Abstract
Objectives
Worldwide, lung ultrasound (LUS) was utilized to assess coronavirus disease 2019 (COVID‐19) patients. Often, imaging protocols were however defined arbitrarily and not following an evidence‐based approach. Moreover, extensive studies on LUS in post‐COVID‐19 patients are currently lacking. This study analyses the impact of different LUS imaging protocols on the evaluation of COVID‐19 and post‐COVID‐19 LUS data.
Methods
LUS data from 220 patients were collected, 100 COVID‐19 positive and 120 post‐COVID‐19. A validated and standardized imaging protocol based on 14 scanning areas and a 4‐level scoring system was implemented. We utilized this dataset to compare the capability of 5 imaging protocols, respectively based on 4, 8, 10, 12, and 14 scanning areas, to intercept the most important LUS findings. This to evaluate the optimal trade‐off between a time‐efficient imaging protocol and an accurate LUS examination. We also performed a longitudinal study, aimed at investigating how to eventually simplify the protocol during follow‐up. Additionally, we present results on the agreement between AI models and LUS experts with respect to LUS data evaluation.
Results
A 12‐areas protocol emerges as the optimal trade‐off, for both COVID‐19 and post‐COVID‐19 patients. For what concerns follow‐up studies, it appears not to be possible to reduce the number of scanning areas. Finally, COVID‐19 and post‐COVID‐19 LUS data seem to show differences capable to confuse AI models that were not trained on post‐COVID‐19 data, supporting the hypothesis of the existence of LUS patterns specific to post‐COVID‐19 patients.
Conclusions
A 12‐areas acquisition protocol is recommended for both COVID‐19 and post‐COVID‐19 patients, also during follow‐up.
Keywords: artificial intelligence, COVID‐19, lung ultrasound, post‐COVID‐19, SARS‐CoV‐2
Abbreviations
- COVID‐19
coronavirus disease 2019
- LUS
lung ultrasound
- RT‐PCR
reverse transcription polymerase chain reaction
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus 2
During the recent coronavirus disease 2019 (COVID‐19) pandemic, lung ultrasound (LUS) has emerged as a powerful ally for clinicians. In fact, thanks to ultrasound technologies portability, cost‐effectiveness, and safety, LUS has been utilized extensively around the world to assess the condition of the lung in patients suspected or affected by COVID‐19. 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 Specifically, LUS has been utilized to intercept the presence of COVID‐19‐associated interstitial pneumonia, and monitor its evolution. To this end, a variety of imaging protocols and scoring systems have been proposed in the literature. 10 However, fundamental aspects such as the amount and spatial distribution of areas of the chest to be scanned are often defined arbitrarily and not following an evidence‐based approach. Defining the right amount and distribution of scanning areas is of significant importance for LUS, given that ultrasound imaging can only provide local information on the status of the lung surface. Consequently, reducing the scanning areas in order to simplify the examination does impact directly on the extent of the inspected lung surface. This is particularly relevant for COVID‐19, given the patchy distribution of the relevant findings. 11 , 12 , 13 , 14 , 15 , 16 Moreover, extended studies on LUS findings on post‐COVID‐19 patients are currently lacking. Therefore, in this multicenter study we investigate the impact of the amount and distribution of scanning areas on the accuracy of the LUS examination. To this end, we analyzed LUS data acquired on a population of 220 patients. Specifically, a 14‐areas acquisition protocol and a 4‐level scoring system were utilized. The prognostic value of this approach has been investigated through a study conducted at the Fondazione Policlinico San Matteo (Pavia, Italy), and involving 52 patients. 17 Results showed how patients showing a cumulative LUS score (the sum of the scores over the 14 areas scanned) higher than 24 had an almost 6‐fold increase in the odds of worsening. Moreover, we investigated LUS findings variability with respect to the implemented imaging protocol. Five imaging protocols were considered, respectively based on 4, 8, 10, 12, and 14 scanning areas. This approach allows to define the optimal trade‐off between a simple and time‐efficient LUS evaluation (which requires minimizing the number of areas to be scanned) and an accurate LUS examination (which requires maximizing the areas to be scanned).
Of the 220 patients, 100 were COVID‐19 positive at the time they were scanned, while 120 patients were post‐COVID‐19, that is, they were negative to reverse transcription polymerase chain reaction (RT‐PCR) test after being originally diagnosed with COVID‐19 by means of the same test.
To our knowledge an extensive study on LUS patterns on post‐COVID‐19 patients represents by itself a significant novelty to the existing scientific literature. Additionally, we report on results from a longitudinal study on a subgroup of 29 patients. These results are important to investigate the evolution of the lung condition over time, and to verify whether a simplified scanning procedure could be adopted during patients' follow‐up. In conclusion, we also report on the level of prognostic agreement between LUS experts and recently developed AI algorithms, 18 which were trained at implementing the previously introduced scoring system. 2 Specifically, we investigate the performance of the AI, differentiating between COVID‐19 and post‐COVID‐19 data. The AI algorithm discussed in this manuscript was the first DL algorithm to be developed worldwide for the analysis of LUS data from COVID‐19 patients. A detailed technical description of the algorithm can be found in a recently published article. 18 This algorithm was then validated in a multicenter study involving 314,879 images from 82 patients. In that study, the DL performance at scoring LUS videos was compared with that of clinical experts. 19 To our knowledge, this is the only DL algorithm that has had a similar validation (distinguishing frame‐level, video‐level, and exam‐level performance) for the analysis of LUS data from COVID‐19 patients. Results from the multicenter study showed a level of agreement between DL and clinical experts of 85.96% in the stratification between patients at high risk of clinical worsening and patients at low risk. In this new work, we have further extended this validation to data acquired from 220 patients, and distinguished between the performance obtained for COVID‐19 and post‐COVID‐19 patients.
The paper is organized as follows. Firstly, we present the study design and population, then we describe the utilized LUS protocol. Successively, we present the methods used to assess the impact of different scanning areas on the LUS exam's evaluation, and describe the design of the longitudinal study and the methods implemented for the analysis of the prognostic agreement between LUS experts and AI. Next, the results are introduced. Finally, we present the discussion and the conclusion.
Materials and Methods
Study Design and Population
The studied population consists of 220 patients (138 male, 82 female, with ages ranging from 23 to 95 years, and average age equal to 63.0 years). Inclusion criteria were age 18 years or older, confirmed COVID‐19 infection based on the detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) on a reverse transcriptase polymerase chain reaction from a nasopharyngeal swab or bronchoalveolar wash, and a collaborative status allowing them to express informed consent. Patients were excluded if they were not able to express their consent, if they were severely obese (body mass index [BMI] > 35 kg/m2), or if they were affected by heart failure or interstitial lung disease, such as usual interstitial pneumonia or lung fibrosis secondary to rheumatologic disease. Patients’ enrolment was performed, for the acute COVID‐19 patients, at the internal medicine ward (converted in a sub‐intensive COVID ward) for San Matteo and Lodi General Hospital, while post‐COVID‐19 patients from San Matteo Hospital were outpatients. From Gemelli Hospital, acute COVID‐19 patients were enrolled when hospitalized in dedicated wards converted in a sub‐intensive COVID ward, while post‐COVID‐19 inpatients were enrolled in the pulmonology ward.
Of the 220 patients, 100 were diagnosed as COVID‐19 positive by a RT‐PCR swab test, and 120 are post‐COVID‐19 patients (mean days between last positive RT‐PCR swab test and LUS examination equal to 47.85 ± 12.82). Of the 100 COVID‐19 patients, 63 (35 male, 28 female, with ages ranging from 26 to 92 years, and average age equal to 63.72 years) were examined within the Fondazione Policlinico San Matteo (Pavia, Italy), 19 (16 male, 3 female, with ages ranging from 34 to 84 years, and average age equal to 63.95 years) within the Lodi General Hospital (Lodi, Italy), and 18 (8 male, 10 female, with ages ranging from 23 to 95 years, and average age equal to 52.11 years) within the Fondazione Policlinico Universitario Agostino Gemelli (Rome, Italy). Of the 120 post‐COVID‐19 patients, 109 (71 males, 38 females, with ages ranging from 36 to 87 years, and average age equal to 63.20 years) were examined within the Fondazione Policlinico San Matteo, and 11 (8 males, 3 females, with ages ranging from 52 to 89 years, and average age equal to 73.64 years) within the Fondazione Policlinico Universitario Agostino Gemelli. It is important to highlight how the post‐COVID‐19 patients examined at Pavia were scanned during follow‐up and were not hospitalized at the date of LUS exam, whereas the post‐COVID‐19 patients examined at Rome were still hospitalized at the date of LUS exam. As a subgroup of COVID‐19 patients was examined multiple times (on different dates), a total of 253 LUS exams were performed (COVID‐19 positive: 94 at Pavia, 20 at Lodi, 19 at Rome; post‐COVID‐19: 109 at Pavia, 11 at Rome), and 3481 LUS videos acquired (COVID‐19 positive: 1291 from Pavia, 276 from Lodi, 242 from Rome; post‐COVID‐19: 1526 from Pavia, 146 from Rome), consisting of 772,780 frames (COVID‐19 positive: 293,194 from Pavia, 44,288 from Lodi, 29,070 from Rome; post‐COVID‐19: 371,168 from Pavia, 35,060 from Rome). LUS data were acquired by LUS experts with more than 10 years of experience. Andrea Smargiassi, Riccardo Inchingolo, Tiziano Perrone and Francesco Tursi respectively acquired the data collected at the Gemelli, San Matteo and Lodi Hospital.
The data from Pavia have been acquired using a convex probe with an Esaote MyLab Twice scanner, and an Esaote MyLab 50, setting an imaging depth from 5 to 13 cm (depending on the patient) and an imaging frequency from 2.5 to 6.6 MHz (depending on the scanner). The data from Lodi have been acquired using a convex probe with an Esaote Mylab Sigma scanner, and a MindRay TE7, setting an imaging depth from 8 to 12 cm (depending on the patient) and an imaging frequency from 3.5 to 5.5 MHz. The data from Rome have been acquired using both convex and linear probes with an Esaote MyLab 50, an Esaote MyLab Alpha, a Philips IU22, and an ATL Cerbero, setting an imaging depth from 5 to 30 cm (depending on the patient) and an imaging frequency from 3.5 to 10 MHz (depending on the scanner).
Figure 1 shows a Sankey diagram where the distribution of the dataset characteristics is illustrated in detail. As visible, the majority of the data have been acquired with an imaging frequency ranging from 2.5 to 7.5 MHz and an imaging depth from 8 to 12 cm.
Figure 1.
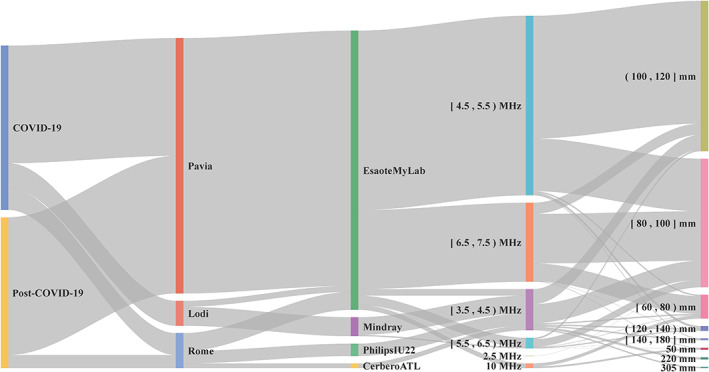
Sankey diagram illustrating the distribution of the dataset characteristics. Square and round brackets are respectively utilized to indicate whether the interval includes or not the endpoints. Data are grouped (from left to right) based on they being from COVID‐19 or post‐COVID‐19 patients, based on the hospital where the data have been collected, on the utilized ultrasound scanner, on the imaging frequency and imaging depth. Frequencies are expressed in Hertz (MHz = 106 Hz) and depths in meters (mm = 10−3 m).
This study was part of a protocol that has been registered (NCT04322487) and received approval from the Ethical Committee of the Fondazione Policlinico Universitario San Matteo (protocol 20200063198), of the Fondazione Policlinico Universitario Agostino Gemelli, Istituto di Ricovero e Cura a Carattere Scientifico (protocol 0015884/20 ID 3117), of Milano area 1, the Azienda Socio‐Sanitaria Territoriale Fatebenefratelli‐Sacco (protocol N0031981). All patients gave informed consent.
LUS Acquisition Protocol
All patients were examined following the standardized acquisition protocol presented by Soldati et al and based on 14 scanning areas. 2 According to this protocol, a score ranging from 0 to 3 was assigned to each video by LUS experts (TP, AS, and FT). 2 Figure 2 shows an example of LUS image for each level of the scoring system. The 4 levels are defined based on the current understanding of ultrasound interaction with lung tissue. Score 0 corresponds to a continuous pleural line with associated horizontal artifacts. These artifacts are generally referred to as A‐lines, and are due to the high reflectivity of the non‐pathological lung surface preventing ultrasound waves to propagate beyond the pleural line. Ultrasound waves are thus scattered multiple times between the lung surface and the probe, giving rise to this particular horizontal pattern. Score 1 signals instead the appearance of the first abnormalities. The pleural line is not continuous anymore and vertical artifacts are visible. We prefer to adopt the general term vertical artifact over a different term generally found in LUS literature (ie, B‐lines). This choice is motivated as to avoid the ambiguity related to the definition of B‐lines. Moreover, the presence, and not the number, of vertical artifacts is considered. In fact, recent clinical studies 20 , 21 showed how the visualization of vertical artifacts is strongly influenced by key imaging settings such as the imaging frequency and bandwidth. Moreover, it is also very important to stress how vertical artifacts are not specific to COVID‐19, and simply signal the presence of local alterations along the lung surface. Their appearance during an ultrasound exam is considered to be due to the formation, along the lung surface, of channels accessible to ultrasound, which can indeed be generated in many pathological states of the lung once volumes originally filled by air are occupied by media that are acoustically much more similar to the intercostal tissue (eg, water, blood, and tissue). 20 , 21 Score 2 is associated with the appearance of small‐to‐large consolidated areas. Differently that with horizontal and vertical artifacts, consolidations are not artifacts, but anatomical findings that appear as hypoechoic areas (darker areas) along the lung surface. The loss of echogenicity of the consolidated areas is a reflection of the loss of aeration and signal the transition of these areas toward acoustic properties similar to soft tissue. They thus signal deaeration. Below the consolidations, vertical artifacts are generally found. The latter are most likely associated with the presence of areas not yet fully deaerated. Ultimately, score 3 is associated with the presence of large, extended (>50% of the pleural line) vertical artifacts (sometimes referred to White Lung in the literature), with or without large consolidations.
Figure 2.
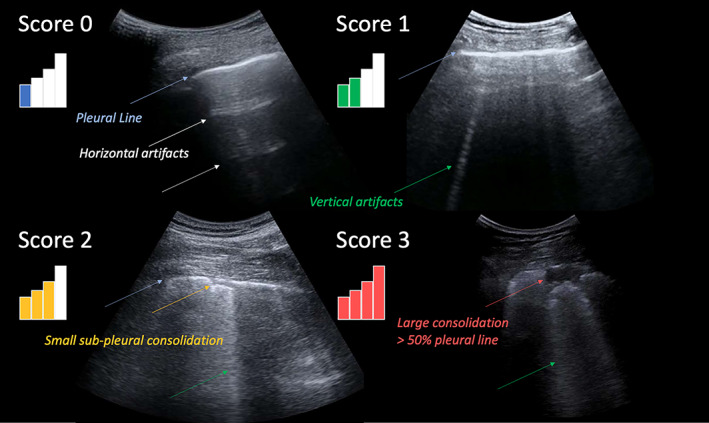
Typical LUS image associated with each level of the scoring system. A higher score is associated with a higher level of deaeration of the lung surface explored by ultrasound. A higher score is thus intended to signal a worsening of the status of the lung surface. 2 Relevant patterns are indicated by color‐coded arrows. The displayed images were acquired with a convex probe.
Impact of Different Scanning Areas on Exam's Evaluation
Consistently with a previous study, 22 we classified each exam according to the highest score (from 0 to 3) assigned to the corresponding 14 LUS videos. Then, different subgroups of scanning areas were considered to reevaluate the worst score of each exam and compare the obtained value with the worst score obtained by the reference protocol (14 scanning areas). 2 , 22 Finally, we computed the percentage of agreement by summing the number of exams sharing the same worst score from the reference protocol (named system 4) 2 and dividing it by the total number of exams. 22 Given the presence of two significantly different groups (ie, COVID‐19 patients and post‐COVID‐19 patients), this evaluation was separately performed for each group.
Firstly, we analyzed the level of agreement by separately considering only the anterior (named 11, 12, 13, 14 2 ), lateral (named 7, 8, 9, 10 2 ), and posterior (named 1, 2, 3, 4, 5, and 6 2 ) areas. 11 , 22 Secondly, we evaluated the level of agreement for three different protocols based on 4 (named system 1), 8 (named system 2), and 12 (named system 3) scanning areas. 10 , 22 Specifically, system 1 is based on scanning areas 7, 9, 12, and 14, system 2 on scanning areas from 7 to 14, system 3 on scanning areas 1, 3, 4, 6 and from 7 to 14, whereas the reference system (system 4) is based on all the scanning areas (from 1 to 14). 2 , 22 Moreover, given the impact of posterior areas in the exam's evaluation, 11 , 22 three modified versions of system 4 (based on 10 areas instead of 14) were evaluated. 22 In particular, these three modified versions were obtained by considering all the anterior and lateral scanning areas (from 7 to 14) together with the basal posteriors (1 and 4), middle posteriors (2 and 5), or apical posteriors (3 and 6). 22
Longitudinal Study
We performed a longitudinal study with a subgroup of 29 COVID‐19‐positive patients (15 males, 14 females, with ages ranging from 39 to 92 years, and average age equal to 67.55 years) that underwent LUS exams twice (in different dates; days between the first and second LUS exams equal to 6.93 ± 5.44), to evaluate how the score assigned to each area of system 4 changes between the two exams. Specifically, we computed, for each patient, the difference between the score assigned to each scanning area at the first LUS exam and at the second LUS exam, which will be referred to as Δ. Therefore, the values of Δ range from −3 to 3, where a negative value represents a worsening of the patient in the considered scanning area, whereas a positive value represents an improvement of the patient in that scanning area. Then, we computed, for each patient, the mean value of Δ by averaging the Δ values obtained for each scanning area, and the minimum and maximum values of Δ (similarly obtained). Hence, we obtained an error bar for each patient, where its length is given by the distance between the minimum Δ value and the maximum Δ value. A long error bar with a mean value in the middle would represent a heterogeneous change of Δ with respect to the different scanning areas, thus highlighting the necessity to scan all the 14 areas every time a new LUS exam is required. In contrast, a short error bar would represent a homogeneous change of Δ with respect to the different scanning areas, thus suggesting the possibility to scan only a subgroup of areas and implicitly predict the scores of the other areas.
Prognostic Evaluation
The prognostic value of the reference acquisition protocol and scoring system 2 has been recently evaluated based on the cumulative score, that is, the sum of the scores on each of the 14 scanning areas. 17 As the score for each LUS video ranges from 0 to 3, the cumulative score ranges from 0 to 42. Specifically, when the exam‐based cumulative score (also called sum of scores) is greater than 24, the patient is considered at high risk of clinical worsening, whereas, when the exam‐based cumulative score is less than or equal to 24, the patient is considered at low risk. This threshold follows from the results of a study conducted at the Fondazione Policlinico San Matteo (Pavia, Italy), and involving 52 patients. 17 This strategy can therefore help the stratification between patients at high risk of clinical worsening and patients at low risk. 17
In this part of the study, we assessed the capability of recently developed artificial intelligence (AI) algorithms 18 of automatically stratifying patients at high risk of clinical worsening from patients at low risk. We hence compared, for each LUS exam, the cumulative score values obtained from the analysis of the data performed by LUS experts, with that provided by the AI. 19 Specifically, we considered clinicians and AI in prognostic agreement when both the cumulative scores are greater than 24 (high risk of worsening) or less than or equal to 24 (low risk of worsening). To perform this comparison, we needed to classify each video with a single score. However, the AI provided frame‐level labeling. Hence, we used an aggregation technique consisting of assigning to each video the highest score assigned at least at a given percentage of frames (threshold) composing the video. 19 In this work we applied the optimal threshold (1%). 19 This threshold was derived from the analysis of a dataset obtained within a multicenter study, and involving 314,879 images from 82 patients. The technical details of the implementation are described in a previous publication. 19
Results
Impact of Different Scanning Areas on Exam's Evaluation
Figure 3 shows the score distributions for anterior, lateral, and posterior areas, for LUS exams performed on COVID‐19 patients (Figure 3a) and post‐COVID‐19 patients (Figure 3b). Considering COVID‐19 patients (Figure 3a), the highest percentage of score 0 (28.01%) was observed in the anterior areas, whereas posterior areas show the highest percentage of score 3 (33.71%). Jointly considering score 2 and 3, the percentages for the anterior, lateral, and posterior areas are 42.11, 58.65, and 62.91%, respectively. This result highlights how the highest scores are focused on the lateral and posterior areas. Consequently, the levels of agreement with system 4 for just the anterior, lateral, and posterior areas are 48, 62, and 89%, respectively (Figure 3a). All these results on COVID‐19 patients are consistent with the results achieved by Mento et al. 22 On the other hand, Figure 3b shows how the distributions of scores are different in post‐COVID‐19 patients. Specifically, score 0 is significantly more present (71.46, 54.37, and 68.19% for anterior, lateral, and posterior areas), whereas score 3 is the less frequent (2.50, 4.17, and 5.83% for anterior, lateral, and posterior areas). Given the almost homogeneous distribution of the worst scores (ie, scores 2 and 3), for post‐COVID‐19 patients, the levels of agreement with system 4 for just the anterior, lateral, and posterior areas are 52, 68, and 69%, respectively (Figure 3b).
Figure 3.
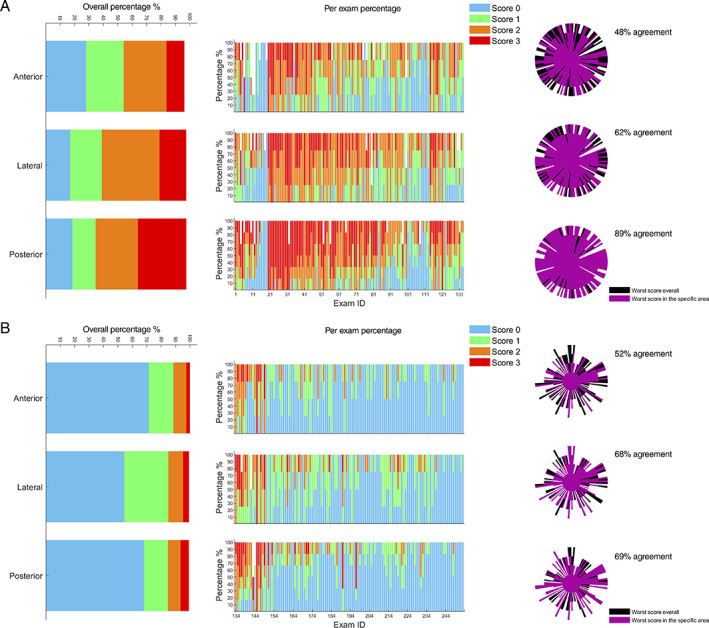
A, Graphs referring to LUS exams performed on COVID‐19 patients; B, graphs referring to LUS exams performed on post‐COVID‐19 patients. The overall distributions of scores, divided per specific area (anterior, lateral, and posterior), are depicted on the left. The percentage of scores assigned for each area and for each exam is depicted in the center. The level of agreement is shown on the right. Each exam is represented by a beam of the polar plot. The score is indicated by the length of a beam. The longer the beam, the higher the score. For further details about the structure of agreement graphs see Smargiassi et al. 11 Each exam was classified according to the worst score. The reference system is system 4 (14 scanning areas).
Figure 4 shows the overall distributions of scores divided per specific area and per each subgroup (ie, acquisition center and kind of patients). It is interesting how the distributions of score 0 and score 3 in COVID‐19 patients are consistent among the three acquisition centers. Moreover, it is clear how in post‐COVID‐19 patients the percentage of worst scores (ie, scores 2 and 3) is significantly higher when comparing patients that were still hospitalized at the date of LUS exam (Rome) with patients that were not (Pavia) (57.57% vs 10.24% in the posterior areas).
Figure 4.
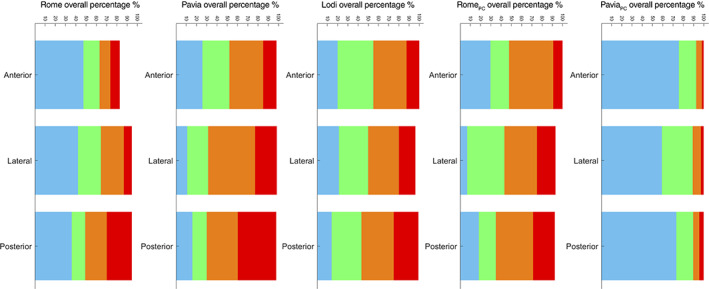
The overall distributions of scores, divided per specific area (anterior, lateral, and posterior) and per each subgroup (from left to the right: COVID‐19 patients of Rome, Pavia, and Lodi, and post‐COVID‐19 patients of Rome and Pavia).
Figure 5 shows how the distributions of scores vary with different systems, for both COVID‐19 (Figure 5‐a) and post‐COVID‐19 (Figure 5b) patients. As introduced in the Materials and Methods section, five systems have been investigated, that is, system 1 (scanned areas, 7, 9, 12, and 14), system 2 (scanned areas, 7–14), system 3 (scanned areas, 1, 3, 4, 6, and 7–14), system 4 (scanned areas, 1–14), and a system based on 10 scanning areas (scanned areas 7–14 plus 2 posterior areas). Figure 5 shows how the trend of score distributions and percentage of agreement is similar when evaluating COVID‐19 patients (Figure 5a) and post‐COVID‐19 patients (Figure 5b). In fact, even though the score distributions of these two groups are significantly different, the levels of agreement with system 4 for systems 1, 2, and 3 are 65, 76, and 98% for COVID‐19 patients (Figure 5a, top right), and 68, 82, and 97% for post‐COVID‐19 patients (Figure 5b, top right). Consistently with a previous study, 22 for this type of analysis the level of agreement was computed by summing the number of patients sharing the same worst score from the reference protocol and dividing it by the total number of patients.
Figure 5.
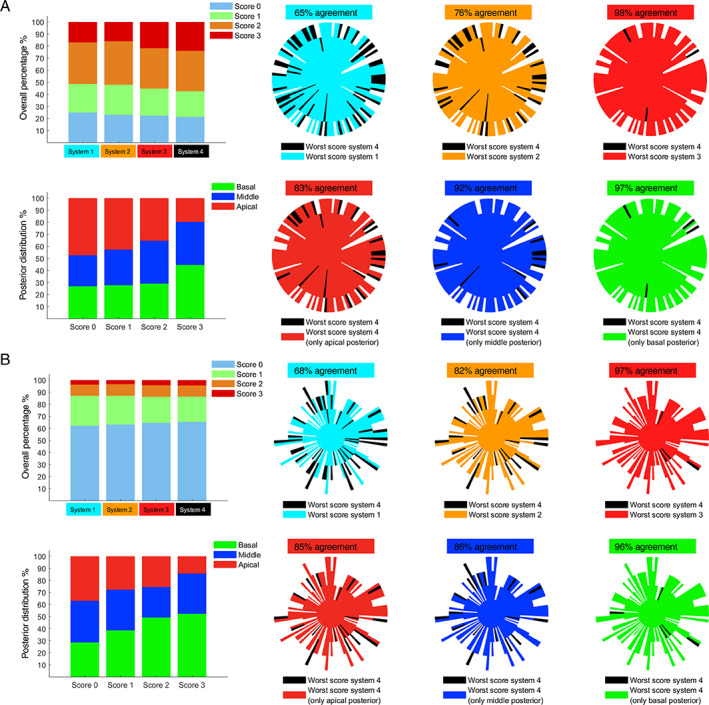
A, Graphs referring to LUS exams performed on COVID‐19 patients; B, graphs referring to LUS exams performed on post‐COVID‐19 patients. On the top left of (A) and (B) the overall distributions of scores considering the four systems are shown, and, on the top right of (A) and (B), the level of agreement between systems 1, 2, and 3 with respect to system 4 is depicted. Each exam is represented by a beam of the polar plot. The score is indicated by the length of a beam. The longer the beam, the higher the score. For further details about the structure of agreement graphs see Smargiassi et al. 11 On the bottom left of (A) and (B) the distributions of each score in the posterior areas (basal, middle, and apical) are shown, and, on the bottom right, the level of agreement between the 3 modified versions of system 4 (10 zones instead of 14: ie, all of the anterior and lateral areas together with apical posteriors, middle posteriors, or basal posteriors) with respect to system 4 is shown.
Moreover, also the distributions of scores in the posterior areas show a similar trend when comparing COVID‐19 patients (Figure 5a, bottom left) and post‐COVID‐19 patients (Figure 5b, bottom left). This is translated in consistent levels of agreement between system 4 and the modified systems 4 (10 areas instead of 14) when looking at COVID‐19 patients (Figure 5a, bottom right) and post‐COVID‐19 patients (Figure 5b, bottom right).
Specifically, the levels of agreement between system 4 and the modified systems 4 are 83, 92, and 97% (COVID‐19 patients), and 85, 86, and 96% (post‐COVID‐19 patients), when the scanned posterior areas were the apical, middle, and basal, respectively.
Longitudinal Study
As shown in Figure 6 (left), Δ values are generally heterogeneously distributed within each patient. As a consequence, the error bars are generally long (55.17% of error bars have a length equal to or greater than 3), with the mean values focalized in the center of each bar (Figure 6, right). It is noticeable that most of the mean values of Δ are positive (65.52%), meaning that most of the patients were recovering from the disease.
Figure 6.
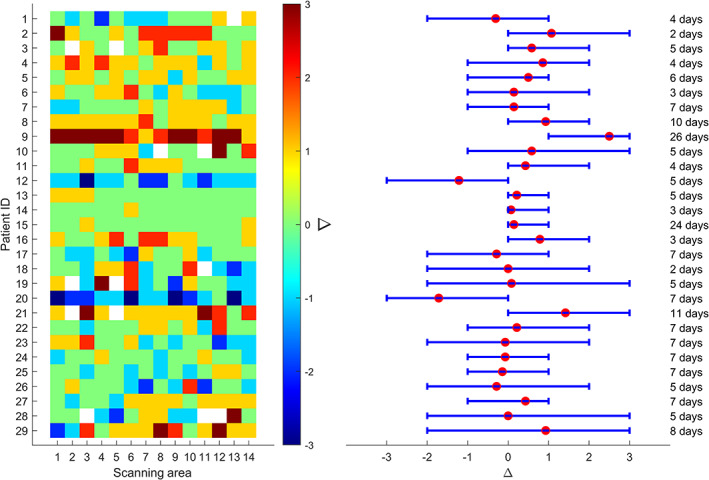
The values of Δ for each scanning area (x‐axis) and for each patient (y‐axis) that was scannied twice (on different dates) are depicted on the left. The 29 patients involved in this longitudinal study are numbered on the y‐axis from 1 to 29. The white squares indicate the absence of the measurement. On the right side, the mean value of Δ for each patient is depicted with a red point, whereas the lower and upper bounds of each error bar represent the minimum and maximum Δ of each patient, respectively. The temporal distance (days) between the two LUS exams is indicated on the right.
Prognostic Evaluation
Figure 7 shows how the prognostic agreement between AI and clinicians is higher in COVID‐19 patients than in post‐COVID‐19 patients. Specifically, the prognostic agreement is 80.45% for COVID‐19 patients (exam ID from 1 to 133) and 72.50% for post‐COVID‐19 patients (exam ID from 134 to 253). It is important to highlight how the AI models were trained on LUS data from COVID‐19 positive patients.
Figure 7.
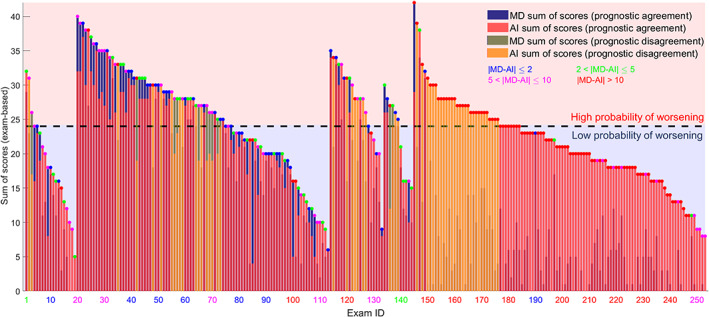
The exam‐based sum of scores for each LUS exam are depicted. MD exam‐based scores and AI exam‐based scores are depicted in blue and red bars, respectively. Each exam is colored (colored points above each bar) in blue, green, purple, and red, depending on the disagreement interval. The bars highlighted in yellow represent the LUS exams where the prognostic evaluation of MD and AI differs. The dark dashed line indicates a cumulative score of 24, which defines the prognostic threshold. The five subgroups of exams have been divided as follows: COVID‐19 patients from Rome (exam ID 1–19), Pavia (exam ID 20–113), and Lodi (exam ID 114–133), post‐COVID‐19 patients from Rome (exam ID 134–144), and Pavia (exam ID 145–253).
As introduced in the Materials and Methods section, the prognostic agreement was calculated assessing the capability of recently developed AI algorithms 18 of automatically stratifying patients at high risk of clinical worsening from patients at low risk. We hence compared, for each LUS exam, the cumulative score values (the sum of the scores over the 14‐areas investigated) obtained from the analysis of the data performed by LUS experts, with that provided by the AI. 19
For post‐COVID‐19 patients, it is plausible to assume that there may be a moment in time in which the recovery process of the damaged lung tissue produces LUS patterns which are not fully compatible with those obtained from healthy or acute patients. This hypothesis could explain the different performance of the AI models on post‐COVID‐19 patients.
Discussion
In this study, we report on new results related to the application of a standardized LUS imaging protocol and scoring system, which was developed to assess LUS data from patients affected by COVID‐19. The objectives of this study are multiple. First, to determine whether a simplified LUS imaging protocol could accurately capture and characterize the sonographic appearance of pleural and sub‐pleural alterations in COVID‐19 and post‐COVID‐19 patients. Standardization and evidence‐based results are in fact fundamental with LUS, since one of the most important limitations of this type of exam is that it relies on qualitative and subjective interpretations of LUS videos, which are scored depending on the presence of relevant imaging patterns. Although standardization cannot remove subjectivity completely, it can help reducing it by defining how many areas need to be scanned and where, as well as detailing the range of key imaging parameters which should be utilized to acquire the data.
Beyond the extended dataset (220 patients), one of the novelties of this study are the comparison between data from COVID‐19 and post‐COVID‐19 patients. From Figure 3, it is clear how post‐COVID‐19 patients present lower scores compared to COVID‐19 patients. This is in line with the expectations, given the prevalence of nonhospitalized patients in the post‐COVID‐19 group.
From Figure 4, it is also interesting to note how the score distribution for COVID‐19 patients was very similar among the different centers. Moreover, a clear difference could be observed between hospitalized and nonhospitalized post‐COVID‐19 patients, with the latter subgroup displaying lower scores.
From Figure 5, we can observe how, for both COVID‐19 and post‐COVID‐19 patients, the optimal trade‐off in terms of amount of scanning areas is 12. Specifically, a level of agreement of 98 and 97% was respectively found when comparing the results with a 14‐areas scanning protocol. Moreover, for both patients' populations, the worst scores are found in the basal posterior areas.
The second objective is to study whether the acquisition protocol could be further simplified during follow‐up.
From Figure 6, although these results were obtained on a limited number of patients (29), it seems that it is not possible to derive the general evolution of the lung condition from a subset of areas. In fact, no strong correlation is found among values of Δ obtained over different areas. This implies that a 12‐areas acquisition protocol is recommended also during follow‐up.
The third objective is to investigate the performance of recently developed AI models to automatically assess LUS videos according to the above‐introduced scoring system. AI models, especially when equipped with explainability mechanisms (which guarantee the possibility for a user to understand the decision made by the AI), can in fact further reduce the subjectivity of the evaluation process by providing a baseline evaluation. Moreover, automatic scoring algorithms can execute the analysis in a shorter time and relentlessly. They can thus be of great support for clinicians in case significant amount of data need to be analyzed in a short period of time.
From Figure 7, it is possible to showcase the potential as well as the limitations of AI models when applied to the analysis of LUS data. In fact, whether a good level of agreement was obtained between the AI and LUS experts in the evaluation of data from COVID‐19 patients, the agreement was significantly reduced for post‐COVID‐19 patients. This is consistent with the fact that the employed AI models were trained only on data from COVID‐19 patients. Once again, these results show how important it is to limit the application of AI models to the analysis of data consistently with the characteristic of the training dataset.
When presenting LUS findings, it is important to remember the current limitations of ultrasound technology when applied to the lungs. First of all, its low specificity. LUS patterns, as those investigated in this manuscript for COVID‐19, are in fact nonspecific. It is thus fundamental not to misinterpret LUS as a tool applicable to diagnose COVID‐19. Differently, it is applicable to evaluate the state of the lung and follow its evolution over time. It should also be acknowledged that ultrasound technology can only assess the surface of the lung, as the presence of air inhibits the exploration of deeper regions, unless the loss of aeration is significantly extended and reaches the surface of the lung. Another limitation of LUS, as performed through the analysis of data acquired with clinical scanners, is its intrinsic qualitative nature. In fact, although a numerical scoring system can be developed which associates specific patterns to a number, these approaches cannot be considered truly quantitative. To do that, measurable physical quantities with the power to characterize the alterations along the lung surface should be identified, and dedicated ultrasound methods designed around the peculiar properties of lung tissue should be developed. Research in this direction is emerging, 20 , 21 but further and extensive clinical studies are necessary to define and validate the potential of these methodologies with respect to their reproducibility, accuracy, and specificity.
Conclusion
In conclusion, the proposed scoring system is applicable to assess COVID‐19 and post‐COVID‐19 patients. For both COVID‐19 and post‐COVID‐19 patients, a 12‐areas acquisition protocol is confirmed as the optimal trade‐off between a time‐efficient and accurate LUS examination procedure. Moreover, the worst scores are confirmed to be found in the basal posterior areas for both patients' populations. As for what concerns follow‐up studies, it appears not to be possible to simplify the acquisition process, as no clear correlation was found among the score evolution across different areas. Finally, LUS data obtained from COVID‐19 and post‐COVID‐19 seem to display differences which are capable of confusing AI models that were not trained on post‐COVID‐19 data. This opens interesting questions on the existence of specific patterns associated to post‐COVID‐19 patients. Research in this direction will be the focus of future studies.
Acknowledgement
Open Access Funding provided by Universita degli Studi di Trento within the CRUI‐CARE Agreement.
This work was supported by the European Institute of Innovation and Technology (project UltraOn, EIT Digital 2020) and the Fondazione Valorizzazione Ricerca Trentina (grant 1, COVID‐19 2020).
Authors declare no conflict of interest.
References
- 1. Soldati G, Smargiassi A, Inchingolo R, et al. Is there a role for lung ultrasound during the COVID‐19 pandemic? J Ultrasound Med 2020; 39:1459–1462. 10.1002/jum.15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Soldati G, Smargiassi A, Inchingolo R, et al. Proposal for international standardization of the use of lung ultrasound for patients with COVID‐19. J Ultrasound Med 2020; 39:1413–1419. 10.1002/jum.15285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Poggiali E, Dacrema A, Bastoni D, et al. Can lung US help critical care clinicians in the early diagnosis of novel coronavirus (COVID‐19) pneumonia? Radiology 2020; 295:E6–E6. 10.1148/radiol.2020200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lomoro P, Verde F, Zerboni F, et al. COVID‐19 pneumonia manifestations at the admission on chest ultrasound, radiographs, and CT: single‐center study and comprehensive radiologic literature review. Eur J Radiol Open 2020; 7:100231. 10.1016/j.ejro.2020.100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nouvenne A, Ticinesi A, Parise A, et al. Point‐of‐care chest ultrasonography as a diagnostic resource for COVID‐19 outbreak in nursing homes. J Am Med Dir Assoc 2020; 21:919–923. 10.1016/j.jamda.2020.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yasukawa K, Minami T. Point‐of‐care lung ultrasound findings in patients with COVID‐19 pneumonia. Am J Trop Med Hyg 2020; 102:1198–1202. 10.4269/ajtmh.20-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xing C, Li Q, Du H, Kang W, Lian J, Yuan L. Lung ultrasound findings in patients with COVID‐19 pneumonia. Crit Care 2020; 24:174. 10.1186/s13054-020-02876-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Peng Q‐Y, Wang X‐T, Zhang L‐N, Chinese Critical Care Ultrasound Study Group (CCUSG) . Findings of lung ultrasonography of novel corona virus pneumonia during the 2019–2020 epidemic. Intensive Care Med 2020; 46:849–850. 10.1007/s00134-020-05996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duclos G, Lopez A, Leone M, Zieleskiewicz L. “No dose” lung ultrasound correlation with “low dose” CT scan for early diagnosis of SARS‐CoV‐2 pneumonia. Intensive Care Med 2020; 46:1103–1104. 10.1007/s00134-020-06058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Allinovi M, Parise A, Giacalone M, et al. Lung ultrasound may support diagnosis and monitoring of COVID‐19 pneumonia. Ultrasound Med Biol 2020; 46:2908–2917. 10.1016/j.ultrasmedbio.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smargiassi A, Soldati G, Torri E, et al. Lung ultrasound for COVID‐19 patchy pneumonia: extended or limited evaluations? J Ultrasound Med 2020; 40:521–528. 10.1002/jum.15428. [DOI] [PubMed] [Google Scholar]
- 12. Yuan M, Yin W, Tao Z, Tan W, Hu Y. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS One 2020; 15:e0230548. 10.1371/journal.pone.0230548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mento F, Perrone T, Fiengo A, et al. Limiting the areas inspected by lung ultrasound leads to an underestimation of COVID‐19 patients' condition. Intensive Care Med 2021; 47:811–812. 10.1007/s00134-021-06407-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yoon SH, Lee KH, Kim JY, et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID‐19): analysis of nine patients treated in Korea. Korean J Radiol 2020; 21:494–500. 10.3348/kjr.2020.0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID‐19): a pictorial review. Eur Radiol 2020; 30:4381–4389. 10.1007/s00330-020-06801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus Disease‐19 (COVID‐19): relationship to duration of infection. Radiology. 2020; 295:200463. 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Perrone T, Soldati G, Padovini L, et al. A new lung ultrasound protocol able to predict worsening in patients affected by severe acute respiratory syndrome coronavirus 2 pneumonia. J Ultrasound Med 2020; 40:1627–1635. 10.1002/jum.15548. [DOI] [PubMed] [Google Scholar]
- 18. Roy S, Menapace W, Oei S, et al. Deep learning for classification and localization of COVID‐19 markers in point‐of‐care lung ultrasound. IEEE Trans Med Imaging 2020; 39:2676–2687. 10.1109/TMI.2020.2994459. [DOI] [PubMed] [Google Scholar]
- 19. Mento F, Perrone T, Fiengo A, et al. Deep learning applied to lung ultrasound videos for scoring COVID‐19 patients: a multicenter study. J Acoust Soc Am 2021; 149:3626–3634. 10.1121/10.0004855. [DOI] [PubMed] [Google Scholar]
- 20. Demi L, Demi M, Prediletto R, Soldati G. Real‐time multi‐frequency ultrasound imaging for quantitative lung ultrasound—first clinical results. J Acoust Soc Am 2020; 148:998–1006. [DOI] [PubMed] [Google Scholar]
- 21. Mento F, Soldati G, Prediletto R, Demi M, Demi L. Quantitative lung ultrasound spectroscopy applied to the diagnosis of pulmonary fibrosis: first clinical study. IEEE Trans Ultrasonics Ferroelectr Freq Control 2020; 67:2265–2273. [DOI] [PubMed] [Google Scholar]
- 22. Mento F, Perrone T, Macioce VN, et al. On the impact of different lung ultrasound imaging protocols in the evaluation of patients affected by coronavirus disease 2019. J Ultrasound Med 2020; 40:2235–2238. 10.1002/jum.15580. [DOI] [PMC free article] [PubMed] [Google Scholar]


