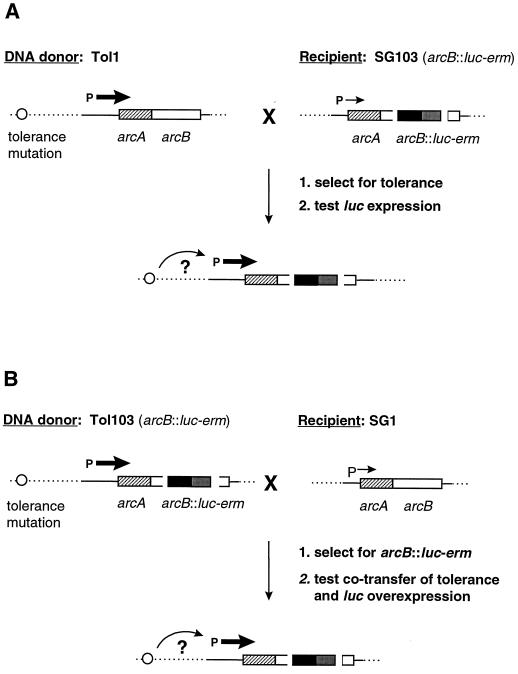
First we tested whether transformation of tolerance from Tol1 was always associated with arc deregulation (Fig. 6A). The parent SG1 strain containing an arcB::luc-erm transcriptional fusion (strain SG103 in Table 1) was transformed either with Tol1 or with control (SG2 [Rfr]) DNA and cycled two times with penicillin. Five independent experiments were performed. In each of them, five independent colonies were isolated after penicillin cycling and tested both for tolerance and for arc deregulation. All of the 25 colonies isolated from the five Tol1-transformed cultures had become tolerant, and all of them expressed luciferase constitutively. In contrast, none of five control colonies isolated from control-transformed cultures was tolerant, and none of them overexpressed luciferase. Thus, transformation of tolerance from Tol1 DNA was indeed associated with arc deregulation.
FIG. 6.
Genetic transformation strategies used to determined the functional and physical linkage between the tolerance mutation of Tol1 and the deregulation of arc. (A) Strategy to test whether the acquisition of tolerance from Tol1 was always associated with arc deregulation. SG103 competent cells (Table 1) were transformed with Tol1 DNA, selected for tolerance by penicillin cycling, and tested for luciferase activity. (B) Strategy to evaluate the genetic linkage between the tolerance mutation and the arc locus by determining the frequency of cotransformation of tolerance and the arcB::luc-erm insert into the SG1 parent. SG1 competent cells were transformed with DNA from Tol103 cells (Table 1). Transformants were selected for Emr and screened both for luciferase activity and for tolerance. The genes are represented by the boxes. P represents the arc operon promoter. Horizontal arrows represent the transcription activity. The circle indicates the putative location of the tolerance mutation, and the curved arrow indicates its cis or trans effect with respect to the arc locus.
Second, we evaluated the genetic linkage between the tolerance mutation of Tol1 and the arc locus (Fig. 6B). SG1 parent cells were transformed with DNA from Tol1 containing the arcB::luc-erm transcriptional fusion (strain Tol103 in Table 1). Transformants were selected for arcB::luc-erm acquisition on erythromycin-containing plates (transformation rate, 10−3) and screened for concomitant tolerance and luciferase overexpression. Cotransformation of arcB::luc-erm and tolerance would indicate physical proximity of the two markers.
Emr transformants (108) were isolated and contained a functional luc gene, as assessed by light emission in the postexponential growth phase. In 105 of 108 (97.8%) of them, the arc locus was regulated and light was expressed only in the postexponential growth phase. In 3 of 108 (2.8%) of them, however, arc was deregulated and light was constitutively expressed. When rechallenged with penicillin, these three transformants were fully tolerant. As a control, three Emr colonies that were randomly picked among transformants expressing low levels of light activity were killed by penicillin. Thus, the arcB::luc-erm insert and the tolerance marker were cotransformed in 2.8% of cases. This was compatible with the concomitant transformation of two separate markers (congression). Moreover, the rate of tolerance cotransformation was compatible with that of a point mutation.
Localization of the tolerance mutation of Tol1 on a specific chromosomal fragment.
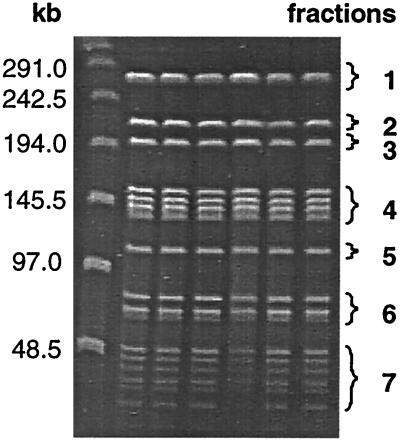
The Tol1 chromosome was digested with SmaI and subjected to PFGE separation. The fragments were purified and tested either separately or together for transformation of tolerance into SG1 recipients containing the arcB::luc-erm fusion (strain SG103 in Table 1) (Fig. 7; Table 3). The cultures transformed with the pooled fragments converted to tolerance at the third penicillin cycle. Likewise, the cultures transformed with fragment 3 converted to tolerance after three cycles. In contrast, none of the other SmaI fragments conferred tolerance (Fig. 7; Table 3). Again, all tolerant transformants expressed luciferase constitutively. Fragment 3, containing the tolerance-transforming activity, was ca. 200 kb in size. Probing with internal arcA and arcB fragments indicated that this DNA band also contained the arc operon (data not shown).
FIG. 7.
Mapping of the tolerance mutation on the Tol1 chromosome (see Table 3). Tol1 DNA was digested with the restriction enzyme SmaI. The resulting fragments were separated by PFGE, extracted from the gel, and utilized alone or pooled together to transform SG103 cells carrying the arcB::luc-erm transcriptional-fusion (Table 1). The PFGE pattern and the various fractions used for transformation are shown. Tolerant transformants were selected after three penicillin cycles. All cultures were tested in parallel for increased luciferase activity.
TABLE 3.
Mapping of the tolerance mutation of Tol1a
| PFGE fractions and genotype
|
Transformation of Smr marker | No. of penicillin cycles | Conversion to Tol+ cells | Luciferase activity | |
|---|---|---|---|---|---|
| DNA donor | Recipient cells | ||||
| Fraction 1 | SG103 (arcB::luc-erm) | NDb | 3 | No | − |
| Fraction 2 | SG103 (arcB::luc-erm) | ND | 3 | No | − |
| Fraction 3 | SG103 (arcB::luc-erm) | ND | 3 | Yes | ++c |
| Fraction 4 | SG103 (arcB::luc-erm) | ND | 3 | No | − |
| Fraction 5 | SG103 (arcB::luc-erm) | ND | 3 | No | − |
| Fraction 6 | SG103 (arcB::luc-erm) | ND | 3 | No | − |
| Fraction 7 | SG103 (arcB::luc-erm) | ND | 3 | No | − |
| Pooled fractions | SG103 (arcB::luc-erm) | 10−3–10−4 | 3 | Yes | ++ |
See the text and the legend to Fig. 7 for a description of the experiment.
ND, not determined.
On a scale from + to +++.
Frequency of arc deregulation in additional tolerant mutants selected with penicillin.
Since only one of the four original mutants analyzed in Fig. 1 was fully characterized, we reassessed whether arc deregulation was frequently associated with tolerance by repeating the original penicillin selection with cells containing the luc transcriptional fusion system. Ten independent cultures of SG103 (containing the arcB::luc-erm insert) (Table 1) were cycled with penicillin as in the original experiment. It took nine cycles for these cultures to convert to tolerance. In 6 of 10 (60%) of them, conversion to tolerance was associated with constitutive arc expression. Ten colonies from each of these cultures were isolated and retested for tolerance and light emission. Each of them reproduced the phenotype of the cycled cultures. Thus, selection of tolerance by penicillin was frequently associated with arc deregulation. However, other tolerance mutations not associated with arc deregulation also existed.
DISCUSSION
The present experiments examined the phenotypic and genetic alterations arising in penicillin-tolerant mutants of S. gordonii. This organism was used as a model because (i) it has a low penicillin MIC, (ii) it is extensively killed by the drug, (iii) it is naturally competent, and (iv) it is a human endocarditis pathogen. Moreover, unlike pneumococci, S. gordonii is not lysed during beta-lactam therapy. This allows an analysis that is more restricted to autolysis-independent mechanisms of penicillin-induced killing and penicillin tolerance (13, 25).
One mutant (Tol1) was fully characterized. Tol1 had unaltered penicillin-binding proteins but contained increased amounts of two cellular polypeptides that were more than 80% similar to the Adi and Oct proteins encoded by the arc operon of several other bacteria (7, 23, 32, 40).
The present results indicated that these two proteins were also encoded by a unique arc operon in S. gordonii. However, although the arc operon was deregulated in Tol1, specific inactivation of this locus did not yield any phenotype. The absence of a direct effect of this locus on tolerance was also supported by the absence of mutations in the partial arc nucleotide sequence of Tol1. On the other hand, it was possible that deregulation of this locus was an indirect sign of a more profound cellular alteration associated with tolerance. Indeed, a clear association was established between arc deregulation and tolerance by using a luciferase-based transcriptional fusion system. In parent cells, arc expression was repressed during exponential growth and up-regulated during postexponential growth. In Tol1, arc expression was constitutive. Transformation of the tolerance phenotype with Tol1 DNA was always accompanied by a parallel deregulation of arc in the recipient cells. Moreover, appraising the genetic distance between the tolerance mutation(s) and the arc locus indicated that the two determinants were physically distant. This suggested that the tolerance mutation might act in trans rather than in cis with regard to arc. Eventually, the tolerance mutation(s) was located on a specific chromosomal fragment and transformed at the rate of a point mutation in DNA backcross experiments.
Several clues suggested that tolerance could result from some global deregulation in the Tol1 mutant. First, the mutation was likely to involve a trans-acting factor influencing the expression of the arc operon. Since the proper arc gene products did not affect tolerance, altered transactivation of this particular locus was probably not the primary purpose of the tolerance mutation. It is likely that the mutation also affected other genes that were more important for tolerance and that arc deregulation was only revealing the alteration.
Second, deregulation of arc was observed not only in Tol1 but also in 6 of 10 (60%) additional tolerant cultures selected by penicillin. Since these mutants emerged in separate tubes, they resulted from separate genetic events. The fact that most of them had altered arc regulation suggested, again, the existence of a common pathway important for penicillin survival.
Third, tolerance was not specific for penicillin but extended to several related and unrelated drugs. This indicated that some central yet relatively nonspecific protective system was operating in these derivatives.
It is noteworthy that in parent S. gordonii, the arc operon was repressed during exponential growth and up-regulated in the postexponential phase. This corresponds to the beginning of phenotypic tolerance, with which slow-growing bacteria can survive beta-lactams and other bactericidal drugs (39). In contrast, this locus was deregulated in the majority of tolerant mutants, suggesting a possible relationship between growth phase, gene regulation, and tolerance. A hint as to such a relation came from the sequence homology with the arginine deiminase operon of other bacteria (17, 41). This locus is controlled by catabolic repression, which responds to carbohydrate starvation and is implicated in arginine degradation and in production of ATP and NH3 (3, 5). Since starvation is linked to the growth rate and the growth rate is linked to phenotypic tolerance, it is tempting to speculate on a possible relationship between genes regulated by catabolic repression and antibiotic tolerance.
A further clue indicating such a possibility was the fact that the S. gordonii arc operon contained a homologue of a catabolic regulatory element (CRE) in its regulatory region. In gram-positive bacteria, catabolic repression is driven both by a cis-acting CRE sequence, located between 100 and 140 bp upstream of the regulated locus (18), and by an as yet undetermined transacting factor. CRE has a consensus sequence of 14 bp, which we were able to find upstream of both the S. pyogenes sagp and S. pneumoniae arc operons. In S. gordonii, a putative CRE sequence, containing only two mismatches, was also found at position 428 of the sequence, 51 bp upstream of the first ATG. Therefore, the S. gordonii arc operon might indeed be regulated by catabolic repression.
Based on the present results, we postulate that the tolerance mutation(s) present in Tol1 and other tolerant mutants affects some kind of global regulatory mechanism of the cell that is important for surviving treatment with penicillin and other antibiotics. The link between tolerance and arc deregulation provides a reporter system that will help track down more central elements of this mechanism. The very likely presence of a CRE element upstream of this operon is a hint as to the possible type of alteration present in Tol1. The fact that some central regulatory system might affect the bactericidal effect of antibiotics is compatible with several observations, including that killing by beta-lactams is modulated by the agr (accessory gene regulator) locus in S. aureus (11) and that VncS-VncR pneumococcal mutants are tolerant to several unrelated kinds of antibiotics (31).
Deregulation of the arc operon was frequently observed in tolerant derivatives selected during penicillin cycling. Moreover, one of the tolerant mutants described herein (Tol1) caused penicillin treatment to fail in rats with experimental endocarditis (9). Since the selective pressure used in these experiments mimicked sequential penicillin treatment, the resulting mutations are likely to be of clinical relevance. Physical characterization of these mutations and their putative link to CRE-regulated loci are the focus of ongoing studies.
ACKNOWLEDGMENTS
This work was supported by grants 32-040836.94 and 32-052501.97 from the Swiss National Funds for Scientific Research.
We thank H. Henry for help in protein biochemistry and Marlyse Giddey for technical support and gratefully acknowledge the gift of the luciferase gene by A. Podbielsky.
REFERENCES
- 1.Bergan T, Oydvin B. Cross-over study of penicillin pharmacokinetics after intravenous infusions. Chemotherapy. 1974;20:263–279. doi: 10.1159/000221816. [DOI] [PubMed] [Google Scholar]
- 2.Buchrieser C, Brosch R, Rocourt J. Use of pulsed field gel electrophoresis to compare large DNA-restriction fragments of Listeria monocytogenes strains belonging to serogroups 1/2 and 3. Int J Food Microbiol. 1991;14:297–304. doi: 10.1016/0168-1605(91)90121-5. [DOI] [PubMed] [Google Scholar]
- 3.Casiano-Colon A, Marquis R E. Role of the arginine deiminase system in protecting oral bacteria and an enzymatic basis for acid tolerance. Appl Environ Microbiol. 1988;54:1318–1324. doi: 10.1128/aem.54.6.1318-1324.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J D, Morrison D A. Cloning of Streptococcus pneumoniae DNA fragments in Escherichia coli requires vectors protected by strong transcriptional terminators. Gene. 1987;55:179–187. doi: 10.1016/0378-1119(87)90278-2. [DOI] [PubMed] [Google Scholar]
- 5.Curran T M, Lieou J, Marquis R E. Arginine deiminase system and acid adaptation of oral streptococci. Appl Environ Microbiol. 1995;61:4494–4496. doi: 10.1128/aem.61.12.4494-4496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daneo-Moore L, Fletcher S H, Massida O, Pittaluga F. Penicillin tolerance in Enterococcus hirae ATCC 9790. In: Actor P, Daneo-Moore L, Higgins M L, Salton M R J, Shokman G D, editors. Antibiotic inhibition of bacterial cell surface assembly and function. Washington, D.C.: American Society for Microbiology; 1987. pp. 628–635. [Google Scholar]
- 7.Degnan B A, Palmer J M, Robson T, Jones C E D, Fischer M, Glanville M, Mellor G D, Diamond A G, Kehoe M A, Goodacre J A. Inhibition of human peripheral blood mononuclear cell proliferation by Streptococcus pyogenes cell extracts is associated with arginine deiminase activity. Infect Immun. 1998;66:3050–3058. doi: 10.1128/iai.66.7.3050-3058.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dowson C G, Coffey T J, Spratt B G. Origin and molecular epidemiology of penicillin binding-protein-mediated resistance to beta-lactam antibiotics. Trends Microbiol. 1994;2:361. doi: 10.1016/0966-842x(94)90612-2. [DOI] [PubMed] [Google Scholar]
- 9.Entenza J M, Caldelari I, Glauser M P, Francioli P, Moreillon P. Importance of genotypic and phenotypic tolerance in the treatment of experimental endocarditis due to Streptococcus gordonii. J Infect Dis. 1997;175:70–76. doi: 10.1093/infdis/175.1.70. [DOI] [PubMed] [Google Scholar]
- 10.Entenza J M, Caldelari I, Glauser M P, Moreillon P. Efficacy of levofloxacin in the treatment of experimental endocarditis caused by viridans group streptococci. J Antimicrob Chemother. 1999;44:775–786. doi: 10.1093/jac/44.6.775. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto D F, Bayles K W. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J Bacteriol. 1998;180:3724–3726. doi: 10.1128/jb.180.14.3724-3726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghuysen J M. Serine beta-lactamases and penicillin-binding proteins. Annu Rev Microbiol. 1991;45:37–67. doi: 10.1146/annurev.mi.45.100191.000345. [DOI] [PubMed] [Google Scholar]
- 13.Giesbrecht P, Kersten T, Maidhof H, Kruger D, Blumel P, Grob H, Wecke J. A novel, “hidden” penicillin-induced death of staphylococci at high drug concentration, occurring earlier than murosome-mediated killing processes. Arch Microbiol. 1994;161:370–383. doi: 10.1007/BF00288946. [DOI] [PubMed] [Google Scholar]
- 14.Gutmann L, Tomasz A. Penicillin-resistant and penicillin-tolerant mutants of group A streptococci. Antimicrob Agents Chemother. 1982;22:128–136. doi: 10.1128/aac.22.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock R E W. Role of porins in outer membrane permeability. J Bacteriol. 1987;169:929–933. doi: 10.1128/jb.169.3.929-933.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handwerger S, Tomasz A. Antibiotic tolerance among clinical isolates of bacteria. Rev Infect Dis. 1985;7:368–386. doi: 10.1093/clinids/7.3.368. [DOI] [PubMed] [Google Scholar]
- 17.Hueck C J, Hillen W. Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria? Mol Microbiol. 1995;15:395–401. doi: 10.1111/j.1365-2958.1995.tb02252.x. [DOI] [PubMed] [Google Scholar]
- 18.Hueck C J, Hillen W, Saier M H J. Analysis of a cis-active sequence mediating catabolite repression in gram-positive bacteria. Res Microbiol. 1994;145:503–518. doi: 10.1016/0923-2508(94)90028-0. [DOI] [PubMed] [Google Scholar]
- 19.Kanaoka M, Negoro T, Kawanaka C, Agui H, Nabeshima S. Streptococcal antitumor protein: expression in Escherichia coli cells and properties of the recombinant protein. Agric Biol Chem. 1991;55:743–750. [PubMed] [Google Scholar]
- 20.Kim K S, Kaplan E L. Association of penicillin tolerance with failure to eradicate group A streptococci from patients with pharyngitis. J Pediatr. 1985;107:681–684. doi: 10.1016/s0022-3476(85)80392-9. [DOI] [PubMed] [Google Scholar]
- 21.Langen H, Takacs B, Evers S, Berndt P, Lahm H W, Wipf B, Gray C, Fountoulakis M. Two-dimensional map of the proteome of Haemophilus influenzae. Electrophoresis. 2000;21:411–429. doi: 10.1002/(SICI)1522-2683(20000101)21:2<411::AID-ELPS411>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 22.Liu H H, Tomasz A. Penicillin tolerance in multiply drug-resistant natural isolates of Streptococcus pneumoniae. J Infect Dis. 1985;152:365–372. doi: 10.1093/infdis/152.2.365. [DOI] [PubMed] [Google Scholar]
- 23.Maghnouj A, de Sousa C, Stalon V, Vander W C. The arcABDC gene cluster, encoding the arginine deiminase pathway of Bacillus licheniformis, and its activation by the arginine repressor argR. J Bacteriol. 1998;180:6468–6475. doi: 10.1128/jb.180.24.6468-6475.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matushek M G, Bonten M J, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDowell T D, Lemanski C L. Absence of autolytic activity (peptidoglycan nicking) in penicillin-induced nonlytic death in a group A streptococcus. J Bacteriol. 1988;170:1783–1788. doi: 10.1128/jb.170.4.1783-1788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreillon P, Markiewicz Z, Nachman S, Tomasz A. Two bactericidal targets for penicillin in pneumococci: autolysis-dependent and autolysis-independent killing mechanisms. Antimicrob Agents Chemother. 1990;34:33–39. doi: 10.1128/aac.34.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreillon P, Tomasz A. Penicillin resistance and defective lysis in clinical isolates of pneumococci: evidence for two kinds of antibiotic pressure operating in the clinical environment. J Infect Dis. 1988;157:1150–1157. doi: 10.1093/infdis/157.6.1150. [DOI] [PubMed] [Google Scholar]
- 28.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 29.Novak R, Braun J S, Charpentier E, Tuomanen E. Penicillin tolerance genes of Streptococcus pneumoniae: the ABC-type manganese permease complex Psa. Mol Microbiol. 1998;29:1285–1296. doi: 10.1046/j.1365-2958.1998.01016.x. [DOI] [PubMed] [Google Scholar]
- 30.Novak R, Cauwels A, Charpentier E, Tuomanen E. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol. 1999;181:1126–1133. doi: 10.1128/jb.181.4.1126-1133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Novak R, Henriques B, Charpentier E, Normark S, Tuomanen E. Emergence of vancomycin tolerance in Streptococcus pneumoniae. Nature. 1999;399:590–593. doi: 10.1038/21202. [DOI] [PubMed] [Google Scholar]
- 32.Ohtani K, Bando M, Swe T, Banu S, Oe M, Hayashi H, Shimizu T. Collagenase gene (colA) is located in the 3′-flanking region of the perfringolysin O (pfoA) locus in Clostridium perfringens. FEMS Microbiol Lett. 1997;146:155–159. doi: 10.1111/j.1574-6968.1997.tb10186.x. [DOI] [PubMed] [Google Scholar]
- 33.Pozzi G, Musmanno R A, Lievens P M, Oggioni M R, Plevani P, Manganelli R. Method and parameters for genetic transformation of Streptococcus sanguis Challis. Res Microbiol. 1990;141:659–670. doi: 10.1016/0923-2508(90)90060-4. [DOI] [PubMed] [Google Scholar]
- 34.Sabath L D, Wheeler N, Laverdiere M, Blazevic D, Wilkinson B J. A new type of penicillin resistance of Staphylococcus aureus. Lancet. 1977;i:443–447. doi: 10.1016/s0140-6736(77)91941-9. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sugai M, Ooku K, Akiyama T, Inoue S, Iseda S, Miyake Y, Suginaka H. Suppression of penicillin-induced lysis of Staphylococcus aureus by cibacron blue 3G-A. FEMS Microbiol Lett. 1991;64:151–154. doi: 10.1016/0378-1097(91)90586-y. [DOI] [PubMed] [Google Scholar]
- 37.Sugai M, Yamada S, Nakashima S, Komatsuzawa H, Matsumoto A, Oshida T, Suginaka H. Localized perforation of the cell wall by a major autolysin: atl gene products and the onset of penicillin-induced lysis of Staphylococcus aureus. J Bacteriol. 1997;179:2958–2962. doi: 10.1128/jb.179.9.2958-2962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomasz A, Albino A, Zanati E. Multiple antibiotic resistance in a bacterium with suppressed autolytic system. Nature. 1970;227:138–140. doi: 10.1038/227138a0. [DOI] [PubMed] [Google Scholar]
- 39.Tuomanen E. Phenotypic tolerance: the search for beta-lactam antibiotics that kill nongrowing bacteria. Rev Infect Dis. 1986;8(Suppl. 3):279–291. doi: 10.1093/clinids/8.supplement_3.s279. [DOI] [PubMed] [Google Scholar]
- 40.Woods G L, Washington J A. Antimicrobial susceptibility tests: dilution and disk diffusion methods. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 1327–1341. [Google Scholar]
- 41.Zúñiga M, Champomier-Verges M, Zagorec M, Pérez-Martínez G. Structural and functional analysis of the gene cluster encoding the enzymes of the arginine deiminase pathway of Lactobacillus sake. J Bacteriol. 1998;180:4154–4159. doi: 10.1128/jb.180.16.4154-4159.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]