Abstract
The role of respiratory superinfections in patients with coronavirus disease 2019 (COVID‐19) pneumonia remains unclear. We investigated the prevalence of early‐ and late‐onset superinfections in invasively ventilated patients with COVID‐19 pneumonia admitted to our department of intensive care medicine between March 2020 and November 2020. Of the 102 cases, 74 (72.5%) received invasive ventilation and were tested for viral, bacterial, and fungal pathogens on Days 0–7, 8–14, and 15–21 after the initiation of mechanical ventilation. Approximately 45% developed one or more respiratory superinfections. There was a clear correlation between the duration of invasive ventilation and the prevalence of coinfecting pathogens. Male patients with obesity and those suffering from chronic obstructive pulmonary disease and/or diabetes mellitus had a significantly higher probability to develop a respiratory superinfection. The prevalence of viral coinfections was high, with a predominance of the herpes simplex virus (HSV), followed by cytomegalovirus. No respiratory viruses or intracellular bacteria were detected in our cohort. We observed a high coincidence between Aspergillus fumigatus and HSV infection. Gram‐negative bacteria were the most frequent pathogen group. Klebsiella aerogenes was detected early after intubation, while Klebsiella pneumoniae and Pseudomonas aeruginosa were related to a prolonged respiratory weaning.
Keywords: bronchoalveolar lavage, coronavirus disease 2019, invasive ventilation, respiratory coinfections, severe acute respiratory syndrome coronavirus 2
Key points
In our cohort, approximately 45% of the invasively ventilated COVID‐19 patients developed a respiratory bacterial, viral, and/or fungal superinfection within 3 weeks after intubation. The most prevalent group of pathogens were Gram‐negative bacteria.
1. INTRODUCTION
The severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a novel β‐coronavirus, identified to consist the main pathogenic agent of coronavirus disease 2019 (COVID‐19), an ongoing pandemic, challenging the public health system worldwide. SARS‐CoV‐2 primarily affects the respiratory system and causes viral pneumonia, which can lead to the development of acute respiratory distress syndrome (ARDS). The involvement of extrapulmonary organs, during the course of the disease, has been also described in the literature. 1 , 2 , 3
Beyond the pathogenesis of SARS‐CoV‐2, respiratory coexisting infections with viruses, bacteria, and fungi have been reported, but their role in diagnosis, clinical presentation, and prognosis of the disease remains unclear. 4 Preliminary evidence shows that microbial copathogens may increase the morbidity and mortality of COVID‐19, especially in critically ill patients. Thus, the difficulty to adjudicate in the presence of a respiratory superinfection, the clinical relevance of the identified microorganisms, and the antimicrobial treatment remains challenging.
Clinical deterioration, elevated inflammatory markers, and bilateral radiological infiltrates may lead to a misperception regarding the presence of a respiratory copathogen and they should, therefore, utilize as an impulse to initiate a comprehensive diagnostic workout with sampling, rather than as an indicator of an underlying superinfection. 5 The consequence is that empirical antimicrobial therapy is systematically initiated until microbiological documentation of coinfecting pathogens is available. A meta‐analysis of studies, reporting antimicrobial prescribing in SARS‐CoV‐2 infected patients, demonstrated a wide use of broad‐spectrum antibacterials in more than 70% of the COVID‐19 cases, despite a paucity of evidence for bacterial coinfection, and no antimicrobial stewardship interventions were described. 6
As controversial reports appeared in the literature, the factor timing gained importance, regarding the clinical relevance of laboratory‐confirmed coinfecting pathogens in COVID‐19 patients and the implication of therapeutic measurements. 7 Some authors reported early‐onset coinfections in SARS‐CoV‐2‐infected patients, 8 , 9 while others identified late‐onset infections. 10 , 11 However, the starting time‐point until a coexisting infection has been identified (beginning of symptoms, admission to the hospital, admission to the intensive care unit [ICU]) and the nomenclature, used in several studies, have been extremely heterogenic, making it difficult to compare their results. The purpose of our study is to investigate the role of timing and differentiate between early‐ and late‐onset respiratory superinfections in an invasively ventilated COVID‐19 population.
According to the US Centers for Disease Control and Prevention (CDC), a superinfection is “an infection following a previous infection especially when caused by microorganisms that are resistant or have become resistant to the antibiotics used earlier,” while a coinfection is one occurring concurrently with the initial infection, the difference being purely temporal. 12 These are the definitions that will be used in the current manuscript.
We present the prevalence and consequences of respiratory coinfections in a consecutive cohort of patients receiving invasive ventilation in Germany's largest department of intensive care medicine between March 2020 and November 2020.
2. METHODS
2.1. Study population
In this retrospective study, 102 laboratory‐confirmed COVID‐19 cases, admitted to the department of intensive care medicine of the university medical center Hamburg‐Eppendorf in Germany, were enrolled consecutively between March 9, 2020 and November 16, 2020. Of the 102 cases, 74 (72.5%) received invasive ventilation and the remaining 28 cases (27.5%) were treated with noninvasive ventilation or oxygen therapy via high‐flow nasal cannula and thus excluded from the study (Figure 1). In all the invasively ventilated patients, the prevalence of viral, bacterial, and fungal pathogens was retrospectively registered in three sequential time periods: on Days 0–7 after intubation, on Days 8–14 under uninterrupted mechanical ventilation, and during the third week (Days 15–21), if a prolonged invasive ventilation was necessary.
Figure 1.
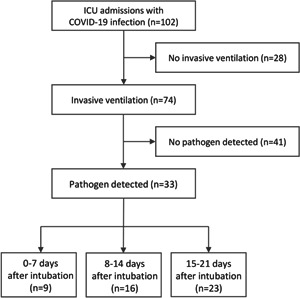
Flow chart of the study population. COVID‐19, coronavirus disease 2019; ICU, intensive care unit
2.2. Microbiological testing
2.2.1. SARS‐CoV‐2‐PCR
On admission to the ICU, SARS‐CoV‐2 polymerase chain reaction (SARS‐CoV‐2‐PCR) was performed, to confirm the diagnosis. Nasopharyngeal swabs were used for spontaneously breathing patients and endotracheal aspirate samples for those being already mechanically ventilated on their admission to the ward.
2.2.2. Respiratory samples
The respiratory samples were selected either as routine aspirates during a closed endotracheal suction, according to the standards of the clinic, or as bronchoalveolar lavage (BAL) during a bronchoscopy. Because of the high transmission risk of the COVID‐19 infection, a bronchoscopy was only conducted when the following criteria were fulfilled: 1. appearance of a relevant clinical deterioration with amelioration of the respiratory insufficiency and/or pneumogenic sepsis and 2. newly emerged infiltrates in the radiological examination of the chest (conventional radiology and/or computed tomography).
2.2.3. Samples testing
All respiratory samples were processed according to internal standards to allow the detection of conventional bacteria and slow‐growing pathogens, such as mycobacteria and fungi. All samples underwent galactomannan test for the detection of mold infections. In addition, the diagnostic panel included a multiplex nested PCR assay that could simultaneously detect different respiratory pathogens: influenza A virus (H1N1, H3N2, and H5N1), influenza B virus, parainfluenza virus types 1, 2, 3, 4a, and 4b, respiratory syncytial virus A and B, human rhinoviruses, human enteroviruses, human metapneumoviruses, Mycoplasma pneumonia, Chlamydophila pneumonia, Legionella pneumophila and adenoviruses (A to F). 13 Cytomegalovirus (CMV), Epstein–Barr virus, varicella‐zoster virus, and herpes simplex virus type 1 and 2 (HSV‐1 and HSV‐2) were tested separately.
3. RESULTS
Sixty‐eight (66.7%) of the total 102 patients were male and 34 (33.3%) were female, with a median age of 62.9 years. The median body mass index was 28.2 (range 17.5–44.5). The main pre‐existing comorbidities, except overweight and obesity, were respiratory diseases (n = 29), diabetes mellitus (n = 37), and underlying immunosuppression (n = 32). The most prevalent respiratory comorbidities were active cigarette smoking, chronic obstructive pulmonary disease (COPD), bronchial asthma, and obstructive sleep apnea/obesity hypoventilation syndrome (Table 1). Of the patients with diabetes mellitus, noninsulin‐dependent diabetes mellitus type 2 was the most prevalent group (n = 19), followed by insulin‐dependent diabetes mellitus type 2 (n = 12), undiagnosed diabetes mellitus (n = 5), and diabetes mellitus type 1 (n = 1). Underlying immunosuppressive conditions were chronic therapy with immunosuppressive drugs (corticosteroids, methotrexate, and lymphodepleting therapeutics) (n = 7), acute myeloid or lymphatic leukemia (n = 9), lymphoma (n = 4), active neoplasia of solid organs (n = 4), and kidney or bone marrow transplantation (n = 8). Of the patients, 68.9% of the invasively ventilated cases developed moderate to severe ARDS, 59.4% underwent prone positioning, and 25.6% required a venovenous extracorporeal membrane oxygenation. Overall, 34% were treated with steroids on admission and 94% received an empirical antibiotic therapy.
Table 1.
Prevalence of respiratory comorbidities in the study population
| Prevalence of respiratory comorbidities | |
|---|---|
| Active cigarette smoking | 12 (11.7%) |
| Chronic obstructive pulmonary disease | 6 (5.6%) |
| Bronchial asthma | 5 (4.9%) |
| Obstructive sleep apnea/obesity hypoventilation syndrome | 6 (5.6%) |
| Sarcoidosis | 1 (0.9%) |
| Bronchiectasis | 2 (1.9%) |
| Emphysema | 1 (0.9%) |
| Pleural asbestosis | 1 (0.9%) |
| History of pulmonary embolism | 2 (1.9%) |
| History of pneumonia | 2 (1.9%) |
| History of pulmonary surgery | 1 (0.9%) |
Approximately 45% of the invasively ventilated patients with COVID‐19 pneumonia were identified to have a respiratory bacterial, viral, and/or fungal superinfection in at least one of the sequential study periods. Of the overall 33 patients with a respiratory superinfection, 9 had one or more pathogens within 7 days after intubation, 16 on Days 8–14 after intubation, and 23 after 2 weeks (on Days 15–21) under mechanical ventilation (Figure 1). In our cohort, the male sex, obesity, COPD, and diabetes mellitus were associated with a significantly increased risk for a respiratory superinfection, whereas bronchial asthma and underlying malignancy were associated with a decreased risk.
All patients (8/8 cases) who got extubated within a week and almost 60% of the patients with successful weaning within 2 weeks (7/12 cases) had no pathogen detected in their samples. Seventy‐five percent of the patients who died within a week (6/8 cases) and 66% of the patients who died within 2 weeks (8/12 cases) had no pathogen detected in the BAL.
Overall, in 12 out of 77 positive samples (15.6%) Aspergillus fumigatus was present (Table 2). The prevalence of viral coinfections was high (27 out of 77 positive samples), with a predominance of HSV, followed by CMV. No respiratory viruses or intracellular bacteria could be detected in our cohort. We observed a high coincidence between A. fumigatus and HSV infection (5 out of 12 A. fumigatus samples were also positive to HSV). The bacterial pathogens were the most frequent in our study population (38 out of 77 positive samples) (Figure 2) and Gram‐negative bacteria was the most predominant pathogen group (Table 2). Klebsiella aerogenes was detected in the first week after intubation and was linked to a favorable outcome, while Klebsiella pneumoniae appeared later in the clinical course and was related to a prolonged respiratory weaning. The same trend was observed for Pseudomonas aeruginosa, which appeared almost exclusively in samples taken after 2 weeks on mechanical ventilation and was linked to a prolonged respiratory weaning.
Table 2.
Specific pathogens in respiratory samples during the study period
| Type of pathogen | Days 0–7 after intubation | Days 8–14 after intubation | Days 15–21 after intubation | Positive samples |
|---|---|---|---|---|
| A. fumigatus | 2 | 5 | 5 | 12 |
| HSV | 2 | 5 | 12 | 19 |
| CMV | 2 | 2 | 4 | 8 |
| A. baumanii complex | 2 | 3 | 5 | |
| C. koseri | 1 | 1 | ||
| E. cloacae complex | 1 | 5 | 6 | |
| E. coli | 1 | 1 | ||
| K. aerogenes | 3 | 1 | 1 | 5 |
| K. pneumoniae | 2 | 4 | 6 | |
| P. aeruginosa | 1 | 6 | 7 | |
| Pseudomonas spp. | 1 | 1 | ||
| S. aureus | 1 | 1 | ||
| S. aureus (MRSA) | 1 | 1 | ||
| S. maltophilia | 1 | 2 | 3 | |
| Viridans group streptococci | 1 | 1 |
Abbreviations: CMV, cytomegalovirus; HSV, herpes simplex virus; MRSA, methicillin‐resistant Staphylococcus aureus.
Figure 2.
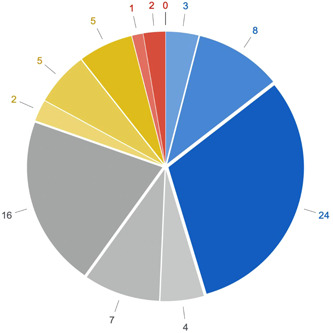
Representation of pathogen types over time. Numbers indicate the numbers of identified pathogens according to the specific type. Color coding: Gram‐negative pathogens are displayed in blue, viral pathogens in gray, fungal pathogens in yellow, and Gram‐positive pathogens in red. Lightest shades code for Days 0–7, medium shades for Days 8–14, and darkest shades for Days 15–21.
4. DISCUSSION
In our cohort, approximately 45% of the invasively ventilated COVID‐19 patients developed a respiratory bacterial, viral, and/or fungal superinfection within 3 weeks after intubation. This is surprisingly higher in comparison to a meta‐analysis by Lansbury et al., 14 who reported a rate of around 14% (95% confidence interval 5%–26%). This difference could be associated with the extensive spectrum of pathogens identified in our study, as long as to the inclusion of exclusively invasively ventilated ICU patients, who underwent the most severe form of illness and may be, therefore, more susceptible to infections, in comparison to the mixed (invasively and noninvasively ventilated) ICU population of the meta‐analysis.
There is a clear correlation between the duration of invasive ventilation and the prevalence of coinfecting pathogens, as described before. 10 , 11 Gram‐negative bacteria, mostly Enterobacteriaceae, were the predominant pathogen group. Interestingly, K. aerogenes was frequently detected in the first week after intubation and was linked to a favorable outcome, while K. pneumoniae and P. aeruginosa, appeared later in the clinical course and were related to a prolonged respiratory weaning.
All patients, who could be successfully weaned within a week, and almost 60% of the patients, who could be weaned within 2 weeks, had no pathogen detected in their samples. This could be a manifestation of a sufficient immunological barrier of the respiratory tract in this collective, as the pulmonary damage was not that prominent as in the prolonged ventilated group. On the other hand, the lack of superinfections may have contributed to the favorable clinical course, in comparison to the patients presented with weaning failure, although according to a meta‐analysis by Melsen et al. 15 the overall attributable mortality of ventilator‐associated pneumonia is 13%, with higher rates in surgical patients with a mid‐range severity score at admission and our study population was consisting exclusively of medical patients with a high‐range severity score at admission.
Another interesting finding was that 6 out of 8 patients, who died within a week, and 8 out of 12 patients, who died on Days 8–14, had no pathogen detected in their respiratory samples. The majority of those patients experienced a severe state of septic shock with unknown origin and had a sterile BAL in the bronchoscopy. This state of septic shock, possibly attributed to SARS‐CoV‐2 itself, without any evidence of other responsible microorganisms, has been already described in former studies. 16 The lack of superinfections in critically ill COVID‐19 patients with a high case fatality has been suggested to be a significant parameter linked to the underestimated prevalence of superinfections in the entire COVID‐19 ICU population. 7
The high prevalence of Staphylococcus aureus, described elsewhere as the main pathogen identified in early‐onset bacterial coinfection, 8 , 9 was not confirmed in our study. Staphylococcus aureus is considered to be a community‐acquired coexisting pathogen. According to the CDC definition, infections identified 48 h after hospital admission should be referred to as hospital‐acquired and those within 48 h as community‐acquired. 12 As the great majority of the ICU patients included in our study was admitted to the hospital several days before their admission to the ICU, no community‐acquired infections were expected in this collective.
The evidence of A. fumigatus was present in more than 15% of the overall positive samples. The prevalence of Herpesviridae was even higher, reaching more than 35% of the positive samples, with a predominance of HSV, followed by CMV. We observed a high coincidence between A. fumigatus and HSV infection (5 out of 12 A. fumigatus samples were also positive to HSV). No respiratory viruses or intracellular bacteria could be detected in our cohort. Due to inconsistent definitions and diagnostic criteria of invasive aspergillosis in non‐neutropenic critically ill patients, the actual prevalence of this entity in our study population is yet unclear. 17 , 18 The inclusion of BAL fluid galactomannan as an additional entry criterion may increase the diagnostic sensitivity for invasive aspergillosis in ICU patients. 19 The coincidence of A. fumigatus with reactivated Herpesviridae is probably a manifestation of an inadequate local and systemic immune response, rather than an active superinfection, requiring a targeted antimicrobial treatment.
A limitation of our study could be the high percentage of patients, who already received broad‐spectrum antibiotics on their admission to the ICU. More than 94% were under an empirical antibiotic therapy before the initiation of the mechanical ventilation and 34% were treated with steroids. This could have influenced the microbiological consistency of the bronchial aspirates. A comparative study, after the adjustment of the guidelines, 20 may elucidate possible differences in this trend. Certainly, multicenter studies with a larger number of subjects are needed to verify and improve our results.
Our manuscript highlights the importance of timing in the development of respiratory superinfections in severe COVID‐19 cases. There is a clear correlation between the duration of invasive ventilation and the prevalence of coinfecting pathogens. However, we could not differentiate if the duration of mechanical ventilation was the reason for the presence of respiratory superinfections, in the terms of ventilator‐associated pneumonia, or the opposite. Specifically designed studies on host–pathogen interaction mechanisms may shed more light on the pathogenic background of this clinical observation.
Furthermore, we could not identify if respiratory coinfections may significantly influence the clinical outcome of critically ill patients with COVID‐19 pneumonia. We believe that respiratory superinfections mainly consist of an indicator of an insufficient immunological response, rather than clinically relevant ongoing infections, requiring an anti‐microbial treatment. However, the initiation of a targeted treatment remains the greatest challenge for clinicians being active in the field. The holistic evaluation of clinical and paraclinical parameters of those patients may improve their personalized management.
CONFLICT OF INTERESTS
S. K. received research support from Ambu, Daiichi Sankyo, ETView Ltd, Fisher & Paykel, Pfizer, and Xenios. He also received lecture fees from Astra, C. R. Bard, Baxter, Biotest, Cytosorbents, Daiichi Sankyo, Fresenius, Gilead, Mitsubishi Tanabe Pharma, MSD, Pfizer, Philips, and Zoll. He received consultant fees from Bayer, Fresenius, Gilead, MSD, and Pfizer. D. W. reports personal consultant and lecture fees from AMEOS, Advanz (formerly Correvio), Eumedica, EUSA, Gilead, Kite, Lilly, MSD, Novartis, Pfizer, and Shionogi. The remaining authors declare no conflict of interests.
ETHICS STATEMENT
The current study was based on the anonymized COVID‐19 patients' register, treated with SARS‐CoV‐2‐induced respiratory insufficiency, in the department of intensive care medicine of the University Medical Center Hamburg‐Eppendorf in Germany. The study protocol and the analysis of patients' data were approved by the ethics committee of the Medical Association of Hamburg (ID Number: WF‐051/20).
AUTHOR CONTRIBUTIONS
Maria Paparoupa, Razaz Aldemyati, and Kevin Roedl analyzed the patient's data and wrote the manuscript, Hannes Roggenkamp, Benjamin Berinson, Dominik Nörz, and Flaminia Olearo performed the microbiological and molecular testing of the patients' samples, Stefan Kluge, Geraldine de Heer, and Dominic Wichmann reviewed the manuscript and provided consultation, regarding intellectual argumentation. All authors read and approved the final version of the manuscript.
ACKNOWLEDGEMENT
Open Access funding enabled and organized by Projekt DEAL.
Paparoupa M, Aldemyati R, Roggenkamp H, et al. The prevalence of early‐ and late‐onset bacterial, viral, and fungal respiratory superinfections in invasively ventilated COVID‐19 patients J Med Virol. 2022;94:1920‐1925. 10.1002/jmv.27548
Maria Paparoupa and Razaz Aldemyati contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Puelles VG, Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS‐CoV‐2. N Engl J Med. 2020;383:590‐592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaltschmidt B, Fitzek ADE, Schaedler J, et al. Hepatic vasculopathy and regenerative responses of the liver in fatal cases of COVID‐19. Clin Gastroenterol Hepatol. 2021;19:1726‐1729.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Roedl K, Jarczak D, Drolz A, et al. Severe liver dysfunction complicating course of COVID‐19 in the critically ill: multifactorial cause or direct viral effect? Ann Intens Care. 2021;11:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X, Liao B, Cheng L, et al. The microbial coinfection in COVID‐19. Appl Microbiol Biotechnol. 2020;104:7777‐7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Verroken A, Scohy A, Gérard L, Wittebole X, Collienne C, Laterre PF. Co‐infections in COVID‐19 critically ill and antibiotic management: a prospective cohort analysis. Crit Care. 2020;24:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rawson TM, Moore LSP, Zhu N, et al. Bacterial and fungal coinfection in individuals with coronavirus: a rapid review to support COVID‐19 antimicrobial prescribing. Clin Infect Dis. 2020;71:2459‐2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bassetti M, Kollef MH, Timsit JF. Bacterial and fungal superinfections in critically ill patients with COVID‐19. Intens Care Med. 2020;46:2071‐2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhu X, Ge Y, Wu T, et al. Co‐infection with respiratory pathogens among COVID‐2019 cases. Virus Res. 2020;285:198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elabbadi A, Turpin M, Gerotziafas GT, Teulier M, Voiriot G, Fartoukh M. Bacterial coinfection in critically ill COVID‐19 patients with severe pneumonia. Infection. 2021;49:1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hughes S, Troise O, Donaldson H, Mughal N, Moore LSP. Bacterial and fungal coinfection among hospitalized patients with COVID‐19: a retrospective cohort study in a UK secondary‐care setting. Clin Microbiol Infect. 2020;26:1395‐1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dudoignon E, Caméléna F, Deniau B, et al. Bacterial pneumonia in COVID‐19 critically ill patients: a case series. Clin Infect Dis. 2021;72:905‐906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feldman C, Anderson R. The role of co‐infections and secondary infections in patients with COVID‐19. Pneumonia (Nathan). 2021;13:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lam WY, Yeung AC, Tang JW, et al. Rapid multiplex nested PCR for detection of respiratory viruses. J Clin Microbiol. 2007;45:3631‐3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lansbury L, Lim B, Baskaran V, Lim WS. Co‐infections in people with COVID‐19: a systematic review and meta‐analysis. J Infect. 2020;81:266‐275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melsen WG, Rovers MM, Groenwold RH, et al. Attributable mortality of ventilator‐associated pneumonia: a meta‐analysis of individual patient data from randomised prevention studies. Lancet Infect Dis. 2013;13:665‐671. [DOI] [PubMed] [Google Scholar]
- 16. Antinori S, Galimberti L, Milazzo L, Ridolfo AL. Bacterial and fungal infections among patients with SARS‐CoV‐2 pneumonia. Infez Med. 2020;28:29‐36. [PubMed] [Google Scholar]
- 17. Bassetti M, Giacobbe DR, Grecchi C, Rebuffi C, Zuccaro V, Scudeller L, FUNDICU investigators . Performance of existing definitions and tests for the diagnosis of invasive aspergillosis in critically ill, adult patients: a systematic review with qualitative evidence synthesis. J Infect. 2020;81:131‐146. [DOI] [PubMed] [Google Scholar]
- 18. Donnelly JP, Chen SC, Kauffman CA, et al. Revision and update of the consensus definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin Infect Dis. 2020;71:1367‐1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schroeder M, Simon M, Katchanov J, et al. Does galactomannan testing increase diagnostic accuracy for IPA in the ICU? A prospective observational study. Crit Care. 2016;20:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kluge S, Janssens U, Welte T, Weber‐Carstens S, Marx G, Karagiannidis C. Empfehlungen zur intensivmedizinischen Therapie von Patienten mit COVID‐19 [Recommendations for critically ill patients with COVID‐19]. Med Klin Intensivmed Notfmed. 2020;115:175‐177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


