Abstract
The rapid spread of the Delta variant of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) became a serious concern worldwide in summer 2021. We examined the copy number and variant types of all SARS‐CoV‐2‐positive patients who visited our hospital from February to August 2021 using polymerase chain reaction (PCR) tests. Whole genome sequencing was performed for some samples. The R.1 variant (B.1.1.316) was responsible for most infections in March, replacing the previous variant (B.1.1.214); the Alpha (B.1.1.7) variant caused most infections in April and May; and the Delta variant (B.1.617.2) was the most prevalent in July and August. There was no significant difference in the copy numbers among the previous variant cases (n = 29, median 3.0 × 104 copies/µl), R.1 variant cases (n = 28, 2.1 × 105 copies/µl), Alpha variant cases (n = 125, 4.1 × 105 copies/µl), and Delta variant cases (n = 106, 2.4 × 105 copies/µl). Patients with Delta variant infection were significantly younger than those infected with R.1 and the previous variants, possibly because many elderly individuals in Tokyo were vaccinated between May and August. There was no significant difference in mortality among the four groups. Our results suggest that the increased infectivity of Delta variant may be caused by factors other than the higher viral loads. Clarifying these factors is important to control the spread of Delta variant infection.
Keywords: COVID‐19, Delta variant, melting curve analysis, SARS‐CoV‐2, viral load
Highlights
There was no significant difference in copy numbers among the previous variant cases, R.1 variant cases, Alpha variant cases, and Delta variant cases.
This suggests that the increased infectivity of Delta variant may be caused by factors other than the higher viral loads.
1. INTRODUCTION
The rapid spread of the Delta variant (B.1.617.2) of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) became a serious problem worldwide in summer 2021. 1 A precise understanding of the features of the Delta variant is crucial for controlling its spread. Tokyo Medical and Dental University Hospital is located in the center of Tokyo and mainly treats patients with severe coronavirus disease 2019 (COVID‐19) from all over Tokyo. We examined the copy number and variant types of the samples from all COVID‐19 patients who visited our hospital.
We previously reported that the R.1 variant (the sublineage of B.1.1.316) rapidly replaced the previously existing strain, B.1.1.214 (originating from the European lineage), in Tokyo in March 2021. 2 After that, the Alpha variant (B.1.1.7) and the Delta variant prevailed.
Information on SARS‐CoV‐2 variants and patient numbers is provided by the Tokyo Metropolitan Institute of Public Health, 3 but is based on a sampling survey and does not include patient data such as viral loads. In this article, we show the precise viral loads and patient profiles based on our clinical data.
2. MATERIALS AND METHODS
2.1. Patients and samples
Data were collected from COVID‐19 patients whose diagnoses were confirmed by polymerase chain reaction (PCR) tests performed from February 1 to August 31, 2021. Nasopharyngeal swab samples were obtained at the time of outpatient visit or admission. The samples were immersed in test tubes containing 1 ml of phosphate‐buffered saline containing 1% dithiothreitol before analysis.
This study was approved by the Medical Research Ethics Committee of Tokyo Medical and Dental University (approval number: M2020‐004) and was conducted in accordance with the ethical standards of the 1964 Helsinki Declaration.
2.2. Quantitative PCR test
To detect and quantify SARS‐CoV‐2 in the samples, one‐step reverse‐transcription quantitative PCR was performed without viral RNA purification using the 2019 Novel Coronavirus Detection Kit (Shimadzu Corp.) and the QuantStudio 5 Dx Real‐Time PCR System (Thermo Fisher Scientific, Inc.). The copy numbers were estimated from the calibration lines of the concentration‐known standard samples.
2.3. Variant screening PCR test
To determine the SARS‐CoV‐2 variant for each patient, we used melting curve analysis of PCR products. Viral RNA was purified from the swab samples that were positive for SARS‐CoV‐2, using the EZ1 Virus Mini Kit v2.0 and EZ1 advanced XL (QIAGEN). Reverse‐transcription polymerase chain reaction (RT‐PCR) was performed using the LightCycler Multiplex RNA Virus Master (Roche Molecular Systems, Inc.) and primers and probes provided in the VirSNiP SARS‐CoV‐2 kits for Spike N501Y, E484K, and L452R (TIB Molbiol). The melting curve analysis of the PCR products was performed according to the manufacturer's instructions. The 501N, 501Y, 484E, 484K, 484Q, 452R, and 452L types were determined by their melting temperature profiles.
Whole genome sequencing (WGS) was performed to identify the lineages of representative samples using a next‐generation sequencer. Libraries were prepared using the QIAseq SARS‐CoV‐2 Primer Panel Kit (QIAGEN) and sequenced using the MiSeq (Illumina, Inc.). Alignment and variant calling were performed using CLC Genomics Workbench software (QIAGEN).
2.4. Statistical analysis
The difference in copy numbers and ages among the four types of variant cases was evaluated using the Steel–Dwass test. The difference in the rate of intensive care unit (ICU) admission and mortality was evaluated using the χ 2 test.
3. RESULTS
Based on the results of variant screening PCR, we divided the samples into four groups: previous strain (501 N, 484E, and 452L), R.1 variant (501N, 484K, and 452L), Alpha variant (501Y, 484E, and 452L), and Delta variant (501N, 484E, and 452R). WGS was conducted on approximately one‐fifth of samples for each variant group, these results consistently agreed with our grouping, confirming that our process was appropriate. PANGO lineages 4 were determined using the WGS data from the representative samples. The strains identified as 501Y and 484K (Beta or Gamma variant) or 484Q and 452L (Kappa variant) were not found in our study sample.
The sequential transitions of the variants are shown in Figure 1. The R.1 variant replaced the previous variants in March, as previously reported. 2 The Alpha variant represented the majority of infections in April and May. The first Delta variant case in our hospital was identified in mid‐May and the Delta variant was predominant in July and August.
Figure 1.
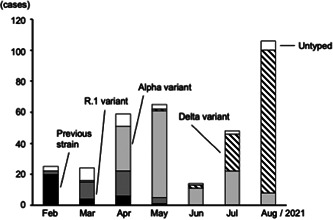
Sequential transition of four types of variant cases determined by PCR‐based melting curve analysis. Untyped bar represents the samples of which variant type was not determined because of that the PCR products were not generated probably due to small copy number. PCR, polymerase chain reaction
The variant types from 20 samples could not be determined as variant screening PCR did not produce any PCR products; this is probably because the samples had a very small copy number (data not shown). Untyped samples (shown as Untyped bar in Figure 1) were excluded from the following analysis.
The clinical profiles of the 288 patients enrolled are shown in Table 1. Comparing the age of each variant group, there was no significant difference between Delta and Alpha cases. However, the Delta variant cases were significantly younger than the R.1 variant cases (p = 5 × 10−5) and the previous strain cases (p = 6 × 10−7).
Table 1.
Clinical profiles of patients with four different variants of SARS‐CoV‐19
| Variant type | Previous (n = 29) | R.1 (n = 28) | Alpha (n = 125) | Delta (n = 106) |
|---|---|---|---|---|
| Age (years) mean ± SD | 68.5 ± 13.8 | 66.9 ± 19.7 | 51.5 ± 15.8 | 47.6 ± 17.6 |
| Male ratio (%) | 70.0 | 57.1 | 72.0 | 64.2 |
| Mortality (number, %) | n = 4, 13.8% | n = 4, 4.3% | n = 4, 3.2% | n = 3, 2.8% |
| ICU admission (%) | 44.8 | 25.0 | 22.4 | 17.0 |
| Viral loads (copies/µl) median | 3.0 × 104 | 2.1 × 105 | 4.1 × 105 | 2.4 × 105 |
Abbreviations: ICU, intensive care unit; SARS‐CoV, severe acute respiratory syndrome coronavirus.
As an indicator of increased severity, the rates of admission to the intensive care unit (ICU) were compared among the four groups. The rate of ICU admission for previous strain cases was significantly higher than that of Delta variant cases (p = 0.022). There were no significant differences among the other variant pairs. With regard to mortality, no significant difference was found between any pair of groups.
The distribution of viral loads as copy numbers in the swab‐soaked samples is shown in Figure 2. There was no significant difference in copy number among the four variant groups. We could not find a clear relationship between copy number and severity (Figure 2–upper panel) and between copy number and generation (Figure 2–lower panel). The copy numbers of patients who died in the Delta variant group seemed high compared with the numbers of those who did not die, however, this finding could not be confirmed as only three patients died in this group.
Figure 2.
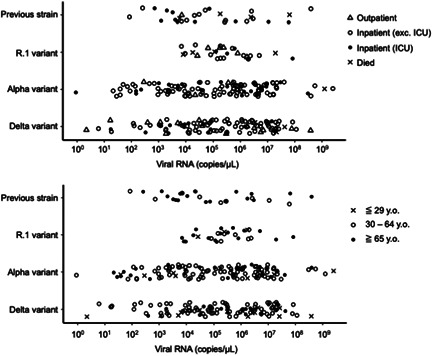
Copy numbers of viral RNA in swab‐soaked samples for four types of SARS‐CoV‐2 variant cases, estimated by reverse transcription‐quantitative PCR. Cases are marked according to the severity (upper panel) and the generation (lower panel). PCR, polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
4. DISCUSSION
The Delta variant has attracted attention for its increased transmissibility. 1 Viral load is thought to be one of the factors that causes increased transmissibility. Li et al. reported that the viral load of Delta variants was more than a thousand times higher than that of the Wuhan strain in the initial wave of 2020 in China. 5 Ong et al. also reported that the Delta variant was associated with lower PCR cycle threshold values compared with the wild‐type in Singapore. 6 Because we had no patients with the Wuhan strains in early 2020, we could not compare the viral load between the Delta variant cases and the Wuhan strain cases.
Contrary to these reports, our data showed that the viral loads of the Delta variant cases were no higher than those of the other variant cases. It should be noted that our data may be biased because our hospital specializes in patients with severe disease and therefore may not reflect the distribution of all cases in Tokyo. However, as this policy did not change over the study period, it is unlikely that it affected the distribution of our data.
The Delta variant was reported to be more frequently detected in the younger generation than other variants, 7 and our data also showed the same tendency. However, it is speculated that this was not due to the nature of Delta variants, but rather to the fact that many elderly individuals (over the age of 65) were vaccinated from May to August in Tokyo, reducing the infection rates of the elderly.
As an indicator of severity, we focused on the rates of ICU admission and mortality. We showed that the Delta variant did not lead to increased severity compared with the other variants. Contrary to our data, Twohig et al. reported the increased severity of the Delta variants compared with the Alpha variants in England. 8
Our study has some limitations. Our data might be affected by the selection bias of the patients mentioned above. Our data included small number of outpatients who did not need hospitalization due to mild symptoms. Some patients were transferred to our hospital due to deterioration from other hospitals after several days of hospitalization. This process meant that more than 1 week may have passed since the onset of illness, which may have affected viral loads in the samples taken at the time of admission to our hospital. In addition, the relatively low mortality of the Delta variant group could be an effect of vaccination in the elderly.
As Bager et al. mentioned, 9 it is difficult to determine causality of the severity across emerging variants and to compare the severity in different studies performed in various countries. This is because social behavior such as vaccination, age distribution, testing, and treatment system are changing and varied. There may also be some differences in genetic factors for immune responses against SARS‐CoV‐2. 10
Our results suggest that the increased infectivity of the Delta variant may not be caused by higher viral loads, but by other factors, such as a higher affinity of the mutated spike proteins to the cells. 11 Clarifying the mechanism is a future task to control the spread of the Delta variant.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Conceived and designed the study: Chihiro Tani‐Sassa, Yumi Iwasaki, Naoya Ichimura, Yoko Nukui, Hiroaki Takeuchi, and Shuji Tohda. Analyzed samples: Chihiro Tani‐Sassa, Yumi Iwasaki, Katsutoshi Nagano, Yuta Takahashi, Sonoka Yuasa, Yuna Takatsuki, Jun Nakajima, and Kazunari Sonobe. Performed WGS: Hiroaki Takeuchi, Kousuke Tanimoto, Yukie Tanaka, and Akinori Kimura. Analyzed data: Naoya Ichimura and Shuji Tohda. Wrote the paper: Shuji Tohda.
ACKNOWLEDGMENTS
The authors would like to thank all the staff involved in COVID‐19 treatment at TMDU hospital. WGS was supported by grant JPMJCR20H2 from JST‐CREST and grant 20nk0101612h0901 from the Japan Agency for Medical Research and Development.
Tani‐Sassa C, Iwasaki Y, Ichimura N, et al. Viral loads and profile of the patients infected with SARS‐CoV‐2 Delta, Alpha, or R.1 variants in Tokyo. J Med Virol. 2022;94:1707‐1710. 10.1002/jmv.27479
Chihiro Tani‐Sassa and Yumi Iwasaki contributed equally to this study.
DATA AVAILABILITY STATEMENT
The data and protocol of RT‐PCR are available from the corresponding author upon request.
REFERENCES
- 1. Campbell F, Archer B, Laurenson‐Schafer H, et al. Increased transmissibility and global spread of SARS‐CoV‐2 variants of concern as at June 2021. Euro Surveill. 2021;26:2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nagano K, Tani‐Sassa C, Iwasaki Y, et al. SARS‐CoV‐2 R.1 lineage variants that prevailed in Tokyo in March 2021. J Med Virol. 2021;93:6833‐6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tokyo Metropolitan Institute of Public Health. SARS‐CoV‐2 variant screening tests at TMIPH. Accessed October 20, 2021. http://www.tokyo-eiken.go.jp/lb_virus/mutation/
- 4.PANGO lineages database.Latest epidemiological lineages of SARS‐CoV‐2. Accessed October 20, 2021. https://cov-lineages.org
- 5. De Crop E, Delgat L, Nuytinck J, Halling RE, Verbeken A. Viral infection and transmission in a large, well‐traced outbreak caused by the SARS‐CoV‐2 Delta variant. Preprint medRxiv. 2021;7:133‐164. 10.1101/2021.07.07.21260122 [DOI] [Google Scholar]
- 6. Ong SWX, Chiew CJ, Ang LW, et al. Clinical and virological features of SARS‐CoV‐2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis. 2021. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Delahoy MJ, Ujamaa D, Whitaker M, et al. Hospitalizations associated with COVID‐19 among children and adolescents—COVID‐NET, 14 States, March 1, 2020‐August 14, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1255‐1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Twohig KA, Nyberg T, Zaidi A, et al, COVID‐19 genomics UK (COG‐UK) consortium . Hospital admission and emergency care attendance risk for SARS‐CoV‐2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect Dis. 2021. Published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bager P, Wohlfahrt J, Rasmussen M, Albertsen M, Krause TG. Hospitalisation associated with SARS‐CoV‐2 delta variant in Denmark. Lancet Infect Dis. 2021;21:1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Downes DJ, Cross AR, Hua P, et al. Identification of LZTFL1 as a candidate effector gene at a COVID‐19 risk locus. Nat Genet. 2021;53:1606‐1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu Y, Liu J, Johnson BA, et al. Delta spike P681R mutation enhances SARS‐CoV‐2 fitness over Alpha variant. bioRxiv. 2021. 10.1101/2021.08.12.456173 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data and protocol of RT‐PCR are available from the corresponding author upon request.


